Abstract
Objective:
Genetic heterogeneity is common in many neurologic disorders. This is particularly true for the hereditary ataxias where at least 36 disease genes or loci have been described for spinocerebellar ataxia and over 100 genes for neurologic disorders that present primarily with ataxia. Traditional genetic testing of a large number of candidate genes delays diagnosis and is expensive. In contrast, recently developed genomic techniques, such as exome sequencing that targets only the coding portion of the genome, offer an alternative strategy to rapidly sequence all genes in a comprehensive manner. Here we describe the use of exome sequencing to investigate a large, 5-generational British kindred with an autosomal dominant, progressive cerebellar ataxia in which conventional genetic testing had not revealed a causal etiology.
Methods:
Twenty family members were seen and examined; 2 affected individuals were clinically investigated in detail without a genetic or acquired cause being identified. Exome sequencing was performed in one patient where coverage was comprehensive across the known ataxia genes, excluding the known repeat loci which should be examined using conventional analysis.
Results:
A novel p.Arg26Gly change in the PRKCG gene, mutated in SCA14, was identified. This variant was confirmed using Sanger sequencing and showed segregation with disease in the entire family.
Conclusions:
This work demonstrates the utility of exome sequencing to rapidly screen heterogeneous genetic disorders such as the ataxias. Exome sequencing is more comprehensive, faster, and significantly cheaper than conventional Sanger sequencing, and thus represents a superior diagnostic screening tool in clinical practice.
Genetic heterogeneity is common in inherited cerebellar ataxias with over 36 genetic loci known to cause spinocerebellar ataxia (SCA) and over 100 genes that primarily present with ataxia (http://neuromuscular.wustl.edu/ataxia/aindex.html). Only a relatively few hereditary ataxia syndromes are associated with distinctive clinical features that indicate a particular ataxia gene. Thus, even neurologists specialized in the care of patients with cerebellar disease are frequently left with long lists of putative genes when dealing with such a patient. Screening such a long list of genes is costly, as well as time-consuming, and genetic testing for some of the rarer and difficult to sequence genes is not commercially available. The net result is that a genetic diagnosis cannot be achieved in at least 40% of ataxia cases, even though a genetic cause is heavily suspected.1
Recently developed genomic techniques provide the opportunity to screen large proportions of the genome in a short time frame. Exome sequencing, which targets the coding portion of the genome, offers an affordable strategy to comprehensively sequence all genes in the human genome.2,3
Here we report our genetic analysis of a large British kindred with autosomal dominant cerebellar ataxia. After excluding the most common genes responsible for heritable cerebellar ataxia by standard screening methods, we performed exome sequencing to find the causative gene. In doing so, we demonstrate that exome sequencing is an efficient and cost-effective method for screening heterogeneous diseases such as ataxia.
METHODS
Samples.
We identified a 5-generation family with autosomal dominant SCA (19 affected, 15 unaffected individuals). Twenty family members were examined by a neurologist specialized in ataxias and 2 investigated for acquired causes of ataxia (H.H.). In patient VI-14, the available diagnostic ataxia gene screening was carried out for SCA1, SCA2, SCA3, SCA6, SCA7, SCA8, SCA10, SCA11, SCA12, SCA15/16, SCA17, POLG1, and DRPLA and a CGH array to reveal large structural changes.4–16
Standard protocol approvals, registrations, and patient consents.
All examined family members gave written informed consent and the study was approved by the appropriate institutional review board (IRB/ethics 06/N076).
Sequencing.
Exome sequencing was performed in one affected individual (IV-14) using NimbleGen Sequence Capture technology (2.1 Human Exome v1.0) according to the manufacturer's instructions (Roche NimbleGen, Madison, WI). The enriched libraries underwent 50 base pair, paired-end sequencing on a HiSeq2000 next-generation sequencing platform (Illumina, San Diego, CA). The sequence data were aligned to the reference human genome (UCSC hg18) and variant calling used the Genome Analysis Toolkit (Broad Institute, Cambridge, MA). After filtering PCR duplicates, previously reported variants (http://browser.1000genomes.org/index.html; http://www.ncbi.nlm.nih.gov/snp, build 130) and synonymous variants were removed. For potential disease-causing variants protein alteration was predicted using SIFT (http://sift.jcvi.org/), presence excluded in 221 neurologically healthy control exomes, and the variant was confirmed using Sanger sequencing (see appendix e-1 on the Neurology® Web site at www.neurology.org for details). A total of 36 known ataxia genes were analyzed for sequencing quality in the exome data from 16 samples as described in e-Methods.
RESULTS
Clinical characteristics.
The pedigree was consistent with an autosomal dominant inheritance pattern (figure e-1). Twenty family members were examined, of which 10 showed signs of cerebellar ataxia (table e-1). Acquired causes of ataxia were excluded by history and investigations in 2 individuals (VI-10, VI-14). Mean age at symptom onset was 37 years (range 15–68 years), and the clinical presentation was a relatively pure cerebellar ataxia predominantly affecting lower limb coordination and speech.17 The age at symptom onset varied significantly within the family; ataxia was only revealed on neurologic examination in some patients. Symptom progression was slow with an overall benign disease course. Two affected members had unilateral mild ptosis (IV-14, IV-10), and one patient had concomitant mild parkinsonism (IV-7). Individual V-9 rapidly developed a very severe ataxia during pregnancy at the age of 24 years (24 weeks gestational age); she plateaued after delivery and has remained severely affected since. She was considered phenotypically different from other family members, as detailed in the supplementary data.
Sequencing.
We sequenced the exome of patient IV-14, which provided >10-fold coverage in 87% of her exome. We identified 22,119 variants of which 1,061 were novel (844 homozygous, 217 heterozygous). In this list, we noted a novel c.76A>G mutation in exon 1 of the PRKCG gene, which is known to cause SCA14.18 The PRKCG gene had previously been analyzed using DHPLC-based WAVE analysis and found to be negative. This variant results in a change from the polar basic amino acid arginine to a hydrophobic, uncharged glycine residue at codon 26 (p.Arg26Gly). This variant is not reported in db-SNP or the 1,000 genome project and is absent in 221 control exomes. Sanger sequencing confirmed the mutation and demonstrated disease segregation in all investigated family members (n = 19; figure e-1), giving a lod score of 5.7 for the mutation. Patient V-9 with a different ataxia phenotype did not carry the mutation.
Ataxia locus screening.
To assess the power of exome sequencing in detecting mutations, we investigated 36 known recessive and dominant ataxia genes for coverage and sequence alignment in the exome data generated for 16 patients (table e-2). Of these 36 genes, 3 were not captured by the NimbleGen Sequence Capture Kit, and in an additional 5 genes the disease-causing variants were noncoding and therefore not targeted in the capture array. Furthermore, we found that the exome data had multiple alignment errors and poor coverage in the region of the known CAG and other disease-causing repeats. The length of expanded coding or noncoding repeats therefore cannot be assessed accurately in the SCAs using exome sequencing.
The other 21 ataxia genes showed good overall coverage with an average read depth above 10 in all of these genes and above 30 in 17 out of 21. Pathogenic mutations in these genes are predominantly reported to be coding nucleotide substitutions, small indels, and splice site mutations, which can all be reliably detected by exome sequencing.
DISCUSSION
We used exome sequencing to identify the causative mutation in a large British family with a pure autosomal dominant cerebellar ataxia, the most heterogeneous group of inherited ataxia. We had previously excluded 13 known ataxia genes for which routine screening tests were available. Exome sequencing data analysis revealed a novel mutation in the SCA14 gene (PRKCG) consistent with this phenotype and the mutation showed complete disease segregation among the 19 members of the family screened. This novel c.76A>G (p.Arg26Gly) mutation is predicted to be protein damaging and occurs in a highly conserved nucleotide (figure 1C and figure 2). We therefore conclude that the p.Arg26Gly variant is disease-causing in this ataxia family.
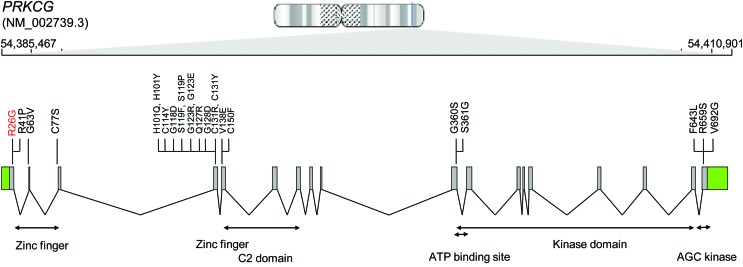
Figure 1. PRKCG gene and mutations.
Shown is the PRKCG locus on chromosome 19. Previously reported nonsynonymous mutations are indicated using black font and the novel p.Arg26Gly mutation is indicated in red.
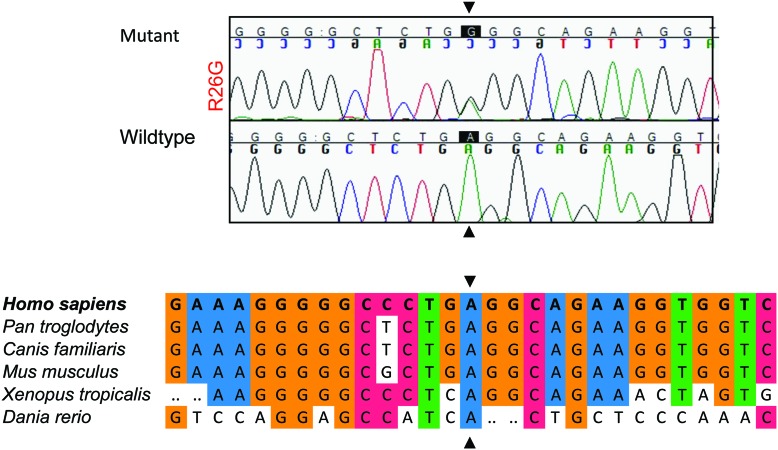
Figure 2. The novel p.Arg26Gly PRKCG mutation.
An electropherogram of the c.76A>G (p.Arg26Gly) mutation in patient IV-14 and sequence conservation plots at the mutated site across different species are displayed.
Exome sequencing reveals a large number of variants (∼20,000) normally present in any individual. Trying to identify a disease-causing mutation in such a large pool of variants is a major challenge. Filtering out variants reported as nonpathogenic in databases such as dbSNP is able to reduce the number of potential mutations to several hundreds but also harbors the risk of excluding pathogenic mutations present in nonmanifesting individuals. To further restrict the list of potential mutations linkage data from large informative kindreds is often necessary.
A potential application of exome sequencing is in medical diagnostics for the screening of heterogeneous genetic diseases such as genetic ataxias. A large number of known disease-causing genes can be investigated in a single experiment. Specifically looking in such candidate genes greatly reduces the list of variants discovered by exome sequencing and will often allow the accurate diagnosis in a single individual as we have shown in our SCA14 patient.
Thus the method of exome sequencing offers a potential alternative to currently available diagnostic sequencing tests. Here we were able to show that the majority of ataxia genes were well covered by this technology. The ability to screen for mutations with exome sequencing does vary depending mainly on the mutational mechanism. Coding nucleotide substitutions, splice site changes, and small indels will be covered19,20 but by way of design, exome sequencing does not routinely sequence noncoding regions and is therefore not suitable for investigating intronic variants unless in close proximity of a targeted exon. However, custom-made libraries could be designed to target these regions. Repetitive sequence stretches, such as triplet repeats, cannot be determined accurately, though advances in sequencing technologies with longer read lengths may resolve this technical issue in the future.
We demonstrated the utility of exome sequencing to rapidly screen a large number of disease-causing genes in a heterogeneous disease such as ataxia by identifying a novel mutation in the PRKCG gene known to cause SCA14. The majority of known ataxia genes can be investigated with this method, and therefore it harbors great potential as a tool for rapid and comprehensive screening of such patients. For a clinical diagnostic application, however, sensitivity and specificity of exome sequencing would still have to be validated in a larger ataxia cohort. Furthermore, we anticipate that costs of exome sequencing will continue to decrease as next-generation sequencing becomes more widely available (currently, ∼$1,500 per patient for exome data production and analysis) and thus the method will be more cost-efficient than a series of individual gene tests, each costing between hundreds and several thousand dollars. As this happens, exome sequencing will become an increasingly important and more widely available test in the investigation of even the most heterogeneous disorders.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the patients and family who supported this work. They also thank the following for grant support: The Medical Research Council (H.H. and N.W.), The MSA Trust (A.S.), Ataxia UK (H.H.), and The Wellcome Trust. This work was undertaken at UCLH/UCL, which received a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme.
GLOSSARY
- SCA
spinocerebellar ataxia
Footnotes
AUTHOR CONTRIBUTIONS
This study was designed and funding obtained by H.H., A.B.S., J.H., H.J.F., and D.H., H.H., A.S., N.W.W., and J.B. collected samples and assessed patients clinically. A.S., S.W.S., and J.O.J. conducted experiments. J.R.B., A.S., S.W.S., B.J.T., A.T., J.D., and V.P. performed data analysis. The manuscript was written by H.H., A.S., S.W.S., and B.J.T.
DISCLOSURE
A. Sailer, S. Scholz, J.R. Gibbs, A. Tucci, J. Johnson, N. Wood, V. Plagnol, H. Hummerich, J. Ding, J. Brown, D. Hernandez, J. Hardy, and H. Federoff report no disclosures. B. Traynor received research support from the ALS Association, The Packard Center for ALS Research, Microsoft Research, Federazione Italiana Giuoco Calcio (FIGC), and The Myasthenia Gravis Foundation. A. Singleton and H. Houlden report no disclosures. Go to Neurology.org for full disclosures.
REFERENCES
- 1. Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol 2010; 9: 885– 894 [DOI] [PubMed] [Google Scholar]
- 2. Johnson JO, Mandrioli J, Benatar M, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2010; 68: 857– 864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009; 461: 272– 276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Orr HT, Chung MY, Banfi S, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet 1993; 4: 221– 226 [DOI] [PubMed] [Google Scholar]
- 5. Houlden H, Johnson J, Gardner-Thorpe C, et al. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat Genet 2007; 39: 1434– 1436 [DOI] [PubMed] [Google Scholar]
- 6. Kawaguchi Y, Okamoto T, Taniwaki M, et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet 1994; 8: 221– 228 [DOI] [PubMed] [Google Scholar]
- 7. Pulst SM, Nechiporuk A, Nechiporuk T, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet 1996; 14: 269– 276 [DOI] [PubMed] [Google Scholar]
- 8. Zhuchenko O, Bailey J, Bonnen P, et al. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat Genet 1997; 15: 62– 69 [DOI] [PubMed] [Google Scholar]
- 9. David G, Abbas N, Stevanin G, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet 1997; 17: 65– 70 [DOI] [PubMed] [Google Scholar]
- 10. Nakamura K, Jeong SY, Uchihara T, et al. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum Mol Genet 2001; 10: 1441– 1448 [DOI] [PubMed] [Google Scholar]
- 11. Koob MD, Moseley ML, Schut LJ, et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat Genet 1999; 21: 379– 384 [DOI] [PubMed] [Google Scholar]
- 12. Matsuura T, Yamagata T, Burgess DL, et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat Genet 2000; 26: 191– 194 [DOI] [PubMed] [Google Scholar]
- 13. Holmes SE, O'Hearn EE, McInnis MG, et al. Expansion of a novel CAG trinucleotide repeat in the 5′ region of PPP2R2B is associated with SCA12. Nat Genet 1999; 23: 391– 392 [DOI] [PubMed] [Google Scholar]
- 14. van de Leemput J, Chandran J, Knight MA, et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet 2007; 3: e108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koide R, Ikeuchi T, Onodera O, et al. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA). Nat Genet 1994; 6: 9– 13 [DOI] [PubMed] [Google Scholar]
- 16. Zeviani M, Servidei S, Gellera C, et al. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature 1989; 339: 309– 311 [DOI] [PubMed] [Google Scholar]
- 17. Harding AE. The clinical features and classification of the late onset autosomal dominant cerebellar ataxias: a study of 11 families, including descendants of the “the Drew family of Walworth. ” Brain 1982; 105: 1– 28 [DOI] [PubMed] [Google Scholar]
- 18. Yabe I, Sasaki H, Chen D-H, et al. Spinocerebellar ataxia type 14 caused by a mutation in protein kinase C gamma. Arch Neurol 2003; 60: 1749– 1751 [DOI] [PubMed] [Google Scholar]
- 19. Asan NFN, Xu Y, Jiang H, et al. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol 2011; 12: R95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clark MJ, Chen R, Lam HYK, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol 2011; 29: 908– 914 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.