Background: Raf kinase inhibitor protein (RKIP) is a regulator of several distinct kinases, including Raf1 and G protein-coupled receptor kinase 2 (GRK2).
Results: Protein kinase C-mediated phosphorylation of RKIP triggers dimer formation of RKIP, which enables RKIP to switch specificity between Raf1 and GRK2.
Conclusion: Phosphorylation-dependent dimerization of RKIP coordinates specific interactions with Raf1 and GRK2.
Significance: Control switches in a kinase regulator permit specific control of multiple kinase signaling pathways and their downstream functions.
Keywords: G Protein-coupled Receptors (GPCR), MAP Kinases (MAPKs), Raf, Serine Threonine Protein Kinase, Signal Transduction, GRK2, RKIP
Abstract
Proteins controlling cellular networks have evolved distinct mechanisms to ensure specificity in protein-protein interactions. Raf kinase inhibitor protein (RKIP) is a multifaceted kinase modulator, but it is not well understood how this small protein (21 kDa) can coordinate its diverse signaling functions. Raf1 and G protein-coupled receptor kinase (GRK) 2 are direct interaction partners of RKIP and thus provide the possibility to untangle the mechanism of its target specificity. Here, we identify RKIP dimer formation as an important mechanistic feature in the target switch from Raf1 to GRK2. Co-immunoprecipitation and cross-linking experiments revealed RKIP dimerization upon phosphorylation of RKIP at serine 153 utilizing purified proteins as well as in cells overexpressing RKIP. A functional phosphomimetic RKIP mutant had a high propensity for dimerization and reproduced the switch from Raf1 to GRK2. RKIP dimerization and GRK2 binding, but not Raf1 interaction, were prevented by a peptide comprising amino acids 127–146 of RKIP, which suggests that this region is critical for dimer formation. Furthermore, a dimeric RKIP mutant displayed a higher affinity to GRK2, but a lower affinity to Raf1. Functional analyses of phosphomimetic as well as dimeric RKIP demonstrated that enhanced dimerization of RKIP translates into decreased Raf1 and increased GRK2 inhibition. The detection of RKIP dimers in a complex with GRK2 in murine hearts implies their physiological relevance. These findings represent a novel mechanistic feature how RKIP can discriminate between its different interaction partners and thus advances our understanding how specific inhibition of kinases can be achieved.
Introduction
The Raf kinase inhibitor protein (RKIP)3 modulates and controls crucial intracellular signaling networks, including the signaling cascades of Raf/MEK/ERK, NFκB, glycogen synthase kinase-3β, and G protein-coupled receptors (GPCRs) (1–5). RKIP belongs to the evolutionarily conserved phosphatidylethanolamine-binding protein (PEBP) family, which is characterized by the ability to bind phospholipids in vitro (6, 7). This protein family is ubiquitously expressed within a panoply of organisms, including humans, yeast, bacteria, and plants (8). RKIP regulates a variety of physiological processes such as differentiation, cell cycle, apoptosis, and contractile activity of cardiomyocytes (9–12).
The structure of the RKIP/PEBP proteins is characterized by a central β-sheet surrounded by more variable β-strands and α-helices (12–14). These secondary structural elements are connected by long loops. Most members of this protein family, particularly mammalian members of this family, possess a ligand-binding pocket formed by highly conserved amino acid residues. This pocket displays high affinity for small anionic groups, e.g. phosphates, and is implicated in the binding of RKIP to Raf1 (14–18). Due to this binding pocket and due to the ability of RKIP to interfere with various signaling cascades and cellular functions, RKIP has been suggested as a major modulator of kinases involved in signal transduction (12, 19).
Only Raf1, MEK1, and ERK2, which belong to the mitogen-activated protein (MAP) kinase cascade, and GRK2, a kinase involved in GPCR desensitization and signaling specificity, have so far been identified as direct interaction partners of RKIP (1, 2, 4). These proteins are ubiquitously expressed kinases with a prominent role in the regulation of cellular functions and, thus, need to be tightly regulated. The Raf/MEK/ERK cascade is involved in cancerogenesis and in development of cardiac hypertrophy (21, 22). This cascade coordinates gene expression, differentiation, proliferation, and cell survival. RKIP overexpression in cells decreased Raf1-induced transformation and transcription, whereas down-regulation of endogenous RKIP had the opposite effects (1). Because reduced RKIP expression in some types of tumors has been shown to trigger metastasis and tumor progression, RKIP is discussed as a prognostic marker in prostate and colorectal cancer (9, 23).
GRKs catalyze the phosphorylation of cytoplasmic serine and threonine residues of activated GPCRs and thereby initiate recruitment of β-arrestins and subsequent receptor desensitization and internalization (24). In mammalian cells, GRK2 is a major feedback inhibitor of GPCRs and has been shown to be implicated among others in psychiatric disorders, immune diseases, and cardiovascular diseases (25–27). Binding of RKIP to GRK2 inhibits GRK2 kinase activity, which leads to enhanced GPCR signaling. Down-regulation of RKIP in cardiomyocytes attenuated β-adrenergic signaling and contractile activity (4).
The ligand binding pocket of RKIP has been shown to be of particular importance for the Raf1/RKIP interaction (14, 17, 18). Under basal conditions RKIP is bound to Raf1 and the release of RKIP from Raf1 is triggered by protein kinase C (PKC)-mediated phosphorylation of RKIP at serine 153 (4, 28). Phosphorylation of Ser-153 also increases the affinity of RKIP toward its substrate GRK2 (4). However, it is still unclear how this single phosphorylation can cause such a dramatic change of binding specificity in such a small protein.
Here we report that phosphorylation of RKIP by PKC facilitates RKIP dimerization and that dimerization of RKIP is necessary for the switch of RKIP from Raf1 to GRK2. Our results reveal phosphorylation-dependent dimerization of RKIP as an important regulatory mechanism for the binding of RKIP to its different substrates.
EXPERIMENTAL PROCEDURES
Antibodies for Immunodetection of Proteins
For immunoblotting we used the following antibodies directed against: c-Myc tag (sc-789, Santa Cruz), ERK1/2 (9102, Cell Signaling), green fluorescent protein (GFP; G1544, Sigma), GRK2 (sc-13143, Santa Cruz), His tag (2366, Cell Signaling), phospho-ERK1/2 (9101, Cell Signaling), phospho-RKIP (Ser-153; pRKIP(S153)) (2445-1, Epitomics; sc-32622, Santa Cruz), phospho-β2-adrenergic receptor (Ser-355/356; pβ2AR(S355/6)) (sc-22191, Santa Cruz), Raf1 (610152, BD Biosciences), and RKIP (sc-5423 and sc-33184, Santa Cruz). For immunoprecipitation we used an antibody directed against the FLAG tag (F3165, Sigma). As secondary antibodies we used peroxidase-conjugated anti-mouse (115035003, Dianova), anti-rabbit (111035144, Dianova), and anti-goat antibodies (sc-2020, Santa Cruz).
Chemicals and Materials
We used [γ-32P]ATP (Hartmann), bisindolylmaleimide I (GFX; Merck), ECL Plus Western blotting detection reagent (GE Healthcare), sulfo-ethylene glycolbis(succinimidylsuccinate) (sEGS; AppliChem), isoprenaline hydrochloride (Iso; Tocris/Biozol), 4× Laemmli buffer (Roth), nickel-nitrilotriacetic acid-agarose (Ni-NTA-agarose; Qiagen), paraformaldehyde (PFA, AppliChem), polyvinylidene difluoride (PVDF) membrane, protein A-Sepharose (GE Healthcare), protein phosphatase 1 (PP1; New England Biolabs), and 12-O-tetradecanoylphorbol 13-acetate (TPA; AppliChem). For in vitro phosphorylation of RKIP we used PKCδ (Merck). For SDS-PAGE we used as standards peqGOLD protein marker 3® (Peqlab) and polypeptide marker (Bio-Rad).
cDNA Constructs
cDNAs coding for c-Myc- and FLAG-tagged rat wild-type RKIP and all mutants or fragments of RKIP were generated by polymerase chain reaction using HA-rRKIP (4) as template. Constructs were subcloned into pcDNA3 expression vector (Invitrogen) using the restriction sites HindIII and XbaI. cDNAs coding for N-terminal His6-tagged rRKIP (His-RKIP) and RKIP mutants were subcloned into the expression vector pET3c (Novagen) using restriction sites NdeI and BamHI. Plasmids for HA-Raf1 and GRK2 have been described previously (4). The correct sequence of all constructs was confirmed by DNA sequencing.
Cell Culture and Transient Transfection
Human embryonic kidney 293 (HEK293) cells and HEK293 cells with stable overexpression of β2-adrenergic receptors (β2AR) were cultured as previously described (4). 5 h after seeding, cells were transfected with the indicated cDNAs using the calcium phosphate precipitation method (29). 24 h later, cells were serum-starved. Assays were performed 40 h after transfection.
Immunoprecipitation Assays and Immunoblotting
Cells were stimulated with TPA (1 μm, 5 min) as indicated, collected in ice-cold phosphate-buffered saline (PBS, 137 mm NaCl, 2.7 mm KCl, 1.5 mm KH2PO4, 10.1 mm Na2HPO4) supplemented with protease inhibitors (10 μg/ml of soybean trypsin inhibitor, 1 mm benzamidine, 1 mm phenylmethylsulfonyl fluoride) and phosphatase inhibitors (50 mm sodium fluoride, 5 mm sodium pyrophosphate, 1 mm sodium vanadate, 3 mm sodium azide), and sonicated. After centrifugation (14,000 × g, 4 °C, 15 min), the supernatant was incubated with anti-FLAG antibodies bound to Protein A-Sepharose for 2 h at 4 °C. Immunoprecipitated proteins were separated by SDS-PAGE, transferred to PVDF membranes, and visualized using the indicated antibodies similarly as described previously (29).
Protein Purification of RKIP, GRK2, and Rhodopsin
His-RKIP was expressed in Escherichia coli and purified by metal-affinity chromatography using Ni-NTA-agarose (4). GRK2 was expressed in Spodoptera frugiperda (Sf9) cells infected with baculoviruses encoding bovine GRK2 and purified via SP-Sepharose and heparin-Sepharose chromatography as described previously (4). Rhodopsin was isolated from rod outer segments (29).
In Vitro Phosphorylation of RKIP by PKCδ and Subsequent Cross-linking
Purified His-RKIP was incubated with recombinant PKCδ in kinase buffer (20 mm HEPES (pH 7.4), 10 mm MgCl2, 0.1 mm CaCl2, 3 mm β-mercaptoethanol, 500 μm ATP) at 30 °C. The reaction was started by addition of TPA (25 μm). After 30 min, proteins were cross-linked as follows: the reaction was carried out in nonnucleophilic buffer (20 mm HEPES (pH 7.4), 2 mm EDTA) with paraformaldehyde (PFA, 0.6% (w/v), 10 min, 28 °C). The reaction was stopped with 4× Laemmli buffer. Proteins were separated by SDS-PAGE and detected by subsequent Western blot analyses.
Cross-linking Experiments
For cross-linking experiments, HEK293 cells were stimulated with TPA (1 μm, 5 min) and collected in ice-cold PBS supplemented with protease and phosphatase inhibitors (see above) and PFA (0.75% (w/v)). After 7 min of incubation at room temperature, cells were centrifuged (1,700 × g, 3 min at room temperature) and the reaction was stopped by addition of 1.25 m glycine (dissolved in PBS) to the cell pellet. Cells were centrifuged again and the pellet was lysed in lysis buffer (50 mm Tris, 150 mm NaCl, 1% (v/v) Nonidet P-40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 1 mm EDTA). Lysates were sonicated and cell debris was removed by centrifugation (14,000 × g, 30 min, 4 °C). For cross-linking experiments in native tissue, we perfused hearts of wild-type C57BL/6 mice with PBS or PBS supplemented with GFX (1.5 μm) for 3 min followed by perfusion with 4% (w/v) PFA in PBS as cross-linking compound. To analyze RKIP phosphorylation levels, hearts were perfused with PBS or PBS supplemented with GFX (1.5 μm) without subsequent PFA treatment. Left ventricles were placed in 600 mm Tris (dissolved in PBS, pH 7.4) for 10 min (4 °C) and homogenized in the above mentioned lysis buffer. Cell debris was removed by centrifugation (14,000 × g, 15 min, 4 °C). Cross-linking experiments with purified RKIP and GRK2 were performed at a RKIP:GRK2 ratio of 5:1 in nonnucleophilic buffer (20 mm HEPES (pH 7.4), 2 mm EDTA) using sEGS (0.3 mm, 15 min, room temperature). The reaction was quenched with Tris buffer (pH 7.4) at a final concentration of 50 mm. Protein complexes were separated by SDS-PAGE and visualized by Western blotting using antibodies directed against anti-His, anti-Myc, or anti-GRK2 as indicated.
Phosphorylation Assays
The kinase activity of GRK2 was assessed in vitro by phosphorylation of rhodopsin in a total volume of 50 μl buffer (20 mm HEPES (pH 7.4), 2 mm EDTA, 10 mm MgCl2) containing 5 nm GRK2, 400 nm rhodopsin, 50 μm [γ-32P]ATP, and 3 μm wild-type His-RKIP, RKIPSK153/7EE, or RKIPΔ143–146. Rhodopsin activation was initiated by light and phosphorylation proceeded for 10 min at room temperature. Phosphorylated rhodopsin was separated by SDS-PAGE and assessed by autoradiography and phosphorimaging analysis.
Phosphorylation of β2AR was determined in HEK293 cells with stable expression of GFP-tagged β2AR. Cells were transiently transfected with GRK2 and Myc-tagged wild-type RKIP or RKIP mutants and stimulated with isoprenaline (1 μm, 5 min) as indicated. Subsequently, cells were lysed using a buffer containing 50 mm Tris (pH 7.4), 300 mm NaCl, 5 mm EDTA, 1% (v/v) Triton X-100, 10 μg/ml of soybean trypsin inhibitor, 1 mm benzamidine, 1 mm phenylmethylsulfonyl fluoride, 50 mm sodium fluoride, 5 mm sodium pyrophosphate, 1 mm sodium vanadate, and 3 mm sodium azide. After SDS-PAGE, phosphorylated receptors were visualized by immunoblotting using anti-pβ2AR(S355/6) antibodies. Phosphorylation of ERK1/2 was analyzed under the above mentioned conditions using anti-pERK1/2 antibodies.
Dephosphorylation of RKIP
Left ventricles of murine hearts (C57BL/6) were homogenized in dephosphorylation buffer (50 mm HEPES (pH 7.5), 100 mm NaCl, 2 mm DTT, 0.01% Brij 35, 50 μg/ml of aprotinin, 50 μg/ml of leupeptin, 0.5 mm PMSF, 1 mm benzamidine) and cell debris was removed by centrifugation (14,000 × g, 15 min, 4 °C). Heart lysate (100 μg) was incubated with 20 units of PP1 for 30 min at 30 °C. The reaction was stopped by the addition of 4× Laemmli buffer and RKIP phosphorylation was analyzed by Western blot.
Statistical Analyses
Statistical significance between groups was analyzed by one-way analysis of variance followed by the Bonferroni test as post hoc test using the GraphPad software (San Diego, CA). Differences were considered significant with p < 0.05. All data are shown as mean ± S.E. For additional experimental procedures, see supplemental “Experimental Procedures.”
RESULTS
PKC-mediated Phosphorylation of RKIP at Ser-153 Promotes RKIP-RKIP Dimerization
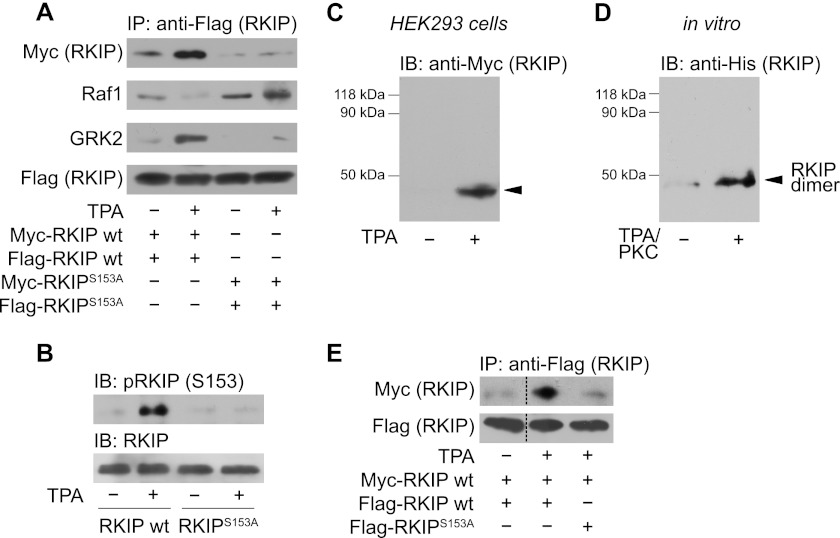
Previous studies have shown that PKC-mediated phosphorylation of RKIP at Ser-153 is necessary for the dissociation of RKIP from Raf1 and binding of RKIP to GRK2 (4, 14, 28). It is known that protein phosphorylation not only affects recognition by other proteins but also induces protein self-association, which can generate new and more diverse binding sites (30). To analyze whether self-association of RKIP supports the switch between different substrates, we performed co-immunoprecipitation assays in HEK293 cells overexpressing differentially tagged RKIP constructs, i.e. FLAG- and Myc-RKIP wild-type (wt) or phosphorylation-deficient FLAG- and Myc-RKIPS153A, respectively. These RKIP constructs were overexpressed at similar expression levels as endogenous RKIP protein (supplemental Fig. S1A). PKC activation with TPA significantly enhanced self-association of RKIP as shown by immunoprecipitation of FLAG-RKIP and subsequent detection of co-immunoprecipitated Myc-RKIP by Western blot analysis. In contrast, TPA treatment was not able to induce self-association of RKIPS153A (Fig. 1A, upper panel). Western blot analysis of corresponding cell lysates with phospho-RKIP(Ser153) antibodies confirmed efficient phosphorylation of wild-type RKIP upon TPA stimulation but not of RKIPS153A (Fig. 1B). Phosphorylation and self-association of RKIP occurred concomitantly with the release of RKIP from Raf1 and its association with GRK2 (Fig. 1A). These experiments show that PKC switches the substrate specificity of RKIP from Raf1 to GRK2, and that this switch is associated with significant self-association of RKIP.
FIGURE 1.
PKC-mediated phosphorylation of RKIP at Ser-153 promotes RKIP-RKIP self-association. A, immunoprecipitation (IP) of wild-type FLAG-tagged RKIP (wt) and phospho-deficient FLAG-RKIPS153A from lysates of HEK293 cells and subsequent immunoblot analysis of co-immunoprecipitated Myc-RKIP wild-type (wt), Myc-RKIPS153A, Raf1, or GRK2. Cells were treated with TPA (1 μm, 5 min) as indicated. B, immunoblot analysis (IB) of Ser-153 (S153) phosphorylation of RKIP in the respective cell lysates of A. C, cross-linking experiments in HEK293 cells expressing wild-type Myc-RKIP. Cells were stimulated with TPA (1 μm, 5 min) as indicated and treated with paraformaldehyde (PFA; 0.75% (w/v), 7 min). Cell lysates were analyzed for RKIP complexes by immunoblotting. D, in vitro cross-linking experiments with purified His-RKIP using PFA (0.6% (w/v), 10 min). Cross-linking was performed before or after phosphorylation of His-RKIP by recombinant PKCδ (30 min, 30 °C). E, immunoprecipitation of FLAG-RKIP wild-type and FLAG-RKIPS153A from lysates of HEK293 cells treated without or with TPA (1 μm, 5 min) and subsequent Western blot analysis for co-immunoprecipitated wild-type Myc-RKIP. n = 4–7 independent experiments.
Because co-immunoprecipitation experiments cannot distinguish between RKIP dimers and RKIP oligomers, we next performed cross-linking experiments using PFA as cross-linking agent both in living cells (Fig. 1C) and with purified proteins (Fig. 1D). Cells overexpressing Myc-tagged RKIP were stimulated with TPA followed by treatment with PFA and cell lysis. Western blot analyses with antibodies directed against the Myc tag revealed a band of about 42 kDa for wild-type RKIP after TPA treatment (Fig. 1C and supplemental Fig. S1B), which matches the calculated molecular mass of RKIP dimers. Higher-order oligomers of RKIP were neither detected after cross-linking with PFA (Fig. 1, C and D) nor after cross-linking with agents comprising longer spacers (e.g. sEGS, data not shown). Similarly, cross-linking experiments with purified His-tagged wild-type RKIP showed an increased formation of RKIP dimers upon phosphorylation with recombinant PKCδ, but again without detectable quantities of higher-order RKIP oligomers (Fig. 1D). These in vitro experiments indicate that RKIP dimerization originates from a direct RKIP-RKIP interaction, which is only dependent on phosphorylation and does not require other components.
To determine whether one or both RKIP molecules have to be phosphorylated for dimer formation, we performed co-immunoprecipitation experiments using FLAG-RKIPS153A and Myc-RKIP wild-type (wt). In contrast to FLAG-RKIP wild-type, phosphorylation-deficient FLAG-RKIPS153A was not able to assemble with Myc-RKIP wild-type in response to TPA stimulation (Fig. 1E). RKIP dimer formation thus requires that both RKIP molecules are phosphorylated at Ser-153.
Taken together, phosphorylation-induced RKIP dimerization is dependent on Ser-153 phosphorylation and occurs concomitantly with the release of Raf1 from RKIP and association of RKIP with GRK2. This may indicate that RKIP dimerization influences the interaction of RKIP with its substrates.
Phosphomimetic RKIP (RKIPSK153/7EE) Possesses High Propensity for Dimerization and GRK2 Binding
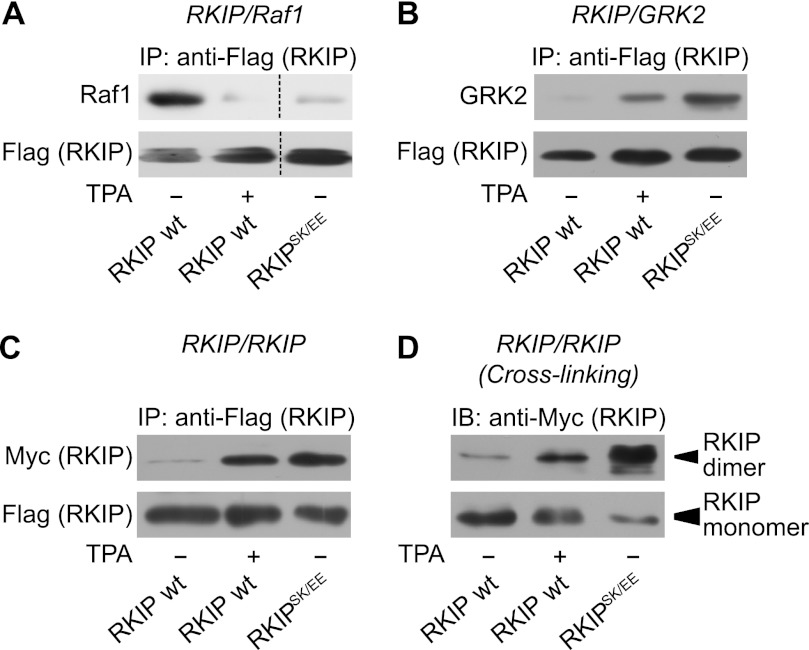
Next, we generated a mutant of RKIP, which mimics PKC phosphorylation on Ser-153 to further evaluate the impact of phosphorylation on dimerization and substrate binding of RKIP. In some cases, replacing target amino acids with negatively charged amino acids such as glutamate or aspartate can mimic transfer of phosphate groups to phosphorylation sites. However, mutation of Ser-153 of RKIP to either glutamate (RKIPS153E) (Ref. 28 and data not shown) or aspartate (RKIPS153D) (data not shown) did not result in a functional phosphomimetic mutant, which would be expected to bind to GRK2, but not to Raf1 (4, 28). Assuming that one negative charge may not be sufficient to mimic the di-ionic phosphate group, we inserted a second negative charge in close proximity. Because Ser-153 is located within an α-helix (8), we inserted in the mutant RKIPS153E an additional glutamate at position 157 to align the inserted charges with similar orientation according to one α-helical turn of ∼4 amino acids (31). Of note, these two newly introduced negatively charged amino acids are located in the corresponding positions in the rape homolog of RKIP (19). Binding of the phosphomimetic mutant RKIPSK153/7EE (RKIPSK/EE) to Raf1 was clearly reduced under basal conditions compared with wild-type RKIP (Fig. 2A). In contrast, RKIPSK153/7EE co-immunoprecipitated GRK2 at least as efficiently as wild-type RKIP after TPA treatment (Fig. 2B). RKIP constructs were overexpressed at similar expression levels as endogenous RKIP protein (supplemental Fig. S2). These experiments indicate that the RKIP mutant, RKIPSK153/7EE, indeed mimics Ser-153 phosphorylation of RKIP with respect to Raf1 and GRK2 binding. To analyze the dimerization properties of RKIPSK153/7EE, we performed co-immunoprecipitation assays with FLAG- and Myc-tagged RKIPSK153/7EE constructs. As shown in Fig. 2C, RKIPSK153/7EE displayed enhanced propensity for dimerization in the absence of TPA. This was further validated in cross-linking experiments in HEK293 cells transfected with Myc-tagged RKIPSK153/7EE (Fig. 2D), again there was no evidence of higher-order oligomers (data not shown). Interestingly, the proportion of monomeric RKIPSK153/7EE under basal conditions was less than that of wild-type RKIP after TPA stimulation (Fig. 2D, lower panel, blot with lower exposure).
FIGURE 2.
Phosphomimetic RKIP has high propensity for dimerization and GRK2 binding. A–C, co-immunoprecipitation assays using HEK293 cells transiently transfected with FLAG- and Myc-tagged wild-type RKIP (wt) or with FLAG- and Myc-tagged RKIPSK153/7EE (RKIPSK/EE) as well as with Raf1 and GRK2. Cells were stimulated with TPA (1 μm, 5 min) as indicated. After immunoprecipitation (IP) of the FLAG-tagged RKIP constructs, co-immunoprecipitated Raf1 (A), GRK2 (B), and Myc-RKIP (C) were visualized by immunoblot analysis with the indicated antibodies. D, cross-linking experiments with PFA (0.75% (w/v), 7 min) in HEK293 cells transfected with wild-type Myc-RKIP or Myc-RKIPSK153/7EE (RKIPSK/EE). Cells were stimulated with TPA (1 μm, 5 min) as indicated. Protein complexes were separated by SDS-PAGE and detected with anti-Myc antibodies. The upper panel shows dimeric RKIP (∼42 kDa) and the lower panel shows monomeric RKIP (∼21 kDa; lower film exposure). n = 4–6 independent experiments.
Mutant RKIP Lacking Amino Acids 143–146 Has a High Propensity for Dimerization and GRK2 Binding
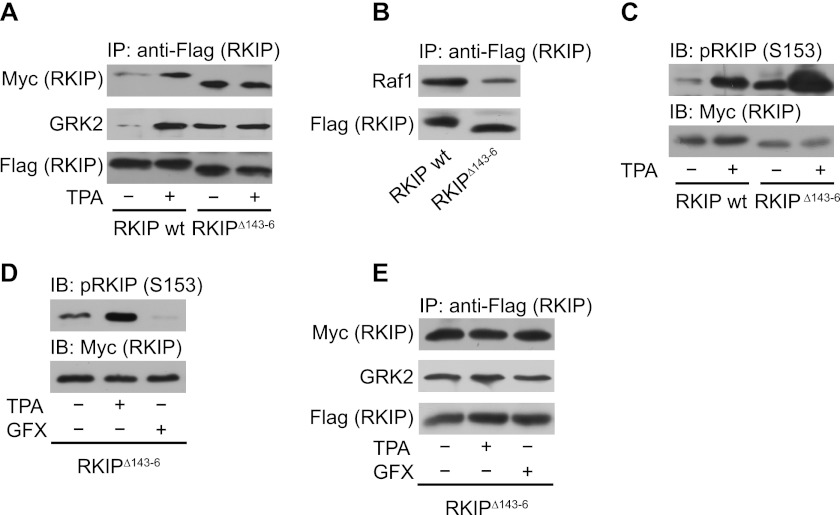
It has been suggested that a loop structure (“loop 127–150”), which is located close to the Ser-153 containing α-helix, might be involved in the RKIP dimer interface (8). We performed mutational analyses to investigate the impact of this particular region. We generated (i) deletion mutants, which lacked 4 or 6 consecutive amino acids all through the loop and a deletion mutant lacking the entire loop (amino acids 127–150) and (ii) a point mutant in which we exchanged several of the charged amino acids to neutral residues. Almost all of these mutants could not be used for interaction studies because they were either not expressed or precipitated unspecifically. However, the deletion of amino acids 143–146 (RKIPΔ143–146) at the C-terminal end of the loop generated a mutant with high propensity for dimerization in the absence of PKC stimulation as shown by co-immunoprecipitation (Fig. 3A) and by cross-linking experiments in HEK293 cells (supplemental Fig. S3A). RKIP constructs were overexpressed at similar expression levels as endogenous RKIP protein (supplemental Fig. S3B). Intriguingly, the corresponding loop of PEBP in Antirrhinum centroradialis is generally 5 amino acids shorter than the corresponding loop of mammalian PEBPs (19), and this RKIP homolog crystallizes as a dimer. This further supports our observation that shortening of this particular loop is favorable for RKIP dimerization. RKIPΔ143–146 displayed a lower affinity to Raf1 (Fig. 3B), whereas its affinity to GRK2 was comparable with that of phosphorylated wild-type RKIP (Fig. 3A). Thus, RKIPΔ143–146 showed similar properties as the phosphomimetic RKIPSK153/7EE. Because RKIPΔ143–146 was already highly phosphorylated in quiescent cells (Fig. 3C), we analyzed whether increased dimer formation and GRK2 binding of this mutant was due to increased RKIPΔ143–146 phosphorylation. However, prevention of RKIPΔ143–146 phosphorylation by the PKC inhibitor bisindolylmaleimide I (GFX) (Fig. 3D), did not affect RKIPΔ143–146 dimerization and GRK2 binding (Fig. 3E). These findings suggest that dimeric RKIP does not require Ser-153 phosphorylation to bind to GRK2. This hypothesis was further supported by the fact that dimerization and GRK2 binding of RKIPΔ143–146 did not further increase in response to TPA treatment (Fig. 3, A and E, and supplemental Fig. S3A). Because RKIPΔ143–146 dimerized under basal conditions (and after PKC inhibitor treatment), we hypothesize that RKIP phosphorylation might induce a conformational change triggering dimerization, which is mimicked by the dimeric RKIP mutant RKIPΔ143–146.
FIGURE 3.
RKIP lacking amino acids 143–146 has a high propensity for dimerization and GRK2 binding. A and B, immunoprecipitation (IP) of wild-type FLAG-RKIP (wt) or FLAG-RKIPΔ143–146 (RKIPΔ143–6) from lysates of HEK293 cells treated with TPA (1 μm, 5 min) as indicated. Co-precipitated wild-type Myc-RKIP (A), Myc-RKIPΔ143–146 (A), GRK2 (A), and Raf1 (B) were detected by Western blot analysis with the respective antibodies. The lower molecular mass of the deletion mutant RKIPΔ143–146 is visible due to the usage of 17% (w/v) acrylamide SDS-PAGE gels. C, immunoblot analysis (IB) of the respective cell lysates of A with anti-pRKIP (Ser-153, S153) antibodies. D, immunoblot analysis of Ser-153 phosphorylation of overexpressed RKIPΔ143–146 in HEK293 cells pretreated with either TPA (1 μm, 5 min) or bisindolylmaleimide I (GFX; 3 μm, 2 h) as indicated. E, immunoprecipitation of FLAG-RKIPΔ143–146 from cells pretreated with either TPA (1 μm, 5 min) or GFX (3 μm, 2 h) as indicated with subsequent immunoblot analysis of co-immunoprecipitated Myc-RKIPΔ143–146 or GRK2. n = 4–7 independent experiments.
Dimerization of RKIP Is Necessary for the Switch of RKIP from Raf1 to GRK2
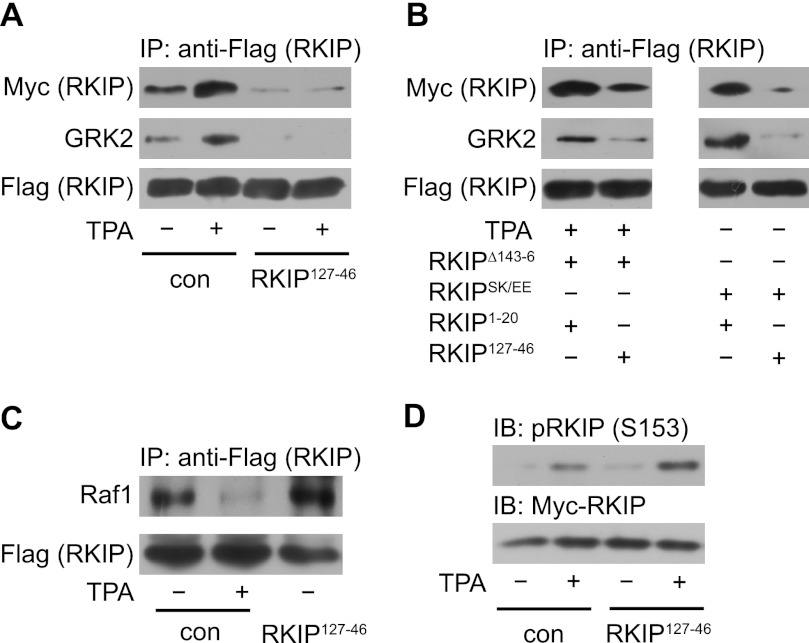
To further investigate the effect of dimerization on RKIP-GRK2 association and on RKIP/Raf1 dissociation, we used peptides containing the loop sequence of the putative dimer interface to competitively interfere with RKIP dimerization. These peptides comprised amino acids 127–146 of rat RKIP, i.e. most of the sequence of the loop 127–150. Co-expression of the RKIP fragment RKIP127–146 as well as longer RKIP fragments, i.e. RKIP1–150 and RKIP84–150, prevented PKC-induced dimerization of wild-type RKIP (Fig. 4A and supplemental Fig. S4, A–C) as well as of RKIPSK153/7EE and RKIPΔ143–146 in HEK293 cells (Fig. 4B). As controls, we used either mock-transfected cells or cells transfected with N-terminal RKIP fragments, which did not contain the loop sequence (supplemental Fig. S4, A (RKIP1–20) and C (RKIP1–67)). In accordance with our previous results, prevention of RKIP dimerization by loop 127–150 was accompanied by inhibition of PKC-induced RKIP-GRK2 assembly (Fig. 4, A and B, and supplemental Fig. S4, A–C), whereas Raf1 binding to RKIP was not affected (Fig. 4C). Of note, the fragment RKIP127–146 did not inhibit PKC-mediated RKIP phosphorylation on Ser-153 (Fig. 4D). We confirmed the expression of the peptides by reverse transcriptase (RT) PCR (RKIP1–20 and RKIP127–146, supplemental Fig. S4D), dot-blot analyses (RKIP127–146 and RKIP84–150, supplemental Fig. S4E), or Tricine-SDS-PAGE (RKIP1–150 and RKIP84–150, supplemental Fig. S4F). These results identify loop 127–150 of RKIP as the dimer interface and they further show that dimerization of RKIP is a mandatory feature for the switch of RKIP from Raf1 to GRK2.
FIGURE 4.
Dimerization is necessary for the switch of RKIP from Raf1 to GRK2. A, immunoprecipitation (IP) of wild-type FLAG-RKIP from cells treated without or with TPA (1 μm, 5 min), which were either co-transfected with empty vector (con) or with the RKIP fragment containing amino acids 127–146 (RKIP127–146, RKIP127–46) of RKIP. Co-precipitated Myc-RKIP wild-type and GRK2 were visualized with the indicated antibodies. B, immunoprecipitation of FLAG-RKIPΔ143–146, RKIPΔ143–6 (left panels) or FLAG-RKIPSK153/7EE (RKIPSK/EE, right panels) from cells, which were co-transfected with RKIP fragments containing amino acids 1–20 (RKIP1–20) or 127–146 (RKIP127–146) of RKIP. Cells transfected with FLAG-RKIPΔ143–146 were treated with TPA (1 μm, 5 min) to induce maximal RKIP phosphorylation for fair comparison with phosphomimetic RKIPSK153/7EE. Co-precipitated Myc-RKIPΔ143–146 or RKIPSK153/7EE and GRK2 were visualized with the indicated antibodies. C, immunoprecipitation of wild-type FLAG-RKIP from cells treated without or with TPA (1 μm, 5 min), which were either co-transfected with empty vector (con) or with RKIP127–146. Co-precipitated Raf1 was visualized with anti-Raf1 antibodies. D, immunoblot analyses (IB) of cell lysates used in A and C analyzed with indicated antibodies. n = 3–5 independent experiments.
High Propensity for RKIP Dimerization Prevents Raf1 Inhibition and Translates into Efficient GRK2 Inhibition
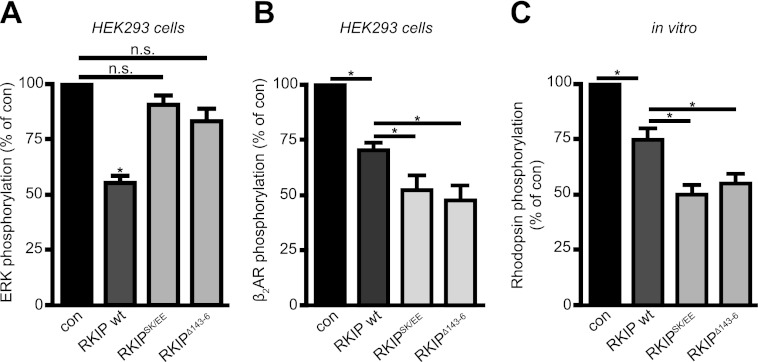
To test the functionality of the phosphomimetic RKIPSK153/7EE and the dimeric mutant RKIPΔ143–146, we analyzed their capability to inhibit Raf1 and GRK2. For these analyses wild-type RKIP, RKIPSK153/7EE, and RKIPΔ143–146 were expressed at similar expression levels as endogenous RKIP in these cells (supplemental Fig. S5A). To analyze Raf1 inhibition, we monitored the activation of its effector kinases ERK1/2 in HEK293 cells using antibodies directed against activated ERK1/2 (pErk1/2(TEY)). Unlike wild-type RKIP, RKIPSK153/7EE and RKIPΔ143–146 did not display a significant inhibitory effect on Raf1 (Fig. 5A and supplemental Fig. S5B). These results are in line with the reduced binding affinities of phosphomimetic RKIPSK153/7EE and dimeric RKIPΔ143–146 to Raf1 (Figs. 2A and 3B). To ensure that the enhanced binding affinity of RKIPSK153/7EE and RKIPΔ143–146 to GRK2 (Figs. 2B and 3A) indeed translates into enhanced GRK2 inhibition, we monitored GRK2-mediated β2AR phosphorylation in HEK293 cells with stable overexpression of GFP-tagged β2AR using phosphospecific β2AR antibodies (pβ2AR(S355/6)) (Fig. 5B and supplemental Fig. S5C). Because these antibodies might also detect β2AR phosphorylation by other kinases, we overexpressed GRK2 to minimize background phosphorylation due to other kinases (supplemental Fig. S5D). All RKIP proteins significantly inhibited β2AR phosphorylation in response to isoproterenol stimulation. The inhibition of β2AR phosphorylation by RKIPSK153/7EE and RKIPΔ143–146 was even more efficient than by wild-type RKIP, which suggests that dimerization leads to more efficient GRK2 inhibition (Fig. 5B and supplemental Fig. S5C). In the absence of isoproterenol, β2AR phosphorylation was too weak to detect potential differences between wild-type RKIP and mutants RKIPSK153/7EE and RKIPΔ143–146 (data not shown). These experiments demonstrate that RKIPSK153/7EE and RKIPΔ143–146 are functional in terms of GRK2 inhibition.
FIGURE 5.
High propensity for RKIP dimerization prevents Raf1 inhibition and translates into efficient GRK2 inhibition. A, quantification of ERK1/2 phosphorylation in HEK293 cells with stable expression of GFP-tagged β2AR co-transfected with empty vector (con), wild-type Myc-RKIP (wt), Myc-RKIPSK153/7EE (RKIPSK/EE), or Myc-RKIPΔ143–146 (RKIPΔ143–6). ERK1/2 phosphorylation was detected with phospho-specific pERK1/2 antibodies. The graph shows ERK1/2 phosphorylation as % of the values in the absence of RKIP. n = 7 independent experiments; *, p < 0.01. B, quantification of β2AR phosphorylation induced by isoproterenol (Iso; 1 μm, 5 min) in HEK293 cells with stable expression of GFP-tagged β2AR and co-transfected GRK2 and empty vector (con), wild-type Myc-RKIP (wt), Myc-RKIPSK153/7EE (RKIPSK/EE), or Myc-RKIPΔ143–146. Receptor phosphorylation was detected with phosphospecific antibodies directed against serine residues 355 and 356 in β2AR. The graph shows isoproterenol-stimulated β2AR phosphorylation as % of the values in the absence of RKIP. n = 5–6 independent experiments; *, p < 0.05. C, quantification of GRK2-mediated phosphorylation of purified rhodopsin in the absence (con) or presence of purified His-RKIP wild-type, His-RKIPSK153/7EE, or His-RKIPΔ143–146. Rhodopsin phosphorylation was assessed using [γ-32P]ATP and quantified by phosphorimaging analysis. The graph shows light-stimulated rhodopsin phosphorylation as % of the values in the absence of RKIP. n = 15 independent experiments; *, p < 0.05. Data are mean ± S.E.
To further substantiate our hypothesis that RKIP dimerization is not only important for GRK2 binding but also for GRK2 inhibition, we next compared purified His-RKIP wild-type, RKIPSK153/7EE, and RKIPΔ143–146 with regard to their ability to inhibit GRK2 in an in vitro receptor phosphorylation assay, i.e. rhodopsin phosphorylation. Of note, no PKC was present in this assay, which could induce RKIP dimerization. Indeed, inhibition of GRK2-mediated rhodopsin phosphorylation was more pronounced in the presence of the “dimeric” mutants, RKIPSK153/7EE or RKIPΔ143–146, than in the presence of wild-type RKIP (Fig. 5C). This experiment demonstrates that the increased propensity for dimer formation indeed enhances GRK2 binding as well as GRK2 inhibition.
GRK2 Preferentially Binds to RKIP Dimers in Vitro
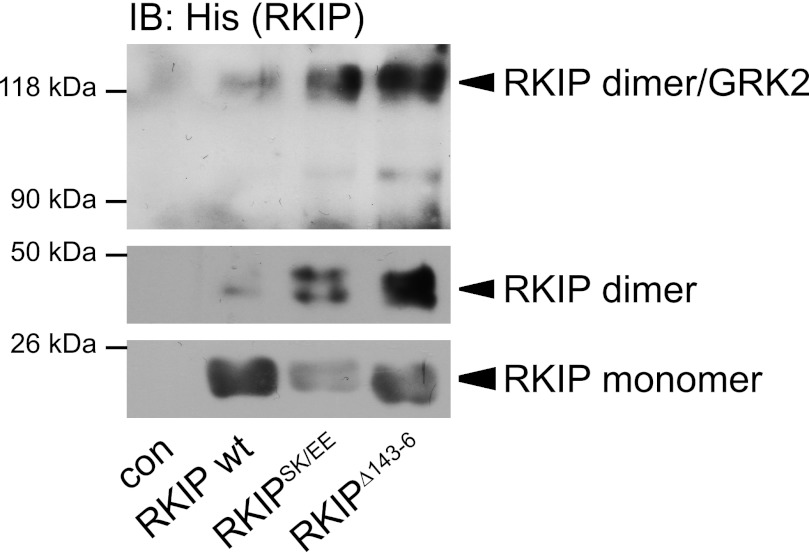
To validate that GRK2 binds to RKIP dimers, we performed cross-linking experiments with purified GRK2 and His-RKIP wild-type, His-RKIPSK153/7EE, or His-RKIPΔ143–146 using sEGS as cross-linking agent. Antibodies directed against the His tag revealed strong immunoreactive bands at ∼21 (molecular mass of RKIP monomers), ∼42 (RKIP dimers), and ∼122 kDa (Fig. 6). The signal at ∼122 kDa corresponds to the molecular mass of a complex consisting of dimeric RKIP (42 kDa) and GRK2 (80 kDa). Interestingly, there was almost no signal detectable at ∼101 kDa, which would refer to a complex of monomeric RKIP with GRK2. Analogous to previous experiments, RKIPSK153/7EE and RKIPΔ143–146 had a higher propensity for dimer formation and GRK2 binding as shown by an increased signal at ∼42 and ∼122 kDa and a decreased signal at ∼21 kDa compared with wild-type RKIP (Fig. 6). Thus, this experiment further sustains our hypothesis that GRK2 preferentially binds to RKIP dimers.
FIGURE 6.
GRK2 preferentially binds to RKIP dimers in vitro. Cross-linking experiments without (con) or with purified wild-type His-RKIP (wt), His-RKIPSK153/7EE (RKIPSK/EE), or His-RKIPΔ143–146 (RKIPΔ143–6) and purified GRK2 using sulfo-EGS as cross-linking agent (0.3 mm, 15 min). RKIP complexes were separated by SDS-PAGE and visualized with anti-His antibodies. The positions of molecular mass standards are marked on the left. The lower panel shows a film with shorter exposure. n = 5 independent experiments.
Detection of ∼122 kDa Complex Consisting of GRK2 and RKIP in Mouse Hearts
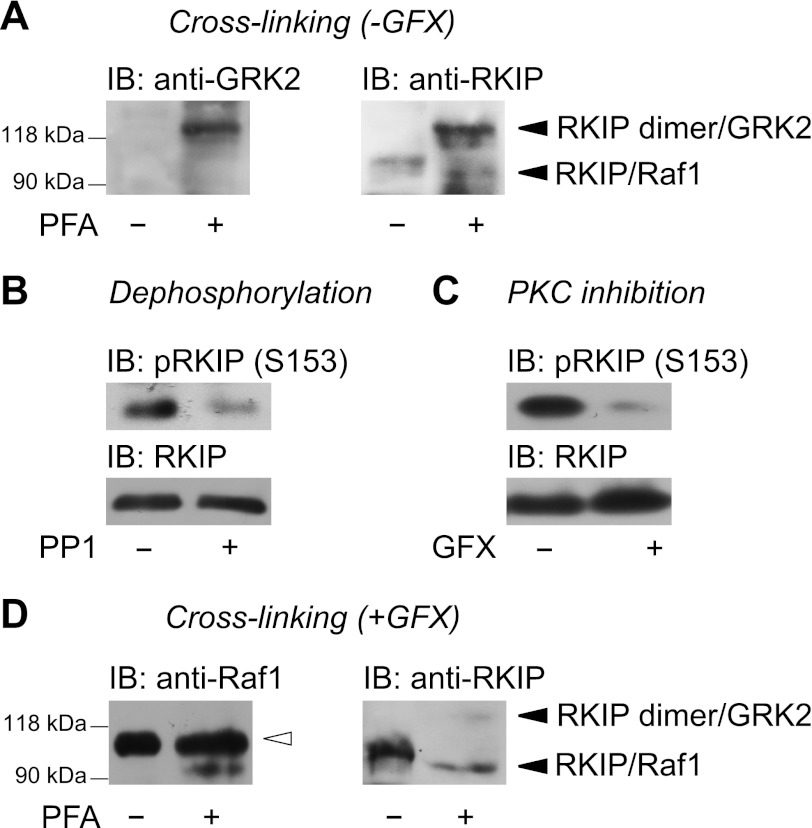
Because GRK2 is of particular interest for cardiac function, we analyzed whether RKIP dimers could potentially be of interest for the regulation of GRK2 function in the heart. Hence, we investigated if a complex of dimeric RKIP with GRK2 can be detected in cardiac tissue. We perfused mouse hearts with PFA as cross-linking compound and analyzed the heart lysates for GRK2-RKIP complexes by Western blot. Intriguingly, antibodies directed against GRK2 as well as antibodies directed against RKIP detected a prominent band of ∼122 kDa after cross-linking with PFA (Fig. 7A). Because RKIP phosphorylation is a prerequisite for RKIP dimerization, we verified that RKIP was phosphorylated in these heart samples. For this analysis, heart lysates were incubated with a recombinant phosphatase, PP1. Analysis of RKIP phosphorylation using phosphospecific pRKIP(Ser153) antibodies revealed a significant reduction of RKIP phosphorylation after PP1 treatment (Fig. 7B). Interestingly, immunoblot analysis of RKIP after cross-linking with PFA revealed an additional faint band at ∼94 kDa (Fig. 7A). To analyze whether this band may represent the RKIP-Raf1 complex, we perfused mouse hearts with GFX before cross-linking to reduce PKC-induced RKIP phosphorylation. GFX treatment indeed reduced RKIP phosphorylation as shown by immunoblot analysis (Fig. 7C). Under these conditions, antibodies directed against Raf1 detected a band at ∼94 kDa, which might represent the RKIP-Raf1 complex, and a very prominent cross-reacting band above (Fig. 7D). In line with the Raf1 immunoblot, antibodies directed against RKIP clearly detected a major band at ∼94 kDa (Fig. 7D) underlining the hypothesis that this band represents the RKIP-Raf1 complex. Unlike non-GFX-treated hearts (Fig. 7A), heart lysates of GFX-treated hearts showed only a faint 122-kDa (“RKIP-RKIP-GRK2”) band (Fig. 7D). Taken together, these experiments suggest that PKC-mediated phosphorylation of RKIP in native tissue also triggers dimer formation of endogenous RKIP and, thus, enables RKIP to switch from Raf1 to GRK2. The detection of RKIP dimers in a complex with GRK2 in native cardiac tissue implies a physiological relevance of RKIP dimers in the heart.
FIGURE 7.
Detection of a ∼122-kDa complex consisting of GRK2 and RKIP in mouse hearts. A, mouse hearts were perfused with the cross-linking agent PFA (4% (w/v) in PBS, 7 min) and lysed. Protein complexes were separated by SDS-PAGE and subjected to Western blot with the indicated antibodies. The positions of molecular mass standards are marked on the left. n = 5 independent experiments. B, immunoblot analysis of RKIP phosphorylation in murine heart lysates treated without or with protein phosphatase 1 (PP1; 20 units of PP1/100 μg of lysate; 30 min, 30 °C) for dephosphorylation using antibodies directed against pRKIP(S153). n = 4 independent experiments. C, perfusion of mouse hearts with the PKC inhibitor GFX (1.5 μm; 3 min) and lysed. RKIP phosphorylation was analyzed by Western blot analysis. n = 4 independent experiments. D, mouse hearts were perfused with GFX (1.5 μm; 3 min) followed by perfusion with PFA (4% (w/v) in PBS, 7 min). Protein complexes were separated by SDS-PAGE and subjected to Western blot with the indicated antibodies. The positions of molecular mass standards are marked on the left. Open arrow indicates a nonspecific anti-Raf1 immunoreactive band. n = 4 independent experiments.
DISCUSSION
The number of signaling cascades known to be modulated by RKIP is constantly increasing. Until Yeung et al. (1) discovered the inhibitory effect of RKIP on the Raf/MEK/ERK cascade, no physiological role of this protein was known. Meanwhile, the inhibition of Raf1 by RKIP has been shown to be implicated in cell transformation (1) and the control of the cell cycle (32) and, thus, tumor progression and metastasis (33). Shortly after the discovery of RKIP-mediated Raf1 inhibition, the inhibitory effect of RKIP on NFκB signaling was discovered (2) and is discussed as one possible mechanism for the chemoresistance of tumor types with low or no RKIP expression (34). RKIP may also influence cell division and apoptosis by interference with glycogen synthase kinase 3β signaling (3). Furthermore, RKIP was suggested to be involved in GPCR signaling (5) and was eventually identified as a physiological and direct inhibitor of GRK2 (4), which is an important regulator of GPCR signaling. RKIP-mediated GRK2 inhibition is involved in cross-regulation of GPCRs (35), neuronal differentiation (36), and cardiomyocyte contractility (4). Taken together, RKIP controls a complex variety of signaling networks in mammalian cells.
It is still a matter of discussion, how such a small protein of 21 kDa can retain control over several different signaling cascades. The only well characterized regulatory mechanism known so far, which controls alternate binding of RKIP to its targets, is the PKC-mediated phosphorylation of RKIP at Ser-153. This phosphorylation releases RKIP from Raf1 and leads to the association of RKIP with GRK2 (4). In general, protein phosphorylations offer flexible communication between interaction partners implicated in gene expression, regulation of receptor activity, and other crucial cellular functions. They promote conformational changes and alter surface properties of proteins and may thereby affect protein-protein interactions, activation/inhibition of enzyme activity, or self-association. Self-association of proteins can increase protein stability and generate new binding sites and thus alter protein function (37–43). Surprisingly, homo-oligomerization as an efficient mechanism to control protein-protein interactions has thus far not been taken into consideration for RKIP. Although putative dimers and higher-order oligomers of RKIP have been observed by Bollengier et al. (44) in human brain lysates and in crystal structures of human RKIP, a bacterial and a plant homolog of RKIP (8, 15, 19), the existence or physiological implications of RKIP dimers or oligomers in mammalian cells have not been investigated under physiological conditions.
Here, we show that (i) RKIP indeed exists as a dimer in intact cells (Fig. 1), (ii) RKIP preferentially forms dimers and not higher oligomers (Figs. 1–3 and supplemental Figs. S1 and S3), (iii) the RKIP-RKIP interaction is of a direct nature (Figs. 1D and 6), (iv) RKIP dimerization is post-translationally regulated by PKC-mediated phosphorylation at Ser-153 (Figs. 1 and 2), and (v) PKC-mediated RKIP dimerization also occurs in native tissue (Fig. 7). Furthermore, our experiments identify a loop structure (loop 127–150) at the surface of RKIP as part of the dimer interface (Fig. 4 and supplemental Fig. S4). This interface is located in immediate proximity to the PKC-phosphorylation site, which may alter the conformation of the loop and may therefore trigger dimerization after PKC-mediated phosphorylation. Perturbations within the loop structure were detected in NMR studies with a mutant of RKIP, which at least partially imitates a phosphorylation at this site (RKIPS153E) (14). Further observations by others strengthen our assumption that this loop represents the interface for RKIP dimerization: loops in general often participate in protein-protein interactions (45, 46); particularly, loops with a relatively high fraction of polar, charged residues, as well as glycines and prolines have been suggested to be involved in protein dimerization (37, 47). Indeed, loop 127–150 contains eight charged and eight polar amino acids, as well as two glycines and two prolines of 24 amino acids; Banfield et al. (8) have generated crystals of dimeric hRKIP and suggested this loop as a putative dimer interface. Others have reported that differences in this loop region participate in opposing effects on flowering of two proteins in Arabidopsis thaliana with high similarity to RKIP (48). Our results, these observations, and the fact that the loop is highly conserved among mammalian RKIP isoforms (12) all suggest that this loop plays an important role in the regulation of protein interactions of RKIP.
To study the impact of RKIP dimerization on its target specificity, we analyzed the switch of RKIP from Raf1 to GRK2. Even though there is a growing number of interaction partners, there are only a few, i.e. Raf1, MEK1, ERK2, and GRK2, of which direct interactions using recombinant, purified proteins have been established. Of these direct interaction partners, a mechanistic understanding may be especially interesting for Raf1 and GRK2, because only for these two inhibition of their kinase activity by RKIP has been shown with an IC50 value in the low micromolar range (∼5 versus ∼0.5 μm, respectively) (1, 4). Our experiments show that phosphorylation-induced dimerization is a prerequisite for RKIP to inhibit GRK2. More precisely, (i) experiments with wild-type RKIP and phosphorylation-deficient RKIPS153A and phosphomimetic RKIPSK153/7EE showed that dimerization occurred concomitantly with the release of RKIP from Raf1 and the binding of RKIP to GRK2 (Figs. 1 and 2); (ii) studies with a dimeric RKIP mutant (RKIPΔ143–146; Fig. 3 and supplemental Fig. S3), and with peptides competitively interfering with RKIP dimerization (Fig. 4 and supplemental Fig. 4) substantiated the hypothesis that the actual binding interface of GRK2 and RKIP is formed by RKIP dimers; (iii) experiments with the dimeric RKIP mutant (RKIPΔ143–146) further show a reduced affinity of dimeric RKIP for Raf1, which suggests that RKIP dimerization causes the release of Raf1 from RKIP (Fig. 3); (iv) in vitro experiments with purified RKIP showed that RKIP dimers are important structural components for GRK2 binding (Figs. 1 and 6); (v) cross-linking experiments in heart tissue showed that PKC-mediated RKIP phosphorylation promotes the complex formation between dimeric RKIP and GRK2 (Fig. 7). The physiological monomer to dimer ratio is very difficult to determine (40). In the case of RKIP dimerization, activation and deactivation of PKC, and most likely also of phosphatases may dynamically alter the monomer to dimer ratio depending on cellular triggers. The finding that the equilibrium is shifted toward monomers in the presence of the loop peptide RKIP127–146 (Fig. 4, A and B, and supplemental Fig. S4, A–C), supports the hypothesis of a dynamic nature of RKIP dimers. Interestingly, RKIP dimerization also seems to be controlled by a threshold mechanism because both RKIP molecules need to be phosphorylated for dimer formation (Fig. 1E).
To prove the functional relevance of RKIP dimers for GRK2 inhibition, we analyzed the effect of phosphomimetic RKIPSK153/7EE and dimeric RKIPΔ143–146 on the inhibition of GRK2-mediated phosphorylation of β2ARs in cells as well as on GRK2-mediated rhodopsin phosphorylation in vitro. Because both mutants, characterized by high propensity for dimerization and for GRK2 binding, are more efficient GRK2 inhibitors than wild-type RKIP (Fig. 5, B and C), these experiments supported the functional impact of RKIP dimers. In line with our hypothesis, that dimerization releases RKIP from Raf1, phosphomimetic RKIPSK153/7EE and dimeric RKIPΔ143–146 hardly interfered with Raf1-mediated ERK1/2 activation (Fig. 5A). Interestingly, the difference in GRK2 inhibition between wild-type RKIP and mutants RKIPSK153/7EE and RKIPΔ143–146 was even more pronounced in vitro. This is most likely due to the absence of PKC-induced phosphorylation of wild-type RKIP under these conditions. GRK2 inhibition by wild-type RKIP (Fig. 5 and Lorenz et al. (4)) in vitro may be explained by the existence of a certain proportion of RKIP dimers under these conditions as shown by cross-linking experiments (Figs. 1 and 6). The tendency of RKIP to dimerize under these conditions may be fostered by relatively high protein concentrations used in in vitro assays (39).
The detection of the RKIP-RKIP-GRK2 complex in murine cardiac tissue suggests a physiological relevance of RKIP dimers in the heart (Fig. 7A). Of note, GRK2 inhibition is of special interest in cardiovascular research because GRK2 activity and expression are significantly increased in heart failure patients (49). Although it is not yet entirely clear whether enhanced GRK2 activity in heart failure is an adaptive or maladaptive response, GRK2 inhibition is extensively discussed as a promising strategy in heart failure therapy (50, 51). This hypothesis is supported by a number of studies in which GRK2 expression and activity in cells or transgenic animals was altered (27). Ablation of GRK2 in cardiac myocytes prevented heart failure progression after myocardial infarction in mice (52), whereas overexpression of GRK2 caused cardiac dysfunction in vivo (53). So far, only RKIP has been described as a physiological inhibitor of GRK2 with an impact on cardiomyocyte function. RKIP interacts with the N terminus of GRK2, a region, that is important for receptor/GRK2 interaction. This interaction is highly specific, because RKIP does not regulate cardiac GRK5 or PKA. RKIP-mediated GRK2 inhibition increases cAMP signaling and enhances contractility of isolated neonatal cardiomyocytes (4). Despite significant efforts to develop small molecule GRK2 inhibitors, a specific and applicable GRK2 inhibitor is still not available (20, 27). The phosphomimetic and the dimeric mutants of RKIP, RKIPSK153/7EE, and RKIPΔ143–146, are promising tools to study the RKIP/GRK2 interface and to understand how RKIP achieves specific and efficient inhibition of GRK2.
Taken together, our study gives insights in how RKIP may be able to control several different kinases. We propose that dimerization is an important mechanistic feature for the substrate specificity of RKIP. We identify PKC-mediated phosphorylation of RKIP at Ser-153 as a prerequisite for RKIP dimerization, which is in turn essential for the binding of RKIP to its substrate GRK2. It will be interesting to study the implication of RKIP dimers in the context of other RKIP targets to shed light on the question how such a multifunctional protein can control several different signaling cascades.
Acknowledgments
We thank Ulrike Zabel, Heide-Marie Roth, Maria Hirschbeck, and Bodo Sander for helpful discussions and Martina Fischer and Christian Dees for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft and the Bundesministerium fuer Bildung und Forschung.

This article contains supplemental “Experimental Procedures” and Figs. S1–S5.
- RKIP
- Raf kinase inhibitor protein
- βAR
- β-adrenergic receptor
- GFX
- bisindolylmaleimide I
- GPCR
- G protein coupled receptor
- GRK2
- G protein-coupled receptor kinase 2
- GFP
- green fluorescent protein
- MAPK
- mitogen-activated protein kinase
- MEK
- mitogen-activated protein kinase and extracellular-regulated kinase kinase
- NFκB
- nuclear factor κ-light chain enhancer of activated B-cells
- PEBP
- phosphatidylethanolamine-binding protein
- PFA
- paraformaldehyde
- sEGS
- sulfo-ethylene glycolbis(succinimidylsuccinate)
- TPA
- 12-O-tetradecanoylphorbol 13-acetate
- PP1
- protein phosphatase 1
- Ni-NTA
- nickel-nitrilotriacetic acid
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Yeung K., Seitz T., Li S., Janosch P., McFerran B., Kaiser C., Fee F., Katsanakis K. D., Rose D. W., Mischak H., Sedivy J. M., Kolch W. (1999) Suppression of Raf-1 kinase activity and MAP kinase signaling by RKIP. Nature 401, 173–177 [DOI] [PubMed] [Google Scholar]
- 2. Yeung K. C., Rose D. W., Dhillon A. S., Yaros D., Gustafsson M., Chatterjee D., McFerran B., Wyche J., Kolch W., Sedivy J. M. (2001) Raf kinase inhibitor protein interacts with NF-κB-inducing kinase and TAK1 and inhibits NF-κB activation. Mol. Cell Biol. 21, 7207–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Al-Mulla F., Bitar M. S., Al-Maghrebi M., Behbehani A. I., Al-Ali W., Rath O., Doyle B., Tan K. Y., Pitt A., Kolch W. (2011) Raf kinase inhibitor protein RKIP enhances signaling by glycogen synthase kinase-3β. Cancer Res. 71, 1334–1343 [DOI] [PubMed] [Google Scholar]
- 4. Lorenz K., Lohse M. J., Quitterer U. (2003) Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature 426, 574–579 [DOI] [PubMed] [Google Scholar]
- 5. Kroslak T. (2001) Human phosphatidylethanolamine-binding protein facilitates heterotrimeric G protein-dependent signaling. J. Biol. Chem. 276, 39772–39778 [DOI] [PubMed] [Google Scholar]
- 6. Bernier I., Tresca J. P., Jollès P. (1986) Ligand-binding studies with a 23-kDa protein purified from bovine brain cytosol. Biochim. Biophys. Acta 871, 19–23 [DOI] [PubMed] [Google Scholar]
- 7. Vallée B. S., Tauc P., Brochon J. C., Maget-Dana R., Lelièvre D., Metz-Boutigue M. H., Bureaud N., Schoentgen F. (2001) Behavior of bovine phosphatidylethanolamine-binding protein with model membranes. Evidence of affinity for negatively charged membranes. Eur. J. Biochem. 268, 5831–5841 [DOI] [PubMed] [Google Scholar]
- 8. Banfield M. J., Barker J. J., Perry A. C., Brady R. L. (1998) Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure 6, 1245–1254 [DOI] [PubMed] [Google Scholar]
- 9. Zeng L., Imamoto A., Rosner M. R. (2008) Raf kinase inhibitory protein (RKIP). A physiological regulator and future therapeutic target. Expert Opin. Ther. Targets 12, 1275–1287 [DOI] [PubMed] [Google Scholar]
- 10. Kolch W. (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6, 827–837 [DOI] [PubMed] [Google Scholar]
- 11. Keller E. T., Fu Z., Brennan M. (2004) The role of Raf kinase inhibitor protein (RKIP) in health and disease. Biochem. Pharmacol. 68, 1049–1053 [DOI] [PubMed] [Google Scholar]
- 12. Granovsky A. E., Rosner M. R. (2008) Raf kinase inhibitory protein. A signal transduction modulator and metastasis suppressor. Cell Res. 18, 452–457 [DOI] [PubMed] [Google Scholar]
- 13. Serre L., Pereira de Jesus K., Zelwer C., Bureaud N., Schoentgen F., Bénédetti H. (2001) Crystal structures of YBHB and YBCL from Escherichia coli, two bacterial homologues to a Raf kinase inhibitor protein. J. Mol. Biol. 310, 617–634 [DOI] [PubMed] [Google Scholar]
- 14. Granovsky A. E., Clark M. C., McElheny D., Heil G., Hong J., Liu X., Kim Y., Joachimiak G., Joachimiak A., Koide S., Rosner M. R. (2009) Raf kinase inhibitory protein function is regulated via a flexible pocket and novel phosphorylation-dependent mechanism. Mol. Cell Biol. 29, 1306–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Serre L., Vallée B., Bureaud N., Schoentgen F., Zelwer C. (1998) Crystal structure of the phosphatidylethanolamine-binding protein from bovine brain. A novel structural class of phospholipid-binding proteins. Structure 6, 1255–1265 [DOI] [PubMed] [Google Scholar]
- 16. Shemon A. N., Heil G. L., Granovsky A. E., Clark M. M., McElheny D., Chimon A., Rosner M. R., Koide S. (2010) Characterization of the Raf kinase inhibitory protein (RKIP) binding pocket. NMR-based screening identifies small-molecule ligands. PLoS ONE 5, e10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rath O., Park S., Tang H. H., Banfield M. J., Brady R. L., Lee Y. C., Dignam J. D., Sedivy J. M., Kolch W., Yeung K. C. (2008) The RKIP (Raf-1 kinase inhibitor protein) conserved pocket binds to the phosphorylated N-region of Raf-1 and inhibits the Raf-1-mediated activated phosphorylation of MEK. Cell Signal 20, 935–941 [DOI] [PubMed] [Google Scholar]
- 18. Yeung K., Janosch P., McFerran B., Rose D. W., Mischak H., Sedivy J. M., Kolch W. (2000) Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the Raf kinase inhibitor protein. Mol. Cell. Biol. 20, 3079–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Banfield M. J., Brady R. L. (2000) The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 297, 1159–1170 [DOI] [PubMed] [Google Scholar]
- 20. Thal D. M., Yeow R. Y., Schoenau C., Huber J., Tesmer J. J. (2011) Molecular mechanism of selectivity among G protein-coupled receptor kinase 2 inhibitors. Mol. Pharmacol. 80, 294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lorenz K., Schmitt J. P., Vidal M., Lohse M. J. (2009) Cardiac hypertrophy. Targeting Raf/MEK/ERK1/2 signaling. Int. J. Biochem. Cell Biol. 41, 2351–2355 [DOI] [PubMed] [Google Scholar]
- 22. Maurer G., Tarkowski B., Baccarini M. (2011) Raf kinases in cancer roles and therapeutic opportunities. Oncogene 30, 3477–3488 [DOI] [PubMed] [Google Scholar]
- 23. Keller E. T., Fu Z., Yeung K., Brennan M. (2004) Raf kinase inhibitor protein. A prostate cancer metastasis suppressor gene. Cancer Lett. 207, 131–137 [DOI] [PubMed] [Google Scholar]
- 24. Pierce K. L., Premont R. T., Lefkowitz R. J. (2002) Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 [DOI] [PubMed] [Google Scholar]
- 25. Ribas C., Penela P., Murga C., Salcedo A., García-Hoz C., Jurado-Pueyo M., Aymerich I., Mayor F. (2007) The G protein-coupled receptor kinase (GRK) interactome. Role of GRKs in GPCR regulation and signaling. Biochim. Biophys. Acta 1768, 913–922 [DOI] [PubMed] [Google Scholar]
- 26. Gurevich E. V., Tesmer J. J., Mushegian A., Gurevich V. V. (2012) G protein-coupled receptor kinases. More than just kinases and not only for GPCRs. Pharmacol. Ther. 133, 40–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rengo G., Lymperopoulos A., Leosco D., Koch W. J. (2011) GRK2 as a novel gene therapy target in heart failure. J. Mol. Cell Cardiol. 50, 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Corbit K. C., Trakul N., Eves E. M., Diaz B., Marshall M., Rosner M. R. (2003) Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J. Biol. Chem. 278, 13061–13068 [DOI] [PubMed] [Google Scholar]
- 29. Eichmann T., Lorenz K., Hoffmann M., Brockmann J., Krasel C., Lohse M. J., Quitterer U. (2003) The amino-terminal domain of G-protein-coupled receptor kinase 2 is a regulatory Gβγ binding site. J. Biol. Chem. 278, 8052–8057 [DOI] [PubMed] [Google Scholar]
- 30. Dayhoff J. E., Shoemaker B. A., Bryant S. H., Panchenko A. R. (2010) Evolution of protein binding modes in homo-oligomers. J. Mol. Biol. 395, 860–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cohen C., Parry D. A. (1986) α-helical coiled coils - a widespread motif in proteins. Trends Biochem. Sci. 11, 245–248 [Google Scholar]
- 32. Eves E. M., Shapiro P., Naik K., Klein U. R., Trakul N., Rosner M. R. (2006) Raf kinase inhibitory protein regulates aurora B kinase and the spindle checkpoint. Mol. Cell 23, 561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keller E. T., Fu Z., Brennan M. (2005) The biology of a prostate cancer metastasis suppressor protein. Raf kinase inhibitor protein. J. Cell Biochem. 94, 273–278 [DOI] [PubMed] [Google Scholar]
- 34. Al-Mulla F., Bitar M. S., Feng J., Park S., Yeung K. C. (2012) A new model for Raf kinase inhibitory protein induced chemotherapeutic resistance. PLoS One 7, e29532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang J., Mahavadi S., Sriwai W., Grider J. R., Murthy K. S. (2007) Cross-regulation of VPAC2 receptor desensitization by M3 receptors via PKC-mediated phosphorylation of RKIP and inhibition of GRK2. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G867–874 [DOI] [PubMed] [Google Scholar]
- 36. Hellmann J., Rommelspacher H., Mühlbauer E., Wernicke C. (2010) Raf kinase inhibitor protein enhances neuronal differentiation in human SH-SY5Y cells. Dev. Neurosci. 32, 33–46 [DOI] [PubMed] [Google Scholar]
- 37. Hashimoto K., Panchenko A. R. (2010) Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states. Proc. Natl. Acad. Sci. U.S.A. 107, 20352–20357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nishi H., Hashimoto K., Panchenko A. R. (2011) Phosphorylation in protein-protein binding. Effect on stability and function. Structure 19, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marianayagam N. J., Sunde M., Matthews J. M. (2004) The power of two. Protein dimerization in biology. Trends Biochem. Sci. 29, 618–625 [DOI] [PubMed] [Google Scholar]
- 40. Nooren I. M., Thornton J. M. (2003) Structural characterization and functional significance of transient protein-protein interactions. J. Mol. Biol. 325, 991–1018 [DOI] [PubMed] [Google Scholar]
- 41. Khokhlatchev A. V., Canagarajah B., Wilsbacher J., Robinson M., Atkinson M., Goldsmith E., Cobb M. H. (1998) Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 93, 605–615 [DOI] [PubMed] [Google Scholar]
- 42. Lorenz K., Schmitt J. P., Schmitteckert E. M., Lohse M. J. (2009) A new type of ERK1/2 autophosphorylation causes cardiac hypertrophy. Nat. Med. 15, 75–83 [DOI] [PubMed] [Google Scholar]
- 43. Anderson E., Quartararo C., Brown R. S., Shi Y., Yao X., Cole J. L. (2010) Analysis of monomeric and dimeric phosphorylated forms of protein kinase R. Biochemistry 49, 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bollengier F., Beeckmans S., Mahler A., Kanarek L. (1989) Further physicochemical characterization of the novel human brain protein h3. J. Neurochem. 52, 1123–1126 [DOI] [PubMed] [Google Scholar]
- 45. Akiva E., Itzhaki Z., Margalit H. (2008) Built-in loops allow versatility in domain-domain interactions. Lessons from self-interacting domains. Proc. Natl. Acad. Sci. U.S.A. 105, 13292–13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Crasto C. J., Feng J. (2001) Sequence codes for extended conformation. A neighbor-dependent sequence analysis of loops in proteins. Proteins 42, 399–413 [DOI] [PubMed] [Google Scholar]
- 47. Jones S., Thornton J. M. (1995) Protein-protein interactions. A review of protein dimer structures. Prog. Biophys. Mol. Biol. 63, 31–65 [DOI] [PubMed] [Google Scholar]
- 48. Ahn J. H., Miller D., Winter V. J., Banfield M. J., Lee J. H., Yoo S. Y., Henz S. R., Brady R. L., Weigel D. (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25, 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ungerer M., Böhm M., Elce J. S., Erdmann E., Lohse M. J. (1993) Altered expression of β-adrenergic receptor kinase and β1-adrenergic receptors in the failing human heart. Circulation 87, 454–463 [DOI] [PubMed] [Google Scholar]
- 50. Lohse M. J., Engelhardt S., Eschenhagen T. (2003) What is the role of β-adrenergic signaling in heart failure? Circ. Res. 93, 896–906 [DOI] [PubMed] [Google Scholar]
- 51. Belmonte S. L., Blaxall B. C. (2011) G protein-coupled receptor kinases as therapeutic targets in cardiovascular disease. Circ. Res. 109, 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Raake P. W., Vinge L. E., Gao E., Boucher M., Rengo G., Chen X., DeGeorge B. R., Jr., Matkovich S., Houser S. R., Most P., Eckhart A. D., Dorn G. W., 2nd, Koch W. J. (2008) G protein-coupled receptor kinase 2 ablation in cardiac myocytes before or after myocardial infarction prevents heart failure. Circ. Res. 103, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koch W. J., Rockman H. A., Samama P., Hamilton R. A., Bond R. A., Milano C. A., Lefkowitz R. J. (1995) Cardiac function in mice overexpressing the beta-adrenergic receptor kinase or a β ARK inhibitor. Science 268, 1350–1353 [DOI] [PubMed] [Google Scholar]