Background: “Designer” nucleosomes are key tools to investigate functions of histone modifications.
Results: Histone H2Aub inhibits PRC2 methylation of histone H3 Lys-27 but not Dot1L methylation of H3 Lys-79.
Conclusion: The position of nucleosome monoubiquitylation affects the specificity and direction of cross-talk with histone methyltransferases.
Significance: Cross-talk between histone modifications and chromatin enzymatic activities may be an important mechanism for generating distinct biological outputs.
Keywords: Chromatin, Chromatin Histone Modification, Chromatin Regulation, Histone Methylation, Ubiquitylation, Histone Monoubiquitylation, Methyltransferases
Abstract
It is well established that chromatin is a destination for signal transduction, affecting many DNA-templated processes. Histone proteins in particular are extensively post-translationally modified. We are interested in how the complex repertoire of histone modifications is coordinately regulated to generate meaningful combinations of “marks” at physiologically relevant genomic locations. One important mechanism is “cross-talk” between pre-existing histone post-translational modifications and enzymes that subsequently add or remove modifications on chromatin. Here, we use chemically defined “designer” nucleosomes to investigate novel enzymatic cross-talk relationships between the most abundant histone ubiquitylation sites, H2AK119ub and H2BK120ub, and two important histone methyltransferases, Dot1L and PRC2. Although the presence of H2Bub in nucleosomes greatly stimulated Dot1L methylation of H3K79, we found that H2Aub did not influence Dot1L activity. In contrast, we show that H2Aub inhibited PRC2 methylation of H3K27, but H2Bub did not influence PRC2 activity. Taken together, these results highlight how the position of nucleosome monoubiquitylation affects the specificity and direction of cross-talk with enzymatic activities on chromatin.
Introduction
Chromatin is the physiologically relevant form of eukaryotic genomes and thereby the biological substrate for DNA-templated processes, including transcription, mitosis, meiosis, and DNA replication and repair. The mononucleosome is the fundamental repeating unit of chromatin and is composed of ∼147 bp of DNA wrapped around an octamer of histone proteins, two each of histones H3, H4, H2A, and H2B (1–3). The diversity of processes regulated by chromatin is mirrored in the immense structural and compositional complexity of chromatin. Notably, histone proteins are extensively post-translationally modified by acetylation, phosphorylation, and, central to this study, monoubiquitylation and methylation (4, 5).
Many individual histone post-translational modifications (PTMs)2 have been studied in depth, but we are only beginning to understand the importance of combinations of PTMs in indexing eukaryotic genomes and specifying phenotypic changes in cell state. Recently, several genome-wide profiling studies of histone modifications revealed combinations of PTMs strongly associated with genome architecture and different chromatin states, including transcriptional status (6–8). These data elicit the question of how PTMs are coordinately regulated to generate meaningful combinations of “marks” at physiologically appropriate genomic locations.
An important mechanism of coordination is histone PTM “cross-talk” whereby the installation or removal of a histone modification is influenced by pre-existing histone PTMs. In landmark papers for trans-histone cross-talk, monoubiquitylation of H2B Lys-120 (H2Bub) was shown to be an in vivo requisite for methylation of H3 Lys-4 and Lys-79 (H3K4me and H3K79me, respectively) (9–12). As H3K79me and especially H3K4me are strongly associated with transcriptionally active genes (13, 14), these in vivo cross-talk studies firmly implicated H2Bub in active transcription.
Although crucial first steps, these yeast genetic studies were not able to rule out indirect effects as the cause of the observed striking ubiquitin-methylation cross-talk. However, a subsequent study using chemically defined ubiquitylated mononucleosomal substrates and purified methyltransferase (MTase) conclusively showed that H2Bub can directly stimulate H3K79 methylation by Dot1L (disruptor of telomeric silencing 1-like) (15). Similarly elegant in vitro cross-talk studies were performed with an H3K4 MTase, Set1 (SET domain-containing protein 1) (16). Furthermore, an evolutionarily conserved motif in Set1 required for H2Bub cross-talk with H3K4me in vitro and in vivo was recently identified.3
Despite the remarkable progress in understanding the mechanistic role of H2Bub in transcriptional regulation (17–20), the function of the predominant ubiquitylated species in metazoan chromatin, H2A monoubiquitylated at Lys-119 (H2Aub), remains less well understood. Studies of H2Aub function have been slowed in part by its restriction mostly to metazoans (unlike H2Bub, which is present in yeast), where histone genetics is not tractable. However, the discovery of the major E3 ligase for H2Aub in mammalian systems, Ring1B, a member of the PRC1 (Polycomb repressive complex 1) family (21), opened the door to studies implicating H2Aub in developmentally regulated transcriptional repression. Recently, using nucleosomes reconstituted with endogenous H2A or H2Aub, Nakagawa et al. (22) showed that deubiquitylation of H2Aub allowed for efficient methylation of H3K4 by the MTase MLL3. This study provided mechanistic support for H2Aub involvement in transcriptional repression. Furthermore, it revealed that H2Aub, in addition to H2Bub, participates in enzymatic cross-talk, albeit of an inhibitory nature.
Here, we employed designer mononucleosomes carrying site-specifically installed monoubiquitin to investigate novel direct cross-talk pathways with MTases for H3K79 and H3K27 (see Fig. 1). As noted above, H2Bub and H2Aub have opposing influences on methylation of H3K4 in vitro. Our specialized nucleosomal reagents allowed us to test whether H2Aub also functions antagonistically to H2Bub in cross-talk with another MTase involved in active transcription, Dot1L. Additionally, given the established direct enzymatic cross-talk between H2Bub and MTases implicated in active transcription, it has been hypothesized that H2Aub may directly stimulate MTases involved in transcriptional repression, in particular PRC2. Using chemically defined mononucleosomes, we tested whether PRC2-mediated H3K27 methylation activity is sensitive to the presence of H2Aub or H2Bub in nucleosomal substrates.
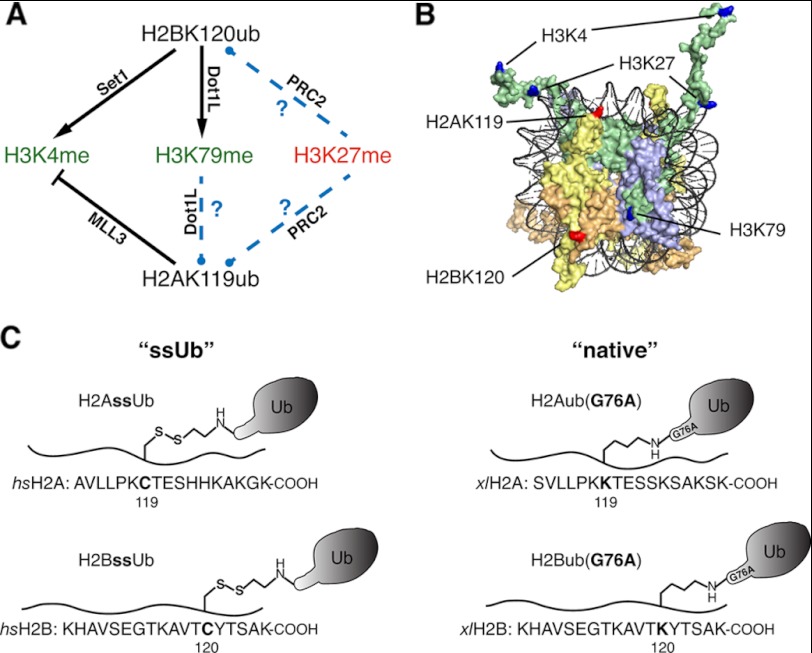
FIGURE 1.
Enzymatic cross-talk between histone ubiquitylation and methylation using designer monoubiquitylated histones. A, the black lines drawn between histone monoubiquitylation and histone methylation sites indicate validated direct enzymatic cross-talk between the indicated PTMs, with the relevant enzyme indicated. Standard and blunt-headed arrows signify enzymatic stimulation and inhibition, respectively, by the modification at the origin of the arrow. Novel cross-talk relationships tested in this study are indicated by dashed blue lines. Methylation states strongly associated with active or repressed transcription are colored green and red, respectively. B, surface rendition of mononucleosome structure (Protein Data Bank code 1KX5). DNA is colored black, and histones H3, H4, H2A, and H2B are colored green, lavender, yellow, and orange, respectively. Lysines site-specifically modified in this study by monoubiquitylation are indicated in red. Lysines colored blue are methylated and are targets for direct enzymatic cross-talk relationships with ubiquitylated histones. C, the four monoubiquitylated histones generated for this study are represented in schematic form. Amino acid sequences of H2A and H2B C termini are shown below a wavy line representing the peptide backbone. Left, ssUb refers to Homo sapiens (hs) H2AssUb and H2BssUb generated by disulfide-mediated ubiquitylation at position 119 in H2A and position 120 in H2B. Right, native refers to Xenopus laevis (xl) H2Aub(G76A) and H2Bub(G76A) generated by semisynthetic expressed protein ligation.
EXPERIMENTAL PROCEDURES
Recombinant Histone Expression and Purification
Recombinant human histones H3.2(C110A), H4, H2A.a4, H2A.a4(K119C), H2B.k, and H2B.k(K120C) were expressed as His6-tagged fusion proteins in Escherichia coli and purified as described previously (23). β-Mercaptoethanol-H2A(K119C) and β-mercaptoethanol-H2B(K120C) adducts formed during purification and tobacco etch virus/rhinovirus 3C cleavage steps were reduced with 20 mm tris(2-carboxyethyl)phosphine at pH 8.5 and purified by reverse-phase HPLC before use in H2AssUb or H2BssUb ligation reactions. Careful removal of reducing agents by extensive dialysis and/or HPLC is crucial for all histone preparations because the eventual disulfide bonds in H2AssUb and H2BssUb are extremely sensitive to reducing agents (see Fig. 2D). Recombinant Xenopus histones were prepared as described previously (15).
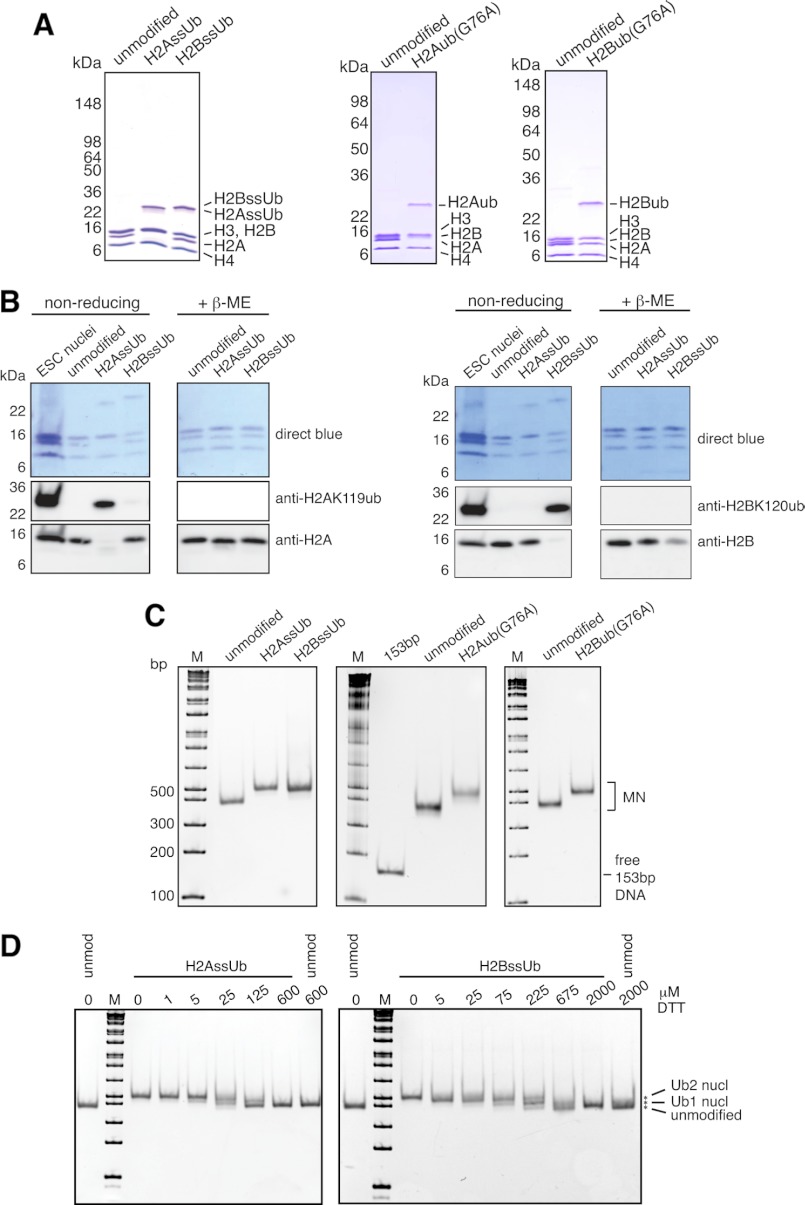
FIGURE 2.
Characterization of designer octamers and mononucleosomes. A and B, octamers assembled from unmodified and monoubiquitylated histones were separated by SDS-PAGE and characterized by Coomassie Blue staining (A) or Western blotting with the indicated antibodies (B). Nuclei from mouse embryonic stem cells (ESC) were used to mark the migration positions of endogenous H2AK119ub and H2BK120ub. β-ME, β-mercaptoethanol. C, human unmodified, H2AssUb, H2BssUb, H2Aub(G76A), and H2Bub(G76A) octamers were assembled into mononucleosomes with 153-bp (601) positioning sequence DNA and separated by native 5% PAGE. Gels were stained with ethidium bromide to confirm the absence of free 153-bp DNA and to assess the structural homogeneity of the assembled mononucleosome (MN) pools. The marker (M) is the 1-kb Plus ladder from New England Biolabs. D, unmodified (unmod nucl), H2AssUb, and H2BssUb mononucleosomes were adjusted to the indicated DTT concentrations and incubated at 37 °C for 3 h before separation by native 5% PAGE and stained with ethidium bromide to assess the stability of the disulfide bond between histone and ubiquitin.
H2AssUb and H2BssUb Preparation
Disulfide-directed ubiquitylation of human histones H2A(K119C) and H2B(K120C) (see Fig. 1C) was performed as described previously (24) with the following alteration. Here, the ubiquitin-aminoethanethiol (Ub-SH) adduct, rather than the histone cysteine sulfhydryl, was activated with 2,2′-dithiobis(5-nitropyridine). Briefly, an asymmetric disulfide between the C-terminal sulfhydryl of Ub-SH and 2,2′-dithiobis(5-nitropyridine) was generated by incubation of 2,2′-dithiobis(5-nitropyridine) and Ub-SH in a 3:1 mixture of acetic acid and water. After 18–24 h at 25 °C, the reaction was dialyzed against water to precipitate unreacted 2,2′-dithiobis(5-nitropyridine), and the Ub-SH-5-nitro-2-pyridinesulfenyl product was purified by C18 reverse-phase HPLC. Purified Ub-SH-5-nitro-2-pyridinesulfenyl was mixed at ∼3:1 molar ratio with histone (either H2A(K119C) or H2B(K120C)) in a small volume of 6 m guanidine HCl and 1 m HEPES (pH 6.9), and the reaction was stopped with 0.1% TFA after 10 min. The resulting H2AssUb and H2BssUb were purified by C18 reverse-phase HPLC. Note that Invitrogen NuPAGE precast gels reduce the disulfide bond between Ub-SH and histone H2A(K119C) and H2B(K120C), but Bio-Rad TGX gels leave the disulfide intact.
H2Bub/H2Aub(G76A) Preparation
Xenopus H2Bub(G76A)was prepared by semisynthetic expressed protein ligation (see Fig. 1C) as described previously (25). Xenopus H2Aub(G76A) was prepared using an adapted semisynthetic expressed protein ligation strategy which will be described in detail elsewhere.4 These ubiquitylated histones contain a normal isopeptide bond linkage but harbor a G76A mutation within ubiquitin (referred to herein as “native”).
Octamer Preparation
Octamers were assembled by combining the appropriate purified histones at ∼2 mg/ml in denaturing buffer (50 mm MOPS (pH 7), 6 m guanidine HCl, 10% glycerol, and 5 mm EDTA) and dialyzed overnight into high salt buffer (10 mm MOPS (pH 7), 2 m NaCl, 10% glycerol, and 0.5 mm EDTA). Folded octamers were purified from soluble material by FPLC with Superdex 200 (GE Healthcare) and stored at −20 °C in 50% glycerol. Xenopus H2Bub(G76A) octamers were prepared as described previously (15).
Nucleosome Positioning Sequence Preparation and Mononucleosome Assembly
DNA fragments for mononucleosome assemblies were prepared by EcoRV digest of 32× 153-bp tandem repeats of the 601 positioning sequence (26) cloned into pUC19 (a kind gift from Kyle Chiang). Plasmid backbone fragments were precipitated with 7.5% PEG 6000 at 500 mm NaCl, and soluble 153-bp fragments were purified with several phenol/chloroform extractions and ethanol-precipitated. Mononucleosomes were prepared by gradient dialysis as described previously (23), followed by dialysis into low salt buffer (10 mm Tris (pH 7.5), 10m M NaCl, 1 mm EDTA, and 10% glycerol). Assembly quality was assessed by native 5% PAGE and ethidium bromide staining.
Dot1L MTase Assays
FLAG-tagged human Dot1L was purified from a baculovirus expression system as described previously (15, 25). Typical assays combined 4.1 pmol of mononucleosomes, 0.14 pmol of FLAG-tagged human Dot1L, and 15 pmol of S-[3H]adenosyl-l-methionine (1 μCi of 3H) in 12 μl of reaction buffer (20 mm Tris (pH 8.0), 10 mm NaCl, 5 mm MgCl2, and 1 mm EDTA) at 30 °C. Activity was assessed by fluorography and/or scintillation counting.
PRC2 MTase Assays
Recombinant PRC2 complex (Ezh2, FLAG-Eed3, Suz12, and RbAP48) was purified using a baculovirus expression system as described previously (27). Typical assays combined 200 ng of nucleosomal DNA, 20 ng of Ezh2 in PRC2 complex, and 15 pmol of S-[3H]adenosyl-l-methionine (1 μCi of 3H) in 12 μl of reaction buffer (20 mm Tris (pH 8.0), 10 mm NaCl, 2.5 mm MgCl2, 1 mm EDTA, and 0.5 mm DTT) at 30 °C. Activity was assessed by fluorography and/or scintillation counting.
Antibodies
The antibodies used were anti-H2A (Millipore 07-146), anti-H2B (Proteintech), anti-H2AK119ub (22), and anti-H2BK120ub (Cell Signaling Technology 5546).
RESULTS
Design and Characterization of Designer Mononucleosomes
The inherent heterogeneity of chromatin purified from endogenous sources has historically obstructed efforts to establish direct enzymatic cross-talk relationships between histone PTMs. Ideal substrates for cross-talk studies require robust and specific installation of modifications on otherwise unmodified histones (28). Recently, several groups have developed chemical strategies for the site-specific installation of histone PTMs, including monoubiquitylation (29). These chemically defined modified histones can be combined with histones expressed recombinantly in E. coli, devoid of PTMs, to reconstitute so-called designer nucleosomes.
We utilized two complementary methods to quantitatively install monoubiquitin at position 119 in H2A and position 120 in H2B (Fig. 1C): disulfide-mediated (ssUb) (24) and expressed protein ligation (native) (25). These approaches are more specific than in vitro enzymatic ubiquitylation and generate histone-ubiquitin linkages that differ minimally from the native linkage (24, 25). Furthermore, H2BssUb and native H2Bub function indistinguishably from H2Bub with the native linkage in Dot1L MTase stimulation (24, 25). For our studies, unmodified and ubiquitylated histones were assembled into designer octamers of six types: human unmodified, H2AssUb, and H2BssUb and Xenopus unmodified, H2Bub(G76A), and H2Aub(G76A) (Fig. 2, A and B).
Relative to other prevalent histone PTMs, monoubiquitylation is an extremely bulky modification (itself being ∼60% of the molecular weight of histones H2A and H2B). For our enzymatic cross-talk studies, we sought to eliminate the possibility that ubiquitylation-dependent changes in array conformation could alter the accessibility of a target lysine for subsequent methylation. Therefore, we reconstituted our octamers into designer mononucleosomes using a 153-bp sequence containing the 601 strong positioning sequence (26). The structural homogeneity of assembled mononucleosomes was assessed by native PAGE electrophoresis and ethidium bromide staining (Fig. 2C).
Anticipating that the disulfide linkage in our ssUb nucleosomes could present challenges for biochemical studies given the frequent necessity of reducing conditions for enzymatic activity, we performed titrations with DTT to ascertain whether low concentrations of reducing agent would leave the histone-ubiquitin disulfide bond appreciably intact. However, the mobility of H2AssUb and H2BssUb nucleosomes on native PAGE gels indicated significant disulfide reduction even at 25 μm DTT (Fig. 2D). Additionally, we found H2AssUb mononucleosomes to be ∼3 times more sensitive to DTT than their H2BssUb counterparts. This is reminiscent of the differential accessibility of H2AK119ub and H2BK120ub to trypsin: H2AK119ub in endogenous chromatin is trypsin-sensitive, whereas H2BK120ub is resistant (30, 31).
H2Bub, but Not H2Aub, Participates in Cross-talk with Dot1L, MTase for H3K79
Monoubiquitylation of H2BK120 is a potent stimulator of Dot1L-mediated methylation of H3K79 (15). Interestingly, monoubiquitylation at several other positions on the nucleosome axial surface, and even on the radial edge, also dramatically stimulated Dot1L MTase activity (Fig. 3A) (24, 32). Taken together, these studies reveal significant positional plasticity in the histone ubiquitylation site for Dot1L MTase stimulation. The location of H2AK119ub, on the opposite end of the nucleosome axial face, and its ∼10-fold greater abundance than H2BK120ub in vivo (33, 34) make H2Aub an interesting test case for Dot1L plasticity to stimulation by histone ubiquitylation.
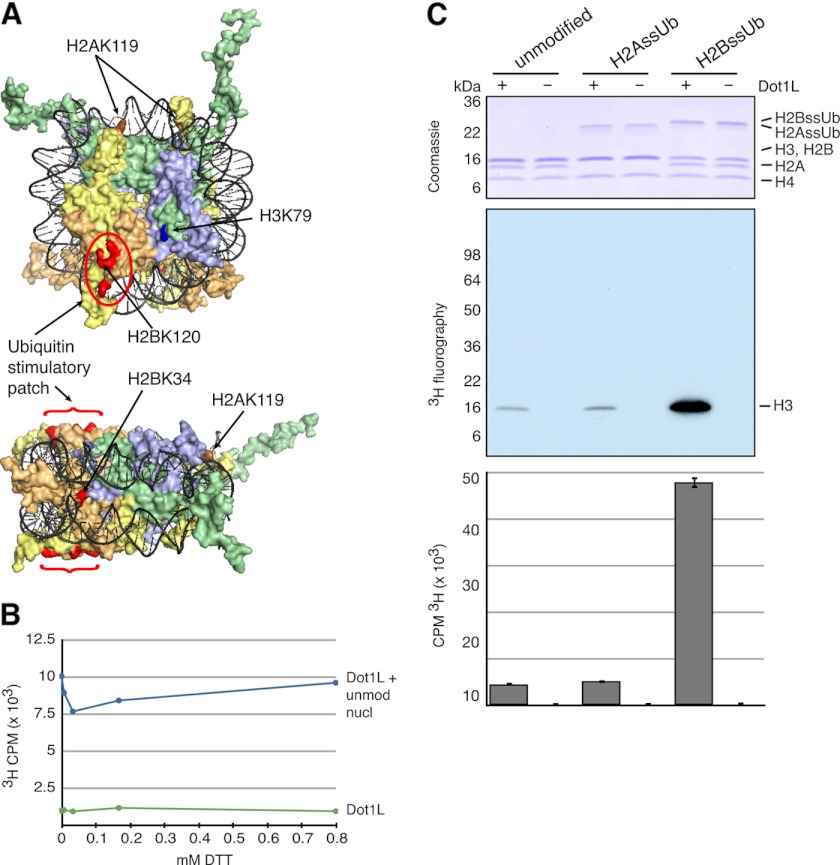
FIGURE 3.
H2Aub does not affect Dot1L MTase activity on mononucleosomes. A, surface rendition of mononucleosome structure shown in axial and radial views (Protein Data Bank code 1KX5). DNA is colored black, and histones H3, H4, H2A, and H2B are colored green, lavender, yellow, and orange, respectively. The Dot1 methylation site (H3K79) is colored dark blue. Positions that when ubiquitylated strongly stimulate Dot1L activity are colored red. Located at the opposite side of the nucleosome axial face from H2BK120, H2AK119 is colored brown. B, DTT titration of Dot1L MTase activity on unmodified mononucleosomes (unmod nucl) as quantitated by scintillation counting of incorporated 3H. C, Dot1L MTase activity was tested on human unmodified and ssUb mononucleosomes as indicated. A representative experiment is shown. Three-fourths of each reaction was separated by SDS-PAGE and Coomassie Blue-stained (upper panel), and activity was assessed by fluorography (middle panel). Activity was also assessed by scintillation counting of incorporated 3H in one-fourth of each reaction (lower panel). Error bars indicate the S.D. of multiple scintillation counts of the same reaction.
Dot1L MTase activity was undiminished under nonreducing conditions (Fig. 3B), which allowed the use of ssUb nucleosomes as substrates for in vitro MTase cross-talk assays. As reported previously (24), Dot1L methylation of H3 is greatly stimulated by nucleosomal H2BssUb. In striking contrast, the incorporation of H2AssUb into an otherwise unmodified nucleosome had no appreciable impact on Dot1L-mediated methylation (Fig. 3C). On the basis of these experiments, we conclude that plasticity in monoubiquitin stimulation of Dot1L does not extend to the most abundant ubiquitylated histone in metazoans, H2AK119ub. In addition to deepening our understanding of Dot1L MTase regulation in vitro, this result supports the view that differential functions of H2Aub and H2Bub exist in vivo with respect to Dot1L and likely other chromatin-remodeling activities.
PRC2, MTase for H3K27, Is Modestly Inhibited by H2Aub but Is Unaffected by H2Bub in Mononucleosomal Substrates
Previous studies have shown several MTases involved in active transcription to be remarkably sensitive to monoubiquitylated nucleosomal substrates (15, 16, 32). However, to our knowledge, there are no reports testing transcriptionally repressive MTases in direct cross-talk experiments with histone ubiquitylation. For a transcriptionally repressive MTase, we chose to investigate the regulation of PRC2, the MTase for H3K27, because of its important role in cellular differentiation, stem cell identity, X-inactivation, cell cycle regulation, and cancer (35).
PRC2 target genes have been mapped in a variety of cellular contexts, and in many cases, these genes can be either methylated at H3K27, H3K4, or both depending on the particular cellular/developmental context (36, 37). In fact, the balance between H3K4me and H3K27me at gene promoters may be an important mechanism of transcriptional regulation (38, 39). Others have shown that the activity of MTases for H3K4 are stimulated or repressed by H2Bub and H2Aub, respectively (16, 22). Given the opposing function of H3K4me and H3K27me in transcriptional regulation, we wondered whether H2Aub and H2Bub also directly and antagonistically regulate PRC2-mediated methylation of H3K27.
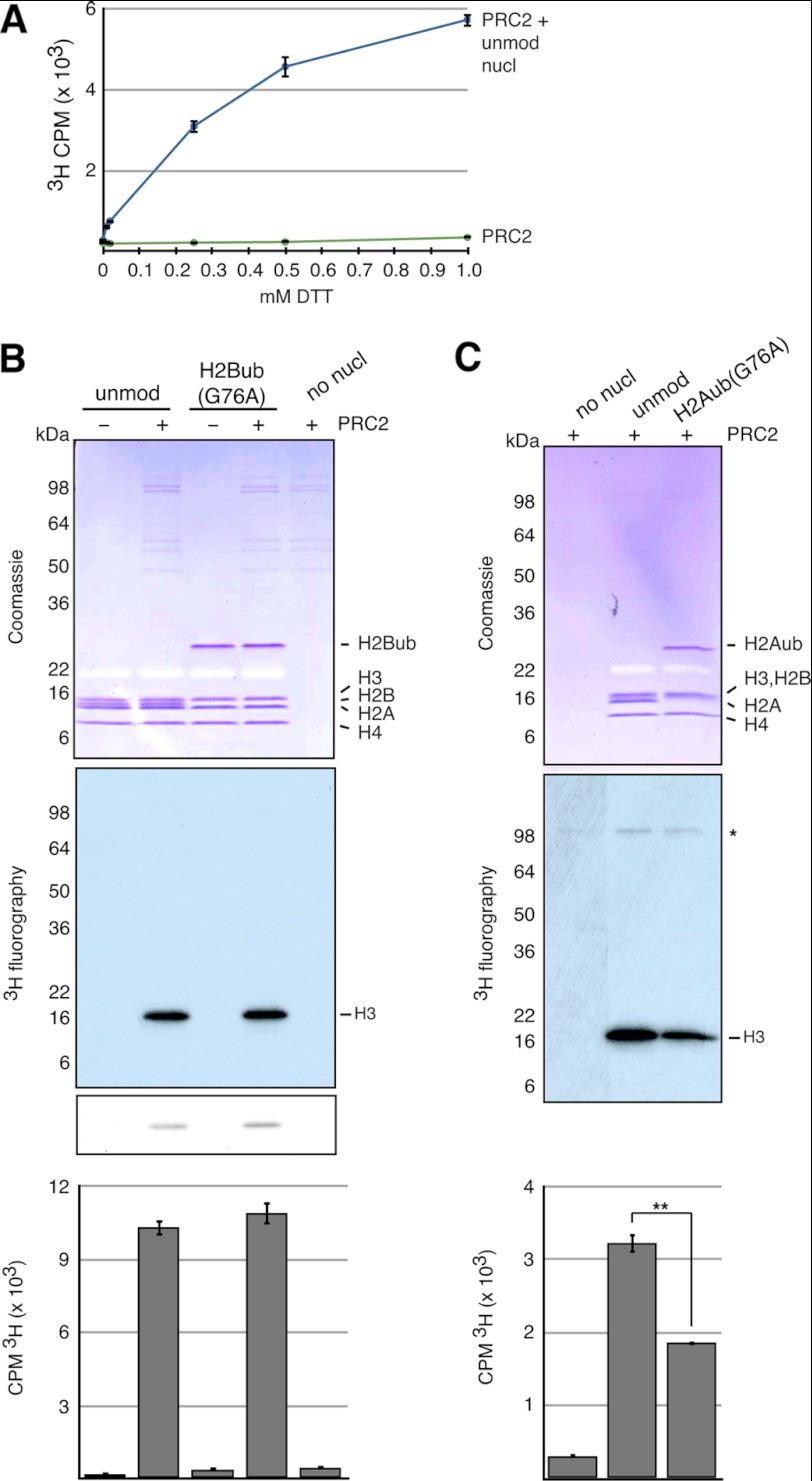
As reducing agents were required for PRC2 activity (Fig. 4A), we turned to native ubiquitylated mononucleosomes for our enzymatic cross-talk studies. Unlike Set1 and Dot1L MTases, PRC2 MTase activity was unaffected by the presence of H2Bub in nucleosomal substrates (Fig. 4B and supplemental Fig. 1). However, H2Aub modestly but reproducibly inhibited PRC2 MTase activity (Fig. 4C). This was an unexpected result because both H3K27me and H2Aub are transduced by PRC2 and PRC1, respectively, with well established roles in gene repression. However, several recent publications provide evidence that the functional link between H2Aub and transcriptional repression is not as clear as was once thought (40–42), and these will be discussed further below.
FIGURE 4.
H2Aub inhibits PRC2 MTase activity on mononucleosomes. A, DTT titration of PRC2 MTase activity on unmodified mononucleosomes (unmod nucl) as quantitated by scintillation counting of incorporated 3H. Error bars represent the S.D. of multiple scintillation counts. B and C, PRC2 MTase activity was tested under reducing conditions on Xenopus unmodified, H2Bub(G76A), and H2Aub(G76A) mononucleosomes. Representative experiments are shown. Reactions were separated by SDS-PAGE and Coomassie Blue-stained, and activity was assessed by fluorography. In B, a short exposure is shown below a longer exposure. In C, automethylation activity is indicated (*). Activity was also assessed by scintillation counting of incorporated 3H into a fraction of each reaction (one-half in A and one-fourth in B). Error bars indicate the S.D. of multiple scintillation counts of the same reaction. In C, **, p < 0.0001 (Student's paired test).
DISCUSSION
Cross-talk between histone PTMs may have evolved as a way to amplify, buffer, and integrate diverse signals impinging on chromatin. Elucidating which PTMs “talk” to each other, which do not, and via what mechanisms is critical in our quest to understand cellular memory, development, and disease. To study direct enzymatic cross-talk relationships between PTMs, the use of highly defined in vitro chromatin templates is critical (28). Here, we utilized chemically defined monoubiquitylated histones to generate a set of mononucleosomal substrates for MTase assays to investigate novel functions of histone ubiquitylation and to shed light on the plasticity/specificity of cross-talk with histone MTases (Fig. 5).
FIGURE 5.
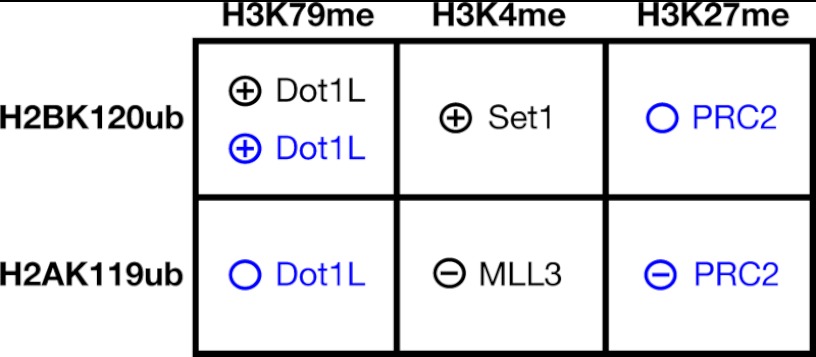
Summary of enzymatic cross-talk relationships between histone monoubiquitylation and methylation. Left, histone monoubiquitylation sites known to directly influence MTase activity directed toward the indicated histone methylation sites (top) are indicated. Inside, the relevant MTase and the type of cross-talk observed are indicated. ⊕, the ubiquitylated histone directly stimulates the indicated MTase; ⊝, inhibition; ○, no detected stimulation or inhibition. The results presented here and those published by others (see text) are colored in blue and black, respectively.
In landmark papers, H2Bub was shown to be an in vivo requisite for methylation of H3K79 by Dot1 (9, 11). Careful subsequent studies using nucleosomes with site-specifically installed monoubiquitin revealed that H2BK120ub directly stimulates Dot1 MTase activity (15). Interestingly, H2BK120ub is not unique in its ability to cross-talk with Dot1L-mediated H3K79 methylation. Ubiquitylation at several other sites on the nucleosome axial face and on the radial edge can also stimulate Dot1L activity (24, 32). From these data, it is clear that Dot1L stimulation is relatively plastic with regard to the position of histone monoubiquitylation. Using a disulfide-mediated approach, we installed monoubiquitin at position 119 of H2A to test whether H2Aub, the most abundant ubiquitylated species in metazoan chromatin, could influence Dot1L MTase activity on mononucleosomes. Our results clearly show that the observed plasticity of the ubiquitylation site for Dot1L stimulation in vitro does not extend to H2Aub (Fig. 3C). Thus, even though H2Aub is a relatively abundant mark in mammalian systems, it seems likely that its biological function is not linked to H3K79 methylation and downstream events.
Most studies of the function of H2Bub in metazoans have focused on its role in transcriptional activation (43). Although the steady-state H2Bub level correlates well with the most highly expressed genes genome-wide (44), in vivo depletion of RNF20 (E3 ligase for H2BK120) and near abolishment of H2Bub revealed a subset of genes repressed by RNF20 (45). Surprisingly, genes repressed by RNF20 were found to be associated with higher levels of H2Bub than genes activated by RNF20. These data raise the possibility that H2Bub may be involved in transcriptional repression in some chromatin contexts.
Given that H2Bub dramatically and directly stimulates the evolutionarily unrelated MTases Dot1 and Set1, we wondered whether the ability of H2Bub to stimulate MTases is restricted to MTases involved in active transcription (e.g. H3K4me and H3K79me) or if it extends to MTases involved in transcriptional repression (such as PRC2). In vitro PRC2 MTase assays revealed no cross-talk, in either a stimulatory or inhibitory direction, between H2Bub and PRC2-mediated H3K27me (Fig. 4B). Based on these results, it is unlikely that putative H2Bub-mediated transcriptional repression, as observed in genome-wide studies (45), is caused by direct stimulation of PRC2 activity. Going forward, it will be interesting to understand at the molecular/structural level how H2Bub selectivity stimulates MTases Dot1L and Set1 but not PRC2.
Although H2Aub is typically understood to cooperate with PRC2-mediated H3K27 methylation in transcriptional repression, we found that H2Aub produced a modest but reproducible inhibition of PRC2 activity in vitro (Fig. 4C). One more trivial explanation for our finding might be that the relatively close proximity of H2AK119ub to H3K27 in the nucleosome (Fig. 1B) may simply sterically hinder PRC2 MTase activity. Although this possibility cannot be ruled out by our studies, recently published data from several groups suggest that the widely accepted link between H2Aub and transcriptional repression may not be so straightforward (40–42).
Eskeland et al. (42) showed that the H2A E3 ligase activity of Ring1B is dispensable for transcriptional repression of Polycomb target genes and for long-range chromatin compaction of Hox gene clusters in vivo. In light of our in vitro results, it would be of interest to know whether cells with catalytically null Ring1B have attenuated or otherwise misregulated activation of Hox genes during differentiation.
Scheuermann et al. (41) identified a novel Polycomb repressive complex in Drosophila with H2A deubiquitinase activity, PR-DUB. Interestingly, they found that H2A deubiquitinase activity is required for proper transcriptional repression of Polycomb target genes in vivo. Furthermore, repression of Polycomb target genes is even more severely compromised if dRing, the E3 ligase for H2Aub, and PR-DUB are simultaneously depleted. Therefore, at least in Drosophila, complexes with directly opposing enzymatic activities on H2A synergistically regulate transcriptional repression. These results suggest that H2Aub in different genomic locations may have opposite influences on transcription or that dynamic cycles of H2A ubiquitylation and deubiquitylation may be required for proper gene repression. We look forward to studies that will investigate how the spatial, temporal, and kinetic control of H2Aub turnover on chromatin contributes to gene regulation. Such studies would greatly inform future in vitro studies using designer chromatin to more closely mimic the dynamism of histone ubiquitylation in vivo.
Finally, Richly et al. (40) identified ZRF1 as an H2Aub-binding protein involved in differentiation-dependent transcriptional activation of Polycomb target genes. If reduced PRC2 activity on Polycomb group-repressed genes is an important early step in pushing the chromatin state toward one more permissive to transcription, then our in vitro cross-talk data may provide another layer of mechanistic explanation for H2Aub involvement in transcriptional derepression.
Although our studies represent a significant advance in the use of designer nucleosomes containing H2Aub versus H2Bub, important uncertainties remain that will need to be addressed in future experiments. In particular, the role of ubiquitin turnover (e.g. due to the cyclic action of ubiquitin ligases and deubiquitinases) is not addressed in our system, nor are potential differences in spatial and temporal control of H2Aub and H3K27me in transcriptional regulation.
We look forward to subsequent studies that will use designer chromatin to explore enzymatic cross-talk relationships between H2Aub and H2Bub with not only MTases but also with other histone-modifying activities. More generally, we expect further study of histone PTM cross-talk relationships to be a fertile approach to understand how combinations of PTMs are coordinately regulated to maintain and affect dynamic changes in chromatin state, such as those that accompany gene expression changes resulting from developmental or environmental signal transduction.
Acknowledgments
We thank L. A. Banaszynski, C. M. Hughes, and J.-A. Kim for critical reading of the manuscript; R. Sadeh for helpful discussions; C. Chatterjee for help and guidance with preparing ssUb histones; and S. Wu and Y. Zhang for the generous contribution of the PRC2 complex.
This work was supported, in whole or in part, by National Institutes of Health Grant 2R01 GM040922-27. This work was also supported by Swiss National Science Foundation Grant PP00P3_133680 and The Rockefeller University.

This article contains supplemental Fig. 1.
J. Kim, J.-A. Kim, C. D. Allis, R. G. Roeder et al., personal communication.
B. Fierz and T. W. Muir, manuscript in preparation.
- PTM
- post-translational modification
- MTase
- methyltransferase
- Ub-SH
- ubiquitin-aminoethanethiol.
REFERENCES
- 1. Kornberg R. D. (1974) Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–871 [DOI] [PubMed] [Google Scholar]
- 2. Van Holde K. E., Allen J. R., Tatchell K., Weischet W. O., Lohr D. (1980) DNA-histone interactions in nucleosomes. Biophys. J. 32, 271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 4. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 5. Tan M., Luo H., Lee S., Jin F., Yang J. S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Z., Zang C., Rosenfeld J. A., Schones D. E., Barski A., Cuddapah S., Cui K., Roh T. Y., Peng W., Zhang M. Q., Zhao K. (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Minoda A., Riddle N. C., Ernst J., Sabo P. J., Larschan E., Gorchakov A. A., Gu T., Linder-Basso D., Plachetka A., Shanower G., Tolstorukov M. Y., Luquette L. J., Xi R., Jung Y. L., Park R. W., Bishop E. P., Canfield T. K., Sandstrom R., Thurman R. E., MacAlpine D. M., Stamatoyannopoulos J. A., Kellis M., Elgin S. C., Kuroda M. I., Pirrotta V., Karpen G. H., Park P. J. (2011) Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471, 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roudier F., Ahmed I., Bérard C., Sarazin A., Mary-Huard T., Cortijo S., Bouyer D., Caillieux E., Duvernois-Berthet E., Al-Shikhley L., Giraut L., Després B., Drevensek S., Barneche F., Dèrozier S., Brunaud V., Aubourg S., Schnittger A., Bowler C., Martin-Magniette M. L., Robin S., Caboche M., Colot V. (2011) Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30, 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Briggs S. D., Xiao T., Sun Z. W., Caldwell J. A., Shabanowitz J., Hunt D. F., Allis C. D., Strahl B. D. (2002) Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418, 498. [DOI] [PubMed] [Google Scholar]
- 10. Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., Johnston M., Shilatifard A. (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277, 28368–28371 [DOI] [PubMed] [Google Scholar]
- 11. Ng H. H., Xu R. M., Zhang Y., Struhl K. (2002) Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277, 34655–34657 [DOI] [PubMed] [Google Scholar]
- 12. Sun Z. W., Allis C. D. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 13. Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. (2002) Active genes are trimethylated at K4 of histone H3. Nature 419, 407–411 [DOI] [PubMed] [Google Scholar]
- 14. Schübeler D., MacAlpine D. M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D. E., O'Neill L. P., Turner B. M., Delrow J., Bell S. P., Groudine M. (2004) The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18, 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McGinty R. K., Kim J., Chatterjee C., Roeder R. G., Muir T. W. (2008) Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453, 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J., Guermah M., McGinty R. K., Lee J. S., Tang Z., Milne T. A., Shilatifard A., Muir T. W., Roeder R. G. (2009) RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henry K. W., Wyce A., Lo W. S., Duggan L. J., Emre N. C., Kao C. F., Pillus L., Shilatifard A., Osley M. A., Berger S. L. (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17, 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717 [DOI] [PubMed] [Google Scholar]
- 19. Tanny J. C., Erdjument-Bromage H., Tempst P., Allis C. D. (2007) Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 21, 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiao T., Kao C. F., Krogan N. J., Sun Z. W., Greenblatt J. F., Osley M. A., Strahl B. D. (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25, 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 22. Nakagawa T., Kajitani T., Togo S., Masuko N., Ohdan H., Hishikawa Y., Koji T., Matsuyama T., Ikura T., Muramatsu M., Ito T. (2008) Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 22, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruthenburg A. J., Li H., Milne T. A., Dewell S., McGinty R. K., Yuen M., Ueberheide B., Dou Y., Muir T. W., Patel D. J., Allis C. D. (2011) Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chatterjee C., McGinty R. K., Fierz B., Muir T. W. (2010) Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol. 6, 267–269 [DOI] [PubMed] [Google Scholar]
- 25. McGinty R. K., Köhn M., Chatterjee C., Chiang K. P., Pratt M. R., Muir T. W. (2009) Structure-activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem. Biol. 4, 958–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lowary P. T., Widom J. (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 [DOI] [PubMed] [Google Scholar]
- 27. Cao R., Zhang Y. (2004) SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15, 57–67 [DOI] [PubMed] [Google Scholar]
- 28. Allis C. D., Muir T. W. (2011) Spreading chromatin into chemical biology. ChemBioChem 12, 264–279 [DOI] [PubMed] [Google Scholar]
- 29. Chatterjee C., Muir T. W. (2010) Chemical approaches for studying histone modifications. J. Biol. Chem. 285, 11045–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Böhm L., Briand G., Sautière P., Crane-Robinson C. (1982) Proteolytic digestion studies of chromatin core histone structure. Identification of limit peptides from histone H2B. Eur. J. Biochem. 123, 299–303 [DOI] [PubMed] [Google Scholar]
- 31. Böhm L., Crane-Robinson C., Sautière P. (1980) Proteolytic digestion studies of chromatin core histone structure. Identification of a limit peptide of histone H2A. Eur. J. Biochem. 106, 525–530 [DOI] [PubMed] [Google Scholar]
- 32. Wu L., Zee B. M., Wang Y., Garcia B. A., Dou Y. (2011) The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in cross-talk with H3 K4 and K79 methylation. Mol. Cell 43, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goldknopf I. L., Busch H. (1977) Isopeptide linkage between non-histone and histone 2A polypeptides of chromosomal conjugate protein A24. Proc. Natl. Acad. Sci. U.S.A. 74, 864–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. West M. H., Bonner W. M. (1980) Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 8, 4671–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Margueron R., Reinberg D. (2011) The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 37. Papp B., Müller J. (2006) Histone trimethylation and the maintenance of transcriptional ON and OFF states by TrxG and PcG proteins. Genes Dev. 20, 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007) Genome regulation by Polycomb and Trithorax proteins. Cell 128, 735–745 [DOI] [PubMed] [Google Scholar]
- 39. Cloos P. A., Christensen J., Agger K., Helin K. (2008) Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22, 1115–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Richly H., Rocha-Viegas L., Ribeiro J. D., Demajo S., Gundem G., Lopez-Bigas N., Nakagawa T., Rospert S., Ito T., Di Croce L. (2010) Transcriptional activation of Polycomb-repressed genes by ZRF1. Nature 468, 1124–1128 [DOI] [PubMed] [Google Scholar]
- 41. Scheuermann J. C., de Ayala Alonso A. G., Oktaba K., Ly-Hartig N., McGinty R. K., Fraterman S., Wilm M., Muir T. W., Müller J. (2010) Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eskeland R., Leeb M., Grimes G. R., Kress C., Boyle S., Sproul D., Gilbert N., Fan Y., Skoultchi A. I., Wutz A., Bickmore W. A. (2010) Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38, 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weake V. M., Workman J. L. (2008) Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–663 [DOI] [PubMed] [Google Scholar]
- 44. Minsky N., Shema E., Field Y., Schuster M., Segal E., Oren M. (2008) Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 10, 483–488 [DOI] [PubMed] [Google Scholar]
- 45. Shema E., Tirosh I., Aylon Y., Huang J., Ye C., Moskovits N., Raver-Shapira N., Minsky N., Pirngruber J., Tarcic G., Hublarova P., Moyal L., Gana-Weisz M., Shiloh Y., Yarden Y., Johnsen S. A., Vojtesek B., Berger S. L., Oren M. (2008) The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 22, 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]