Background: Neurodegenerative diseases involve proteotoxic stress and apoptosis.
Results: NPD1 inhibits proteotoxic stress-induced apoptosis.
Conclusion: NPD1 synthesis is an early response to proteotoxic stress.
Significance: This might be one of the first survival defenses activated in neurodegenerations.
Keywords: Apoptosis, Lipids, Neurodegenerative Diseases, Oxidative Stress, Retinal Degeneration
Abstract
Neurodegenerative diseases share two common features: enhanced oxidative stress and cellular inability to scavenge structurally damaged abnormal proteins. Pathogenesis of polyglutamine (poly(Q)) diseases involves increased protein misfolding, along with ubiquitin and chaperon protein-containing nuclear aggregates. In spinocerebellar ataxia, the brain and retina undergo degeneration. Neuroprotectin D1 (NPD1) is made on-demand in the nervous system and retinal pigment epithelial (RPE) cells in response to oxidative stress, which activates prosurvival signaling via regulation of gene expression and other processes. We hypothesized that protein misfolding-induced proteotoxic stress triggers NPD1 synthesis. We used ARPE-19 cells as a cellular model to assess stress due to ataxin-1 82Q protein expression and determine whether NPD1 prevents apoptosis. Ectopic ataxin-1 expression induced RPE cell apoptosis, which was abrogated by 100 nm docosahexaenoic acid, 10 ng/ml pigment epithelium-derived factor, or NPD1. Similarly, NPD1 was protective in neurons and primary human RPE cells. Furthermore, when ataxin-1 82Q was expressed in 15-lipoxygenase-1-deficient cells, apoptosis was greatly enhanced, and only NPD1 (50 nm) rescued cells from death. NPD1 reduced misfolded ataxin-1-induced accumulation of proapoptotic Bax in the cytoplasm, suggesting that NPD1 acts by preventing proapoptotic signaling pathways from occurring. Finally, NPD1 signaling interfered with ataxin-1/capicua repression of gene expression and decreased phosphorylated ataxin-1 in an Akt-independent manner, suggesting that NPD1 signaling modulates formation or stabilization of ataxin-1 complexes. These data suggest that 1) NPD1 synthesis is an early response induced by proteotoxic stress due to abnormally folded ataxin-1, and 2) NPD1 promotes cell survival through modulating stabilization of ataxin-1 functional complexes and pro-/antiapoptotic and inflammatory pathways.
Introduction
Neurodegenerative diseases are a complex group of progressive disorders with diverse etiologies linked by a common outcome: neuronal cell death. Although neurotoxins (1), vascular deficiencies (2), and mitochondrial failure (3) are proposed factors in neurodegeneration, only genetic abnormalities are unequivocally proven to be the cause of some of these disorders. From this latter group, a subset of nine neurodegenerative disorders emerges with a clear etiology. An error in DNA replication causes some proteins to carry an abnormally large polyglutamine tract. This feature alters the folding, stability, and activity of the protein in which it is contained, affecting cell survival. Spinocerebellar ataxia type 1 (SCA1) is the best characterized disorder from this group. Mutant ataxin-1, containing more than 39 glutamines in its tract, accumulates in the nuclei, causing toxicity in cerebellar Purkinje cells (4–6). Ataxin-1 was widely used as a model of degeneration, and thus its interactions with transcription factors and spliceosome components are well known (7, 8). In addition, compelling evidence shows that not only is the cerebellum compromised, but the retina also undergoes degeneration (9–11), thus providing the basis for applying this model to retinal studies as well.
Protein misfolding contributes to degeneration in the central nervous system. For instance, the abnormal folding of rhodopsin and its subsequent aggregate formation in the retina lead to the increased neurodegeneration observed in retinitis pigmentosa (12–15). Proteostasis encompasses the homeostatic regulation of the protein pool present in a cell (16), and it is maintained through the orchestration of several systems involved in protein synthesis and clearance. In this context, protein homeostasis may be modulated by various mechanisms, including stress-inducible responses (17). Protein misfolding interferes with the equilibrium, inducing cytotoxicity in two ways: 1) through its accumulation or 2) by causing loss of function (16). Misfolded protein accumulation may evoke a stress response called unfolded protein response as a way for the cell to maintain proteostasis (18). Unfolded protein response alleviates this condition; however, if the stress is sustained and the cell cannot cope with the disequilibrium, the system tends toward overproduction of reactive oxygen species (19–21). Reciprocally, oxidative conditions induce not only protein damage but also impairment of normal protein turnover through the inhibition of the ubiquitin/proteosome system, thereby leading to abnormal protein accumulation (22) due to failure of the endoplasmic reticulum-related degradation system. In this sense, oxidative and proteotoxic stresses are intimately related.
Neuroprotectin D1 (NPD12; (10R,17S)-dihydroxydocosa-(4Z,7Z,11E,13E,15Z,19Z)-hexaenoic acid) is a stress-response lipid mediator derived from docosahexaenoic acid (DHA). Oxidative stress stimulation initiates the enzymatic oxygenation of DHA through the activation of 15-lipoxygenase-1 (15-LOX-1) (23). NPD1 enhances survival signaling in retinal pigment epithelial (RPE) cells confronted with oxidative stress by promoting modulation of the activity and content of proteins directly involved in deciding cell fate (24). We reasoned that NPD1 signaling, as in the oxidative stress scenario, may be involved in the unfolded protein response by promoting cell survival. In the present report, we evaluate this prediction in a system known to endogenously produce NPD1: RPE cells constitutively expressing ataxin-1 containing an extended polyglutamine tract.
EXPERIMENTAL PROCEDURES
Reagents and Constructs
A construct that carried an 830-bp cyclooxygenase-2 (COX-2) promoter DNA fragment, which drives luciferase expression, was used previously to study modulation of proinflammatory gene expression. Expression plasmids containing the ataxin-1 forms 30Q, 2Q, and 82Q, as well as the AXH truncated form (ΔAXH), the self-folding AXH domain, and huntingtin 17Q and 72Q, have been described previously (25, 26). The antibodies used for detecting total and phosphorylated forms of ataxin-1 Western blot and immunocytochemistry were obtained from LifeSpan Biosciences (Seattle, WA), Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), Cell Signaling Technology (Danvers, MA), and Abcam (Cambridge, MA), respectively. Anti-Bax was obtained from Cell Signaling Technology. Buffers and supplies for Western blot were obtained from Invitrogen, Bio-Rad, and Sigma. Anti-MAP2 antibody was obtained from Millipore (Temecula, CA). Media and additives were purchased from Invitrogen, and FBS was from Tissue Culture Biologicals Inc. (Seal Beach, CA).
Cell Culture, Transfection, and NPD1/DHA Treatments
ARPE-19 cells (a human RPE cell line) were plated and grown in DMEM/F-12 containing 10% FBS and 1× penicillin/streptomycin at 37 °C, 5% CO2, 99% relative humidity for 24 h. Silenced 15-LOX-1 cells (from the stable silenced cell line 15-LOX-1d), described in detail elsewhere (23), were treated like their mother cell line (ARPE-19) with the addition of Geneticin (500 μg/ml). Primary human RPE cultures were performed using human eye cups, provided by the National Disease Research Interchange, following a modification of a previously described protocol (27). In brief, the vitreous humor and neural retina were removed. Squares of 5 × 5 mm2 containing the RPE, choroid, and sclera were cut and placed on Petri dishes. RPE cells were allowed to grow outside the graft for 3 weeks before being transferred to flasks with MEM containing 10% fetal calf serum, 5% newborn calf serum, nonessential amino acids, 4 mm glutamine, amphotencin B (0.5 μg/ml), and gentamicin (10 μg/ml). Identification of RPE cells was verified by staining with anti-RPE65, anti-MiTF, and anti-peropsin in a 1:250 dilution (Abcam). Rat cortical neuronal culture was performed following the method described in detail elsewhere (28). ARPE-19 cells were transfected using FuGene 6 (Roche Applied Science). Human RPE and rat cortical primary cultures were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's directions. The FuGene 6/plasmid ratio was 3:1, and Lipofectamine 2000/plasmid ratio was 5:2. FuGene 6·plasmid transfection complexes were left in the culture for 24 h, whereas Lipofectamine ones were removed after 5 h of incubation to prevent cell death due to reagent toxicity. The optimal amount of plasmids used for ARPE-19 and neuronal cultures was 2 μg/ml, and the optimal amount was 1 μg/ml for hRPE primary culture. Under these conditions transfection yielded 80–90% in ARPE-19 (supplemental Fig. 1), 90–98% in hRPE primary culture (supplemental Fig. 2), and 22–41% in rat neuronal culture (supplemental Figs. 4 and 13). Differences were attributed to cell viability after transfection. For instance, transfection efficiency in hRPE primary cells was decreased by adding a second expression vector from 98 to 69% (supplemental Fig. 3). For this reason, we chose to make parallel transfection controls using GFP in separate wells. In each case, those experiments that did not meet the transfection efficiency ranges established for each type of cells were discarded. Cultured cells were transfected with expression constructs containing wild type, mutant, and truncated forms of ataxin-1 open reading frame. The expression of ataxin-1 was evaluated by Western blot analysis (supplemental Fig. 11) and semiquantitative conventional RT-PCR to ensure even expression of the constructs using the primers 5′-ccaacatgggcagtctgag-3′ (forward) and 5′-tggacgtactggttctgctg-3′ (reverse) for ataxin-1 and 5′-gagtcaacggatttggtcgt-3′ (forward) and 5′-ttgattttggagggatctcg-3′ (reverse) for GAPDH using the following PCR protocol: 1 cycle of 5 min at 95 °C; 25 cycles of 15 s at 95 °C, 30 s at 55 °C, 30 s at 72 °C; and 1 cycle of 5 min at 72 °C. An example test is provided in supplemental Fig. 5B. Transfected cells were followed for a period of 72 h by Western blot and immunocytochemistry to determine the periods of incubation and treatment (supplemental Fig. 5A). Cells were serum-starved for 8 h before treatment. NPD1 (50 nm) or DHA (100 nm) plus PEDF (10 ng/ml) or brain-derived neurotrophic factor (BDNF; 10 ng/ml) were added to the culture medium dropwise and incubated for different time periods as indicated in each experiment.
Immunocytochemistry, Hoechst Staining, and TUNEL Assay
Immunocytochemistry assays were performed in 8-well slide chambers, and the remaining experiments were performed in 6-well plates. Medium was removed, and cells were washed once in 1× PBS and fixed in 4% paraformaldehyde in phosphate buffer for 20 min and then cold methanol for 15 min. Cells were further permeabilized with Triton X-100 buffer, and nonspecific epitopes were blocked in 10% normal serum and 1% bovine serum albumin (BSA) in 1× PBS for 1 h at room temperature. Immunostaining was performed by incubating the cells with the corresponding primary antibody at proper dilution for 2 h. Secondary antibodies conjugated with Alexa Fluor 594, 555, or 488 or fluorescein isothiocyanate (FITC) were used for 1 h at room temperature, and nuclei were stained with DAPI or Hoechst for 10 min. TUNEL staining was performed using the DeadEnd fluorometric TUNEL system (Promega, Madison, WI) following the manufacturer's directions. Caspase activation was measured using anti-activated caspase-3 antibody (Millipore, Temecula, CA), which recognizes the 17-kDa fragment of cleaved caspase-3. Slides were mounted in Vectashield fluorescent medium (Vector Laboratories, Burlingame, CA) and analyzed using a Zeiss LSM 510 Meta confocal laser-scanning microscope or Axioplan 2 deconvolution microscope. Images were obtained and processed using SlideBook 4.2 and 5.0 software (Intelligent Imaging Innovations Inc., Denver, CO). For apoptotic ratio determinations, a validated Hoechst staining protocol was used (24). Briefly, cells were fixed with 4% paraformaldehyde for 20 min and then methanol for 15 min, washed with PBS, incubated with 2 μm Hoechst reagent in PBS (Promega, Madison, WI) at room temperature for 15 min, and then photographed with a DIAPHOT 200 microscope (Nikon, Melville, NY) with fluorescent optics. Images were recorded by a color-chilled 3CCD camera (Hamamatsu, Bridgewater, NJ). Apoptotic cells presented hyperpyknotic stains, whereas healthy ones were normopyknotic. Both types of cells were counted using ImageJ 1.42 (National Institutes of Health), using two threshold values to differentiate by intensity. In addition to intensity, two other criteria were used to discriminate apoptotic from normal and dividing cells: 1) nuclear size (distinctively small in apoptotic cells) and 2) circularity (apoptotic nucleus appears usually in a perfect circular shape as opposed to normal (elliptical) or mitotic (irregular)). The ratio of hyperpyknotic cells over the total was calculated for nine randomly chosen fields per sample obtained from three independent wells per experiment. Each experiment was repeated at least three times to confirm the findings. In the experiments involving early stages of apoptosis, we assessed the total number of cells per field and counted them to ensure unbiased observations (supplemental Fig. 6).
Human COX-2 and Capicua Binding Site Promoter Induction
RPE cells were transfected for 48 h with 5 μg of human COX-2 luciferase construct (830 bp) alone or capicua binding site-Luc+/− in addition to the plasmids containing the expression constructs for ataxin-1 30Q, 82Q, and 2Q. The medium was removed, and fresh medium was added. Cells were serum-starved for 8 h before treatment with NPD1 (50 nm), DHA/PEDF (50 nm/10 ng), or DHA/BDNF (50 nm/10 ng) for 24, 48, or 144 h. Cells were harvested and homogenized to perform luciferase assay. The internal standard used was constitutively expressed β-galactosidase in the capicua expression experiment. For COX-2 promoter activity measurement as well as for every other experiment, the transfection efficiency was measured in separate wells. A typical control experiment is shown in supplemental Fig. 1. The samples were read using a Thermo Scientific Appliskan luminescence plate reader.
Lipidomic Analysis
The lipid extraction was performed using Folch's method (29). Briefly, medium and cells were collected separately, and 5 μl of internal standard (prostaglandin D2-d4, 0.01 μg/μl) was added to each sample. Cells were extracted with 1 ml of methanol. Lipid extracts were removed from plates and collected in test tubes containing chloroform to yield a ratio of 2:1 chloroform/methanol. The medium was centrifuged, and 1 ml was collected into 9 ml of cold chloroform/methanol (1:1). Protein precipitates were then separated by centrifugation at 1,000 × g (30 min, 4 °C). Lipids were extracted from cells and medium. Eluates were concentrated on a nitrogen stream evaporator and resuspended in 100 μl of methanol before MS analysis. Samples were loaded to a liquid chromatography-tandem mass spectrometer (LC-TSQ Quantum; Thermo Scientific Co., Waltham, MA) installed with a Pursuit 5-μm C18 column (100 mm × 2.1 mm; Thermo Scientific Co.) and eluted in a linear gradient (100% solution A (40:60:0.01 methanol/water/acetic acid, pH 3.5) to 100% solution B (99.99:0.01 methanol/acetic acid)) at a flow rate of 300 μl/min for 45 min. LC effluents were diverted to an electrospray ionization probe on a TSQ Quantum triple quadrupole mass spectrometer. Lipid standards (Cayman Chemical Company, Ann Arbor, MI) were used for tuning and optimization and to create calibration curves. The instrument was set on full-scan mode to detect parent ions and selected reaction for quantitative analysis, to detect product ions, simultaneously. The selected parent/product ions (m/z) and collision energy (v) obtained by running on negative ion detection mode were 359/153/22 for NPD1 and 355/275/22 for prostaglandin D2-d4 (used as an internal standard).
Pull-down Experiments and Western Blot Analysis
Immunoprecipitation was performed using agarose beads coupled to protein A/G following the manufacturer's recommendations (Santa Cruz Biotechnology, Inc.). Briefly, the volume of lysate corresponding to 500 μg of total protein was initially cleared by incubating it with normal serum along with the agarose beads. The primary antibody against total ataxin-1 was incubated with the precleared sample for 1 h, and the agarose beads were added. The binding to the column proceeded at 4 °C for 16 h. The beads were then washed five times with 1× PBS and heated with Laemmli loading buffer for 5 min at 95 °C and loaded in a NuPAGE precast gel. As in the case of the Western blots, the gel was run in an Invitrogen X-Cell running system using MOPS buffer. The transference took place in the iBLOT system following the manufacturer's instructions. Primary antibodies were hybridized for 16–48 h at 4 °C, washed, and incubated for 1 h with the secondary antibody at room temperature. The membranes were developed using ECL (GE Healthcare) following the manufacturer's instructions.
Statistical Analysis
The data collected from biological replicates were subjected to one-factor analysis of variance, and differences were compared pairwise using Student's t test.
RESULTS
NPD1 Protects ARPE-19 Cells from Ataxin-1 82Q-induced Apoptosis
To address the effects of NPD1 on misfolded protein stress, we used the ARPE-19 cell line expressing the 82-glutamine form of ataxin-1 (ataxin-1 82Q) as a model. Ataxin-1 82Q was first noticeable 24 h after transfection (supplemental Fig. 5A) with a concomitant increase in the nuclear Ser-776-phosphorylated form, as evidenced by immunostained cells (supplemental Figs. 7, 8, 9 (A and B), and 10), and reached higher levels at 48 h (supplemental Figs. 7–9).
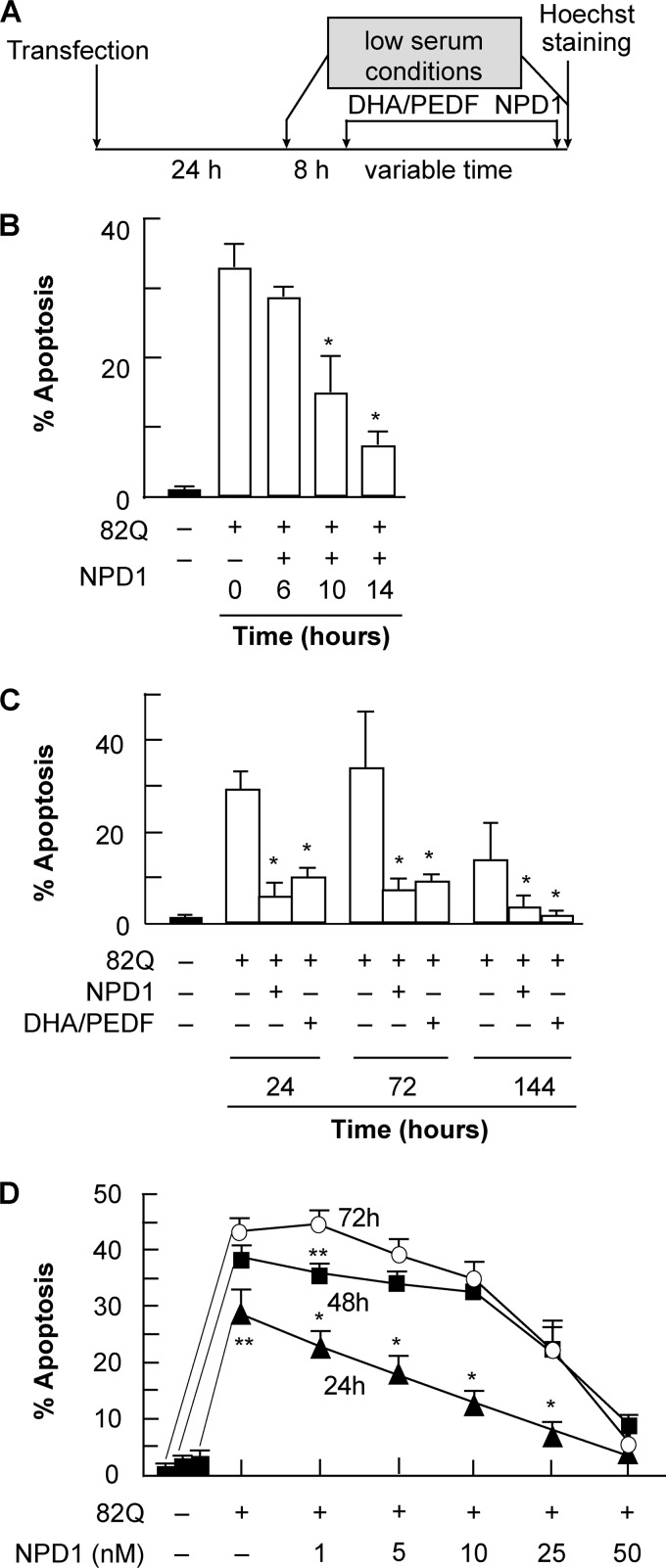
To assess whether or not NPD1 prevents ataxin-1 82Q-induced apoptosis, ARPE-19 cells expressing the mutant protein were treated with NPD1 (50 nm) and its precursor, DHA (100 nm) (Fig. 1A). PEDF was added as an agonist of NPD1 synthesis. In the short term, the minimum time required for NPD1 to decrease the apoptosis percentage by half was 10 h, and the time necessary to reach minimal levels was 14 h (Fig. 1B). Ataxin-1 82Q expression increased cell death to 30%, and both NPD1 and DHA/PEDF addition reduced the levels to 5 and 7%, respectively; there were no significant differences between the two treatments at any time point (Fig. 1C).
FIGURE 1.
NPD1 prevents ataxin-1 82Q-induced apoptosis in ARPE-19 cells. A, time line showing the experimental design. ARPE-19 cells, transfected with an expression construct containing ataxin-1 82Q, were treated with 50 nm NPD1 for 0, 6, 10 and 14 h (B); NPD1 (50 nm) or DHA (100 nm) along with PEDF (10 ng/ml) for 24, 72, and 144 h (C); or 1, 5, 10, 25, or 50 nm NPD1 at three different time points, 24, 48, and 72 h (D). Apoptosis percentage was calculated by dividing the number of pyknotic cells by the total count of cells (see “Experimental Procedures” for a detailed description of the technique). Solid bars, control groups; empty bars, treated groups. Results are represented by averages ± S.D. (error bars). **, p < 0.005; *, p < 0.0005.
We evaluated the dynamics of NPD1 action by building a dose-response curve at three different time points, representing three stages of proteotoxicity, which was provided by a sustained source of misfolded ataxin-1. However, continuous expression of the protein after 48 h induced maximum levels of apoptosis that were only reversed by the addition of the highest dose tested. NPD1 showed a dose-dependent linear effect at 24 h. This complex kinetic trend is compatible with an elevated turnover promoted by NPD1 in a dose-dependent manner. Half of the protective activity was accomplished at low dose (5 nm) by 24 h, but for 48 and 72 h, only the 25 nm dose was effective, suggesting a threshold effect for NPD1 (Fig. 1D). The maximum level of protection was reached at 50 nm NPD1 regardless of the amount of protein accumulated (Fig. 1D).
Altogether, these results suggest that low concentrations of NPD1 are effective when the levels of the toxic protein are low, whereas higher concentrations of NPD1 are required to counteract apoptosis when it reaches higher levels of accumulation; the response to the chronic stimulus then becomes time-independent.
NPD1 Promotes Survival in Neurons and Primary RPE Cells Undergoing Proteotoxic Stress
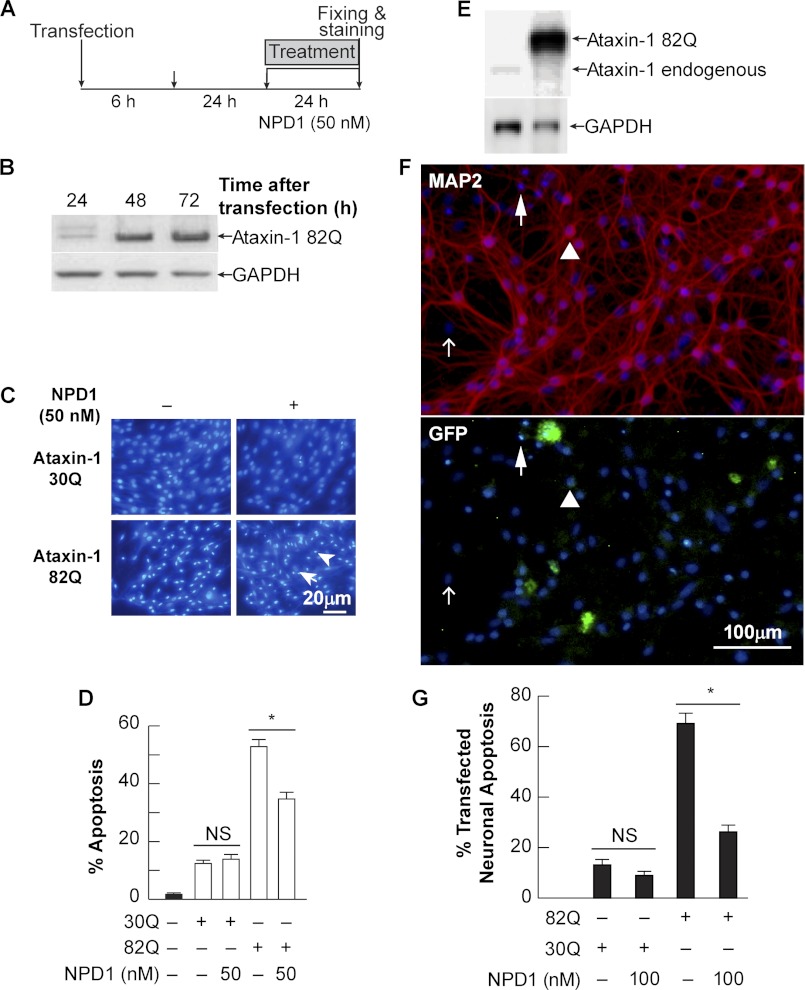
In order to validate and expand the use of ARPE-19 cells as a model to investigate the effects of ataxin-1 and NPD1, proteotoxic stress was induced in human RPE and rat neuronal primary cultures. In these two scenarios, we predicted that NPD1 would rescue cells from apoptosis as well. Cells were transfected with the 82Q and wild type (30Q) forms of ataxin-1 following the protocol depicted in Fig. 2A. The expression of the mutated form peaked its maximum accumulation at 72 h in hRPE cells and neurons (Fig. 2, B and E). As in ARPE-19, neurons and hRPE cells showed differential morphology between positive and negative Hoechst nuclei (Fig. 2, C and F). Positive Hoechst staining was characterized by high intensity, size reduction, and high circularity, which are distinctive features of the apoptotic shrinking nuclei (24). Furthermore, neurons were co-transfected using the construct and GFP expression plasmid to pinpoint those neurons that were transfected and underwent apoptosis, as opposed to those that were transfected but showed a normal morphology (Fig. 2F, arrow and arrowhead). 50 nm NPD1 prevented the apoptosis induced from the expression of mutated ataxin-1 by 52.8 ± 2.5% to 34.7 ± 2.3% in human RPE cells (Fig. 2, C and D), whereas 100 nm NPD1 reduced the cell death by 62.3% in rat neuronal culture (Fig. 2G). These results support the idea that NPD1 prevents a common cellular toxic event that occurs during ataxin-1 82Q-induced proteotoxic stress and gave us the grounds to study these effects in the ARPE-19 cell line.
FIGURE 2.
NPD1-induced neuroprotection in human RPE primary culture and neurons undergoing ataxin-1 82Q-triggered proteotoxic stress. A, time line depicting the protocol used to induce proteotoxic stress in human RPE primary cells and rat neuronal culture. B and E, representative immunoblots showing overexpression of ataxin-1 82Q 24, 48, and 72 h post-transfection. C, representative images showing Hoechst-positive cells. Apoptotic nuclei were selected following three criteria: high intensity (emission at 480-nm wavelength), nuclei size, and circularity. Arrows and arrowheads indicate living and apoptotic cells, respectively. D, quantification of Hoechst-positive nuclei in hRPE cells and neurons. *, p < 0.0005. F, representative images of Hoechst and MAP2 immunostaining (top) and GFP of the primary neuronal culture co-transfected with the construct expressing ataxin-1 82Q and GFP expression plasmid used to produce the results shown in G. Open arrow, non-neuronal cell; arrow, Hoechst-, GFP-, and MAP2-positive cell; arrowhead, GFP- and MAP2-positive cell. NS, not significant.
NPD1 Counteracts Proapoptotic and Proinflammatory Signaling Mediated by Ataxin-1 82Q
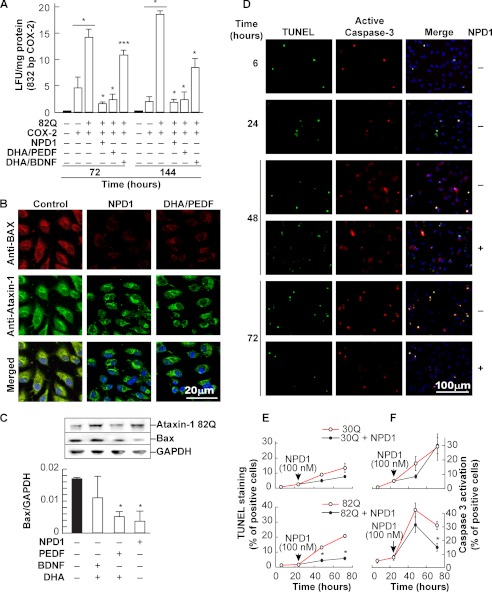
NPD1 promotes survival through modulation of the inflammatory and apoptotic signaling in ARPE-19 and hRPE cells undergoing oxidative stress (23, 24, 30, 31). COX-2 promoter activity was measured as a marker of inflammation (24) in ARPE-19 cells to test the prediction that NPD1 signaling leads to the lessening of proinflammatory events triggered by proteotoxic stress. After 72 h of continuous expression of the ataxin-1 82Q construct, cells showed an increase in the COX-2 promoter-induced luciferase, evidenced by its augmented activity (Fig. 3A). When either NPD1 or DHA/PEDF was present, there was a severe reduction in COX-2 promoter activity, suggesting that DHA is acting through its derivative NPD1 to counteract proinflammatory signaling triggered by proteotoxic stress (Fig. 3A).
FIGURE 3.
NPD1 blocks ataxin-1 82Q-induced apoptotic signaling. A, COX-2 promoter activity measured by the means of the luciferase reporter gene assay in human RPE cells expressing ataxin-1 82Q and treated with either NPD1 (50 nm), a combination of DHA/PEDF (50 nm/10 ng), or DHA/BDNF (50 nm/10 ng) for 72 and 144 h. B, representative fields obtained by confocal microscopy of ARPE-19 cells expressing ataxin-1 82Q treated or not with NPD1 (50 nm) or DHA (100 nm)/PEDF(10 ng/ml) using anti-Bax (red) and anti-ataxin-1 (green) antibodies. Nuclei are shown in blue. Solid and empty bars, control groups and treated groups, respectively. C, Bax expression assessed by Western blot analysis in cells expressing ataxin-1 82Q (top) and treated with NPD1 (50 nm) and DHA (100 nm) along with PEDF or BDNF (10 ng/ml each). D, representative images acquired to quantify activated caspase-3 and TUNEL-positive cells in E and F. Anti-activated caspase-3 antibody detected the 17-kDa fragment of the cleaved caspase-3 selectively. E and F, quantification of TUNEL and activated caspase-3-positive cells. Time-dependent ratio of the positive and total cells was assessed in 30Q- and 82Q-transfected ARPE-19 cells. NPD1 (100 nm) was added or not 24 h post-transfection. Bars, averages ± S.D. (error bars). *, p < 0.05; **, p < 0.005; ***, p < 0.0005.
Polyglutamine-expanded forms of ataxin-3 and -7 induce the up-regulation of Bax in cerebellar neurons as a part of programmed cell death caused by poly(Q) cytotoxicity (32, 33). NPD1 and its precursor, DHA, were shown to reduce the increase of Bax triggered by oxidative stress stimulus (24). To address whether or not NPD1 affects Bax content in ARPE-19 cells expressing ataxin-1 82Q, the levels of cytoplasmic protein were assessed as a marker of programmed cell death. Immunocytochemistry revealed an increase in the cytoplasmic Bax elicited by the expression of ataxin-1 82Q that was suppressed by NPD1 or a combination of DHA/PEDF. These findings were then confirmed by Western blot analysis (Fig. 3C). In addition, other markers of apoptosis were tested in the presence and absence of NPD1. After 24 h of transfection using the wild type and the mutant constructs, the NPD1 or vehicle was added, and apoptotic caspase-3 activation and DNA fragmentation were tested by means of immunocytochemistry and TUNEL assays (Fig. 3, D–F). Caspase-3 activation showed a peak signal 48 h after transfection, independent of NPD1 addition. Differences became significant only 48 h after treatment (Fig. 3F), supporting the values obtained for Hoechst staining (Fig. 1D). However, the differences in the DNA fragmentation between treated and non-treated cells became significant at least 24 h after NPD1 was applied (Fig. 3E). These data suggest not only that DHA/NPD1-mediated cell survival involves the down-regulation of the proapoptotic protein Bax as an early event (Fig. 3, B and C) but also that the bioactive lipid acts through resolution of the caspase-dependent apoptotic pathway (Fig. 3F). This resulted in decreased DNA fragmentation (Fig. 3E), which occurs at a later stage in the developing programmed cell death. For these two events, the effect of NPD1 followed a different time pattern. Altogether, these results show that NPD1 signaling results in inflammation resolution upon the stress caused by the mutant protein and prevents the resulting cell death by counteracting apoptotic signaling.
NPD1 Decreases Capicua (CIC)-mediated Repression
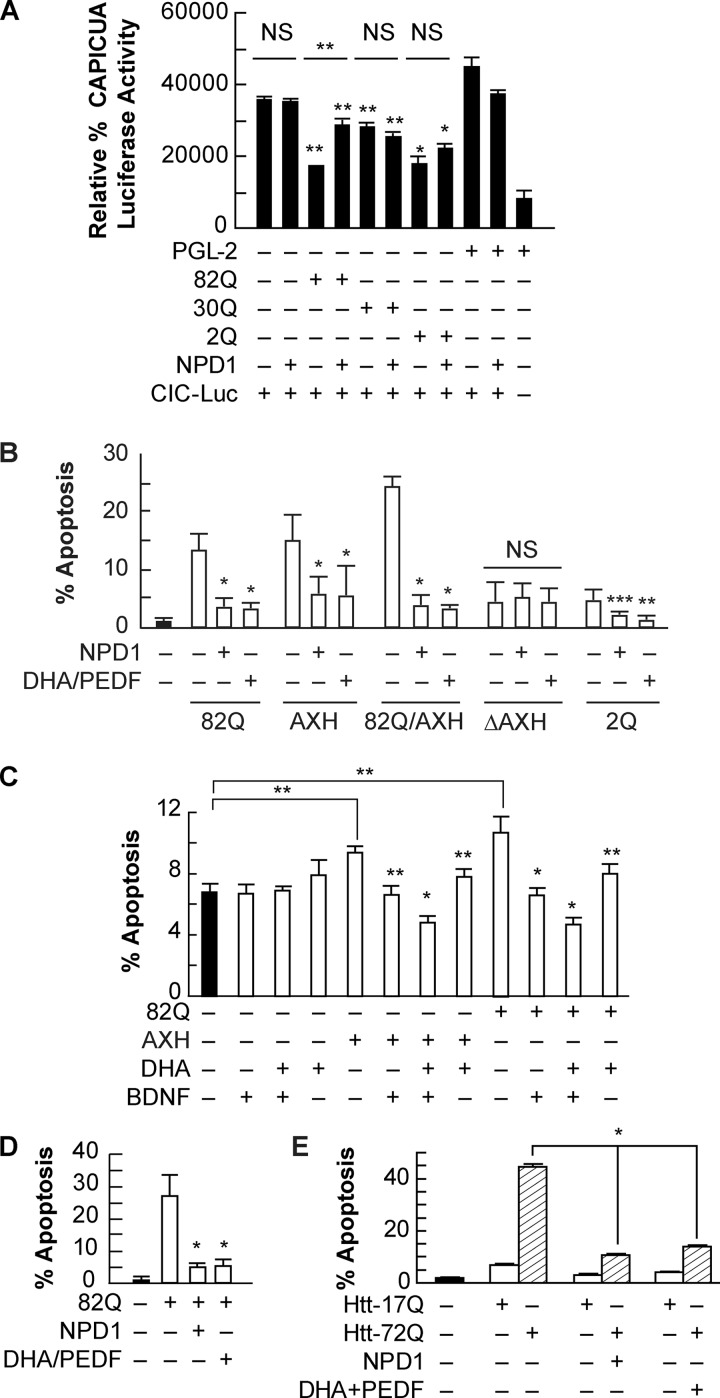
It was proposed that the pathogenic mechanism involves partial loss of function of ataxin-1 and the toxicity induced by the extended poly(Q) tract (8). The abnormally long polyglutamine tract in ataxin-1 alters CIC repressor activity upon binding (34). To explore the role of NPD1 in the activity of the CIC repressor complex and whether or not this is related to the content of glutamine in ataxin-1, we measured its activity using a luciferase reporter assay for the CIC binding site in cells expressing the 30Q, 82Q, or 2Q form of ataxin-1. NPD1 relieved 79% of the repression in the transcription induced by overexpression of ataxin-1 82Q (Fig. 4A). Intriguingly, the polyglutamine truncated (2Q) and the wild type (30Q) forms of ataxin-1 exerted significant repression on the CIC-luciferase construct, but NPD1 did not modulate this effect. These data suggest that NPD1 is only active upon stress when the toxic form of the protein is present and that it promotes homeostasis by balancing the complex formation, decreasing CIC-DNA binding activity, and thus reducing CIC-induced repression (Fig. 4A).
FIGURE 4.
NPD1 prevents ataxin-1 82Q-dependent apoptosis. A, ARPE-19 cells expressing the CIC-luciferase construct, along with ataxin-1 wild type (30Q), ataxin-1 mutant (82Q), and ataxin-1 without the glutamine tract (2Q), were treated with NPD1 (50 nm) or DHA (100 nm) and PEDF (10 ng/ml) for 24 h. B, ARPE-19 cells were transfected with the above-mentioned constructs plus ataxin-1 containing truncated AXH (ΔAXH) and the self-folding domain alone (AXH) and treated with NPD1 (50 nm) or DHA (100 nm) and PEDF (10 ng/ml) for 16 h. C and D, human RPE primary culture cells were transfected with ataxin-1 82Q or AXH domain and treated with 10 ng/ml BDNF (C) or PEDF (D) in combination with DHA (100 nm). E, 17Q and 72Q huntingtin-expressing cells were treated with NPD1 and DHA (100 nm)/PEDF (10 ng/ml). The percentage of apoptosis is shown for each treatment. Bars, averages ± S.D. (error bars). **, p < 0.05; *, p < 0.005. NS, not significant.
Ataxin-1 AXH Domain Contribution to Cytotoxicity Is Blocked by NPD1
To further investigate whether NPD1 affects regulation of mutant ataxin-1 activity in relationship with its protein structure, the mortality rate of ARPE-19 expressing the AXH self-folding domain and the different forms of ataxin-1 were compared in the presence and the absence of NPD1. AXH by itself enhanced apoptosis at comparable levels of the mutant 82Q, and both NPD1 and DHA/PEDF treatment promoted survival (Fig. 4B). The same pattern was observed in human RPE primary cell culture (Fig. 4, C and D). The co-expression of AXH and 82Q forms further augmented cell death, suggesting that two different sources of toxicity are supported by the mutant protein. Both NPD1 and DHA reduced apoptosis levels. In addition, ΔAXH and 2Q expression did not significantly affect cell death induction, presumably because these two forms of ataxin-1 lack the active domain AXH and contain a lower number of glutamines, respectively (Fig. 4B). These results suggest a strong contribution of the two features to the toxicity of the mutant protein and highlight the stress-driven activity of NPD1.
Other neurotrophic factors besides PEDF are able to enhance NPD1 synthesis in RPE cells (30). For instance, BDNF alone improved the survival of human RPE cells expressing 82Q or AXH constructs to the same extent as DHA, but when added together, BDNF and DHA induced a reduction in cell death of about 50% compared with the controls (Fig. 4C). BDNF did not help to prevent inflammation as PEDF did when added along with DHA. This suggests that both neurotrophic factors act through different pathways and thus provides evidence for additive effects (Fig. 4, C and D).
NPD1 also prevented cell death induced by the extended poly(Q) tract contained in other proteins, such as huntingtin (Fig. 4E). This evidence supports the role of NPD1 in preventing toxicity elicited by the altered structure of ataxin-1, aside from its deregulated activity. In addition, the excess of the AXH domain alone is capable of causing cytotoxicity in RPE cells, potentiating the damage induced by structure disruption caused by the 82Q extension. Finally, these results show that NPD1 and its precursor, DHA, protect against structure/activity-dependent damage.
Ataxin-1 82Q Protein Induces NPD1 Synthesis
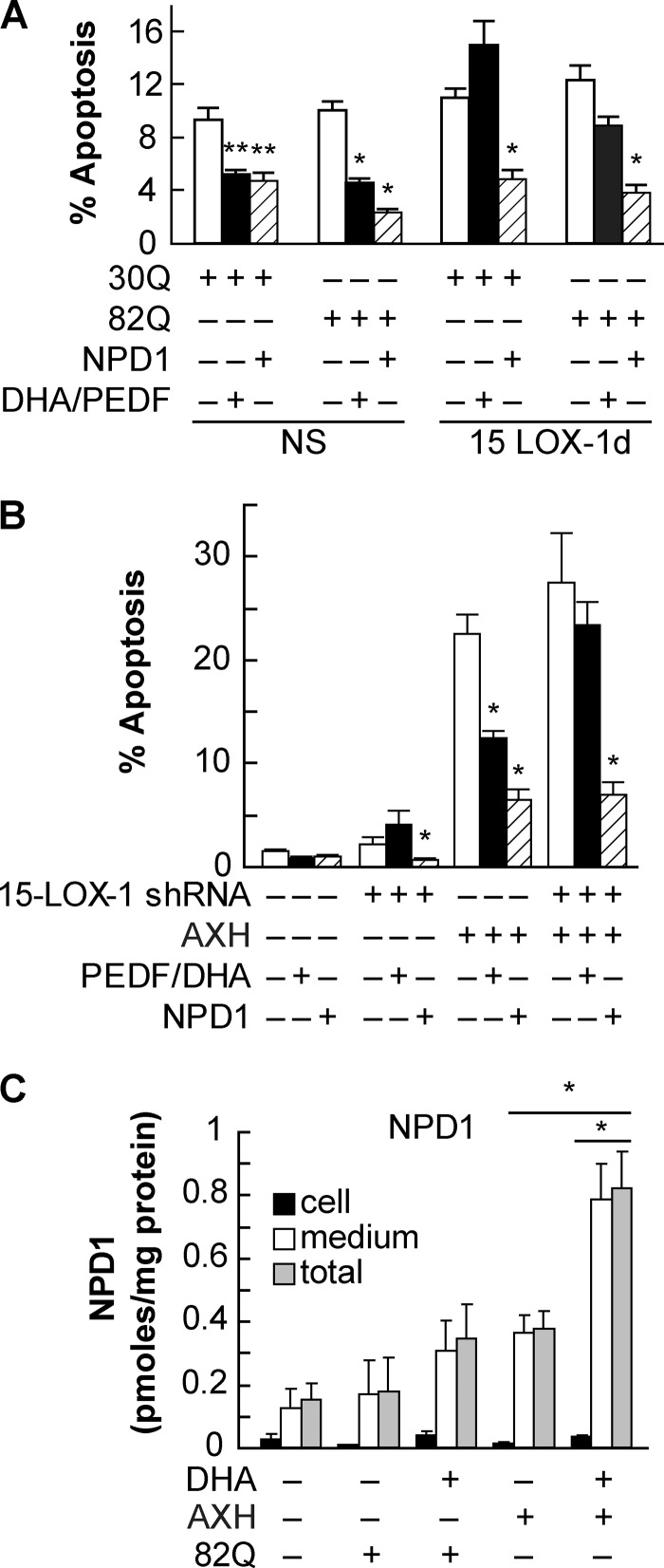
The addition of exogenous NPD1 promoted survival in cells subjected to proteotoxic stress. 15-LOX-1 is the main enzyme involved in the oxygenation of DHA into NPD1 in ARPE-19 cells. Deficiency of this enzyme reduces the ability of these cells to produce NPD1 and therefore to cope with oxidative stress damage, thus increasing their susceptibility to apoptosis (23). To test the prediction that decreased NPD1 synthesis enhances proteotoxic stress-induced apoptosis in ARPE-19 cells, stably 15-LOX-1-silenced cells were transfected using constructs carrying 30Q and 82Q forms of ataxin-1, as well as the AXH domain, to determine the apoptotic ratio in comparison with the nonspecific silenced cells. Nonspecific cells possess normal levels of the enzyme and therefore are able to induce NPD1 synthesis upon proteotoxic stress (23). The percentage of apoptosis was slightly higher in the 15-LOX-1-deficient cells than in the control cells but not enough to be significant (Fig. 5, A and B). On the other hand, the addition of exogenous NPD1 and DHA/PEDF decreased apoptosis in control cells, whereas in deficient cells, DHA/PEDF failed to promote survival (Fig. 5A), suggesting that the action of DHA is achieved through its conversion into NPD1. The same was observed in AXH-transfected deficient cells (Fig. 5B).
FIGURE 5.
Endogenous NPD1 prevents ataxin-1 82Q-induced apoptosis in RPE cells. A and B, stable 15-LOX-1-silenced ARPE-19 cells were transfected with a plasmid containing the wild type (30Q), ataxin-1 (82Q), or AXH domain. Cells were treated for 16 h with DHA (100 nm)/PEDF (10 ng/ml) or NPD1 (50 nm). C, primary human RPE cells were transfected with 82Q or AXH constructs. Levels of cellular (black bars), secreted (white bars), and total (gray bars) NPD1 were measured by LC MS-MS with or without the addition of DHA. NS, not significant. **, p < 0.05; *, p < 0.005.
Oxidative stress triggers NPD1 synthesis and release to provide autocrine/paracrine stimuli that compete with proapoptotic and proinflammatory signaling to promote survival (23, 24, 30, 31, 35). To establish if proteotoxic stress induces endogenous activation of NPD1 synthesis by chronic proteotoxic stress in human primary RPE cell culture, NPD1 was assessed in 1) the postincubation medium to quantify its release and 2) the cells, in response to overexpression of ataxin-1 82Q and AXH (Fig. 4C). As shown previously for oxidative stress (23), NPD1 was found in higher amounts in the medium and in lower amounts intracellularly. Furthermore, there was no significant change in NPD1 in the cellular fraction among treatments. AXH, but not 82Q, triggered substantial NPD1 synthesis and release, as evidenced by NPD1 concentration in the medium. The addition of exogenous DHA further increased NPD1 production (Fig. 5C). These results suggest that the AXH domain induced enough stress, per se, to produce activation of NPD1 synthesis. Also, the increased susceptibility in the silenced cells, due to their inability to produce NPD1 from DHA, implies that proteotoxic stress triggers endogenous NPD1 synthesis as a prosurvival mechanism to counteract proapoptotic signaling induced by cell damage.
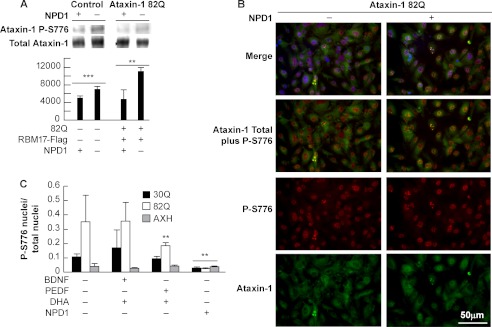
NPD1 Decreases Phosphorylated Ataxin-1
It was proposed that the mechanism that mediates neurodegeneration in SCA1 involves displacement of the complex equilibrium formed by ataxin-1 due to the change in its folding/phosphorylation status (7, 8). To test the prediction that NPD1 modifies the activity of the CIC repressor through modification of ataxin-1 phosphorylation, we immunoprecipitated the protein from ARPE-19 cells incubated in the presence and absence of NPD1. The cells expressing ataxin-1 82Q showed an increase in phosphorylated ataxin-1 in comparison with cells that were not transfected. These observations are in agreement with the immunocytochemical data observed (supplemental Figs. 7–10). Intriguingly, transfected cells that were treated with NPD1 displayed a decrease in phosphoataxin-1 (Fig. 6A) in a pull-down assay using anti-phosphoataxin-1. Furthermore, NPD1 also decreased the phosphorylated ataxin-1 content in non-transfected cells. This observation was confirmed by immunocytochemistry (Fig. 6, B and C). Quantification of the immunocytochemical data showed that DHA/PEDF treatment was less potent in reducing the number of nuclei displaying a positive S-776 phosphoataxin-1 signal than NPD1, and the differences were significant only in cells expressing the 82Q form. DHA/BDNF treatment did not affect the phosphorylation status of ataxin-1 (Fig. 6C). In fact, AXH expression did not induce an increase in phosphoataxin-1 above normal levels; nor did it show modification of the phosphorylation state under any of the treatments (Fig. 6C). Altogether, these results suggest that NPD1 favors clearance of the complex, probably by increasing the dynamics of assembly-disassembly through modulation of ataxin-1 phosphorylation in a way that prevents toxicity induced by the overexpression of the 82Q form of the protein.
FIGURE 6.
NPD1 decreases the content of ataxin-1 phospho-Ser-776. ARPE-19 cells were transfected with the constructs 82Q, 30Q, and AXH with or without a construct expressing RBM17. A, immunoprecipitation using anti-ataxin-1 Ser(P)-776 antibody (P-S776) from ARPE-19 cells expressing the ataxin-1 82Q form and treated with and without NPD1 (50 nm). Bars, densitometric measurement of ataxin-1 Ser(P)-776/total ataxin-1 ratio. B, immunocytochemistry of ARPE-19 cells expressing ataxin-1 82Q and treated with NPD1 using anti-phosphoataxin-1 (red) and anti-ataxin-1 (green). C, percentage of nuclei showing ataxin-1 Ser-776 phosphorylation signal in ARPE-19 cells expressing the constructs 30Q, 82Q, and AXH and treated with NPD1 or DHA and PEDF or BDNF. Values are mean ± S.D. (error bars). ***, p < 0.05; **, p < 0.005; *, p < 0.0005.
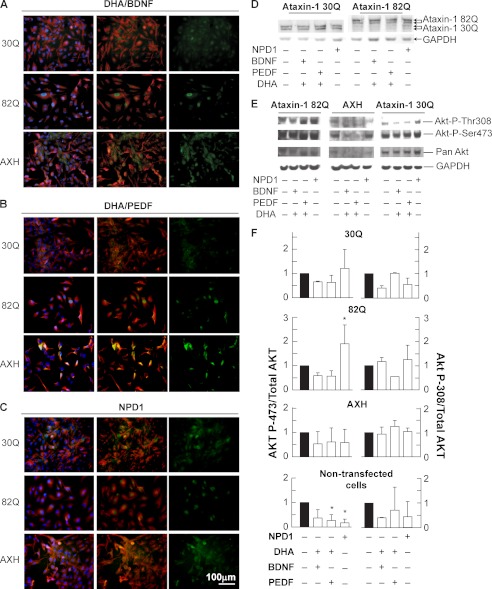
NPD1 Does Not Prevent Akt-mediated Ataxin-1 Phosphorylation
Phosphoataxin-1 cellular amount varies depending on the necessities of cell activity. The final content of phosphoataxin-1 depends on phosphorylation/dephosphorylation ratios (7). Some evidence points to Akt as the kinase responsible for ataxin-1 phosphorylation (36). We investigated this issue by assessing the Akt activation achieved by phosphorylation in threonine 308 and serine 473 as well as its translocation from the membrane to its action site. In control and AXH-expressing cells, the Akt signal was associated with non-nuclear localization, possibly cytoplasmic or close to the membrane (Fig. 7, A–C). When ataxin-1 82Q is expressed in the presence of NPD1 or DHA/PEDF, Akt is translocated to the perinuclear/nuclear space (Fig. 7, B and C). BDNF is a known activator of Akt (37, 38). However, to a lesser extent, DHA/BDNF induced Akt translocation but only in cells expressing the 82Q form (supplemental Fig. 11), suggesting that Akt activation is only accompanied by its translocation when the poly(Q) protein is present. To address Akt phosphorylation upon activation, we performed Western blot assay using the anti-phosphoserine (Ser(P)-473) and threonine (Thr(P)-308) Akt. The ratio of Ser(P)-473 Akt over total Akt was increased significantly only when cells were expressing 82Q and treated with NPD1, suggesting that ataxin-1 phosphorylation takes place although the content of phosphorylated ataxin-1 decreases. This evidence indicates that NPD1 may induce ataxin-1 dephosphorylation and turnover (Fig. 8).
FIGURE 7.
Akt activation and translocation to the nucleus is induced by NPD1 upon expression of ataxin-1 82Q. ARPE-19 cells were transfected with the constructs 82Q, 30Q, and AXH and treated with the lipid messengers and growth factors for 24 h in low serum medium. A–C, immunocytochemistry using anti-pan-Akt (green) and anti-14-3-3 η (red). D, immunoblots and densitometric determination of the expression of ataxin-1 30Q and 82Q. E, representative immunoblots of ARPE-19 cells subjected to the same conditions shown in A–C using anti-phospho-Ser-473 Akt, anti-phospho-Thr-308 Akt, anti-pan-Akt, and GAPDH. F, densitometric analysis of the immunoblot shown in E. *, p < 0.05. Error bars, S.D.
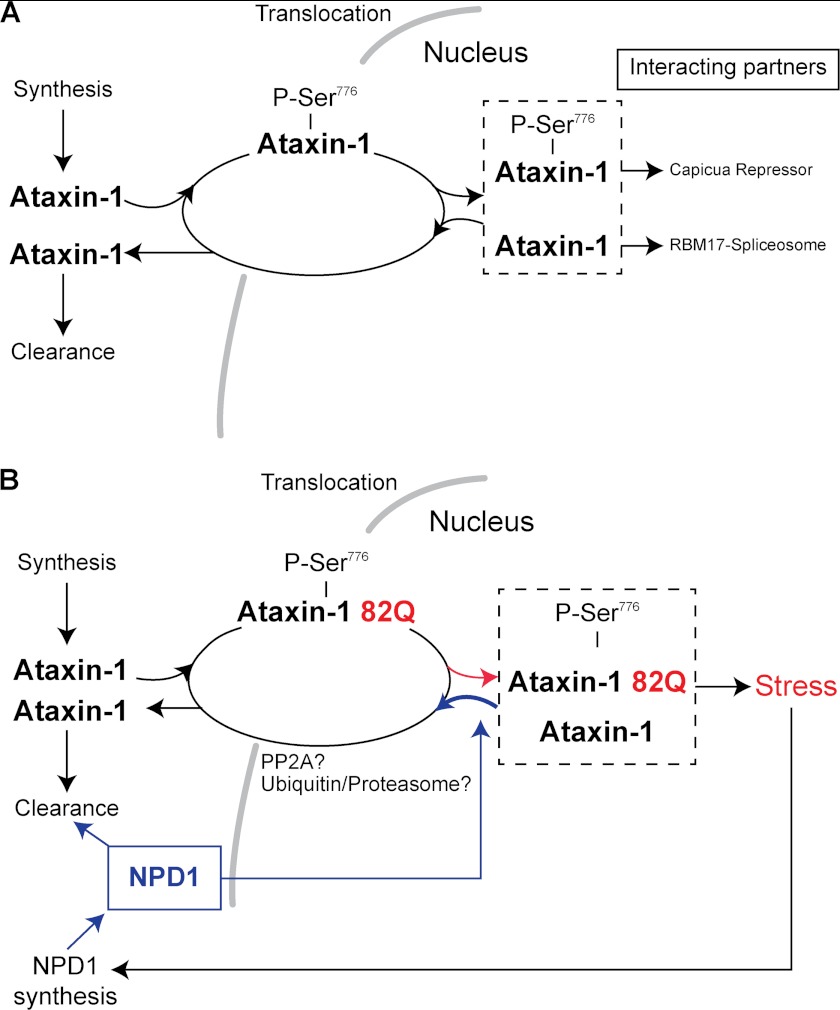
FIGURE 8.
NPD1 mechanism of protection against proteotoxic stress. Schematic representation of the mechanism proposed for NPD1 in RPE cells. A, model extracted from data obtained in Purkinje cells; ataxin-1 corrected folding. B, model in human RPE cells; misfolded ataxin-1 disequilibrium and proposed NPD1 action in human RPE cells.
DISCUSSION
In the present report, we provide evidence in support of the role of NPD1 signaling in promoting an increase in turnover rate that helps clear abnormally folded proteins through a general mechanism. “Proteostasis” is the term coined to describe the events leading to restored balance of the synthesis, activation/deactivation, localization, and degradation of proteins to achieve a dynamic equilibrium of activity that satisfies cell demands in specific conditions. When this equilibrium is disrupted, proteins accumulate and thereby induce formation of inclusion bodies and toxic subproducts. Examples of these imbalances are drusen in age-related macular degeneration, Lewis bodies in Parkinson disease, and amyloid plaques in Alzheimer disease. Although it is not always clear whether protein tangles are the cause or consequence of neurodegeneration, efforts are being made to prevent protein accumulation because it is still considered a toxic factor (16).
SCA1 is an autosomal dominant disorder characterized by progressive loss of motor coordination due to cerebellar and brainstem degeneration (39, 40). Some evidence links SCA1 with decreased visual acuity and visual fields along with impairment of color vision (9). Specifically, a recent report describes a case in which an SCA1 patient displayed pigmentary macular dystrophy, which is commonly encountered in SCA7 patients (11). Amplified glutamine repeats are present in a variety of proteins involved in several neurodegenerative diseases (40). The oversized polyglutamine tracts are proposed to induce misfolding in the proteins in which they are contained. This promotes abnormal accumulation that causes either a loss of function when the protein is inactivated by the tangle or a gain of toxic function when protein degradation fails, resulting in toxic properties that lead to cell death (41). In this report, we showed that ataxin-1 82Q expression induced apoptosis in RPE cells. To a lesser extent, the overexpression of the normal form of ataxin-1 (30Q) also induced apoptosis (Figs. 3 (E and F) and Fig. 5A and supplemental Fig. 12A), although in this case, DHA/PEDF treatment did not affect the outcome, suggesting that overexpression of the normal protein form differs in its toxic effect from the pathological form. A plausible explanation may be that while the function is intact, the wild type protein overflow alters the normal stoichiometry of the complexes, and thus a moderate cytotoxicity occurs; however, when the misfolded form is present, stress-triggered mechanisms are awakened, and these are responsible for activating prosurvival pathways through enhanced NPD1 synthesis (Fig. 5C). In addition, caspase-3 activation was prevented to a lesser extent by NPD1 in apoptosis induced by the wild type constructs. Therefore, only the misfolded poly(Q) ataxin-1 induced stress-dependent, prosurvival signaling mediated by NPD1, which provides evidence for the prohomeostatic properties of this lipid messenger. Altogether, these data suggest that NPD1 counteracts the mechanisms set forward by the proteotoxicity once stress is triggered. This is a desirable property in therapeutic treatments, given that NPD1 does not disrupt mechanisms to prevent apoptosis but instead promotes the establishment of equilibrium (42).
Activity at the CIC binding site was increased by the expression of normal ataxin-1, showing a decreased luciferase activity compared with the control (Fig. 4A). Furthermore, the 2Q form promoted significant repression (Fig. 4A), suggesting that the activity exerted jointly with capicua-binding protein is inversely proportional to the length of the poly(Q) tract. Conversely, the 82Q tract induced increased activity of the repressor released by the addition of NPD1. This apparent contradiction may be explained in terms of the dynamics of the complexes formed that guided regulation of the function. Fig. 8B depicts the proposed relationship with the complex components. We propose that NPD1 increases ataxin-1 turnover, reducing sequestration of the wild type active protein by the 82Q inactive form of ataxin-1. The system used in the present study also expresses the wild type protein (supplemental Fig. 5C) that may, in part, restore the function of the complexes by replacing the faulty protein. Thus, we still observed repression to some extent in the NPD1-treated cells. Following this line of reasoning, in ARPE-19 cells, we found that NPD1 reduced the phosphorylated signal in the nuclei, which was positively correlated with survival of the cells carrying the 82Q construct (Figs. 1 and 6C). Conversely, repressive activity accomplished by the formation of the complex ataxin-1·CBP was decreased by NPD1. In this scenario, the inactive 82Q form was cleared together with the endogenously active wild type one, causing loss of activity to a greater extent than in cells that expressed only the functional wild type protein. This may have caused the decrease in activity observed in Fig. 3B. On the other hand, NPD1 had no effect on the endogenously expressed ataxin-1, in agreement with the results obtained from the 30Q overexpression. This shows that only the presence of the 82Q form triggers a response from NPD1.
Other poly(Q) proteins, such as huntingtin (Htt), are capable of triggering apoptosis and a prosurvival response from NPD1. Htt 72Q expression increased apoptosis in ARPE-19 cells, and NPD1 counteracted it almost completely (Fig. 4E). These observations may support the idea that the poly(Q) tract evokes NPD1 protective actions independent of the specific protein activity that carries it, which leads to the conclusion that NPD1 is a protective mechanism. A candidate process that may be affected by NPD1 is the ubiquitin/proteasome pathway. Compelling evidence links deficits in the proteasome pathway to an increase in pathology severity. In fact, ataxin-1 is polyubiquitinated, and intriguingly, Ube3a-deficient SCA1 mice show a reduction in the nuclear inclusions in the cerebellar Purkinje cells (43). On the other hand, HSP70 overexpression reduces phenotype severity, but nuclear inclusions are increased (44). Another piece of evidence that contributes to the concept of an NPD1-enhanced scavenging system is the dose/time-related dependence, which showed that at 24 h, NPD1 doses of 10 nm or lower did not have a significant effect on cell survival, whereas 50 nm prevented apoptosis even after 72 h (Fig. 1C). The 24 h linearity is consistent with NPD1-enhanced scavenging because this is an accumulation mechanism. Beyond this point, NPD1 is no longer effective at low doses, probably because cells require a stronger activation for clearance mechanisms.
The phosphorylation state is an indicator of the type of activity the protein is performing, as mentioned above (7). We observed a positive temporal correlation between the emerging Ser-776 phosphoataxin-1 nuclear granules (supplemental Figs. 7–10) and apoptosis (Figs. 1B and 3, E and F). An alternative explanation would involve displacement of equilibrium in the complexes. It was described previously that the phosphorylation state determines the activity of the normal protein (7). The phosphorylation on Ser-776 in the normal ataxin-1 protein reduces the interaction with RBM17 (also known as SPF45), displacing the equilibrium of complexes formed in the nucleus from transcription repression toward spliceosomal activity (7, 36). The non-phosphorylated protein may be diverted into the formation of the spliceosome complex, reducing both toxicity and CIC repressive activity. NPD1 favors ataxin dephosphorylation in the nucleus; this notion arises from the observation that NPD1 did not prevent activation of Akt, the main enzyme responsible for the ataxin-1 phosphorylation (Fig. 7D). On the contrary, NPD1 seems to enhance Akt activation, as evidenced by the increase in the Ser-473-phosphorylated form of Akt and its translocation to the nucleus (Fig. 7, B, C, and E). This is in agreement with previous reports (44) demonstrating that the phosphorylation state of Akt is modified by the presence of the poly(Q) form of the protein. Conversely, AXH failed to induce Akt translocation into the nucleus (Fig. 7, A–C, third row) and ataxin-1 phosphorylation, suggesting again that the causes of the AXH toxicity differ from the 82Q ones. Finally, these results support the idea that there are at least two components of toxicity elicited by ataxin-1 82Q; one is related with the disequilibrium in ataxin-1 function, and the other is related with the stress mechanisms set off by the poly(Q) tract itself. NPD1 action may be triggered by the second component to affect the first.
Although phosphorylation at Ser-776 is an important event in ataxin-1 activity, dephosphorylation more than likely seems to be the mechanism for switching ataxin-1 activity type. By the same token, it seems that the presence of a protein phosphatase 2A inhibitor may be involved in this process. Anp32, a potent protein phosphatase 2A inhibitor, was proposed to be involved in SCA1 pathogenesis based on its expression time frame, pattern, and differential interaction with the expanded form of ataxin-1, although this was later suggested to be indirect (44). In agreement with our hypothesis that there is a relationship between NPD1 regulation of phosphorylated and non-phosphorylated ataxin-1 activity, NPD1 decreased the phosphorylated content of ataxin-1 in ARPE-19 cells (Fig. 6, B and C). NPD1 was previously shown to increase protein phosphatase 2A activity (45). As such, NPD1 may counteract protein phosphatase 2A inhibition, allowing the 82Q form to be dephosphorylated and cleared or relocated into the spliceosomal function. The fact that Anp32 was proposed to have a stronger interaction with the expanded form rather than with the wild type ataxin-1 makes this protein an excellent target candidate for NPD1 signaling (46). Furthermore, the expression of the 30Q form in our system affected the levels of phosphorylation but not to the same extent as ataxin-1 82Q. Further research is needed to clarify this issue because another report shows that PPA2 inhibitors do not affect ataxin-1 dephosphorylation (47).
In addition to the expansions in the polyglutamine tract, AXH has an important role in the functionality of ataxin-1. AXH, a self-folding domain present in ataxin-1, is responsible for the protein-protein interactions between ataxin-1 and other transcription factors, such as the capicua homolog CIC protein (33). The sequestration of the complex partners formed by ataxin-1 by its inactive counterpart may be involved in the loss of function observed in SCA1 pathology (48). Brother of ataxin-1 (Boat), another member of the AXH domain-containing protein family, is an example of the proposed loss of function. Boat is an in vivo binding partner of ataxin-1 that is also affected by the malfunction of ataxin-1 82Q (49). Following this line of reasoning, the expression of AXH alone would produce toxic effects by competing with the domains contained by ataxin-1, thereby inducing disassembly of the complexes. Indeed, AXH expression in ARPE-19 cells resulted in increased apoptosis. Furthermore, it aggravated the cytotoxicity induced by ataxin-1 82Q (Fig. 4B). Unlike the sequestration scenario, in which the complexes are formed but are inactive, AXH induces toxicity in this case by increasing disassembly of the complex, thus promoting inactivation of its partners. NPD1 signaling promotes survival by modulating a set of genes that homeostatically control cell fate. NPD1 was capable of reversing the toxicity of both AXH and ataxin-1 82Q in ARPE-19 cells (Fig. 4B).
Finally, the occurrence of ataxin-1 82Q toxicity in human RPE cells (Fig. 2, C and D) confirmed that these cells are susceptible to proteotoxic stress induced by ataxin-1. Accordingly, neurons expressing ataxin-1 82Q were successfully rescued from apoptosis by NPD1 (Fig. 2, H and I), supporting the validity of the model.
Altogether, these results demonstrate that 1) the RPE cellular model depicts highly similar features occurring in Purkinje cells, laying the groundwork for unveiling new pathways and therapeutic approaches, and 2) NPD1 attenuates the deleterious effects of proteotoxic stress caused by extended poly(Q) tract ataxin-1 expression in RPE cells due to the increase in turnover. These findings may lead to new therapeutic applications for DHA and NPD1 for treating and preventing many brain and retinal conditions and diseases, such as age-related macular degeneration and Alzheimer and Parkinson diseases.
Acknowledgments
We thank Dr. Huda Zoghbi for the ataxin-1 and huntingtin expression constructs used in this report and for invaluable input; Stephen Prescott and Dan A. Dixon for the COX-2 promoter construct; Dr. William C. Gordon for advice on TUNEL staining; and Dr. Houhui Xia, Hou Hailong, and Miguel Molina for the rat neuronal cultures.
This work was supported, in whole or in part, by National Institutes of Health (NIH), NEI, Grant EY005121 and NIH, NCRR, Grant RR016816. This work was also supported in part by an unrestricted departmental grant from Research to Prevent Blindness, Inc., New York.

This article contains supplemental Figs. 1–13.
- NPD1
- neuroprotectin D1
- 15-LOX-1
- 15-lipoxygenase-1
- BDNF
- brain-derived neurotrophic factor
- CIC
- capicua
- COX-2
- cyclooxygenase-2
- DHA
- docosahexaenoic acid
- PEDF
- pigment epithelium-derived factor
- RPE
- retinal pigment epithelial
- hRPE
- human RPE
- SCA1
- spinocerebellar ataxia 1.
REFERENCES
- 1. Shaw C. A., Höglinger G. U. (2008) Neurodegenerative diseases. Neurotoxins as sufficient etiologic agents? Neuromolecular Med. 10, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kalaria R. N. (2010) Vascular basis for brain degeneration. Faltering controls and risk factors for dementia. Nutr. Rev. 68, S74–S87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khusnutdinova E., Gilyazova I., Ruiz-Pesini E., Derbeneva O., Khusainova R., Khidiyatova I., Magzhanov R., Wallace D. C. (2008) A mitochondrial etiology of neurodegenerative diseases. Evidence from Parkinson's disease. Ann. N.Y. Acad. Sci. 1147, 1–20 [DOI] [PubMed] [Google Scholar]
- 4. Cummings C. J., Mancini M. A., Antalffy B., DeFranco D. B., Orr H. T., Zoghbi H. Y. (1998) Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 19, 148–154 [DOI] [PubMed] [Google Scholar]
- 5. Kaytor M. D., Warren S. T. (1999) Aberrant protein deposition and neurological disease. J. Biol. Chem. 274, 37507–37510 [DOI] [PubMed] [Google Scholar]
- 6. Dikshit P., Jana N. R. (2008) Role of ubiquitin protein ligases in the pathogenesis of polyglutamine diseases. Neurochem. Res. 33, 945–951 [DOI] [PubMed] [Google Scholar]
- 7. de Chiara C., Menon R. P., Strom M., Gibson T. J., Pastore A. (2009) Phosphorylation of Ser-776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS One. 4, e8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zoghbi H. Y., Orr H. T. (2009) Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J. Biol. Chem. 284, 7425–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abe T., Abe K., Aoki M., Itoyama Y., Tamai M. (1997) Ocular changes in patients with spinocerebellar degeneration and repeated trinucleotide expansion of spinocerebellar ataxia type 1 gene. Arch. Ophthalmol. 115, 231–236 [DOI] [PubMed] [Google Scholar]
- 10. Isashiki Y., Kii Y., Ohba N., Nakagawa M. (2001) Retinopathy associated with Machado-Joseph disease (spinocerebellar ataxia 3) with CAG trinucleotide repeat expansion. Am. J. Ophthalmol. 131, 808–810 [DOI] [PubMed] [Google Scholar]
- 11. Saito Y., Matsumura K., Shimizu S., Ichikawa Y., Ochiai K., Goto J., Tsuji S., Shimizu T. (2006) J. Neurol. Neurosurg. Psychiatry 77, 1293–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu X., Garriga P., Khorana H. G. (1996) Structure and function in rhodopsin. Correct folding and misfolding in two point mutants in the intradiscal domain of rhodopsin identified in retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 93, 4554–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saliba R. S., Munro P. M., Luthert P. J., Cheetham M. E. (2002) The cellular fate of mutant rhodopsin. Quality control, degradation, and aggresome formation. J. Cell Sci. 115, 2907–2918 [DOI] [PubMed] [Google Scholar]
- 14. Mendes H. F., van der Spuy J., Chapple J. P., Cheetham M. E. (2005) Mechanisms of cell death in rhodopsin retinitis pigmentosa. Implications for therapy. Trends Mol. Med. 11, 177–185 [DOI] [PubMed] [Google Scholar]
- 15. Tam B. M., Moritz O. L. (2006) Characterization of rhodopsin P23H-induced retinal degeneration in a Xenopus laevis model of retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 47, 3234–3241 [DOI] [PubMed] [Google Scholar]
- 16. Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Adapting proteostasis for disease intervention. Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 17. Morimoto R. I. (2008) Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomas M., Yu Z., Dadgar N., Varambally S., Yu J., Chinnaiyan A. M., Lieberman A. P. (2005) The unfolded protein response modulates toxicity of the expanded glutamine androgen receptor. J. Biol. Chem. 280, 21264–21271 [DOI] [PubMed] [Google Scholar]
- 19. Haynes C. M., Titus E. A., Cooper A. A. (2004) Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol. Cell 15, 767–776 [DOI] [PubMed] [Google Scholar]
- 20. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 21. Hotamisligil G. S. (2010) Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140, 900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gray D. A., Woulfe J. (2005) Lipofuscin and aging: A matter of toxic waste. Sci. Aging Knowledge Environ. 2005, re1. [DOI] [PubMed] [Google Scholar]
- 23. Calandria J. M., Marcheselli V. L., Mukherjee P. K., Uddin J., Winkler J. W., Petasis N. A., Bazan N. G. (2009) Selective survival rescue in 15-lipoxygenase-1-deficient retinal pigment epithelial cells by the novel docosahexaenoic acid-derived mediator, neuroprotectin D1. J. Biol. Chem. 284, 17877–17882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mukherjee P. K., Marcheselli V. L., Serhan C. N., Bazan N. G. (2004) Neuroprotectin D1. A docosahexaenoic acid-derived docosatriene protects human retinal pigment epithelial cells from oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 101, 8491–8496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koshy B., Matilla T., Burright E. N., Merry D. E., Fischbeck K. H., Orr H. T., Zoghbi H. Y. (1996) Spinocerebellar ataxia type-1 and spinobulbar muscular atrophy gene products interact with glyceraldehyde-3-phosphate dehydrogenase. Hum. Mol. Genet. 5, 1311–1318 [DOI] [PubMed] [Google Scholar]
- 26. Tsuda H., Jafar-Nejad H., Patel A. J., Sun Y., Chen H. K., Rose M. F., Venken K. J., Botas J., Orr H. T., Bellen H. J., Zoghbi H. Y. (2005) The AXH domain of ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell 122, 633–644 [DOI] [PubMed] [Google Scholar]
- 27. Ishida M., Lui G. M., Yamani A., Sugino I. K., Zarbin M. A. (1998) Culture of human retinal pigment epithelial cells from peripheral scleral flap biopsies. Curr. Eye Res. 17, 392–402 [DOI] [PubMed] [Google Scholar]
- 28. Stark D. T., Bazan N. G. (2011) Synaptic and extrasynaptic NMDA receptors differentially modulate neuronal cyclooxygenase-2 function, lipid peroxidation, and neuroprotection. J. Neurosci. 31, 13710–13721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 30. Mukherjee P. K., Marcheselli V. L., Barreiro S., Hu J., Bok D., Bazan N. G. (2007) Neurotrophins enhance retinal pigment epithelial cell survival through neuroprotectin D1 signaling. Proc. Natl. Acad. Sci. U.S.A. 104, 13152–13157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mukherjee P. K., Marcheselli V. L., de Rivero Vaccari J. C., Gordon W. C., Jackson F. E., Bazan N. G. (2007) Photoreceptor outer segment phagocytosis attenuates oxidative stress-induced apoptosis with concomitant neuroprotectin D1 synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 13158–13163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chou A. H., Yeh T. H., Kuo Y. L., Kao Y. C., Jou M. J., Hsu C. Y., Tsai S. R., Kakizuka A., Wang H. L. (2006) Polyglutamine-expanded ataxin-3 activates mitochondrial apoptotic pathway by up-regulating Bax and down-regulating Bcl-xL. Neurobiol. Dis. 21, 333–345 [DOI] [PubMed] [Google Scholar]
- 33. Wang H. L., Yeh T. H., Chou A. H., Kuo Y. L., Luo L. J., He C. Y., Huang P. C., Li A. H. (2006) Polyglutamine-expanded ataxin-7 activates mitochondrial apoptotic pathway of cerebellar neurons by upregulating Bax and downregulating Bcl-xL. Cell. Signal. 18, 541–552 [DOI] [PubMed] [Google Scholar]
- 34. Lam Y. C., Bowman A. B., Jafar-Nejad P., Lim J., Richman R., Fryer J. D., Hyun E.D., Duvick L. A., Orr H. T., Botas J., Zoghbi H. Y. (2006) ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 127, 1335–1347 [DOI] [PubMed] [Google Scholar]
- 35. Marcheselli V. L., Hong S., Lukiw W. J., Tian X. H., Gronert K., Musto A., Hardy M., Gimenez J. M., Chiang N., Serhan C. N., Bazan N. G. (2003) Novel docosanoids inhibit brain ischemia-reperfusion-mediated leukocyte infiltration and pro-inflammatory gene expression. J. Biol. Chem. 278, 43807–43817 [DOI] [PubMed] [Google Scholar]
- 36. Chen H. K., Fernandez-Funez P., Acevedo S. F., Lam Y. C., Kaytor M. D., Fernandez M. H., Aitken A., Skoulakis E. M., Orr H. T., Botas J., Zoghbi H. Y. (2003) Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell 113, 457–468 [DOI] [PubMed] [Google Scholar]
- 37. Azadi S., Johnson L. E., Paquet-Durand F., Perez M. T., Zhang Y., Ekström P. A., van Veen T. (2007) CNTF + BDNF treatment and neuroprotective pathways in the rd1 mouse retina. Brain Res. 1129, 116–129 [DOI] [PubMed] [Google Scholar]
- 38. Wu C. L., Hwang C. S., Chen S. D., Yin J. H., Yang D. I. (2010) Neuroprotective mechanisms of brain-derived neurotrophic factor against 3-nitropropionic acid toxicity. Therapeutic implications for Huntington's disease. Ann. N.Y. Acad. Sci. 1201, 8–12 [DOI] [PubMed] [Google Scholar]
- 39. Zoghbi H. Y., Orr H. T. (2000) Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23, 217–247 [DOI] [PubMed] [Google Scholar]
- 40. Matilla-Duenas A., Goold R., Giunti P. (2008) Clinical, genetic, molecular, and pathophysiological insights into spinocerebellar ataxia type 1. Cerebellum 7, 106–114 [DOI] [PubMed] [Google Scholar]
- 41. Williams A. J., Paulson H. L. (2008) Polyglutamine neurodegeneration. Protein misfolding revisited. Trends Neurosci. 31, 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bazan N. G., Calandria J. M., Serhan C. N. (2010) Rescue and repair during photoreceptor cell renewal mediated by docosahexaenoic acid-derived neuroprotectin D1. J. Lipid Res. 51, 2018–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cummings C. J., Reinstein E., Sun Y., Antalffy B., Jiang Y., Ciechanover A., Orr H. T., Beaudet A. L., Zoghbi H. Y. (1999) Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron 24, 879–892 [DOI] [PubMed] [Google Scholar]
- 44. Cummings C. J., Sun Y., Opal P., Antalffy B., Mestril R., Orr H. T., Dillmann W. H., Zoghbi H. Y. (2001) Overexpression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum. Mol. Genet. 10, 1511–1518 [DOI] [PubMed] [Google Scholar]
- 45. Antony R., Lukiw W. J., Bazan N. G. (2010) Neuroprotectin D1 induces dephosphorylation of Bcl-xL in a PP2A-dependent manner during oxidative stress and promotes retinal pigment epithelial cell survival. J. Biol. Chem. 285, 18301–18308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Chiara C., Menon R. P., Pastore A. (2008) Structural bases for recognition of Anp32/LANP proteins. FEBS J. 275, 2548–2560 [DOI] [PubMed] [Google Scholar]
- 47. Kaytor M. D., Byam C. E., Tousey S. K., Stevens S. D., Zoghbi H. Y., Orr H. T. (2005) A cell-based screen for modulators of ataxin-1 phosphorylation. Hum. Mol. Genet. 14, 1095–1105 [DOI] [PubMed] [Google Scholar]
- 48. Crespo-Barreto J., Fryer J. D., Shaw C. A., Orr H. T., Zoghbi H. Y. (2010) Partial loss of ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis. PLoS Genet. 6, e1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mizutani A., Wang L., Rajan H., Vig P. J., Alaynick W. A., Thaler J. P., Tsai C. C. (2005) Boat, an AXH domain protein, suppresses the cytotoxicity of mutant ataxin-1. EMBO J. 24, 3339–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]