Mutations in over 50 genes have been associated with Charcot-Marie-Tooth (CMT) disease. As new genes continue to be discovered, the task of sequencing each of them individually with traditional techniques becomes increasingly burdensome. In addition, these approaches have disadvantages that may reduce their sensitivity. Exome sequencing uses targeted sequence capture of exons and next-generation sequencing to evaluate the sequence of all exons simultaneously. Here, we report a family with the CMT2C phenotype in whom exome sequencing revealed a novel mutation in the transient receptor potential vanilloid 4 (TRPV4) gene missed by previous Sanger sequencing.1
Methods.
Standard protocol approvals, registrations, and patient consents.
Subjects were evaluated after giving written consent to a National Institute of Neurological Disorders and Stroke Institutional Review Board–approved protocol. DNA extracted from peripheral blood was sequenced on a Genome Analyzer IIx platform (Illumina, CA). Sequence variants were confirmed by Sanger sequencing using newly designed primers.
Cellular studies.
Calcium imaging and cell death assays were performed using transiently transfected HEK293 cells as previously described.1 Statistical significance was determined using 2-tailed unpaired t tests.
Results.
Exome sequencing reveals a novel mutation in a family with CMT2C.
We evaluated 9 family members (figure 1A), 3 of whom exhibited features consistent with the CMT2C phenotype,1,2 including progressive distal limb muscle weakness and atrophy, hoarseness of voice, and stridor on exertion (table e-1 on the Neurology® Web site at www.neurology.org). Nerve conduction studies confirmed an axonal neuropathy with phrenic nerve involvement; laryngoscopy showed reductions in vocal fold opening and closing. Two individuals had scoliosis, but skeletal dysplasia was not evident on bone radiographs. One individual had bilateral sensorineural hearing loss.
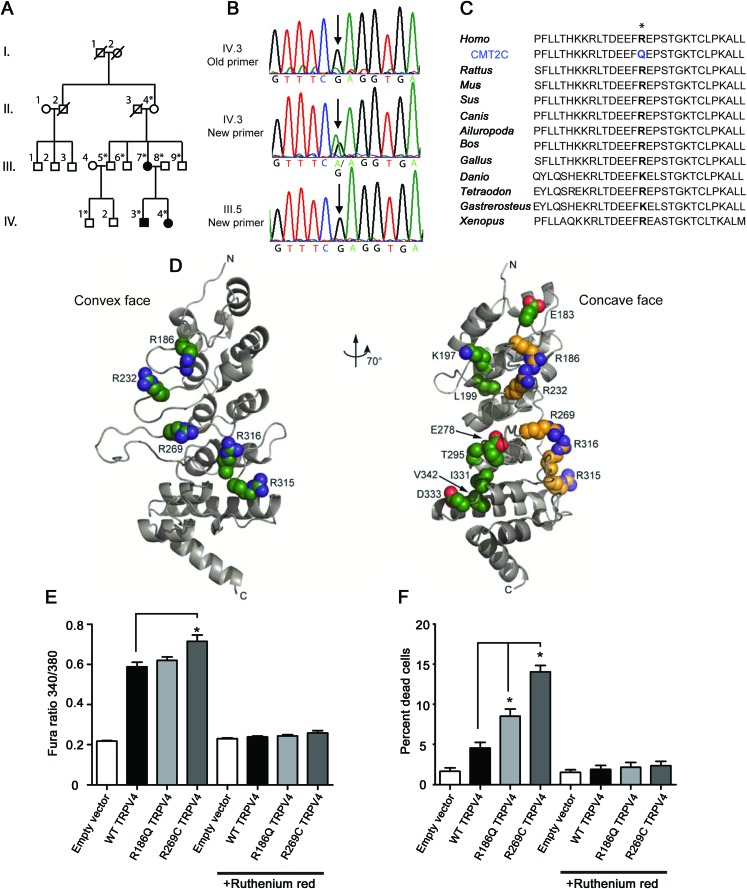
Figure 1. A family with CMT2C has a novel mutation in TRPV4.
(A) Pedigree of the family shows vertical transmission of the disease (white = unaffected, black = affected, *DNA collected). (B) Chromatographs show the c.557G>A mutation initially missed by Sanger sequencing and later identified by exome sequencing in an affected individual. This sequence variant was not apparent using the original primers (top; the “A” variant peak is comparable to the background noise), but became clearly visible when primers that do not contain single nucleotide polymorphisms (SNPs) were used to amplify the genomic DNA (middle). The sequence variant is not present in unaffected individuals (bottom). (C) The R186 residue in TRPV4 is highly conserved across a wide range of species. (D) Like previously described neuropathy-causing TRPV4 mutations, the R186 residue is situated on the convex face of the TRPV4 ankyrin repeat domain (ARD). In contrast, those skeletal dysplasia mutations occurring in the TRPV4 ARD localize to its concave face (at left: green = neuropathy mutations; at right: yellow = neuropathy mutations, green = skeletal dysplasia mutations). (E) Basal calcium levels are modestly increased in HEK293 cells expressing R186Q-TRPV4 and significantly increased in cells expressing R269C-TRPV4, compared to cells expressing WT-TRPV4 for 24 hours. The TRP channel antagonist ruthenium red completely blocks this calcium influx (*p < 0.05, n = 8 per group). (F) Approximately 10% of cells expressing R186Q-TRPV4 and 15% of cells expressing R269C-TRPV4 compared to approximately 5% of cells expressing WT-TRPV4 were dead at 36 hours post-transfection (*p < 0.01, n = 3 per group). Treatment with ruthenium red completely blocked cellular toxicity. Cells were incubated with 2 μM calcein AM and 4 μM EthD-1 for 30 minutes at room temperature and the percentage of dead cells calculated using a Zeiss AxioImager Z1 microscope and AxioVision 4.6 software.
Previous Sanger sequencing failed to identify TRPV4 mutations in this family (figure 1B), designated family 3.1 In the present study, we performed exome sequencing in 1 affected individual and, surprisingly, identified a novel sequence variant (c.557G>A) in TRPV4. No significant variants were identified in other CMT-causing genes. A review of the Sanger sequencing primers used to amplify this region of TRPV4 revealed that the forward and reverse primers each contained a single nucleotide polymorphism (SNP). Further Sanger sequencing with newly designed primers revealed the c.557G>A sequence change in all 3 individuals with CMT2C, but not in the 6 unaffected individuals (figure 1B), nor in 200 controls. This nucleotide change results in an arginine to glutamine substitution at amino acid 186 (R186Q), a highly conserved residue of the TRPV4 protein (figure 1C), a calcium-permeable cation channel. Like previously described neuropathy-causing TRPV4 mutations, R186 is an exposed arginine residue situated on the convex face of the TRPV4 ankyrin repeat domain (ARD) (figure 1D),1,2 a domain mediating regulatory interactions.3
Expression of R186Q-TRPV4 causes cellular toxicity.
Neuropathy-causing TRPV4 mutations (e.g., R269C) have been shown to increase TRPV4 channel activity, leading to elevated intracellular calcium levels and cellular toxicity.1 Here, we examined HEK293 cells expressing wild-type (WT), R186Q, and R269C forms of TRPV4. As previously reported, cells expressing R269C-TRPV4 showed significantly increased basal calcium levels (figure 1E) and cellular toxicity (figure 1F; figure e-1) compared to cells expressing WT-TRPV4. Cells expressing R186Q-TRPV4 showed a slight increase in calcium levels at 24 hours and significantly increased cell death at 36 hours (figure 1, E and F; figure e-1). Both effects were completely abrogated by the TRP channel antagonist ruthenium red (figure 1, E and F), suggesting the R186Q mutation increases constitutive TRPV4 activity.
Discussion.
CMT2C and forms of distal spinal muscular atrophy are caused by dominant missense mutations in TRPV4.1,2,4–6 TRPV4 mutations also cause forms of autosomal dominant skeletal dysplasia.7 Here, we report a novel TRPV4 mutation in a family with the CMT2C phenotype and no evidence of skeletal dysplasia. The localization of the R186Q substitution to the convex face of the TRPV4 ARD, a face on which skeletal dysplasia-causing mutations have not been described, provides further evidence that this region is of particular importance to TRPV4 function in the peripheral nervous system. This mutation was not initially detected by traditional Sanger sequencing, but was identified by exome sequencing, a technology being used increasingly to detect mutations in mendelian diseases. The initial Sanger sequencing primers used to amplify the relevant TRPV4 genomic region overlie known SNPs that may be in linkage disequilibrium with the mutation, likely leading to monoallelic amplification of the WT allele. Exome sequencing technology uses multiple sequence alignments of the same gene fragment, thus avoiding the false negatives that can be encountered during Sanger sequencing.
The identification of the R186Q mutation in TRPV4 by exome sequencing both highlights the importance of the ARD in TRPV4 function in the peripheral nervous system and emphasizes the advantages of rapidly emerging sequencing technologies.
Supplementary Material
Footnotes
Editorial, page 112
Supplemental data at www.neurology.org
Author contributions: Guida Landouré contributed to patient characterization, completed some sequencing, mutagenesis, and cell death assays, and drafted the manuscript. Jeremy M. Sullivan completed cell death assays and cell calcium imaging. Janel O. Johnson performed exome sequencing. Clare H. Munns performed cell calcium imaging. Yijun Shi performed sequencing and cloning. Oumarou Diallo sequenced controls. J. Raphael Gibbs analyzed exome sequencing data. Rachelle Gaudet performed the TRPV4 structural analyses. Christy L. Ludlow contributed to patient recruitment and laryngeal evaluations. Kenneth H. Fischbeck examined patients and interpreted genetic analysis data. Bryan J. Traynor performed exome sequencing. Barrington G. Burnett designed experiments and completed mutagenesis and cell death assays. Charlotte J. Sumner directed the study, examined patients, performed cell death assays, and completed the manuscript.
Acknowledgment: The authors thank the patients and their families for participating in this study; Alice Schindler for aid in patient characterization; Mary Kay Floeter and Tanya Lehky for performing neurophysiologic studies; Jim Nagle and Deborah Kauffmann at the NINDS DNA sequencing facility for help with sequencing; and Stacie Anderson at the NHGRI/DIR Flow Cytometry Core for assistance with cell death analysis.
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
References
- 1. Landouré G, Zdebik AA, Martinez TL, et al. Mutations in TRPV4 cause Charcot-Marie-Tooth disease type 2C. Nat Genet 2010;42:170–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zimoń M, Baets J, Auer-Grumbach M, et al. Dominant mutations in the cation channel gene transient receptor potential vanilloid 4 cause an unusual spectrum of neuropathies. Brain 2010;133:1798–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phelps CB, Wang RR, Choo SS, Gaudet R. Differential regulation of TRPV1, TRPV3, and TRPV4 sensitivity through a conserved binding site on the ankyrin repeat domain. J Biol Chem 2010;285:731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deng HX, Klein CJ, Yan J, et al. Scapuloperoneal spinal muscular atrophy and CMT2C are allelic disorders caused by alterations in TRPV4. Nat Genet 2010;42:165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Auer-Grumbach M, Olschewski A, Papic L, et al. Alterations in the ankyrin domain of TRPV4 cause congenital distal SMA, scapuloperoneal SMA and HMSN2C. Nat Genet 2010;42:160–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen DH, Sul Y, Weiss M, et al. CMT2C with vocal cord paresis associated with short stature and mutations in the TRPV4 gene. Neurology 2010;75:1968–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guilak F, Leddy HA, Liedtke W. Transient receptor potential vanilloid 4: The sixth sense of the musculoskeletal system? Ann NY Acad Sci 2010;1192:404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.