Abstract
DNA repair and transcription process complex nucleic acid structures. The mammalian cell can cross-utilize select components of either pathway to respond to general or special situations arising in either path. These functions comprise activity networks capable of addressing unique requirements for each process. Here, we discuss examples of such networks that are tailored to respond to the demands of both DNA repair and transcription.
Keywords: Chromatin Remodeling, DNA Repair, Post-translational Modification, Transcription, Tumor Suppressor Gene, Orphan Nuclear Receptors, PARP
Introduction
The mammalian genome encodes many damage-responsive elements. It is established that p53, AP-1, and GADD45, for example, among numerous other mammalian proteins, have increased expression following DNA damage (reviewed in Ref. 1). It is not the goal here to examine the regulation of gene expression in response to genome damage or stress; rather, the goal is to note the role of proteins that act in both DNA transcription and repair. Although a protein may have been studied historically for DNA repair or transcription, the two pathways share functions to enact complex cellular processes and may reflect a common processing path.
In the course of our recent NIH-Nuclear Receptor Signaling Atlas (NURSA) multi-investigator project, our own extensive immunoprecipitation/MS study of coregulator complexes involved in transcription and DNA repair revealed substantial numbers of protein-protein interactions that occur between these classes of molecules (2). From overall coverage that extends into ∼40% of the proteome, we recovered >200 gene products that are involved in DNA repair and ∼900 gene products that were annotated as transcriptional regulators by Gene Ontology (GO) and NURSA consortia. In our study alone, strong evidence from reciprocal co-immunoprecipitations suggests that at least 2500 unique pairwise protein associations bridge these major nuclear processes: DNA transcription and DNA repair (2). Thus, this unbiased analysis indicated that proteins long identified as “repair” or “transcription” proteins showed tight interaction. In concert with interactions for proteins previously found to participate in both transcription and repair in small-scale studies, our evidence demonstrates that cross-talk between these two networks clearly occurs at physical and functional levels. In the remainder of this minireview, we summarize seven selected published examples that highlight this conclusion.
Transcription-coupled Repair: CSA and CSB
Early evidence that transcription might be directly involved with DNA repair came from studies of Cockayne syndrome (CS)2 cells demonstrating that there is a prolonged delay in the resumption of RNA synthesis after UV damage in CS cells compared with normal human cells (3, 4). CS cells show sensitivity to UV light compared with normal human fibroblasts, although they have normal nucleotide excision repair (NER) (5).
The resolution of this apparent inconsistency came from observations that DNA repair occurs more rapidly in actively transcribed genes (6). These observations gave rise to the concept of global genomic repair (GGR) and transcription-coupled repair (TCR). TCR occurs on the actively transcribed strand of the genome (reviewed in Ref. 7). The human genes that are needed for TCR were found to be defective in the two complementation groups of CS cells; without the CS factors (CSA and CSB) that couple repair to transcription, repair of the transcribed strand occurs at the slower rate of GGR (8, 9).
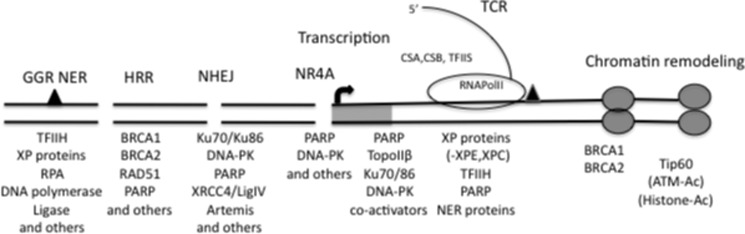
TCR differs from the NER of GGR in the initiation step and in recognition of the DNA damage, with the prototypic damage being the cyclobutane pyrimidine dimer resulting from UV light. Models for TCR assume an arrested RNA polymerase II (RNAPII) when transcription encounters a lesion. The hindrance of the massive transcription apparatus effectively precludes access of the repair proteins. Initiation of repair in TCR depends on recruitment of the CSB protein, which binds to RNAPII and recruits additional factors: CSA, p300, and HMGN1. Transcription factor (TF) IIS associates with arrested RNAPII, as shown by ChIP studies, and appears to stimulate reverse translocation of RNAPII in the presence of CSB (7, 8). CSA is a component of a ubiquitin ligase complex and may promote remodeling of chromatin to assist “bubble” formation (10). In combination with the infrastructure of NER proteins, repair proceeds. However, XPC and XPE proteins, normally required for lesion recognition and for creating a small bubble at the site in NER, are not needed for TCR (11). Overall, although TCR uses the CSA and CSB proteins not needed for normal transcription, the process demonstrates a clear interdependence of transcription and repair and highlights the use of transcription proteins for maintenance of genome stability (Fig. 1).
FIGURE 1.
DNA transcription and repair as an overlapping process. The parallel lines represent the DNA helix with breaks indicated. A given DNA repair process is indicated above the line. Multiple-use proteins are noted below the repair response. Non-transcription repair is to the left of the promoter; the promoter region is represented by the shaded box, and transcription is indicated to the right. RPA, replication protein A; HRR, homologous recombination repair; LigIV, ligase 4; RNAPolII, RNA polymerase II; Ac, acetylated. TFIIH is the transcription preinitiation complex, and TFIIS is the transcription elongation complex.
NER: XPB and XPD, Components of TFIIH
NER deals with bulky adducts in the genome. Primary observations by Cleaver in the 1960s (reviewed in Ref. 12) linked deficiency of NER to the disease xeroderma pigmentosum (XP) and carcinogenesis. Understanding the complex NER pathway in mammals came decades later, awaiting the merger of independent lines of studies involving yeast mutants deficient in repair (reviewed in Refs. 13 and 14), categorization of XP patients into complementation groups that indicated distinct genetic defects (reviewed in Ref. 15), and rodent cell systems with high DNA transformation efficiency (reviewed in Ref. 16). Unexpectedly, when mammalian genes responsible for the defects in XP were identified, two of them were found to be core components of the initiating TFIIH: XPB and XPD (17). Both of these proteins are helicases; XPB proceeds 3′–5′, and XPD proceeds 5′–3′, unwinding the DNA duplex in an ATP-dependent process. At the site of the DNA adduct, XPB and XPD relax the DNA enough for repair to proceed. The XPD helicase activity is required for NER but is dispensable for transcription (18). The other subunits of TFIIH also appear to be utilized for NER. These observations demonstrate that major components of the transcription machinery are specifically required for normal genome maintenance and repair.
Non-homologous End Joining and Regulation of Transcription: DNA-dependent Protein Kinase, Ku70, Ku86, and Topoisomerase IIβ
In contrast to the observations demonstrating that components of transcription act in NER, components of DNA repair also have been shown to be active in transcription. Non-homologous end joining (NHEJ) is a repair response for DNA double-strand breaks (DSBs) that leads to rejoining of DNA strand ends. Defects in NHEJ also produce immune deficiency secondary to failure of V(D)J recombination. NHEJ utilizes the DNA-dependent protein kinase (DNA-PK) catalytic subunit and proteins Ku70 and Ku86, along with other proteins, including XRCC4 (x-ray repair cross-complementing component 4)/ligase 4 (reviewed in Refs. 19–21). DNA-PK is a very abundant nuclear protein in all cell types, and its kinase activity is stimulated 500-fold by DSBs (22).
DNA-PK has also been implicated in the regulation of transcription. We noted a requirement for DNA-PK phosphorylation of the progesterone receptor during transcription at a progesterone-regulated promoter (23), and additional similar evidence has since been reported (24–26). Transcription at endocrine nuclear receptor-binding sites has been shown to require DNA topoisomerase IIβ (topoIIβ) and Ku70/Ku86 in addition to DNA-PK (27). topoIIβ alters the linking number (excess turns of the duplex over the helical structure) of the DNA in units of two, reducing supercoiling via double-strand cleavage and religation, and has been speculated to have a number of roles, including genome stability (28). The function of topoIIβ in regulation of transcription may be relaxation of the supercoil structure at the promoter, facilitating RNAPII access to the site. topoIIβ is required for transcription of several additional genes investigated, including the androgen receptor, retinoic acid receptor, and thyroid receptor genes (27). Because topoIIβ catalyzes incision and resealing, XRCC4/ligase 4 are not required, as would be the case in DSB repair. Thus, transcription initiation differs significantly from NHEJ while using core components of the NHEJ/DSB repair path. These findings demonstrate that proteins known to be required for NHEJ are required for transcription
ADP-ribosylation in Transcription and Repair: Poly(ADP-ribose) Polymerase
Poly(ADP-ribose) polymerase (PARP) is a DNA repair enzyme that was identified almost 50 years ago (29). In humans, there are 18 genes in the family, with PARP-1 being the prototype and most abundant (30). In response to DNA damage, PARP-1 catalyzes attachment of ADP-ribose moieties from NAD+ substrate to proteins in polymers up to a length of 200 bases (“PARylation”). PARP-1 is a nuclear protein present at high concentrations in cells (up to 2 million copies/cell) and attaches to and is stimulated 500-fold by DNA termini or cruciforms (30–33). DNA damage causes PARP-1 to bind to either DNA single-strand breaks or DSBs via its zinc finger motifs, where it may act as a sensor or signal molecule to recruit repair proteins and also PARylate proteins after undergoing extensive post-translational modification (PTM) (34, 35).
Despite the attention to DNA damage, it had been suspected for some time that the function of PARP-1 might not be restricted to DNA repair (36). During the past decade, numerous interactions with other proteins, including DNA-PK, topoisomerases, core histones, histone H1, NF-κB, and p53, have indicated roles for PARP-1 function in chromatin structure, inflammation, and transcription. PARP proteins modulate chromatin structure by PARylation of histones (37). In particular, the association of PARP-1 and DNA-PK suggests a regulatory interaction; PARP can stimulate DNA-PK activity via PARylation, and PARP can be phosphorylated by DNA-PK (38).
PARP-1 enables transcription at promoter sites for estrogen-responsive genes (27). It appears that PARP-1 is part of a corepressor complex with nucleolin, nucleophosmin, and Hsp70 in the non-induced state but is also part of a coactivator complex when the ligand is present (27, 39). ChIP studies indicate a general role for PARP-1 in all transcription, as analysis by genomic microarrays demonstrated that PARP-1 binds to 90% of RNAPII promoters (40). However, it appears that only ∼3–4% of the transcriptome is regulated directly by PARP-1, primarily positively (41).
Given the very high concentration of PARP-1 as a nuclear protein and its utilization of NAD+ as a cofactor, PARP-1 appears to be a major regulator of NAD+ levels in the cell. PARP-1 regulates the activity of the histone deacetylase SIRT1 by NAD+ pool management (42); PARP-1 and SIRT1 are in competition for the available NAD+. In addition, PARP-2 directly affects transcription of SIRT1 as a negative regulator of the SIRT1 promoter (43). Overall, it appears that the PARP-1 and SIRT proteins may counter-regulate both by manipulation of NAD+ pool size and direct promoter modification and by possible bidirectional regulatory ADP-ribosylation.
The findings show that PARP-1, a “repair protein,” is a primary factor in transcription and PTM of proteins. PARP-1 also might control transcription indirectly by modification of chromatin structure and is a central actor in energy metabolism by regulation of NAD+ levels and sirtuin transcription.
Orphan Nuclear Receptors: NR4A
The NR4A orphan nuclear receptors are transcriptional coregulators that do not have known ligands and are constitutively active (44, 45). The NR4A group participates in genome stability, and its members are tumor suppressors (45). Isolation of proteins interacting with NR4A shows that DNA-PK is associated but does not apparently influence the transcriptional activity of NR4A (46). However, phosphorylation by DNA-PK is required for NR4A action in DNA repair. NR4A expression is increased with DNA damage (DSBs) and NR4A localizes to nuclear foci along with numerous repair proteins, dependent on PARP-1 (46). Depletion of NR4A produces increased DSBs after cellular DNA strand breaks caused by inhibition of topoisomerase I (46). However, the DNA-binding domain of NR4A, required for transcription, is not required for DSB response. These findings demonstrate that the NR4A nuclear receptors (transcriptional coregulators) are required for normal DSB repair and must be modified by both DNA-PK and PARP-1 for activation and localization in the DNA repair process.
Protein Acetylation: Tip60
Histone acetylases (HATs) comprise components of histone-remodeling complexes that acetylate histones at specific sites to regulate transcription (reviewed in Refs. 47 and 48). This action constitutes a complex code for regulation of gene activation. Acetylation of histones also occurs after DNA damage, perhaps to allow access or recruit DNA repair enzymes, serving a signaling function (49, 50).
HAT activities have been demonstrated to target proteins other than histones. As an example, the HAT Tip60, an HIV-1 Tat-interactive protein, has a wide range of activities (51, 52). Tip60 is a component of the chromatin-remodeling complex Nu4A and a member of the MYST protein family and acts as a transcriptional coregulator. HAT proteins can acetylate nuclear receptors (47, 53, 54), and Tip60 directly acetylates the androgen receptor, enhancing transcription (55). Thus, Tip60 acts to regulate transcription indirectly as a regulator of chromatin structure and directly as a transcriptional coactivator for specific promoters.
HATs as a class of enzymes also act in genome repair and stability. Tip60 is known to have specific functions in the DNA damage response; it acetylates ATM (ataxia telangiectasia mutated) prior to ATM autophosphorylation, and the acetylation is a required step for the checkpoint and repair functions of ATM (56, 57). This finding demonstrates a direct role in DNA repair for Tip60. We found that Tip60 acts in the repair of interstrand cross-linking (ICL) and that depletion of Tip60 leads to cellular sensitivity to DNA-cross-linking agents. Moreover, Tip60 can promote apoptosis after ICL damage (58, 59). A possible basis for the role in ICL repair might be that Tip60 modulates chromatin structure in the region of DSBs, a function apparently required for ICL repair (52). These findings show that Tip60, a HAT, is active in chromatin remodeling and protein acetylation, regulating transcription, but is also required for DNA repair and normal genome stability.
Tumor Suppressors: BRCA1 and BRCA2
The breast tumor suppressor genes BRCA1 and BRCA2 do not have simple, precisely identified functions but appear to act in genome stability, DNA repair, and regulation of transcription and chromatin structure (60–63). We found that cells depleted of BRCA1 or BRCA2 demonstrate genome instability, even without DNA damage (64, 65). BRCA1 and BRCA2 are transcriptional coregulators, and genomic studies have defined promoter binding (66, 67). Because BRCA1 and BRCA2 are tumor suppressors, it may be postulated that increased transcriptional activity of oncogenes is one basis for carcinogenic effects following mutations causing loss of function of the BRCA1 or BRCA2 protein. BRCA2 is also negatively regulated by PARP, suggesting another mechanism whereby PARP might influence transcription (68).
BRCA1 associates with numerous DNA repair proteins, as shown by the BASC (BRCA1-associated genome surveillance complex) complex, identified by co-immunoprecipitation (69). We also have identified protein interactions of BRCA1 and BRCA2 by mass spectroscopy, defining a set of interactive proteins, minimal endogenous modules (2). Together, these results support roles in DNA repair and genome stability for BRCA1 and BRCA2. Indeed, the BRCA2 gene is part of the Fanconi anemia pathway and has been identified as the FANCD1 gene (70). BRCA1 also acts in the ICL repair response, functioning downstream of the FANCD2-FANCI ubiquitinated dimer (71, 72). These findings demonstrate that the BRCA proteins have protean primary roles in transcriptional regulation and in DNA repair and genome stability.
Summary and Perspective
From the foregoing examples of systems acting in both transcription and genome repair, several points stand out. First, it appears artificial to separate DNA repair and transcription. Rather, they are processes that deal with similar or identical nucleic acid structures and shared mechanisms. These processes might be better thought of as activity networks than pathways. For such networks, some members will be promiscuous and act in other networks. Such second network activity may not require certain functions of the protein needed for the first network. For example, the action of NR4A in DNA repair versus its action in transcription requires differential PTM.
Second, components of modular protein assemblies may act selectively in more than one process of transcription or genome repair. For example, Tip60 can act as a chromatin-remodeling agent or as a protein acetylase, with ATM as the target. For other processes such as NER, substitutions may occur in the protein modules, such as with the XPD function that is needed for the repair activity of TFIIH, but not for transcription.
Third, for modular protein assemblies, specialized proteins may allow the network to execute a select function, as in the case of the CSA and CSB proteins enabling NER components to support TCR activity. In such a circumstance, the specialized pathway protein may eliminate the need for some of the original network components, as, for example, XPC and XPE not being needed for TCR.
Finally, regulation of energy metabolism and transcriptional elements appear to be closely linked. PARP-1 is a node in both processes, interacting with and likely counter-regulating sirtuins and DNA-PK. At the same time, PARP-1 acts selectively in transcription and broadly in PTM of proteins to enable DNA repair and transcription.
This work was supported, in whole or in part, by National Institutes of Health Grant HD8188 (to B. W. O.).
- CS
- Cockayne syndrome
- NER
- nucleotide excision repair
- GGR
- global genomic repair
- TCR
- transcription-coupled repair
- RNAPII
- RNA polymerase II
- TF
- transcription factor
- XP
- xeroderma pigmentosum
- NHEJ
- non-homologous end joining
- DNA-PK
- DNA-dependent protein kinase
- DSB
- double-strand break
- topoIIβ
- topoisomerase IIβ
- PARP
- poly(ADP-ribose) polymerase
- PTM
- post-translational modification
- HAT
- histone acetylase
- ICL
- interstrand cross-linking.
REFERENCES
- 1. Friedberg E. C., Walker G. C., Seide W., Wood R. D., Schultz R. A., Ellenberger T. (2006) DNA Repair and Mutagenesis, 2nd Ed., pp. 817–837, ASM Press, Washington, D.C. [Google Scholar]
- 2. Malovannaya A., Lanz R. B., Jung S. Y., Bulynko Y., Le N. T., Chan D. W., Ding C., Shi Y., Yucer N., Krenciute G., Kim B. J., Li C., Chen R., Li W., Wang Y., O'Malley B. W., Qin J. (2011) Analysis of the human endogenous coregulator complexome. Cell 145, 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mayne L. V., Lehmann A. R. (1982) Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne syndrome and xeroderma pigmentosum. Cancer Res. 42, 1473–1478 [PubMed] [Google Scholar]
- 4. Lehmann A. R., Kirk-Bell S., Mayne L. (1979) Abnormal kinetics of DNA synthesis in ultraviolet light-irradiated cells from patients with Cockayne syndrome. Cancer Res. 39, 4237–4241 [PubMed] [Google Scholar]
- 5. Mayne L. V., Lehmann A. R., Waters R. (1982) Excision repair in Cockayne syndrome. Mutat. Res. 106, 179–189 [DOI] [PubMed] [Google Scholar]
- 6. Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C. (1985) DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40, 359–369 [DOI] [PubMed] [Google Scholar]
- 7. Hanawalt P. C., Spivak G. (2008) Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 [DOI] [PubMed] [Google Scholar]
- 8. Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J. H. (1992) ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne syndrome and preferential repair of active genes. Cell 71, 939–953 [DOI] [PubMed] [Google Scholar]
- 9. Lommel L., Hanawalt P. C. (1991) The genetic defect in the Chinese hamster ovary cell mutant UV61 permits moderate selective repair of cyclobutane pyrimidine dimers in an expressed gene. Mutat. Res. 255, 183–191 [DOI] [PubMed] [Google Scholar]
- 10. Sarker A. H., Tsutakawa S. E., Kostek S., Ng C., Shin D. S., Peris M., Campeau E., Tainer J. A., Nogales E., Cooper P. K. (2005) Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne syndrome. Mol. Cell 20, 187–198 [DOI] [PubMed] [Google Scholar]
- 11. Mellon I., Hanawalt P. C. (1989) Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature 342, 95–98 [DOI] [PubMed] [Google Scholar]
- 12. Cleaver J. E. (2005) Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer 5, 564–573 [DOI] [PubMed] [Google Scholar]
- 13. Friedberg E. C., Bardwell A. J., Bardwell L., Feaver W. J., Kornberg R. D., Svejstrup J. Q., Tomkinson A. E., Wang Z. (1995) Nucleotide excision repair in the yeast Saccharomyces cerevisiae: its relationship to specialized mitotic recombination and RNA polymerase II basal transcription. Philos. Trans. R Soc. Lond. B Biol. Sci. 347, 63–68 [DOI] [PubMed] [Google Scholar]
- 14. Prakash S., Sung P., Prakash L. (1993) DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 27, 33–70 [DOI] [PubMed] [Google Scholar]
- 15. Diderich K., Alanazi M., Hoeijmakers J. H. (2011) Premature aging and cancer in nucleotide excision repair disorders. DNA Repair 10, 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thompson L. H. (1996) Identifying genes and proteins involved in human DNA repair processes using somatic cell and molecular genetics. Prog. Clin. Biol. Res. 395, 175–199 [PubMed] [Google Scholar]
- 17. Schaeffer L., Roy R., Humbert S., Moncollin V., Vermeulen W., Hoeijmakers J. H., Chambon P., Egly J. M. (1993) DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science 260, 58–63 [DOI] [PubMed] [Google Scholar]
- 18. Lehmann A. R. (2001) The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 15, 15–23 [DOI] [PubMed] [Google Scholar]
- 19. Kong X., Shen Y., Jiang N., Fei X., Mi J. (2011) Emerging roles of DNA-PK besides DNA repair. Cell. Signal. 23, 1273–1280 [DOI] [PubMed] [Google Scholar]
- 20. Mladenov E., Iliakis G. (2011) Induction and repair of DNA double-strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat. Res. 711, 61–72 [DOI] [PubMed] [Google Scholar]
- 21. Allen C., Ashley A. K., Hromas R., Nickoloff J. A. (2011) More forks on the road to replication stress recovery. J. Mol. Cell. Biol. 3, 4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meek K., Dang V., Lees-Miller S. P. (2008) DNA-PK: the means to justify the ends? Adv. Immunol. 99, 33–58 [DOI] [PubMed] [Google Scholar]
- 23. Weigel N. L., Carter T. H., Schrader W. T., O'Malley B. W. (1992) Chicken progesterone receptor is phosphorylated by a DNA-dependent protein kinase during in vitro transcription assays. Mol. Endocrinol. 6, 8–14 [DOI] [PubMed] [Google Scholar]
- 24. Wong R. H., Sul H. S. (2009) DNA-PK: relaying the insulin signal to USF in lipogenesis. Cell Cycle 8, 1977–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wong R. H., Chang I., Hudak C. S., Hyun S., Kwan H. Y., Sul H. S. (2009) A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell 136, 1056–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bozulic L., Hemmings B. A. (2009) PIKKing on PKB: regulation of PKB activity by phosphorylation. Curr. Opin. Cell Biol. 21, 256–261 [DOI] [PubMed] [Google Scholar]
- 27. Ju B. G., Lunyak V. V., Perissi V., Garcia-Bassets I., Rose D. W., Glass C. K., Rosenfeld M. G. (2006) A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 [DOI] [PubMed] [Google Scholar]
- 28. Austin C. A., Marsh K. L. (1998) Eukaryotic DNA topoisomerase IIβ. BioEssays 20, 215–226 [DOI] [PubMed] [Google Scholar]
- 29. Chambon P., Weill J. D., Mandel P. (1963) Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid-synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 11, 39–43 [DOI] [PubMed] [Google Scholar]
- 30. Bürkle A. (2005) Poly(ADP-ribose). The most elaborate metabolite of NAD+. FEBS J. 272, 4576–4589 [DOI] [PubMed] [Google Scholar]
- 31. Kim M. Y., Zhang T., Kraus W. L. (2005) Poly(ADP-ribosyl)ation by PARP-1: “PAR-laying” NAD+ into a nuclear signal. Genes Dev. 19, 1951–1967 [DOI] [PubMed] [Google Scholar]
- 32. Kraus W. L. (2008) Transcriptional control by PARP-1: chromatin modulation, enhancer binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kraus W. L., Lis J. T. (2003) PARP goes transcription. Cell 113, 677–683 [DOI] [PubMed] [Google Scholar]
- 34. Krishnakumar R., Kraus W. L. (2010) The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luo X., Kraus W. L. (2012) On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 26, 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cleaver J. E., Morgan W. F. (1991) Poly(ADP-ribose) polymerase: a perplexing participant in cellular responses to DNA breakage. Mutat. Res. 257, 1–18 [DOI] [PubMed] [Google Scholar]
- 37. Tulin A., Naumova N. M., Menon A. K., Spradling A. C. (2006) Drosophila poly(ADP-ribose) glycohydrolase mediates chromatin structure and SIR2-dependent silencing. Genetics 172, 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruscetti T., Lehnert B. E., Halbrook J., Le Trong H., Hoekstra M. F., Chen D. J., Peterson S. R. (1998) Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem. 273, 14461–14467 [DOI] [PubMed] [Google Scholar]
- 39. Ju B. G., Solum D., Song E. J., Lee K. J., Rose D. W., Glass C. K., Rosenfeld M. G. (2004) Activating the PARP-1 sensor component of the Groucho/TLE1 corepressor complex mediates a CaM Kinase IIδ-dependent neurogenic gene activation pathway. Cell 119, 815–829 [DOI] [PubMed] [Google Scholar]
- 40. Krishnakumar R., Gamble M. J., Frizzell K. M., Berrocal J. G., Kininis M., Kraus W. L. (2008) Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319, 819–821 [DOI] [PubMed] [Google Scholar]
- 41. Ogino H., Nozaki T., Gunji A., Maeda M., Suzuki H., Ohta T., Murakami Y., Nakagama H., Sugimura T., Masutani M. (2007) Loss of Parp-1 affects gene expression profile in a genome-wide manner in ES cells and liver cells. BMC Genomics 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bai P., Cantó C., Oudart H., Brunyánszki A., Cen Y., Thomas C., Yamamoto H., Huber A., Kiss B., Houtkooper R. H., Schoonjans K., Schreiber V., Sauve A. A., Menissier-de Murcia J., Auwerx J. (2011) PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 13, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bai P., Canto C., Brunyánszki A., Huber A., Szántó M., Cen Y., Yamamoto H., Houten S. M., Kiss B., Oudart H., Gergely P., Menissier-de Murcia J., Schreiber V., Sauve A. A., Auwerx J. (2011) PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 13, 450–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao Y., Bruemmer D. (2010) NR4A orphan nuclear receptors: transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler. Thromb. Vasc. Biol. 30, 1535–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ramirez-Herrick A. M., Mullican S. E., Sheehan A. M., Conneely O. M. (2011) Reduced NR4A gene dosage leads to mixed myelodysplastic/myeloproliferative neoplasms in mice. Blood 117, 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Malewicz M., Kadkhodaei B., Kee N., Volakakis N., Hellman U., Viktorsson K., Leung C. Y., Chen B., Lewensohn R., van Gent D. C., Chen D. J., Perlmann T. (2011) Essential role for DNA-PK-mediated phosphorylation of NR4A nuclear orphan receptors in DNA double-strand break repair. Genes Dev. 25, 2031–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang C., Tian L., Popov V. M., Pestell R. G. (2011) Acetylation and nuclear receptor action. J. Steroid Biochem. Mol. Biol. 123, 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peng L., Seto E. (2011) Deacetylation of non-histone proteins by HDACs and the implications in cancer. Handb. Exp. Pharmacol. 206, 39–56 [DOI] [PubMed] [Google Scholar]
- 49. Costelloe T., Lowndes N. F. (2010) Chromatin assembly and signaling the end of DNA repair require acetylation of histone H3 on lysine 56. Subcell. Biochem. 50, 43–54 [DOI] [PubMed] [Google Scholar]
- 50. Zhu Q., Wani A. A. (2010) Histone modifications: crucial elements for damage response and chromatin restoration. J. Cell. Physiol. 223, 283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Squatrito M., Gorrini C., Amati B. (2006) Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 16, 433–442 [DOI] [PubMed] [Google Scholar]
- 52. Xu Y., Price B. D. (2011) Chromatin dynamics and the repair of DNA double-strand breaks. Cell Cycle 10, 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang C., Powell M. J., Popov V. M., Pestell R. G. (2008) Acetylation in nuclear receptor signaling and the role of sirtuins. Mol. Endocrinol. 22, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang C., Fu M., Angeletti R. H., Siconolfi-Baez L., Reutens A. T., Albanese C., Lisanti M. P., Katzenellenbogen B. S., Kato S., Hopp T., Fuqua S. A., Lopez G. N., Kushner P. J., Pestell R. G. (2001) Direct acetylation of the estrogen receptor α hinge region by p300 regulates transactivation and hormone sensitivity. J. Biol. Chem. 276, 18375–18383 [DOI] [PubMed] [Google Scholar]
- 55. Sapountzi V., Logan I. R., Robson C. N. (2006) Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 38, 1496–1509 [DOI] [PubMed] [Google Scholar]
- 56. Sun Y., Jiang X., Chen S., Fernandes N., Price B. D. (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 102, 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun Y., Jiang X., Price B. D. (2010) Tip60: connecting chromatin to DNA damage signaling. Cell Cycle 9, 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hejna J., Holtorf M., Hines J., Mathewson L., Hemphill A., Al-Dhalimy M., Olson S. B., Moses R. E. (2008) Tip60 is required for DNA interstrand cross-link repair in the Fanconi anemia pathway. J. Biol. Chem. 283, 9844–9851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hejna J., Bruun D., Pauw D., Moses R. E. (2010) A FANCD2 domain activates Tip60-dependent apoptosis. Cell Biol. Int. 34, 893–899 [DOI] [PubMed] [Google Scholar]
- 60. Mullan P. B., Quinn J. E., Harkin D. P. (2006) The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene 25, 5854–5863 [DOI] [PubMed] [Google Scholar]
- 61. Rosen E. M., Ma Y., Fan S. (2009) BRCA1 cross-talk with hormone receptors. Cancer Treat. Res. 147, 1–20 [DOI] [PubMed] [Google Scholar]
- 62. Rosen E. M., Fan S., Ma Y. (2006) BRCA1 regulation of transcription. Cancer Lett. 236, 175–185 [DOI] [PubMed] [Google Scholar]
- 63. Venkitaraman A. R. (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108, 171–182 [DOI] [PubMed] [Google Scholar]
- 64. Hussain S., Wilson J. B., Medhurst A. L., Hejna J., Witt E., Ananth S., Davies A., Masson J. Y., Moses R., West S. C., de Winter J. P., Ashworth A., Jones N. J., Mathew C. G. (2004) Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum. Mol. Genet. 13, 1241–1248 [DOI] [PubMed] [Google Scholar]
- 65. Bruun D., Folias A., Akkari Y., Cox Y., Olson S., Moses R. (2003) siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair 2, 1007–1013 [DOI] [PubMed] [Google Scholar]
- 66. Gorski J. J., Savage K. I., Mulligan J. M., McDade S. S., Blayney J. K., Ge Z., Harkin D. P. (2011) Profiling of the BRCA1 transcriptome through microarray and ChIP-chip analysis. Nucleic Acids Res. 39, 9536–9548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gorski J. J., James C. R., Quinn J. E., Stewart G. E., Staunton K. C., Buckley N. E., McDyer F. A., Kennedy R. D., Wilson R. H., Mullan P. B., Harkin D. P. (2010) BRCA1 transcriptionally regulates genes associated with the basal-like phenotype in breast cancer. Breast Cancer Res. Treat. 122, 721–731 [DOI] [PubMed] [Google Scholar]
- 68. Wang J., Bian C., Li J., Couch F. J., Wu K., Zhao R. C. (2008) Poly(ADP-ribose) polymerase-1 down-regulates BRCA2 expression through the BRCA2 promoter. J. Biol. Chem. 283, 36249–36256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., Qin J. (2000) BASC, a supercomplex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14, 927–939 [PMC free article] [PubMed] [Google Scholar]
- 70. Howlett N. G., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., Ikeda H., Fox E. A., D'Andrea A. D. (2002) Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297, 606–609 [DOI] [PubMed] [Google Scholar]
- 71. Patel K. J., Joenje H. (2007) Fanconi anemia and DNA replication repair. DNA Repair 6, 885–890 [DOI] [PubMed] [Google Scholar]
- 72. Kee Y., D'Andrea A. D. (2010) Expanded roles of the Fanconi anemia pathway in preserving genome stability. Genes Dev. 24, 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]