Abstract
In order to address neutrophil activation during inflammation we assessed the expression of interleukin 1 receptor type 1 (IL-1R1) following in-vivo extravasation. Extravasated neutrophils were collected from 11 healthy study subjects by a skin chamber technique and compared to neutrophils in peripheral blood. Expression of IL-1R1 was assessed by microarray, quantitative polymerase chain reaction (qPCR), Western blot, flow cytometry, enzyme linked immunosorbent assay (ELISA) and immunoelectron microscopy (iEM). IL-1R1 was induced following extravasation, demonstrated by both gene array and qPCR. Western blot demonstrated an increased expression of IL-1R1 in extravasated leucocytes. This was confirmed further in neutrophils by flow cytometry and iEM that also demonstrated an increased intracellular pool of IL-1R1 that could be mobilized by N-formyl-methionine-leucine-phenylalanine (fMLP). Stimulation of peripheral neutrophils with IL-1 resulted in transcription of NFκB and a number of downstream chemokines and the corresponding chemokines were also induced following in-vivo extravasation. The present results demonstrate that IL-1R1 is induced following extravasation and exists on the neutrophil surface, as well as in a mobile intracellular pool. Furthermore, neutrophils express functional IL-1R1 as demonstrated by the induction of chemokines following IL-1 stimulation. The results indicate a potential role for IL-1 in the activation of neutrophils at inflammatory sites.
Keywords: extravasation, IL-1R1, neutrophils
Introduction
Neutrophils are recruited to sites of inflammation by interaction with adhesion molecules and chemokines, which are expressed on interleukin (IL)-1β- and tumour necrosis factor (TNF)-α-activated endothelium [1]. The process of extravasation primes the neutrophil for innate defence mechanisms such as phagocytosis, generation of reactive oxygen metabolites and release of anti-microbial and proteolytic proteins. Neutrophils can also orchestrate the inflammatory response through the production and release of cytokines and chemokines [2], a process regulated partly by nuclear factor kappa B (NF-κB) [3],[4].
IL-1 is a proinflammatory cytokine that is produced early during inflammation and participates in the acute phase response. IL-1 binds to the IL-1 receptor type 1 (IL-1R1), which transmits a signal to downstream targets, including mitogen activated protein kinases (MAPK), activator protein 1 (AP-1) and NF-κB. Circulating neutrophils express 500–900 IL-1R1 per cell [5]–[7], and internalization of radiolabelled IL-1 indicates that the receptor is biologically active [7]. Although the expression of IL-1R1 on circulating neutrophils increases during sepsis and by in-vitro incubation with granulocyte macrophage colony-stimulating factor (GM-CSF) [8], the expression on extravasated neutrophils has not been assessed thoroughly. A minor up-regulation of IL-1R1 following in-vivo extravasation has been shown previously by gene array [9]. However, the expression of IL-1R1 at protein level has not been assessed, and it remains unclear whether extravasated neutrophils express IL-1R1 and, thus, are receptive for IL-1 at local inflammatory sites.
Activation of NF-κB following in-vivo extravasation, phagocytosis or in-vitro stimulation results in the production and release of chemokines [3],[4],[9]–[13]. Several neutrophil-generated chemokines have been identified that may impact the immune response. Chemokine (C-C motif) ligand 3 (CCL3) [also known as macrophage inflammatory protein (MIP)-1α] and CCL4 (MIP-1β) are chemotactic for monocytes and activated lymphocytes [14], CCL20 (MIP-3α) attracts lymphocytes [15] and chemokine (C-X-C motif) ligand 2 (CXCL2) (MIP-2α) is chemotactic for neutrophils and up-regulated by IL-1 and TNF-α[16]. Hence, because neutrophils dominate in the early inflammatory reaction, they may have a potentially important role in modulating the succeeding inflammatory response by the production of chemokines.
The skin chamber method is useful for studies of leucocyte activation at local inflammatory sites [9],[17]–[19]. Given that extravasation is an important event to modulate activation and function of neutrophils and that IL-1 is a crucial proinflammatory cytokine that can tune these events, we aimed to investigate the expression of IL-1R1 in extravasated neutrophils by use of the skin chamber method.
Materials and methods
Study subjects
Eleven healthy study subjects, nine males and two females, with a median age of 63 (59–67) years, were included for skin chamber experiments, and three healthy blood donors were included for in-vitro assays. Study subjects with known inflammatory or infectious diseases and those given immunosuppressive drugs or warfarin were excluded; median C-reactive protein (CRP) was 1·6 (0·7–5·2) mg/l. All subjects gave informed consent and the study was approved by the ethical committee at the Karolinska University Hospital, Stockholm, Sweden.
The skin chamber technique
This method has been described previously in detail [20]. Briefly, two to five skin blisters were induced on one of the forearms by vacuum (300 mm Hg) and gentle heating (39°C). After 14 h, the wound areas were washed with phosphate-buffered saline (PBS) and the epidermal roofs were removed. Sterilized plastic chambers with an open bottom were mounted over the wounds and filled with autologous serum containing heparin. After 10 h of incubation, the exudates were collected and the chambers were washed with an equal volume of PBS that was added to the collected sample. Total incubation time with the skin chambers was 10 h. The samples were centrifuged, the supernatants were frozen and the cell pellets were dissolved in RPMI-1640 (Hyclone Laboratories Inc., Logan, UT, USA).
Isolation of neutrophils
Skin chamber neutrophils were isolated by density centrifugation for 20 min at 600 g at room temperature using Ficoll Paque (Pharmacia Biotech, Uppsala, Sweden). The pellets, together with a band of activated neutrophils, were collected, pooled and washed with PBS.
Circulating neutrophils were isolated from 20 ml of citrated blood (0·129 M Na-citrate) (Vacutainer; Becton Dickinson, Plymouth, UK). The blood was diluted 1:1 with PBS and Ficoll Paque density centrifugation was performed for 30 min at 400 g in room temperature. The pellets were collected and the erythrocytes were haemolysed by addition of isotonic solution [154 mM NH4Cl, 10 mM KHCO3 and 0·1 mM ethylenediamine tetraacetic acid (EDTA), pH 7·2] at 4°C for 12 min. The samples were centrifuged and fresh lysing solution was added for additional 2 min in order to haemolyse the remaining erythrocytes. The samples were centrifuged again and the neutrophil pellet was washed with PBS. The purity of the neutrophil population was judged by flow cytometry forward and side-scatter.
RNA extraction and microarray analysis
RNA was extracted from the purified neutrophils by use of the Qiagen RNeasy kit, according to the manufacturer's instructions (Qiagen, Solna, Sweden). Microarrays were performed according to the procedure provided by Affymetrix (Affymetrix Inc., Santa Clara, CA, USA).
Analysis of microarray data
Affymetrix Cel files from three healthy study subjects were normalized using GeneChip robust multi-array averaging (GC-RMA) within GeneSpring™ software version 7·3 (Silicon Genetics, CA, USA). Global differences in gene expression between circulating and extravasated neutrophils were distinguished by unsupervised hierarchical cluster analysis. Genes with an altered expression by a factor of 2 or greater which were significant by analysis of variance (anova) (P < 0·05) were considered differentially expressed following extravasation.
Twenty thousand genes were compared; 1029 genes had a higher expression and 1179 genes had a lower expression following extravasation. To identify categories and functional relations among genes, the web-based analysis tools Ingenuity (Ingenuity systems, Redwood City, CA, USA) and Gencards (http://www.genecards.org) were used. The microarray data are accessed online at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi with the accession number GSE35786.
Quantitative polymerase chain reaction (qPCR) validation of gene expression
Analysis by qPCR includes seven study subjects. First-strand cDNA was synthesized from total RNA using SuperScript II reverse transcriptase and oligo(dT)12–18 (Invitrogen Inc., Carlsbad, CA, USA). Using cDNA as a template, TaqMan gene expression assays were performed by means of the FAM dye labelling system (Applied Biosystems, Stockholm, Sweden). Commercially available primers (Table 1) were used (Applied Biosystems) and the qPCR reaction was conducted in a total volume of 20 µl using a fast plate thermocycler ABI 7900 (Applied Biosystems).
Table 1.
TaqMan gene expression probes used for quantitative polymerase chain reaction (qPCR).
| Gene | Assay ID |
|---|---|
| IL-1R1 | Hs00168392_m1 |
| IL-1RN | Hs00893626_m1 |
| IRAK1 | Hs00155570_m1 |
| NF-κB1 | Hs00765730_m1 |
| CCL3 | Hs00234142_m1 |
| CCL4 | Hs99999148_m1 |
| CCL20 | Hs00355476_m1 |
| CXCL2 | Hs00236966_m1 |
| GAPDH | Hs99999905_m1 |
IL-R1, interleukin 1 receptor type 1; IRAK, interleukin-1 receptor associated kinase; NF-kB, nuclear factor kappa B; CCL, chemokine (C-C motif) ligand; CXCL, chemokine (C-X-C motif) ligand; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
The results were normalized against the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and the relative expression was calculated according to instructions provided by Applied Biosystems.
Western blot of IL-1R1
IL-1R1 expression in the total leucocyte pool was analysed by Western blot. Protein was prepared following cell lysis induced by repeated freezing/thawing in a modified RIPA buffer [50 mM Tris-HCL pH 7·5, 150 mM NaCl, 1 mM EDTA, 0·1% Triton X-100, 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulphonate (CHAPS)]. The protein concentration was measured by the Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, USA) and 24 µg of protein was loaded into precasted Novex Tris-glycine, 12% gels (Invitrogen) and separated by electrophoresis. The samples were then transferred onto nitrocellulose membranes (Bio-Rad Laboratories, Sundbyberg, Sweden) by the use of semidry blot. The nitrocellulose membranes were blocked in 5% milk and incubated with a rabbit monoclonal antibody towards IL-1R1, diluted 1:1500 in 5% milk (EP409Y) (Abcam, Cambridge, UK). Following washing with Tris-buffered saline (TBS)-T the membrane was incubated with a secondary antibody conjugated to horseradish peroxidase (HRP), diluted 1:20 000 in TBS-T (ab6721) (Abcam). The membranes were then washed and an HRP substrate was added by the SuperSignal West Pico reagents (Thermo Scientific). CL-XPosure Films (Thermo Scientific) were then exposed to the fluorescent bands. The membranes were stripped by a low pH buffer and reanalysed for the expression of the GAPDH loading control by a monoclonal antibody diluted 1:5000 in 5% milk (EPR1977Y) (Abcam).
Flow cytometry of IL-1R1 and CD66b
Expression of IL-1R1 was analysed in blood and chamber neutrophils from three study subjects. The blood was collected in tubes containing 0·129 M Na-citrate (Vacutainer; Becton Dickinson), divided into 100 µl portions, haemolysed for 5 min at 4°C by an isotonic solution (154 mM NH4Cl, 10 mM KHCO3 and 0·1 mM EDTA, pH 7·2) and then washed with PBS. Half the samples were stimulated for 15 min at 37°C with 0·5 µM formyl-Met-Leu-Phe (fMLP) (Sigma, St Louis, MO, US), diluted in RPMI-1640 with 5% fetal calf serum (FCS). The remaining half of the samples were kept on ice with RPMI-1640 and 5% FCS and are referred to as non-stimulated. Following incubation, the leucocytes were washed with PBS and stained for 30 min on ice with a polyclonal anti-IL-1R1 phycoerythrin (PE)-conjugated antibody (R&D Systems, Abingdon, UK). The leucocytes were then washed and analysed by flow cytometry (Epics Elite, Beckman Coulter Inc., Hialeah, FL, USA). The neutrophil and monocyte populations, respectively, were selected and the percentage of positive cells was determined by gating against the isotype control.
CD66b, a marker for specific and gelatinase granules, was analysed directly by staining with an anti-CD66b fluorescein isothiocyanate (FITC)-conjugated monoclonal antibody (BD Biosciences, San Jose, CA, USA).
Immunoelectron microscopy (iEM) of IL-1R1
Analysis by iEM includes samples from one study subject. Leucocytes from peripheral blood and the skin chamber were fixated in 3% paraformaldehyde in 0·1 M phosphate buffer. An anti-human IL-1R1 polyclonal antibody (Abcam), diluted 1:100, was used and the protocol for iEM is described elsewhere [21].
Measurement of soluble mediators
Soluble mediators were analysed with commercial enzyme-linked immunosorbent assays (ELISAs) (Quantikine immunoassay; R&D Systems), according to the manufacturer's instructions; samples from the seven study subjects analysed by qPCR were included. Lowest detectable value in each analysis was IL-1α: 1·0 pg/ml; IL-1β: 1·0 pg/ml; IL-1R antagonist (IL-1Ra): 6·26 pg/ml; soluble IL-1R2: 10·0 pg/ml; CCL3: 10·0 pg/ml; CCL20: 0·47 pg/ml and CCL4: 11·0 pg/ml and 4·0 pg/ml in serum and skin chamber, respectively. Soluble IL-1R1 was analysed by a commercial ELISA from USCN Life Science Inc. (Wuhan, China) with a lowest detectable value of 0·048 ng/ml.
Gene expression and chemokine release following in-vitro IL-1 stimulation
Peripheral blood from three healthy blood donors was collected in tubes containing 0·129 M Na-citrate (Vacutainer; Becton Dickinson). The neutrophils were isolated as described previously in this paper. Two and 3 million of the purified neutrophils were seeded per well in 12-well cell culture plates in RPMI-1640 with 10% FCS for analysis of gene expression and chemokine release, respectively. Human recombinant IL-1Ra (R&D Systems) was added to one well per donor and plate and the plate was incubated for 15 min at 37°C; the final concentration of IL-1Ra was 1 µg/ml. Recombinant human IL-1α and IL-1β (R&D Systems) were then added in combination, both at the final concentration of 10 ng/ml. The plates were incubated for additional 3 or 5 h at 37°C for the analysis of gene expression and chemokine release, respectively. As a control, one well per donor and plate was incubated with RPMI-1640/FCS alone.
After 3 h of incubation, the neutrophils assessed for gene expression were collected and washed. RLT lysing solution (Qiagen, Solna, Sweden) was added to the wells and thereafter to the collected neutrophil pellets. Analysis by qPCR was conducted as described previously in this paper. Up-regulation of CCL3, CCL4, CCL20, CXCL2, NFκB1 and GAPDH were analysed and the unstimulated sample was used for normalization.
After 5 h of incubation, the neutrophil culture medium was collected and frozen and the release of CCL3 and CCL4 was measured with ELISA, as described previously. The unstimulated sample was used for normalization. Mean recovery following 5 h of incubation was 84% and the amount of apoptotic neutrophils, measured by annexin V and PI, was less than 10% in all wells.
Statistics
Gene-arrays (n = 3) were normalized and analysed as described previously in this paper. Data obtained by qPCR and ELISA (n = 7) were analysed by Wilcoxon's matched-pairs test and data obtained by flow cytometry (n = 3) were analysed by independent t-test. A P < 0·05 was considered significant. Data from gene array, qPCR and flow cytometry are presented as mean ± standard deviation (s.d.) and data from ELISA are presented as median and 25–75% interquartile range.
Results
Quality control
In the peripheral circulation the median leucocyte count was 5·0 (3·6–6·2) 109 cells/µl with 52% (51–55%) granulocytes. The number of leucocytes collected from the skin chamber was 3·0 (1·5–5·1) million with 80% (77–82%) granulocytes. The specific granule protein, CD66b, was analysed by means of neutrophil activation in three healthy study subjects and the mean expression was 6·0 ± 1·9 mean fluorescence intensity (MFI) in peripheral neutrophils and 19·6 ± 4·4 MFI in extravasated neutrophils, indicating cellular degranulation. In addition, following extravasation, the number of apoptotic neutrophils, evaluated by annexin V and PI staining, is less than 10% (unpublished data).
Neutrophils were isolated by density centrifugation and the purity was judged by flow cytometry forward and side-scatter. The median percentage of neutrophils purified from blood was 99·3% (98·7–99·5%) and from the skin chambers 95·7% (95·0–96·0%). Lineage-specific genes from other cells than neutrophils were not detected (Table 2), indicating a pure neutrophil population. The purified RNA had a median optical density ratio 260:280 of 1·87 (1·85–1·89).
Table 2.
Gene-array on lineage specific genes in purified neutrophils.
| Lineage-specific gene | Cell type | Detectable transcript |
|---|---|---|
| G-CSFR | Neutrophils | Yes |
| IL-5RA | Eosinophils/basophils | No |
| M-CSFR | Monocytes/macrophages | No |
| TCR | T lymphocytes | No |
| FGFR1 | Fibroblasts | No |
| FGFR2 | Fibroblasts/epidermal cells | No |
| EGFR | Epidermal cells | No |
| VEGFR | Endothelial cells | No |
| VCAM1 | Endothelial cells | No |
G-CSFR, granulocyte colony-stimulating factor receptor; IL, interleukin; TCR, T cell receptor; FGFR, fibroblast growth factor receptor; EGFR, epidermal growth factor receptor; VEGFR, vascular endothelial growth factor receptor; VCAM, vascular cell adhesion molecule.
Expression of IL-1R1
Neutrophil gene expression following extravasation was assessed by gene array (n = 3) and qPCR (n = 7) and the expression of IL-1R1, interleukin-1 receptor antagonist (IL-1RN) and interleukin-1 receptor associated kinase 1 (IRAK1) is presented in Fig. 1a. IL-1R1 was up-regulated significantly by a factor of 20 assessed by gene array and by a factor of 10 assessed with qPCR, P < 0·05, for both assessments. Minor alterations were seen in IL-1RN and IRAK1.
Fig. 1.

Expression of interleukin 1 receptor type 1 (IL-1R1) on neutrophils from blood and chamber exudate. (a) Gene expression analysed by gene array and quantitative polymerase chain reaction (qPCR). The figure shows changes in gene expression between extravasated and circulating neutrophils, expressed as mean ± standard deviation (s.d.). (b) The expression of IL-1R1 in circulating and extravasated leucocytes assessed by Western blot. (c) Expression of IL-1R1 assessed by flow cytometry. Data by flow cytometry is viewed as the percentage of IL-1R1-positive neutrophils in the peripheral circulation and in the skin chamber, at basal expression and following additional in-vitro N-formyl-methionine-leucine-phenylalanine (fMLP) stimulation, presented as mean ± s.d. Independent t-test was used for comparison between groups. (d,e) Expression of IL-1R1 in peripheral and extravasated neutrophils, respectively, assessed by immunoelectron microscopy (iEM).
Protein expression of IL-1R1 was assessed by Western blot (n = 1), flow cytometry (n = 3) and immunoelectron microscopy (iEM) (n = 1) and soluble IL-1R1 was assessed by ELISA (n = 7). IL-1R1 was first analysed using Western blot of a protein extract from the total leucocyte pool, collected from peripheral blood and the skin chamber. An intense band corresponding to IL-1R1 was detected from extravasated leucocytes (Fig. 1b). From circulating leucocytes, a weak band could be detected only after a long film exposure (data not presented). In order to discriminate the expression of IL-1R1 on neutrophils from that on other leucocytes, the expression was assessed further by flow cytometry (Fig. 1c). Flow cytometry indicated a low expression on circulating neutrophils that could be induced significantly following fMLP stimulation, P = 0·021. Following extravasation, the percentage of neutrophils that had a detectable expression of IL-1R1 increased (P = 0·016) and the expression could be induced further by fMLP (P = 0·005). A similar expression pattern was detected on monocytes, P = not significant (n.s.), between neutrophils and monocytes. Mean expression on circulating monocytes was 2·0 ± 0·5% and following fMLP stimulation 10·2 ± 4·2%. On extravasated monocytes, corresponding numbers were 30·1 ± 15·8% at rest and 26·0 ± 11·1% following fMLP stimulation. The increased expression of IL-1R1 on extravasated neutrophils was confirmed with iEM, which demonstrated an increased expression of IL-1R1 both at the cell surface as well as intracellularly in extravasated neutrophils (Fig. 1e) compared to circulating neutrophils (Fig. 1d). In order to quantify the expression of IL-1R1, the number of receptors was counted and the results are expressed as the median number of IL-1R1 in relation to the µm2 of cytoplasm. In neutrophils from blood, eight sections were calculated and the concentration of IL-1R1 was 0·79 (0·62–1·44) IL-1R1/µm2. In neutrophils from the skin chamber, 13 sections were calculated and the corresponding number was 4·27 (2·83–5·00) IL-1R1/µm2, P = 0·017, between blood and chamber samples, analysed by Wilcoxon's matched-pairs test. As IL-1R1 is cleaved from the surface into a soluble form as part of its regulation, soluble IL-1R1 was analysed in serum and in chamber fluid by ELISA. The concentration of soluble IL-1R1 was 0·34 (0·26–0·48) ng/ml in serum and 5·36 (5·08–7·08) ng/ml in chamber fluid, P = 0·028.
Concentrations of IL-1α, IL-1β, IL-1R antagonist and soluble IL-1R2
The concentrations of IL-1α, IL-1β, IL-1Ra and soluble IL-1R2 were measured in serum and in skin chamber fluid by ELISA. IL-1α and IL-1β were below detection level in serum and are therefore not presented. In the chamber fluid, the concentrations of IL-1α and IL-1β were: 57·9 (42·7–115·6) pg/ml and 200·3 (51·2–382·8) pg/ml, respectively. The level of IL-1Ra was 185·4 (132·9–380·1) pg/ml in serum and 11 735·8 (8211·6–30727·2) pg/ml in the chamber fluid, P = 0·018. The ratio between IL-1Ra and IL-1α in the skin chamber fluid was 189 (112–462) and the corresponding ratio to IL-1β was 132 (53–204). The concentration of soluble IL-1R2 was 11·1 (10·1–13·2) ng/ml in serum and 16·0 (13·8–17·6) ng/ml in chamber fluid, P = 0·018.
Chemokine expression following IL-1 stimulation or extravasation
In order to confirm a functional response by IL-1R1, in-vitro IL-1 stimulations of purified neutrophils were performed, assessing the expression of a number of chemokines regulated by the IL-1R1/NF-κB axis. IL-1 stimulation induced the transcription of CCL3, CCL4, CCL20, CXCL2 and NFκB1 in purified neutrophils and the effect was blocked by pretreatment with IL-1Ra (Fig. 2a). Furthermore, the release of CCL3 and CCL4 into the cell culture medium was detected by ELISA after 5 h of stimulation with IL-1 (Fig. 2b). Data are presented as the ratio between IL-1 stimulated cells and non-stimulated cells, with or without pretreatment with IL-1Ra (n = 3). Up-regulation of the corresponding chemokine genes was also observed in neutrophils following extravasation, assessed by gene array (n = 3) and qPCR (n = 7). Figure 2c shows the increased expression of NFκB1, CCL3, CCL4, CCL20 and CXCL2 in extravasated neutrophils compared to that in circulating neutrophils, P < 0·05, for all markers. In addition, concentrations of CCL3, CCL4 and CCL20, assessed by ELISA (n = 7), were all significantly higher in the skin chamber fluid compared to in serum (Fig. 2d).
Fig. 2.
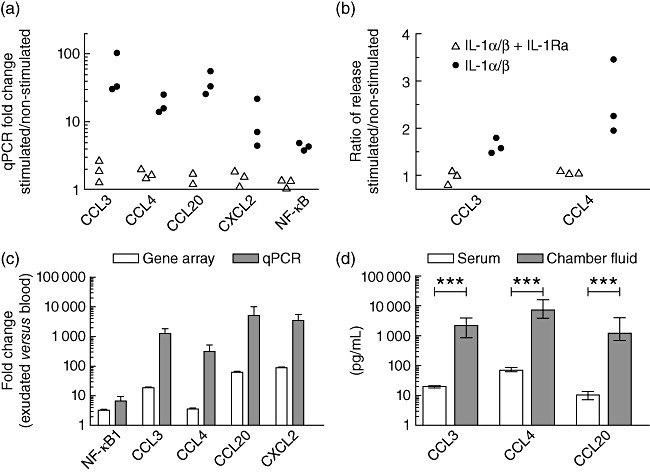
Chemokine expression following interleukin (IL)-1 stimulation and extravasation. (a) Induction of the CCL3, CCL4, CCL20, CXCL2 and NFκB1 genes following 3 h of in-vitro stimulation with IL-1, assessed by quantitative polymerase chain reaction (qPCR). The expression pattern is normalized to cells incubated with cell culture medium alone. (b) Release of CCL3 and CCL4 following 5 h of in-vitro IL-1 incubation in relation to the release following incubation with cell culturing medium alone, assessed by enzyme-linked immunosorbent assay (ELISA). Triangles refer to samples challenged with IL-1 in presence of interleukin-1 receptor antagonist (IL-1Ra) and circles refer to samples treated with IL-1 alone. (c) Up-regulation of the CCL3, CCL4, CCL20, CXCL2 and NFκB1 genes following in-vitro extravasation assessed by gene array and qPCR, presented as mean ± standard deviation. P < 0·05 for all markers, analysed by analysis of variance (anova) (gene array) and Wilcoxon's matched-pairs test (qPCR). (d) Concentrations of CCL3, CCL4 and CCL20 in serum and in the chamber exudate, assessed by ELISA and presented as median and interquartile range, analysed by Wilcoxon's matched-pairs test.
Discussion
In the present study, we demonstrate transcription of IL-1R1 in human neutrophils following extravasation. In addition, expression of IL-1R1 increased following in-vivo extravasation and IL-1R1 could be mobilized further from an intracellular pool to the cell surface by fMLP. A functional response by IL-1R1 was demonstrated by the transcription and release of chemokines following in-vitro IL-1 stimulation. In addition, the corresponding chemokines were transcribed following extravasation and high concentrations were assayed in the skin chamber fluid.
The skin chamber method is well established, and has been used previously to study gene expression in extravasated neutrophils. In line with the present study, Theilgaard-Mönch et al. demonstrated up-regulation of IL-1R1, IL-1β and CCL3 in neutrophils following extravasation [9]. In the present study, a more pronounced up-regulation of IL-1R1 was detected following extravasation. Using gene array, the expression increased 20-fold and using qPCR a 10-fold up-regulation was detected, both indicating that IL-1R1 is transcribed in neutrophils following extravasation.
Increased expression of IL-1R1 in extravasated leucocytes was first confirmed by Western blot. A strong induction of IL-1R1 was detected following extravasation while only a weak band could be detected from circulating leucocytes, indicating translation of IL-1R1 following extravasation. As approximately 80% of the chamber leucocytes were neutrophils the remaining 20%, which were mainly monocytes, could have influenced the result. However, flow cytometry indicated a similar expression of IL-1R1 in neutrophils and monocytes which limits a potential skewing of the results by present monocytes. The number of IL-1R on circulating human neutrophils has been estimated previously as 500–900 receptors per cell [5]–[7], and this relatively low number of receptors may be difficult to detect by conventional immunofluorescence flow cytometry [22]. In line with this notion, we report a low expression of IL-1R1 on circulating neutrophils assessed by flow cytometry. However, the expression increased significantly following extravasation, and could be induced further by the bacterial-related molecule fMLP. Mobilization of additional IL-1R1 following extravasation and the encounter of a bacterial-related stimuli provides an interesting link for neutrophil activation in vivo. The expression of IL-1R1 on circulating and extravasated neutrophils was confirmed further by iEM that demonstrated an increased IL-1R1 on extravasated neutrophils, both at the cell surface and intracellularly. Together, these data indicate that IL-1R1 is expressed at the cell surface of neutrophils and that it can be mobilized from an intracellular compartment following fMLP stimulation. In addition, as part of the regulation, IL-1R1 is cleaved into a soluble form during inflammation, creating a balance between soluble and cell bound receptors. An increased concentration of sIL-1R1 was detected at the interstitial site compared to in serum, which further confirms an increased expression of IL-1R1 at the local inflammatory site.
IL-1 provokes a biological response at low IL-1R1 occupancy and the IL-1 axis is therefore tuned at several levels. Mechanisms for regulation include enzymatic processing of precursor proteins, high concentrations of IL-1Ra and the decoy receptor IL-1R2, as well as regulation of the signal transduction by an IL-1 receptor accessory protein [23]. The biological response by IL-1 can be blocked in vitro by a 100-fold excess of IL-1Ra [24]. Neutrophils have been shown to produce high concentrations of IL-1Ra [25],[26], and it is interesting to speculate that a consecutive production of IL-1Ra by neutrophils might be one way of regulating the IL-1 axis at the inflammatory site. In line with this, we found a strong up-regulation of IL-1RN, assessed by qPCR. The kinetics of IL-1 and IL-1Ra have been studied by inducing endotoxaemia in humans, demonstrating a rise in plasma IL-1Ra which reached a peak concentration, 100-fold greater than IL-1β, 1–2 h after the peak of IL-1β[27]. In the current paper we add information and demonstrate that, at a local aseptic inflammatory site in vivo, IL-1α, IL-1β and IL-1Ra are produced at high concentrations and IL-1Ra is produced at more than 100 times higher than the concentrations of IL-1α and IL-1β.
Signal transduction from IL-1R1 results in nuclear mobilization and activation of NF-κB [11],[28] and the IL-1R1/NF-κB axis plays an important role in the induction of CCL3, CCL4 and CXCL2[12],[13]. In the present paper, a biological response by IL-1R1 was confirmed by in-vitro IL-1 stimulation of peripheral neutrophils, which resulted in up-regulation of NFκB and expression of chemokine genes and the corresponding proteins. Inhibition by IL-1Ra verified an IL-1R1-specific response. Significant up-regulations of CCL3, CCL4, CCL20 and CXCL2 were also detected in extravasated neutrophils compared to circulating neutrophils. However, although activation via IL-1 can stimulate production of these chemokines, it is uncertain whether the observed up-regulation is induced by the extravasation process per se or by IL-1R1 engagement. It is intriguing to speculate that the increased expression of IL-1R1 following extravasation provides a mechanism for sensitizing the extravasated neutrophil towards the IL-1 axis. The concentrations of CCL3, CCL4 and CCL20 were also increased significantly at protein levels in the skin chamber fluid compared to in serum. The data should, however, be interpreted by caution, because the inflammatory milieu in the skin chamber is influenced by both infiltrated leucocytes and residual cells.
It is likely that the increased expression of IL-1R1 on extravasated neutrophils represents newly synthesized receptors. The present data, which show transcription of IL-1R1 following extravasation as well as a higher IL-1R1 expression, assessed by Western blot, iEM, ELISA and flow cytometry, suggest that IL-1R1 is synthesized following extravasation. In addition, the response towards fMLP indicates an interesting link between inflammation and neutrophil encounter of bacterial stimuli. An increased expression of IL-1R1 at the local inflammatory site might impact neutrophil receptiveness towards the inflammatory milieu and thereby tune the succeeding transcriptional programme of the transformed tissue-dwelling neutrophil. It is also interesting to note that the commonly accepted view that neutrophils have a life span of less than 1 day has been challenged recently by Pillay and co-workers [29], and that an increased life span induced by extravasation and by T lymphocytes has been reported [30]. These observations, together with the current data, suggest that neutrophils might also be important modulators of the proinflammatory milieu at time-points beyond degranulation. Previously, a direct link between neutrophil secretion products and monocyte recruitment has been proposed [31]. In the current paper we propose that neutrophils also may provide a link to subsequent recruitment of mononuclear cells at time-points beyond degranulation as a consequence of the increased production of chemokines.
In conclusion, in the present paper we demonstrate that neutrophils express IL-1R1 and that IL-1R1 is induced following extravasation and exists in a mobile intracellular compartment. In addition, neutrophils respond to in-vitro IL-1 stimulation by up-regulation of chemokines, both at gene and protein levels, and the same chemokine genes are also up-regulated in neutrophils following extravasation. We therefore propose that extravasated neutrophils are transformed into chemokine-producing cells and that IL-1 may constitute an important stimulus. Hence, neutrophils that dominate during the early inflammatory reaction may have an immunomodulatory role and tune the inflammatory response towards subsequent recruitment of mononuclear cells.
Acknowledgments
The authors would like to thank Anette Bygden-Nylander for assistance with the skin chamber method and Kjell Hultenby for assistance with iEM. The study was supported by unrestricted grants from Karolinska Institutet, Hesselman Foundation and TERUMO EUROPE NV.
Disclosures
The authors declare no competing economical interests.
References
- 1.Furie MB, McHugh DD. Migration of neutrophils across endothelial monolayers is stimulated by treatment of the monolayers with interleukin-1 or tumor necrosis factor-alpha. J Immunol. 1989;143:3309–17. [PubMed] [Google Scholar]
- 2.Theilgaard-Monch K, Porse BT, Borregaard N. Systems biology of neutrophil differentiation and immune response. Curr Opin Immunol. 2006;18:54–60. doi: 10.1016/j.coi.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Malcolm KC, Arndt PG, Manos EJ, Jones DA, Worthen GS. Microarray analysis of lipopolysaccharide-treated human neutrophils. Am J Physiol Lung Cell Mol Physiol. 2003;284:L663–70. doi: 10.1152/ajplung.00094.2002. [DOI] [PubMed] [Google Scholar]
- 4.McDonald PP, Cassatella MA. Activation of transcription factor NF-kappa B by phagocytic stimuli in human neutrophils. FEBS Lett. 1997;412:583–6. doi: 10.1016/s0014-5793(97)00857-0. [DOI] [PubMed] [Google Scholar]
- 5.Rhyne JA, Mizel SB, Taylor RG, Chedid M, McCall CE. Characterization of the human interleukin 1 receptor on human polymorphonuclear leukocytes. Clin Immunol Immunopathol. 1988;48:354–61. doi: 10.1016/0090-1229(88)90029-3. [DOI] [PubMed] [Google Scholar]
- 6.Borish L, Rosenbaum R, McDonald B, Rosenwasser LJ. Recombinant interleukin-1 beta interacts with high-affinity receptors to activate neutrophil leukotriene B4 synthesis. Inflammation. 1990;14:151–62. doi: 10.1007/BF00917454. [DOI] [PubMed] [Google Scholar]
- 7.Parker KP, Benjamin WR, Kaffka KL, Kilian PL. Presence of IL-1 receptors on human and murine neutrophils. Relevance to IL-1-mediated effects in inflammation. J Immunol. 1989;142:537–42. [PubMed] [Google Scholar]
- 8.Fasano MB, Cousart S, Neal S, McCall CE. Increased expression of the interleukin 1 receptor on blood neutrophils of humans with the sepsis syndrome. J Clin Invest. 1991;88:1452–9. doi: 10.1172/JCI115454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theilgaard-Monch K, Knudsen S, Follin P, Borregaard N. The transcriptional activation program of human neutrophils in skin lesions supports their important role in wound healing. J Immunol. 2004;172:7684–93. doi: 10.4049/jimmunol.172.12.7684. [DOI] [PubMed] [Google Scholar]
- 10.McDonald PP, Bald A, Cassatella MA. Activation of the NF-kappaB pathway by inflammatory stimuli in human neutrophils. Blood. 1997;89:3421–33. [PubMed] [Google Scholar]
- 11.Coldren CD, Nick JA, Poch KR, et al. Functional and genomic changes induced by alveolar transmigration in human neutrophils. Am J Physiol Lung Cell Mol Physiol. 2006;291:L1267–76. doi: 10.1152/ajplung.00097.2006. [DOI] [PubMed] [Google Scholar]
- 12.Calkins CM, Bensard DD, Shames BD, et al. IL-1 regulates in vivo C-X-C chemokine induction and neutrophil sequestration following endotoxemia. J Endotoxin Res. 2002;8:59–67. [PubMed] [Google Scholar]
- 13.Cloutier A, Ear T, Blais-Charron E, Dubois CM, McDonald PP. Differential involvement of NF-kappaB and MAP kinase pathways in the generation of inflammatory cytokines by human neutrophils. J Leukoc Biol. 2007;81:567–77. doi: 10.1189/jlb.0806536. [DOI] [PubMed] [Google Scholar]
- 14.Taub DD, Conlon K, Lloyd AR, Oppenheim JJ, Kelvin DJ. Preferential migration of activated CD4+ and CD8+ T cells in response to MIP-1 alpha and MIP-1 beta. Science. 1993;260:355–8. doi: 10.1126/science.7682337. [DOI] [PubMed] [Google Scholar]
- 15.Baba M, Imai T, Nishimura M, et al. Identification of CCR6, the specific receptor for a novel lymphocyte-directed CC chemokine LARC. J Biol Chem. 1997;272:14893–8. doi: 10.1074/jbc.272.23.14893. [DOI] [PubMed] [Google Scholar]
- 16.Haskill S, Peace A, Morris J, et al. Identification of three related human GRO genes encoding cytokine functions. Proc Natl Acad Sci USA. 1990;87:7732–6. doi: 10.1073/pnas.87.19.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sengelov H, Follin P, Kjeldsen L, Lollike K, Dahlgren C, Borregaard N. Mobilization of granules and secretory vesicles during in vivo exudation of human neutrophils. J Immunol. 1995;154:4157–65. [PubMed] [Google Scholar]
- 18.Follin P, Wymann MP, Dewald B, Ceska M, Dahlgren C. Human neutrophil migration into skin chambers is associated with production of NAP-1/IL8 and C5a. Eur J Haematol. 1991;47:71–6. doi: 10.1111/j.1600-0609.1991.tb00564.x. [DOI] [PubMed] [Google Scholar]
- 19.Follin P, Dahlgren C. A skin chamber technique as a human model for studies of aseptic inflammatory reactions. Methods Mol Biol. 2007;412:333–46. doi: 10.1007/978-1-59745-467-4_22. [DOI] [PubMed] [Google Scholar]
- 20.Thylen P, Lundahl J, Fernvik E, Gronneberg R, Hallden G, Jacobson SH. Impaired monocyte CD11b expression in interstitial inflammation in hemodialysis patients. Kidney Int. 2000;57:2099–106. doi: 10.1046/j.1523-1755.2000.00060.x. [DOI] [PubMed] [Google Scholar]
- 21.Mandic Havelka A, Yektaei-Karin E, Hultenby K, et al. Maternal plasma level of antimicrobial peptide LL37 is a major determinant factor of neonatal plasma LL37 level. Acta Paediatr. 2010;99:836–41. doi: 10.1111/j.1651-2227.2010.01726.x. [DOI] [PubMed] [Google Scholar]
- 22.Zola H. Detection of cytokine receptors by flow cytometry. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im0621s26. Chapter 6: Unit 6.21. [DOI] [PubMed] [Google Scholar]
- 23.Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–32. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arend WP, Welgus HG, Thompson RC, Eisenberg SP. Biological properties of recombinant human monocyte-derived interleukin 1 receptor antagonist. J Clin Invest. 1990;85:1694–7. doi: 10.1172/JCI114622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schröder AK, von der Ohe M, Kolling U, et al. Polymorphonuclear leucocytes selectively produce anti-inflammatory interleukin-1 receptor antagonist and chemokines, but fail to produce pro-inflammatory mediators. Immunology. 2006;119:317–27. doi: 10.1111/j.1365-2567.2006.02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jablonska E, Jablonski J, Piotrowski L, Grabowska Z. IL-1beta, IL-1Ra and sIL-1RII in the culture supernatants of PMN and PBMC and the serum levels in patients with inflammation and patients with cancer disease of the same location. Immunobiology. 2001;204:508–16. doi: 10.1078/0171-2985-00059. [DOI] [PubMed] [Google Scholar]
- 27.Granowitz EV, Santos AA, Poutsiaka DD, et al. Production of interleukin-1-receptor antagonist during experimental endotoxaemia. Lancet. 1991;338:1423–4. doi: 10.1016/0140-6736(91)92725-h. [DOI] [PubMed] [Google Scholar]
- 28.Dunne A, O'Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE. 2003;171:re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 29.Pillay J, den Braber I, Vrisekoop N, et al. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 2010;116:625–7. doi: 10.1182/blood-2010-01-259028. [DOI] [PubMed] [Google Scholar]
- 30.Pelletier M, Micheletti A, Cassatella MA. Modulation of human neutrophil survival and antigen expression by activated CD4+ and CD8+ T cells. J Leukoc Biol. 2010;88:1163–70. doi: 10.1189/jlb.0310172. [DOI] [PubMed] [Google Scholar]
- 31.Soehnlein O, Zernecke A, Eriksson EE, et al. Neutrophil secretion products pave the way for inflammatory monocytes. Blood. 2008;112:1461–71. doi: 10.1182/blood-2008-02-139634. [DOI] [PMC free article] [PubMed] [Google Scholar]


