Abstract
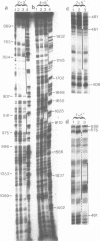
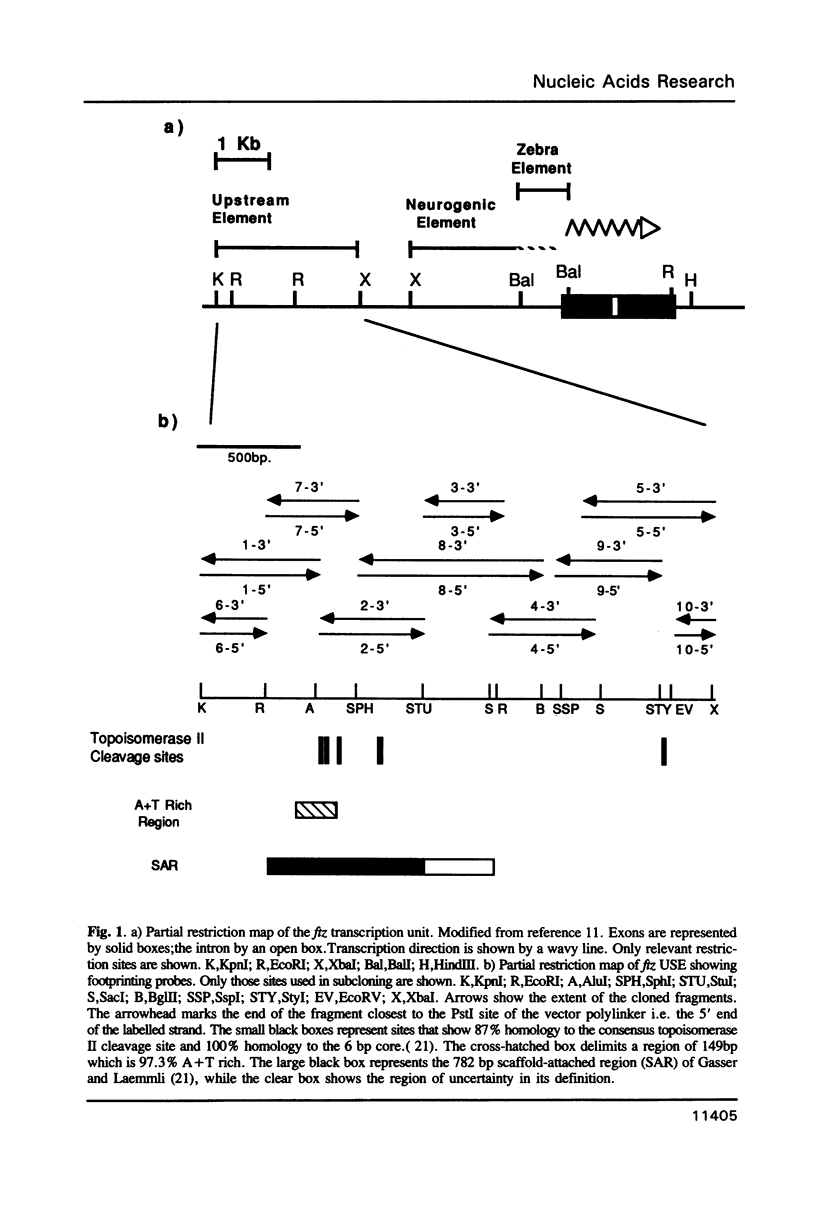
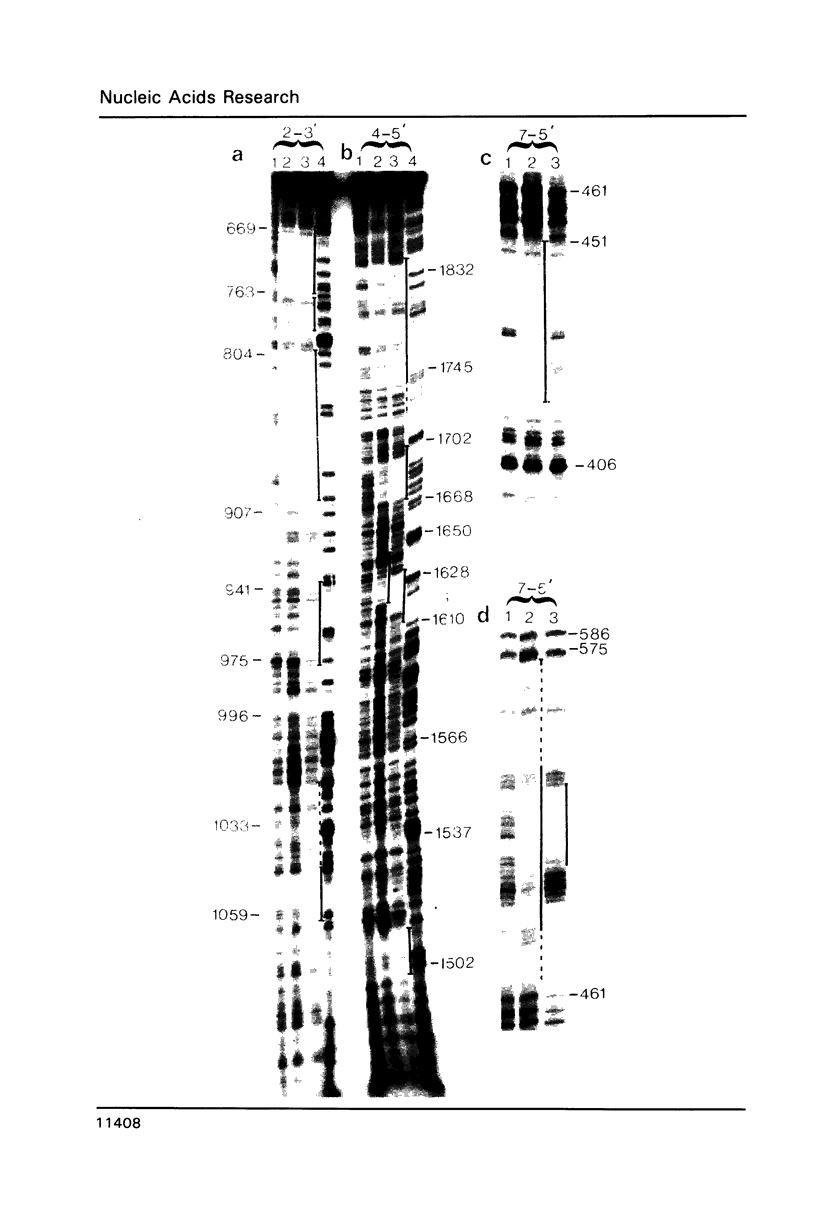
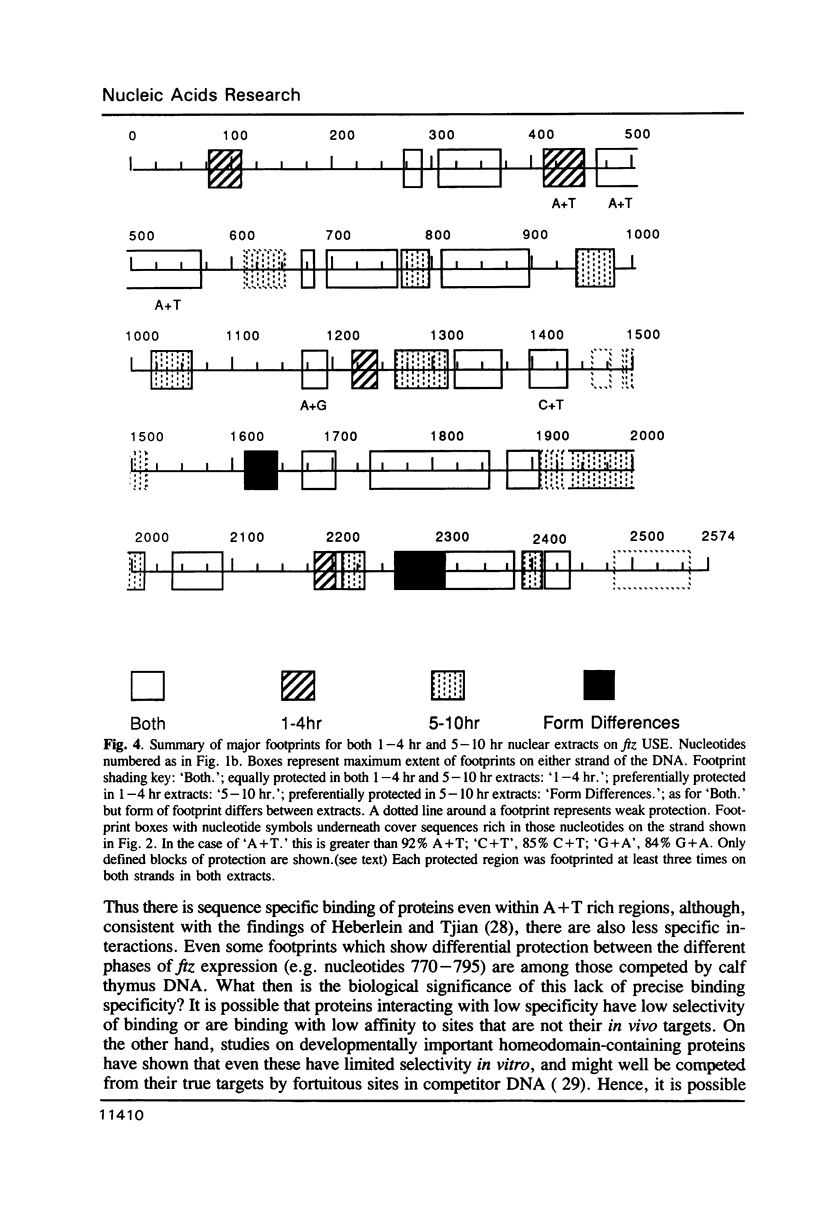
With a view to identifying proteins that regulate the expression of the Drosophila ftz gene we have sequenced its enhancer-like upstream element (USE) and determined the binding sites for embryonic nuclear proteins within this region by in vitro DNAaseI footprinting. We find that greater than 50% of this element is bound by nuclear protein. By footprinting and gel-retardation studies in embryonic extracts from different developmental stages, we have characterised a number of USE/protein complexes whose nature alters in concert with changes in the ftz expression pattern, suggesting that these USE-binding proteins may be involved in the regulation of gene activity. In some cases this suggestion is substantiated by the observation that the protected DNA sequences show homology to the binding sites for ftz regulating DNA-binding proteins such as the pair-rule gene product even-skipped.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987 Sep;101(1):1–22. [PubMed] [Google Scholar]
- Carroll S. B., Scott M. P. Localization of the fushi tarazu protein during Drosophila embryogenesis. Cell. 1985 Nov;43(1):47–57. doi: 10.1016/0092-8674(85)90011-x. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., Scott M. P. Zygotically active genes that affect the spatial expression of the fushi tarazu segmentation gene during early Drosophila embryogenesis. Cell. 1986 Apr 11;45(1):113–126. doi: 10.1016/0092-8674(86)90543-x. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., Winslow G. M., Schüpbach T., Scott M. P. Maternal control of Drosophila segmentation gene expression. Nature. 1986 Sep 18;323(6085):278–280. doi: 10.1038/323278a0. [DOI] [PubMed] [Google Scholar]
- Davidson I., Fromental C., Augereau P., Wildeman A., Zenke M., Chambon P. Cell-type specific protein binding to the enhancer of simian virus 40 in nuclear extracts. Nature. 1986 Oct 9;323(6088):544–548. doi: 10.1038/323544a0. [DOI] [PubMed] [Google Scholar]
- Degelmann A., Hardy P. A., Perrimon N., Mahowald A. P. Developmental analysis of the torso-like phenotype in Drosophila produced by a maternal-effect locus. Dev Biol. 1986 Jun;115(2):479–489. doi: 10.1016/0012-1606(86)90268-x. [DOI] [PubMed] [Google Scholar]
- Desplan C., Theis J., O'Farrell P. H. The Drosophila developmental gene, engrailed, encodes a sequence-specific DNA binding activity. Nature. 1985 Dec 19;318(6047):630–635. doi: 10.1038/318630a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M., Levine M. Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1987 Nov;1(9):981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. The organisation of chromatin loops: characterization of a scaffold attachment site. EMBO J. 1986 Mar;5(3):511–518. doi: 10.1002/j.1460-2075.1986.tb04240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaul U., Jäckle H. Pole region-dependent repression of the Drosophila gap gene Krüppel by maternal gene products. Cell. 1987 Nov 20;51(4):549–555. doi: 10.1016/0092-8674(87)90124-3. [DOI] [PubMed] [Google Scholar]
- Hafen E., Kuroiwa A., Gehring W. J. Spatial distribution of transcripts from the segmentation gene fushi tarazu during Drosophila embryonic development. Cell. 1984 Jul;37(3):833–841. doi: 10.1016/0092-8674(84)90418-5. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Heberlein U., Tjian R. Temporal pattern of alcohol dehydrogenase gene transcription reproduced by Drosophila stage-specific embryonic extracts. Nature. 1988 Feb 4;331(6155):410–415. doi: 10.1038/331410a0. [DOI] [PubMed] [Google Scholar]
- Hiromi Y., Gehring W. J. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1987 Sep 11;50(6):963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- Hiromi Y., Kuroiwa A., Gehring W. J. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985 Dec;43(3 Pt 2):603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- Hoey T., Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988 Apr 28;332(6167):858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Howard K., Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell. 1986 Mar 28;44(6):949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Ingham P. W., Ish-Horowicz D., Howard K. R. Correlative changes in homoeotic and segmentation gene expression in Krüppel mutant embryos of Drosophila. EMBO J. 1986 Jul;5(7):1659–1665. doi: 10.1002/j.1460-2075.1986.tb04409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988 Sep 1;335(6185):25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Pinchin S. M. Pattern abnormalities induced by ectopic expression of the Drosophila gene hairy are associated with repression of ftz transcription. Cell. 1987 Nov 6;51(3):405–415. doi: 10.1016/0092-8674(87)90636-2. [DOI] [PubMed] [Google Scholar]
- Laughon A., Scott M. P. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984 Jul 5;310(5972):25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Ingham P., Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986 Dec 5;47(5):721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Mahoney P. A., Lengyel J. A. The zygotic segmentation mutant tailless alters the blastoderm fate map of the Drosophila embryo. Dev Biol. 1987 Aug;122(2):464–470. doi: 10.1016/0012-1606(87)90310-1. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A., Lawrence P. A. Parasegments and compartments in the Drosophila embryo. Nature. 1985 Feb 21;313(6004):639–642. doi: 10.1038/313639a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980 Oct 30;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Riddihough G., Pelham H. R. An ecdysone response element in the Drosophila hsp27 promoter. EMBO J. 1987 Dec 1;6(12):3729–3734. doi: 10.1002/j.1460-2075.1987.tb02707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Carroll S. B. The segmentation and homeotic gene network in early Drosophila development. Cell. 1987 Dec 4;51(5):689–698. doi: 10.1016/0092-8674(87)90092-4. [DOI] [PubMed] [Google Scholar]
- Shore D., Stillman D. J., Brand A. H., Nasmyth K. A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987 Feb;6(2):461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., LeBowitz J. H., Baldwin A. S., Jr, Sharp P. A. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell. 1988 Feb 12;52(3):415–423. doi: 10.1016/s0092-8674(88)80034-5. [DOI] [PubMed] [Google Scholar]
- Soeller W. C., Poole S. J., Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988 Jan;2(1):68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature. 1988 Mar 17;332(6161):281–284. doi: 10.1038/332281a0. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Lamond A. I., Mace H. A., Berman M. L. RNA polymerase interactions with the upstream region of the E. coli tyrT promoter. Cell. 1983 Nov;35(1):265–273. doi: 10.1016/0092-8674(83)90229-5. [DOI] [PubMed] [Google Scholar]
- Weir M. P., Kornberg T. Patterns of engrailed and fushi tarazu transcripts reveal novel intermediate stages in Drosophila segmentation. Nature. 1985 Dec 5;318(6045):433–439. doi: 10.1038/318433a0. [DOI] [PubMed] [Google Scholar]