Background: The cellular response to DNA damage requires multiple signaling pathways to guarantee genomic stability.
Results: Vaccinia-related kinase 1 (VRK1) is activated by ionizing radiation and required for formation of 53BP1 foci in response to DNA damage.
Conclusion: Human VRK1 is an early step in the cellular response to DNA damage induced by ionizing radiation.
Significance: VRK1 forms part of a novel pathway for DNA protection.
Keywords: DNA Damage Response, Phosphorylation Enzymes, Protein Phosphorylation, Serine-Threonine Protein Kinase, Signal Transduction, 53BP1, VRK1, Ionizing Radiation
Abstract
Cellular responses to DNA damage require the formation of protein complexes in a highly organized fashion. The complete molecular components that participate in the sequential signaling response to DNA damage remain unknown. Here we demonstrate that vaccinia-related kinase 1 (VRK1) in resting cells plays an important role in the formation of ionizing radiation-induced foci that assemble on the 53BP1 scaffold protein during the DNA damage response. The kinase VRK1 is activated by DNA double strand breaks induced by ionizing radiation (IR) and specifically phosphorylates 53BP1 in serum-starved cells. VRK1 knockdown resulted in the defective formation of 53BP1 foci in response to IR both in number and size. This observed effect on 53BP1 foci is p53- and ataxia-telangiectasia mutated (ATM)-independent and can be rescued with VRK1 mutants resistant to siRNA. VRK1 knockdown also prevented the activating phosphorylation of ATM, CHK2, and DNA-dependent protein kinase in response to IR. VRK1 activation in response to DNA damage is a novel and early step in the signaling of mammalian DNA damage responses.
Introduction
When cells encounter stress, such as DNA damage, they activate complex signaling networks that regulate their ability to detect and repair the damage and return to homeostatic equilibrium. These networks integrate a wide variety of signals from inside the cell, transduced through protein kinases (1–3), to ultimately control cell cycle arrest or progression in the case of dividing cells (4). A major risk of DNA damage is the transmission of unrepaired DNA lesions to the progeny (5) that are implicated in aging and cancer (6). In addition to cancer, defective DNA repair is also associated with neurodegenerative diseases (7). Understandably, most research into DNA damage responses has been performed in the context of cell division (5). However, in the human body, most of the cells are in the G0 state and are not dividing at the time of DNA damage. Such responses are particularly important for resting or quiescent cells, such as stem cells and neurons. Thus, the initial response to DNA damage is an individual cell function that does not have to be linked to cell division. Francis Crick (8) predicted that several redundant mechanisms must exist to repair damaged DNA and maintain genome integrity. Since then, several pathways have been identified (1–3).
We hypothesized that cells must rapidly and efficiently sense these DNA lesions before they divide to avoid transmission to their progeny. Because nuclear kinases are capable of rapidly responding to changes in the cell and its environment and integrating diverse stimuli, we suspected that they might be implicated in sensing and triggering downstream signaling that is needed for proper DNA damage repair. The most common pathways in the DNA damage response (DDR)3 implicate protein phosphorylation by different kinases, such as ATM (1), ATR (2), and DNA-PK (3), which have been mostly studied in the context of cell division and cell cycle checkpoints (4). In response to double strand breaks induced by ionizing radiation (IR), 53BP1 scaffold protein is recruited to IR-induced foci, constituting an important marker for monitoring cellular DDR. 53BP1 foci are intermediate steps in DDR activation (9, 10) and are known to be regulated by ATM in response to DNA double strand breaks (DSBs) (11) and by ATR in response to replication stress (12). However, it is also known that the DNA damage response to DNA DSBs can be ATM-independent (13). This suggests that there are alternative kinases participating in DDR induced by ionizing radiation.
DDR signaling induced by different genotoxic agents converge on p53 phosphorylation events (14, 15). Nuclear kinase vaccinia-related kinase 1 (VRK1), a Ser-Thr kinase, phosphorylates p53 at Thr-18 (16–18). This p53 residue is the key residue controlling the binding of p53 with its negative regulator Mdm2. Thr-18 phosphorylation increases binding of p53 to transcriptional cofactors by 3 orders of magnitude (19, 20), whereas p53 with unphosphorylated Thr-18 preferentially binds to ubiquitin ligases, such as Mdm2/Hdm2. Thr-18 phosphorylation plays a threshold role in p53 responses (17, 21), and additional p53 phosphorylations contribute to the specificity of cofactor selection (20). VRK1, which can phosphorylate p53 (16), has been associated with later stages of the DDR and multiple effector mechanisms (18, 22). Human VRK1 is required for entry in the cell cycle (23), and its loss causes a cell cycle arrest in G0 (23). VRK1 phosphorylates histones and promotes chromatin compaction in mitosis (24, 25) as does nucleosomal histone kinase-1 (NHK-1), its Drosophila melanogaster ortholog (26). VRK1 directly interacts with histones, and its kinase activity is regulated by these interactions (27). VRK1 also regulates the proviral DNA preintegration complex of retroviruses (28). VRK1 participation in nucleoprotein complexes can have a dual role either affecting local chromatin structure or alternatively reacting to changes in chromatin, which suggests that VRK1 might function as part of chromatin signaling pathways (29–31). But a kinase responding to changes in chromatin structure should also mediate responses aiming to signal or repair damaged DNA, which also requires chromatin remodeling. VRK1 can thus be a novel protein involved in these processes. This regulatory function is a very suitable role for a kinase that appeared late in evolution, such as VRK1 (32), whose role is more likely to be the coordination of preexisting pathways in higher and more complex organisms. Here we identify a novel role for VRK1 in responding to DSBs that directly connects the signal of damaged DNA to the DDR mediator protein 53BP1 in a p53- and ATM-independent mechanism.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture
A549, H1299, MCF7, GM9607, HT144, and HEK293T cells were from ATCC. Cells were grown as monolayers in DMEM supplemented with 10% fetal calf serum, 2 mm glutamine, 50 μg/ml penicillin, and 50 μg/ml streptomycin. Cells were detached with trypsin-EDTA solution. All cell culture reagents were from Invitrogen.
Plasmids
VRK1 protein was expressed in bacteria from clone pGEX4T-GST-VRK1 (16) or pET23-VRK1-his. GST-53-BP1 fragments in pGEX were from J. Chen (33). GST-p53(1–85) was described previously (16). GST fusion proteins were purified using glutathione-Sepharose 4B (GE Healthcare) and eluted with reduced glutathione. His-tagged proteins were purified using Talon resin (Clontech). Mammalian expression plasmid p-CEFL-HA-VRK1 was described previously (17). Plasmid p-CEFL-HA-VRK1(R391/R393/V394), resistant to si-VRK1–01, was generated by introducing three silent nucleotide changes at the third codon position in residues Arg-391, Arg-393, and Val-394 by site-directed mutagenesis with the QuikChange mutagenesis kit from Stratagene (San Diego, CA). Plasmid p-HA-53BP1 full length was from T. Halazonetis (9) as well as plasmids pCMV-FLAG-CHK2 and pCMV-FLAG-CHK2(A347). Plasmids pBJ1-HA-ATM wild type and kinase-dead were from J. R. Nevins (34). Plasmids pcDNA3-FLAG-ATR wild type and kinase-dead (D2475A) were from R. T. Abraham (35).
Cell Transfections
In experiments in which plasmid constructs were transiently transfected, cells were in exponential growth phase. Twenty-four hours before transfection, cells were plated in P100 dishes at 60–70% confluence at the time of transfection. Plasmid DNAs for mammalian transfections were prepared using the JETStar endotoxin-free maxiprep kit (Genomed GmbH). The specific plasmids used are indicated in the individual experiments. Transfections of the cell lines used in this report have already been reported (25, 36).
RNA Interference
Specific silencing of VRK1 was performed using two different siRNAs: siVRK1-01 (siV1-01) and siVRK1-02 (siV1-02). Both were designed with the SMARTselection algorithm (Dharmacon RNAi Technologies) and obtained from Dharmacon RNAi Technologies. The sequence targets of the two VRK1 siRNA oligos were as follows: siVRK1-01, GAAAGAGAGTCCAGAAGTA; and siVRK1-02, CAAGGAACCTGGTGTTGAA. As a negative control, indicated as siCt in experiments, the ON-TARGETplus siCONTROL Non-targeting siRNA from Dharmacon RNAi Technologies was used. The efficiency of RNAi transfection was determined with siGLO RISC-free siRNA (Dharmacon RNAi Technologies) labeled with a red fluorochrome.
Cells were transfected with the indicated siRNA at a concentration of 20 nm using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. After transfection, cells were processed for specific experiments at the times indicated. Experiments were performed as reported previously (18, 23, 37). For rescue experiments, cells were transfected with the indicated siRNA using Lipofectamine 2000 reagent. After 36 h, cells were retransfected with plasmids using Lipofectamine at a 1:5 ratio (μg of DNA:μl of Lipofectamine). Targeted protein and plasmid expression was analyzed 36 h after the second transfection.
Kinase Assays
In vitro kinase activity of VRK1 was analyzed using recombinant proteins expressed in Escherichia coli BL21 strain in a buffer containing 20 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 0.5 mm DTT, 150 mm KCl, 5 μm ATP, and 5 μCi of (0.1 μm) [γ-32P]ATP (PerkinElmer Life Sciences) in a final volume of 30 μl. As substrates, recombinant proteins also were used as indicated in each individual assay. Reactions were performed at 30 °C for 30 min (38) with mild shaking in a Thermomixer Compact (Eppendorf). Endogenous VRK1 assays were performed under the same conditions using the protein immunoprecipitated from the cell line indicated in the experiment in a final volume of 65 μl. For kinase assays, VRK1 was routinely immunoprecipitated with 1F6 monoclonal antibody (39).
ATM and ATR kinase assays were performed in a buffer containing 10 mm Hepes, pH 7.5, 10 mm MgCl2, 10 mm MnCl2, 50 mm NaCl, 1 mm DTT, 50 μm ATP, and 5 μCi of (0.1 μm) [γ-32P]ATP (33). The CHK2 kinase assay was performed in buffer containing 50 mm Hepes, pH 8.0, 10 mm MgCl2, 2.5 mm EDTA, 1 mm DTT, 10 μm ATP, and 5 μCi of (0.1 μm) [γ-32P]ATP (40).
Reactions were stopped by adding loading buffer (62.5 mm Tris-HCl, pH 6.8, 10% glycerol, 2.3% SDS, 0.1% bromphenol blue, and 5% β-mercaptoethanol) and heating at 100 °C for 5 min. Proteins were separated by SDS-PAGE (25). Gels were either stained with Coomassie Blue or transferred to a PDVF membrane (Immobilon-P, Millipore). Radioactivity was detected by exposure to x-ray film (Fujifilm) or by Molecular Imager FX (Bio-Rad). Control bands were detected with Coomassie Blue, Ponceau staining, or Western blot with a specific antibody as indicated in the individual experiment. Detection of specific phosphorylation in specific residues was performed by Western blot with the indicated phosphospecific antibodies of the corresponding protein.
Antibodies
Human VRK1 was detected by immunofluorescence and in Western blots with monoclonal 1B5 (39) or polyclonal anti-VRK1 from Sigma (HPA000660). Endogenous VRK1 was routinely immunoprecipitated with 1F6 monoclonal antibody (39). 1B5 and anti-VRK1 (Sigma) were used in Western blots at a dilution 1:1000 and by immunofluorescence at 1:50 or 1:100, respectively. 53BP1 was detected by immunofluorescence with either polyclonal 53BP1 (H-300) (Santa Cruz Biotechnology) or monoclonal anti-53BP1 from Upstate at a 1:50 dilution (no differences in focus number or size were found between the two antibodies detecting 53BP1). Antibodies against phospho-53BP1(Ser-25/Ser-29), phospho-p53(Thr-18), phospho-CHK2(Thr-68), CHK2, phospho-CHK1(Ser-317), CHK1, and phospho-Rb(Ser-807/Ser-811) were from Cell Signaling Technology. Antibodies for phospho-ATRIP(Ser-68/Ser-72), ATRIP, phospho-DNA-PK(Ser-2056), and DNA-PK were from Abcam (Cambridge, UK). Antibodies against p53 (DO-1 and Pab-1801 clones), phospho-p53(Ser-15), p27, and cyclin D1 were purchased from Santa Cruz Biotechnology. Antibodies to phospho-ATM(Ser-1981) and ATM were from Calbiochem. Anti-phosphohistone H2AX(Ser-139) was detected with monoclonal antibody JBW301 clone (Millipore). MDC1 was detected with a rabbit polyclonal antibody (Abcam). Other antibodies used were anti-β-actin (Sigma), anti-GST (Santa Cruz Biotechnology), anti-HA (Covance), and anti-FLAG (Sigma). The secondary antibodies used for Western blots, anti-(mouse-HRP) and anti-(rabbit-HRP) were from GE Healthcare. Bands were visualized with an ECL kit (GE Healthcare) followed by exposure to x-ray film (Fujifilm). When indicated, quantification of bands intensity was performed with Quantity One software (Bio-Rad). The secondary antibodies used for immunofluorescences were FluoroLinkCy2 anti-mouse, FluoroLinkCy2 anti-rabbit, FluoroLinkCy3 anti-mouse, and FluoroLinkCy3 anti-rabbit from GE Healthcare.
DNA Damage
Cells were irradiated with doses ranging from 2 to 10 Gy using a Gammacell 1000 Elite irradiator with a 137Cs source. Doxorubicin was used at 1 μm, etoposide was used at 50 μm, and hydroxyurea was used at 2 mm. DNA damage induced by UV-C light was performed with a Stratalinker (Stratagene). The actual UV dose delivered was determined by direct measurement with a radiometer (Spectroline XS-254 nm-UVC, Spectronics Corp., Westbury, NY).
Immunofluorescence Confocal Microscopy and Analysis
Cells were seeded on cover slides, and for HEK293T cells, cover slides were treated with 0.01% polylysine to improve its adherence to the glass. Cells were washed in cold PBS and fixed with 3% paraformaldehyde in PBS for 30 min at room temperature. After fixation, cells were washed with 0.2 m glycine to block the remaining aldehyde groups. Cells were permeabilized with 0.2% Triton X-100 solution in PBS for 30 min and blocked with 1% BSA in PBS for 30 min at room temperature. For the simultaneous detection of two proteins, the incubations with primary antibodies were performed sequentially. Each antibody was diluted in blocking buffer and incubated for 1 h at room temperature. Following antibody incubations, cells were washed three times with PBS for 5 min. Secondary antibodies labeled with Cy2 or Cy3 were diluted in blocking buffer, and incubation was performed for 1 h at room temperature. Finally, nuclei were stained with DAPI (4′,6′-diamidine-2-fenilindol), diluted in PBS, and incubated for 10 min at room temperature. Cells were washed three times for 5 min in PBS. The final wash was in distilled water. Coverslips were mounted on a slide with Mowiol 4-88 (Calbiochem). Subcellular localization was determined by confocal microscopy using a Leica TCS SP5 DMI-6000B confocal microscope using the following lasers: argon (488 nm), diode-pumped solid state (561 nm), and UV diode (405 nm). Fluorescent images were captured with a 63.0× lens zoomed in 1.5–3× with a 1024 × 1024 frame and 600-Hz scanning speed. The following scanner settings were maintained constant throughout all samples examined: pinhole (95.6 μm), laser intensity, and photomultiplier gain and offset. Images were analyzed with Leica LAS AF and NIH ImageJ software.
Immunoprecipitations
Immunoprecipitations were performed using 1.5 mg of total cell extracts. Cells were lysed in the indicated lysis buffer (see below). To avoid nonspecific interactions, cell extracts were preincubated with 30 μl of GammaBind Plus Sepharose beads (GE Healthcare) equilibrated in the same buffer as the extracts and incubated for 1 h at 4 °C with orbital rotation. The beds were removed by centrifugation, and the extract was incubated with the specific antibody indicated in the experiment for 3 h. Afterward, 40 μl of GammaBind Plus Sepharose beads, which had been previously blocked with seralbumin (BSA), were added and incubated for 2–3 h or overnight at 4 °C with rotation. Next the resin was washed several times in lysis buffer before processing for gel loading. The immunoprecipitate was fractionated by SDS-PAGE, and the gel was transferred to a PDVF membrane (Immobilon-P, Millipore) for Western blot analysis (41, 42). For kinase assays, immunoprecipitated proteins were washed several times in the appropriate kinase buffer and then used in kinase assays. Lysis buffer used for VRK1 immunoprecipitation was composed of 50 mm Tris, pH 8.0, 1% Triton X-100, 200 mm NaCl, 5 mm EDTA, 1 mm NaF, and 1 mm sodium orthovanadate plus protease inhibitors (25). Lysis buffer for 53BP1 immunoprecipitation (NETN buffer; Ref. 33) contained 20 mm Tris-HCl, pH 8.0, 0.5% Nonidet P-40, 300 mm NaCl, 1 mm EDTA, 1 mm NaF, and 1 mm sodium orthovanadate plus protease inhibitors. Lysis buffer for ATM immunoprecipitation (43) contained 50 mm Tris, pH 7.5, 150 mm NaCl, 1% Tween 20, 0.5% Nonidet P-40, 50 mm NaF, and 1 mm sodium orthovanadate plus protease inhibitors. Lysis buffer for ATR immunoprecipitation (35) contained 25 mm Hepes, pH 7.4, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 50 mm β-glycerophosphate, 0.5% Triton X-100, 1 mm NaF, and 1 mm sodium orthovanadate plus protease inhibitors.
GST Pulldown Assays
GST pulldown experiments were performed by incubating 3 μg of GST-tagged 53BP1 fragments with 0.1 μg of purified VRK1-His in 250 μl of binding buffer (50 mm Tris-HCl, pH 7.4, 250 mm NaCl, 0.1% Triton X-100, 5 mm EDTA, and 2 mm DTT) for 2 h at 4 °C. GST-tagged 53BP1 fragments along with interacting VRK1 were precipitated by adding glutathione-Sepharose beads. Beads were washed five times with binding buffer, and proteins were resolved by SDS-PAGE. Interacting VRK1 was detected with anti-VRK1 monoclonal antibody (1F6) (39), and GST-53BP1 fragments were detected with anti-GST antibody.
Flow Cytometry Analysis
Cellular DNA content and cell cycle profiles were determined by flow cytometry in a FACSCalibur flow cytometer (BD Biosciences). Data acquisition was carried out with the Cell Quest program (BD Biosciences), and data were analyzed with Paint-a-Gate and ModFit programs (BD Biosciences).
RESULTS
Activation of VRK1 by DNA Damage in Resting Cells
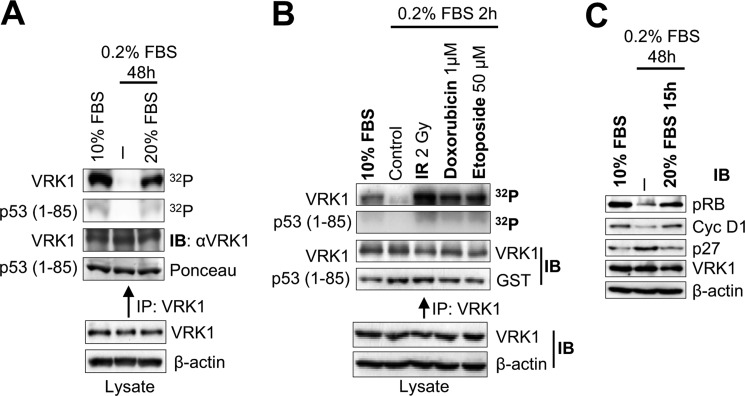
VRK1 has been indirectly linked to the DDR as a p53 kinase (16–18, 22). Because VRK1 phosphorylates p53 at Thr-18 (16–18) and is also implicated in some DDR (18, 22), we hypothesized that VRK1 might also be activated in early steps of cellular DDR. In this work, we determined whether VRK1 can be regulated by DNA damage as an indication of response to altered chromatin structure and determined its consequences on the response to DSBs detected by the formation of 53BP1 foci, a process that is independent of p53 (9). In the first place, we sought to determine whether arresting cells by serum deprivation would affect the activity of VRK1. In the breast human adenocarcinoma MCF7 cell line, we observed that serum deprivation led to loss of VRK1 kinase activity, which was restored upon readdition of serum (Fig. 1A). Next, we determined whether cells arrested and treated with different physical and chemical agents that cause DSBs were able to induce VRK1 activation (Fig. 1B). Cell treatment with IR, the DNA intercalator doxorubicin, and the topoisomerase inhibitor etoposide C caused an 8–12-fold increase in VRK1 kinase activity (Fig. 1B). The activity of VRK1 is known to be insensitive to caffeine (17), an inhibitor of PI3K-related kinases, such as ATM and ATR, and to KU55933, an ATM-specific inhibitor (44). The arrested status of the cell cycle was confirmed by a reduction in phospho-Rb and cyclin D1 and an increase in p27 inhibitor levels (Fig. 1C). The immediate activation of the kinase activity of VRK1 by IR suggested that its role is likely to be that of a sensor protein.
FIGURE 1.
VRK1 kinase activation in response to DNA damage. A, effect of serum withdrawal on the activity of VRK1. Endogenous VRK1 was immunoprecipitated from MCF7 cells either growing in 10% FBS (lane 1), serum-starved for 48 h (lane 2), or serum-starved for 48 h followed by 15 h in 10% FBS (lane 3). VRK1 activity was assayed in vitro for autophosphorylation and phosphorylation of GST-p53(1–85) fragment. B, kinase activity of endogenous VRK1 in response to DSBs. MCF7 cells were serum-starved and then treated with 2 Gy of ionizing radiation, 1 μm doxorubicin, or 50 μm etoposide. Endogenous VRK1 protein was immunoprecipitated and used in an in vitro kinase assay detecting its autophosphorylation and phosphorylation of GST-p53(1–85). C, detection of cell cycle markers by Western blot. MCF7 cells were grown in the presence of serum, serum-starved for 48 h, or serum-starved for 48 h followed by 15 h in serum. Extracts were blotted for phospho-Rb, cyclin D1, p27, VRK1, and actin. IP, immunoprecipitation; IB, immunoblot.
IR Activates VRK1 and Induces Its Association to and Phosphorylation of 53BP1
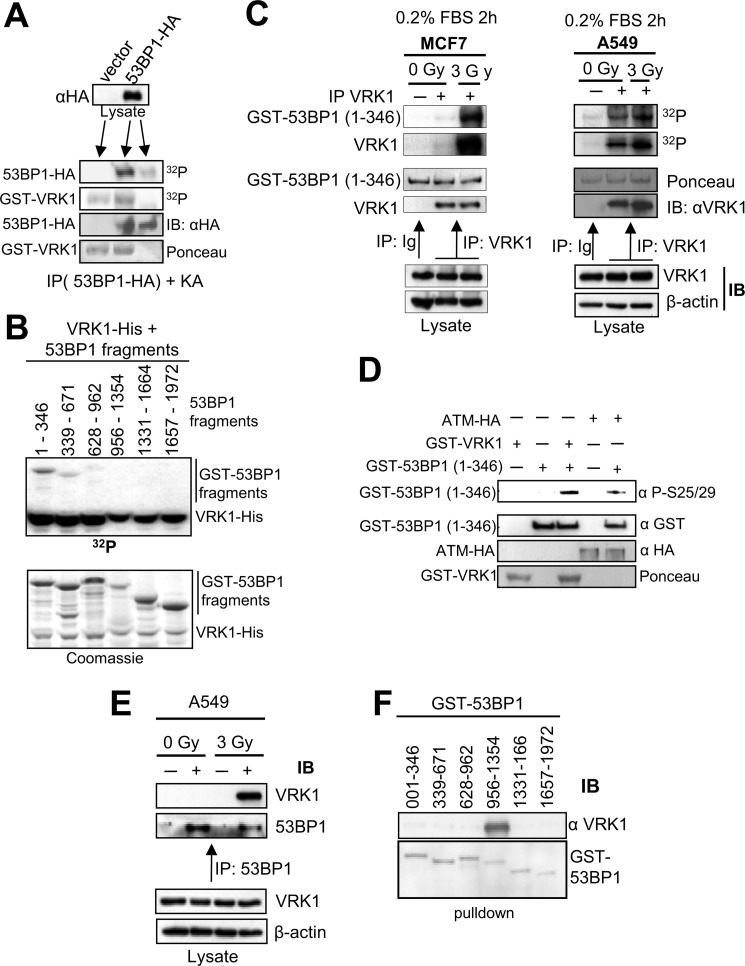
The 53BP1 protein plays an important early role as a scaffold protein in DDR and is required for the assembly of DNA repair foci (9), activation of ATM (45), and downstream regulation of cell cycle checkpoints (46). We hypothesized that 53BP1 is also a phosphorylation substrate of VRK1. To test this possibility, we transfected HEK293T cells (9) with full-length 53BP1 protein, immunoprecipitated 53BP1, and used it as substrate in a VRK1 in vitro kinase assay. Results showed that 53BP1 was phosphorylated by VRK1 (Fig. 2A). To identify the specific target region of this phosphorylation, several GST-53BP1 fusion proteins spanning the complete sequence of 53BP1 (33) were used as substrates in the VRK1 kinase assay. VRK1 strongly phosphorylated the N-terminal region of 53BP1 (residues 1–346) and more weakly phosphorylated the 339–671 and 628–962 regions (Fig. 2B). To further confirm that VRK1 phosphorylates 53BP1, serum-starved MCF7 and A549 cells were irradiated with 3 Gy, and 30 min later, the endogenous VRK1 was immunoprecipitated and tested for its kinase activity with the N-terminal fragment of 53BP1 (residues 1–346) as a substrate. Results showed that endogenous VRK1 phosphorylated 53BP1, and this phosphorylation was enhanced after the activation of VRK1 by γ radiation in both cell lines (Fig. 2C). Finally we identified the residue phosphorylated in 53BP1. For this assay, we used the N-terminal (1–346) 53BP1 region as a substrate of VRK1 or ATM, and we detected the phosphorylation of 53BP1 at Ser-25/29 with a phosphospecific antibody. Significantly, both kinases independently phosphorylated these residues (Fig. 2D), indicating that the 53BP1 protein is targeted by both kinases. Intriguingly, both kinases produce a similar phosphorylation pattern on 53BP1 (33).
FIGURE 2.
Phosphorylation of 53BP1 by VRK1 and IR-induced interaction of VRK1 with 53BP1. A, full-length 53BP1-HA or empty vector was transfected in 293T cells and immunoprecipitated with anti-HA antibody. Immunoprecipitates were used as a substrate of GST-VRK1 in an in vitro kinase assay (25). B, GST-53BP1 fragments encoding different regions of 53BP1 as indicated were tested for phosphorylation by His-VRK1 in an in vitro kinase assay. The gel was exposed to x-ray films to detect phosphorylated proteins. C, starved MCF7 (left) or A549 (right) cells were left unirradiated or exposed to 3 Gy of γ rays. Endogenous VRK1 was immunoprecipitated 30 min after irradiation and assayed for autophosphorylation or phosphorylation of GST-53BP1(1–346) fragment in a kinase assay. Ig represents an immunoprecipitation with a nonspecific antibody (anti-HA). D, specific phosphorylation of 53BP1. In vitro phosphorylation of GST-53BP1(1–346) fragment was assayed either with GST-VRK1 (bacterially expressed) or ATM-HA (prepared by immunoprecipitation from 293T cells). The phosphorylation of 53BP1 was determined by Western blot with an anti-Ser-25/29 phosphospecific antibody (Cell Signaling Technology). E, induction of endogenous VRK1 and 53BP1 protein interaction by ionizing radiation. A549 cells were either left untreated or irradiated with 3 Gy. After 30 min, endogenous 53BP1 was immunoprecipitated using a polyclonal anti-53BP1 antibody. Control immunoprecipitations were performed with a polyclonal antibody (anti-FLAG). Endogenous VRK1 protein was only detected in the 53BP1 immunoprecipitate from irradiated cells. F, mapping the interaction region of 53BP1 with VRK1. Six GST-53BP1 fragments spanning the full-length 53BP1 protein as indicated were incubated with purified VRK1-His and subjected to GST pulldown (25, 74). Proteins were separated by SDS-PAGE followed by immunoblotting with anti-VRK1 antibody (1F6 mAb). IP, immunoprecipitation; IB, immunoblot.
Because phosphorylation requires at least a transient interaction between VRK1 and 53BP1, we also determined whether this interaction could be induced by ionizing radiation. We immunoprecipitated endogenous 53BP1 protein from non-irradiated or irradiated A549 cells, and we determined the presence of VRK1. In irradiated cells, we could observe endogenous VRK1 in the 53BP1 immunoprecipitate forming a complex with endogenous 53BP1, confirming that this interaction is inducible in vivo by IR (Fig. 2E). Because 53BP1 is a large protein with several domains, to assess the specific region of interaction with VRK1, we performed pulldown assays with different GST-53BP1 constructs spanning its complete length. Western blot confirmed that VRK1 interacts with the 53BP1 region comprising residues 956–1354 (Fig. 2F).
Next we determined the sequential activation of VRK1 and 53BP1 focus formation in response to IR by confocal microscopy. The conformational activation of VRK1 reached its peak at 5 min after IR, whereas 53BP1 focus formation reached its maximum between 15 and 30 min after IR in A549 cells (supplemental Fig. S1). Thus, VRK1 activation precedes 53BP1 focus formation.
Loss of VRK1 Results in Defective Formation of 53BP1 Foci in Response to IR
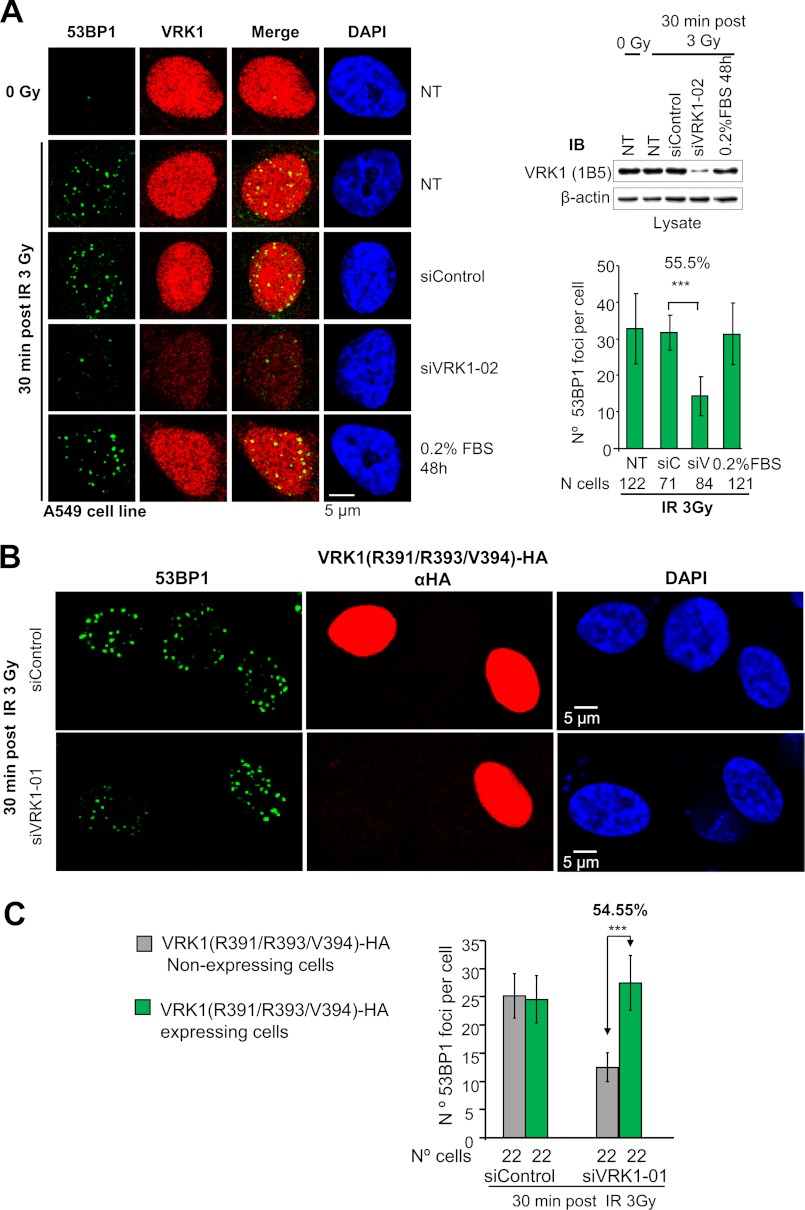
The 53BP1 interaction and its phosphorylation by VRK1 induced by IR suggested that VRK1 might be necessary for the formation of 53BP1 foci in the DSB response (9, 10, 47). Therefore, we determined whether VRK1 knockdown (17, 23) would affect focus formation in DDR by detecting the assembly of 53BP1 into IR-induced foci in A549 cells (9). VRK1 knockdown causes entry of the cells in G0 even in the presence of serum, which cannot revert this cell cycle arrest (23) because VRK1 is required for cyclin D1 gene expression (48). Notably, following irradiation, we observed a decrease in 53BP1 focus formation in siVRK1-02- (Fig. 3A and supplemental Fig. S2A) or siVRK1-01-treated cells (supplemental Fig. S2B) but not in control cells (not transfected), starved cells, and siControl. The formation of foci in starved cells permitted us to rule out the stop in cell cycle as the cause of the effect. Although VRK1 shows a pan-nuclear localization, 53BP1 staining overlaps with VRK1 signal in IR-induced foci (Fig. 3A and supplemental Fig. S2B), which is likely to represent a minor VRK1 subpopulation of the abundant nuclear VRK1 protein. Quantification of 53BP1 foci showed that in irradiated cells with siVRK1-02 or siVRK1-01 knockdown there was a significant reduction in both number and size of 53BP1 foci (Fig. 3A and supplemental Fig. S2B), which suggests that VRK1 is necessary for proper assembly of 53BP1 foci in response to ionizing radiation. A similar effect was also detected in HEK293T (supplemental Fig. S3A), H1299 (p53−/−) (supplemental Fig. S3B), and MCF7 cells (supplemental Fig. S3C), indicating that the effect of VRK1 on 53BP1 focus formation is independent of cell type and p53 status.
FIGURE 3.
Knockdown of VRK1 prevents 53BP1 ionizing radiation-induced focus formation. A, A549 cells were transfected with siRNA oligos targeting VRK1 (siVRK1-02), control oligos (siControl), not transfected (NT), or not transfected and serum-starved (0.2% FBS). Forty-eight hours after transfection, cells were either untreated or treated with 3 Gy. 53BP1 focus formation was determined 30 min after irradiation by confocal microscopy (9). VRK1 was detected with a polyclonal antibody, and 53BP1 was detected with a monoclonal antibody. VRK1 knockdown efficiency was checked by Western blot (top right) (18, 23). Quantification of the number of 53BP1 foci per cell was performed with ImageJ software. Around 100 cells for each condition were analyzed. Means, S.D., and number of cells analyzed are shown in the graph (bottom right). A representative field with several cells to show the phenotype and the cell cycle profile of these cells are shown in supplemental Fig. S2A. A different siRNA induced the same phenotype (supplemental Fig. S2B). ***, p < 0.001. NT, non-transfected control; siC, siControl; siV, siVRK1-02. B, rescue of defective 53BP1 focus formation by an siRNA-resistant VRK1. A549 cells were transfected with siVRK1-01 to knock down endogenous VRK1 or with siControl. After 36 h, cells were retransfected with 2 μg of plasmid HA-VRK1(R391/R393/V394) with three silent substitutions that render the cDNA insensitive to si-VRK1-01. Thirty-six hours after retransfection, cells were irradiated with 3 Gy and left to recover for 30 min. Cells were immunostained with anti-HA monoclonal antibody to identify cells expressing exogenous VRK1. 53BP1 foci were visualized by staining with 53BP1 polyclonal antibody. The efficiency of endogenous VRK1 silencing and expression of siRNA-resistant VRK1 was determined by Western blot (supplemental Fig. S5). C, quantification of the number of 53BP1 foci in the rescue experiment. NIH ImageJ software was used for counting 53BP1 foci. Means of the number of 53BP1 foci per cell and S.D. are represented in the graph. The number of analyzed cells is indicated below. ***, p < 0.001. siV1-1, siVRK1-01; IB, immunoblot. Error bars represent the standard deviation.
Next we performed a rescue of the defective 53BP1 focus phenotype. We transfected A549 cells in which the endogenous VRK1 was previously knocked down with siVRK1-01 with a plasmid expressing a mutated form of VRK1 containing silent nucleotide changes in three codons (Arg-391, Arg-393, and Val-394), which make it insensitive to siRNA-01 (supplemental Fig. S4). We treated cells with 3 Gy and determined the formation of 53BP1 foci. Interestingly, transfection of cells with the resistant VRK1 mutant restored both the number and size of 53BP1 foci (Fig. 3, B and C) to control levels, whereas in cells that did not take up the mutant, the formation of foci was deficient, and the phenotype of siVRK1 was clearly manifested (Fig. 3B). This observation confirmed that VRK1 is a necessary step for the correct formation of 53BP1 foci.
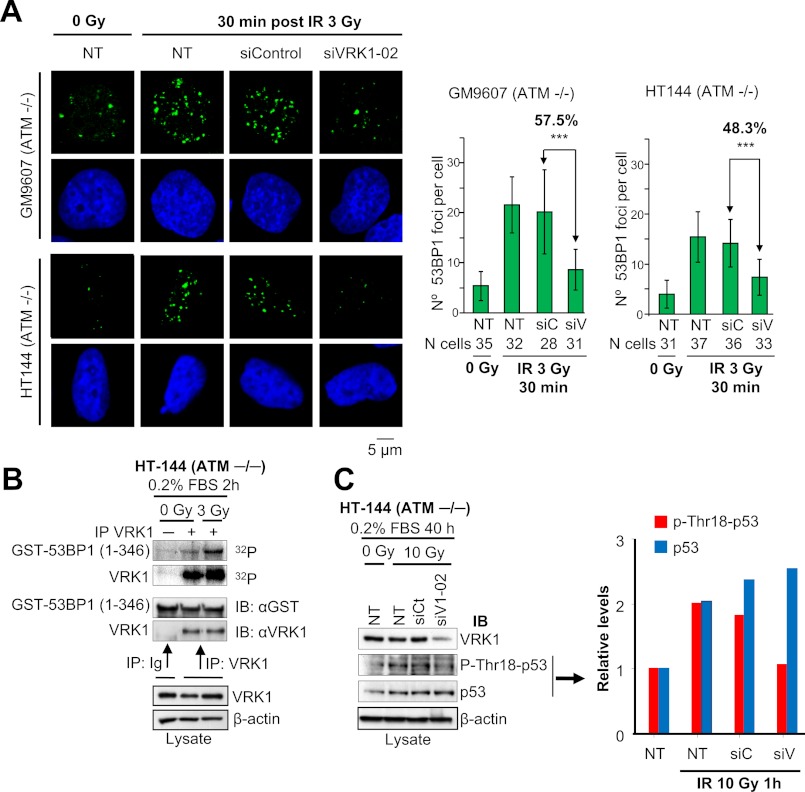
VRK1 Is Activated by IR and Induces 53BP1 Foci in ATM−/− Cells
The response to double strand breaks is mainly mediated by the ATM-CHK2 pathway (49).Therefore, we hypothesized that if VRK1 has an effect on 53BP1 focus formation that is independent of ATM VRK1 knockdown should affect the formation of these foci in ATM-deficient cells. To test this, we knocked down VRK1 in HT144 and GM9607 cells and determined its effect on 53BP1 focus formation in response to IR. These two ATM-deficient cell lines have a high level of background 53BP1 foci, which are increased by IR (Fig. 4A and supplemental Fig. S5). In cells in which VRK1 was knocked down, we were able to identify a significant reduction in size and number of 53BP1 foci (p < 0.001). This result confirms the ATM-independent role of VRK1 in DDR. Next, we determined the activation of VRK1 kinase activity in response to IR in ATM-deficient HT144 cells through autophosphorylation and 53BP1 phosphorylation assays. We observed an increase of phosphorylation in both proteins despite the lack of ATM (Fig. 4B). These data suggested that activation of VRK1 and phosphorylation of 53BP1 are mediated independently of ATM in serum-starved HT144 cells. We also observed that the specific phosphorylation of p53 at Thr-18 was induced by IR in these cells, but it was lost if VRK1 was knocked down (Fig. 4C). Thus, we can conclude that this Thr-18 phosphorylation of p53 is VRK1-dependent and ATM-independent. These results indicated that VRK1 represents an alternative DNA damage response component independent of ATM and upstream of p53 and 53BP1 in DDR.
FIGURE 4.
Defective 53BP1 foci in ATM−/− cell lines by loss of VRK1. A, HT144 and GM9607 cells were transfected with siVRK1-02 or siControl or not transfected (NT). After 72 h, cells were either untreated or exposed to 3 Gy of IR and allowed to recover for 30 min. Cells were immunostained with 53BP1 polyclonal antibody. The quantification of the number of 53BP1 foci per cell in each condition is shown in the graphs. Means and S.D. are represented. The number of cells analyzed is indicated below. Images showing several cells and VRK1 protein levels detected by Western blot are shown in supplemental Fig. S5. ***, p < 0.001. B, activation of VRK1 in response to IR in HT144 cells. Endogenous VRK1 was immunoprecipitated from non-irradiated or 3-Gy irradiated HT144 starved-cells. This immunoprecipitate was used to assess VRK1 autophosphorylation and phosphorylation of GST-53BP1(1–346) in an in vitro kinase assay. Ig is a negative control with a nonspecific antibody (anti-HA). C, phosphorylation of p53 at Thr-18 is dependent on VRK1 in HT144 cells. HT144 cells were transfected with siVRK1-02 or siControl or not transfected (NT). After 24 h, cells were serum-starved for 40 h and then irradiated with 10 Gy of IR or left untreated. Cell lysates were prepared 1 h after irradiation, and Western blots were performed with antibodies to VRK1 (1B5), phospho-Thr-18-p53, p53, and actin. Quantification of the levels of phospho-Thr-18-p53 and total p53 is shown in the graph. IP, immunoprecipitation; IB, immunoblot. Error bars represent the standard deviation.
VRK1 Elimination Prevents Activating Phosphorylations of ATM, CHK2, and DNA-PK
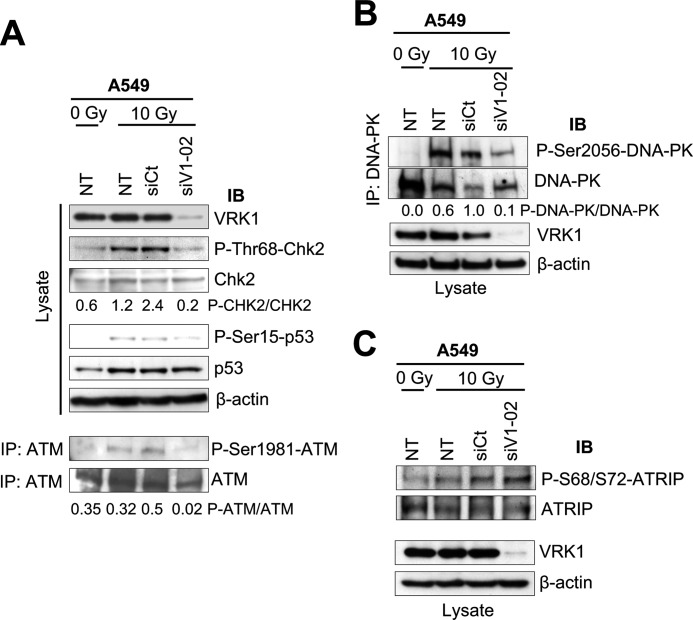
Next we determined whether VRK1 could also affect the efficient activation of ATM-CHK2 in the DDR signaling. We observed that irradiation of A549 cells with γ rays induced activation of ATM and CHK2 as detected by specific phosphorylation of ATM at Ser-1981 (43) and CHK2 at Thr-68 (50, 51). However, these specific phosphorylation events were lost upon knockdown of VRK1 with a reduction greater than 90% when VRK1 was knocked down as detected by the changes in the ratios between phosphorylated and unphosphorylated proteins (Fig. 5A). Thus, VRK1 is also upstream of ATM-CHK2. In these cells, p53 accumulated in response to IR as expected. We observed similar results when we tested the effect of VRK1 knockdown on the activating autophosphorylation of DNA-PK at Ser-2056; there was a reduction in phosphorylation of 90% (52) (Fig. 5B).
FIGURE 5.
VRK1 is an upstream element in DDR. A, effect of VRK1 knockdown on the activation of ATM and CHK2 induced by IR. A549 cells were transfected with siVRK1-02 or siControl oligos or not transfected (NT). After 48 h, cells were treated with 10 Gy or left untreated. Cell lysates were prepared 1 h later and immunoblotted for VRK1 expression (1B5 antibody) and for specific phosphorylation of CHK2 in Thr-68 (anti-phospho-Thr-68-CHK2) and p53 at Ser-15 (anti-phospho-Ser-15-p53). ATM was immunoprecipitated from cell lysates with anti-ATM antibody and immunoblotted for specific phosphorylation at Ser-1981. The ratio of phospho-CHK2 (P-CHK2)/CHK2 and phospho-ATM (P-ATM)/ATM are shown below the corresponding gels. B, effect of VRK1 knockdown on the activation of DNA-PK. Cell lysates were immunoblotted with antibodies to phospho-Ser-2056-DNA-PK, DNA-PK, VRK1 (1B5 antibody), and actin. The ratio of phospho-DNA-PK (P-DNA-PK)/DNA-PK is shown below the gel. C, effect of VRK1 knockdown on ATR-dependent phosphorylation of ATRIP. Immunoblots were performed with antibodies to phospho-Ser-68/Ser-72-ATRIP, ATRIP, VRK1 (1B5 antibody), and actin. IP, immunoprecipitation; IB, immunoblot; siCt, siControl; siV1, siVRK1.
Although ATR plays a secondary role in DDR (53), we also wanted to determine whether VRK1 knockdown had any effect on ATR activation. We observed that the elimination of VRK1 did not affect the phosphorylation of ATRIP at Ser-68/72 (Fig. 5C), a direct target of ATR, although 53BP1 focus formation was impaired under these conditions (Fig. 3A and supplemental Fig. S2). Therefore, we conclude that the ATR signal is not affected by VRK1 in this DSB response and is not implicated in the immediate formation of 53BP1 foci, although it might play a role at later points during the evolution in time of DDR complexes (54). Overall, these results indicate that VRK1, in addition to its direct effect on IR-induced focus assembly, is a novel upstream component in the DDR mediated by the ATM and DNA-PK, but not ATR, signaling pathways. Finally, to rule out the possibility that VRK1, ATM, ATR, and CHK2 were substrates of each other, we studied their in vitro combinations using kinase-dead proteins as targets and p53 as a positive control (supplemental Fig. S6, A–C). None of these active kinases phosphorylated kinase-dead VRK1, nor were their inactive forms phosphorylated by VRK1. These results indicated that these kinases are not directly interacting or phosphorylating each other, but whether they might be forming part of a larger complex cannot be ruled out.
Effect of VRK1 Knockdown on the Formation of γH2AX and MDC1 Foci Induced by IR
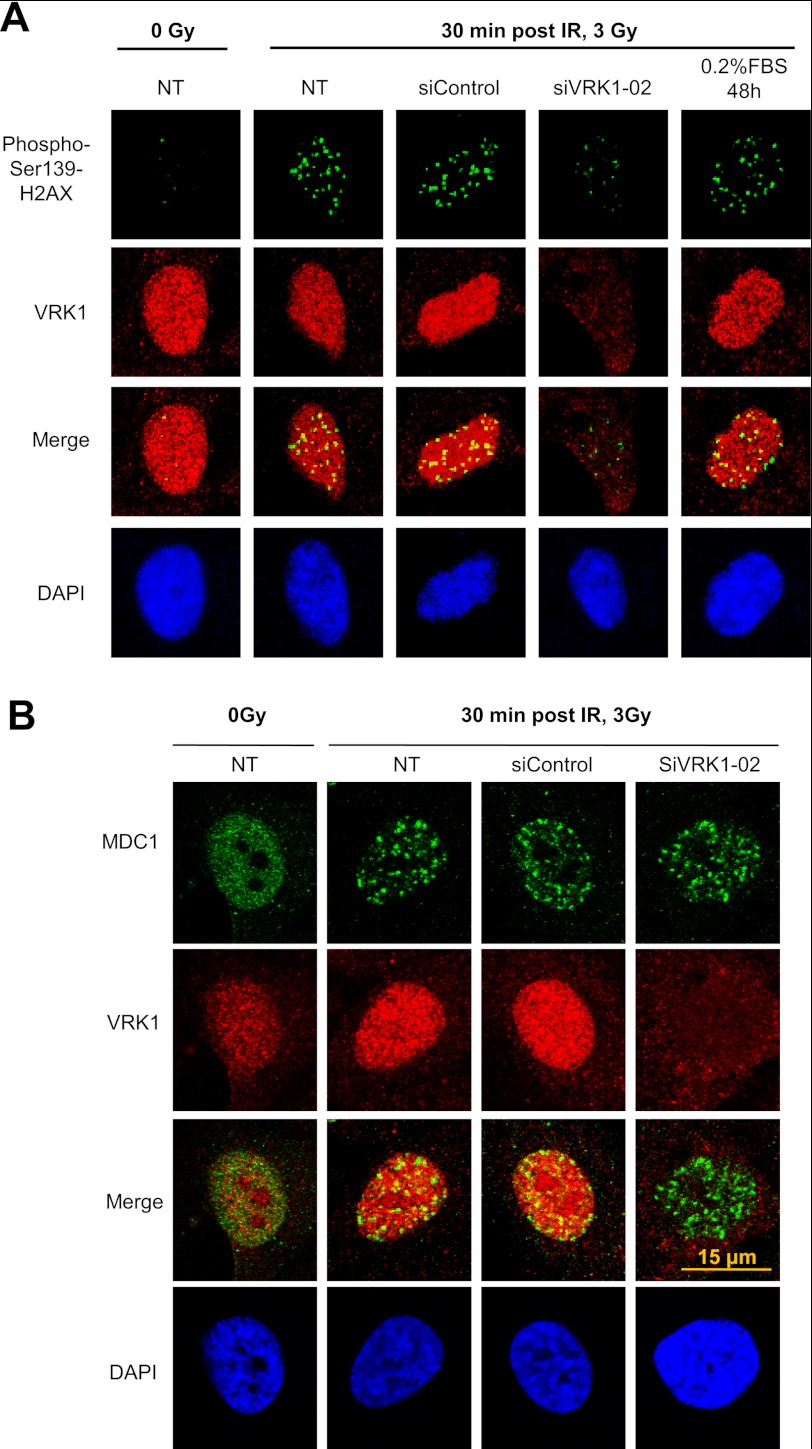
The previous results suggested that the effect of VRK1 is an early event in the response to DDR that is upstream of ATM. To confirm this role, the effect of VRK1 knockdown on the formation of γH2AX, that is the initial event (55), and on MDC1 foci, which are dependent on ATM (56, 57), was determined. MDC1 is required for recruitment of 53BP1 (58). The elimination of VRK1 in A549 cells resulted in defective formation of histone γH2AX (Fig. 6A), consistent with an early role for VRK1 in DDR. But on the other hand, knockdown of VRK1 had no effect on MDC1 foci (Fig. 6B), suggesting that there are alternative routes of focus formation.
FIGURE 6.
Effect of VRK1 knockdown on the formation of histone γH2AX (A) and MDC1 (B) foci in response to IR. A549 cells were transfected with siControl or siVRK1-02 and treated with 3 Gy as indicated under “Experimental Procedures.” The formation of γH2AX foci was determined with monoclonal antibody JBW301, and the formation of MDC1 foci was determined with a polyclonal antibody by confocal microscopy. VRK1 was detected with monoclonal antibody 1B5, which detects a conformational change in its C-terminal region. NT, not transfected.
DISCUSSION
The initial response to DNA damage is likely to be the consequence of an altered chromatin structure. At the site of the DNA damage, specific detection of signaling mechanisms need to be activated according to the type of damage to achieve a proper repair. Until now, most of the studies on DDR have centered on mechanisms operating in the context of cell cycle regulation and its checkpoints to maintain genome integrity. One important observation is that most of an organism's cells are not dividing and that induced DNA damage occurs most frequently in resting cells. This fact supports the survival advantage for additional elements in DDR that might have a redundant role with other already known mechanisms and that are uncoupled from cell cycle regulation. In this work, we have identified that VRK1 is a kinase whose activity is enhanced in response to DNA damage and is a key component involved in channeling a specific response. The induction of VRK1 association with 53BP1 upon IR is consistent with it being both upstream and downstream of IR-induced focus formation. However, because the elimination of VRK1 results in defective 53BP1 focus formation, we can conclude that it has an upstream role in the process, although it may be acting in concert with other known pathways. It is important to note that VRK1 plays a role in defective focus formation even in the absence of ATM. 53BP1 foci are formed even in the absence of ATM, which is a transducer protein in DDR. Furthermore, VRK1 knockdown results in loss of activation of several pathways, including ATM-CHK2 and DNA-PK, as detected by the loss of their specific activating phosphorylation marks. This observation has two clear implications. First, VRK1 is upstream of 53BP1 foci and at the same time is a common component of sensor pathways. Second, the specificity of the response is determined by downstream components in the process. The position of VRK1 upstream of 53BP1 is also consistent with the observation of 53BP1 as an upstream step in ATM activation (45). Furthermore, the observation that VRK1 affects IR-induced focus formation in ATM-defective cells suggests that the VRK1-mediated response is an independent pathway whose activity is required for ATM and DNA-PK, but not ATR, activation (59). These results suggest that VRK1 represents an alternative and perhaps partially redundant pathway of DDR (8), which might be particularly relevant in the context of arrested cells, such as stem cells (60).
The significance of the 53BP1 phosphorylation at residue Ser-25/29 by ATM or VRK1 is that it likely affects its interaction with other proteins in its less characterized N-terminal region. This phosphorylation does not affect DNA repair (61), but it is necessary for 53BP1 binding to the BRCT domains of hPTIP (62, 63), and its abrogation results in reduced CHK2 activation (63). This is consistent with the effect of VRK1 loss on CHK2 activation (Fig. 4D), although mutation of this residue does not affect the ability of the cell to repair damaged DNA (61), suggesting that there are alternative response pathways. VRK1 interacts with the central region, residues 956–1354, of 53BP1, which is outside the Tudor and BRCT domains that induce 53BP1 foci and interact with p53, respectively (45). This 53BP1 region overlaps with a putative binding site for Plk1 (64). VRK1 probably plays different roles during DDR. Initially it is required for the recruitment or assembly of 53BP1 foci, but following its incorporation into these foci, VRK1 probably participates in the regulation or incorporation of additional components in the response pathway that remain to be identified and that are temporally and spatially regulated.
The evidence indicates that VRK1 is likely to play more than one role in DDR. First, it is clearly an early component as indicated by the immediate activation of its kinase activity following irradiation and preceding formation of γH2AX and 53BP1 foci. But the DDR response is dynamic, and roles change with time from damage-sensing to damage repair, and thus downstream signaling might be affected as is the case for phosphorylation of p53, ATM, CHK2, and DNA-PK, all of which are reduced by loss of VRK1. Targeting VRK1 with inhibitors might eventually be a potential aim in chemotherapy, although mechanistically inhibitors might have different roles. In proliferating tumor cells, inhibitors are most likely to function by the effect of VRK1 loss on the cell cycle (23). However, in non-proliferating tumor cells, the effect might be indirect and a consequence of allowing the accumulation of additional genetic damage in resting tumor cells so that when reentering the cell cycle they will be more prone to die because of the accumulated damage. VRK1 is not sensitive to kinase inhibitors (44) due to the structure of its kinase domain (65, 66) and low promiscuity with regard to inhibitors (67).
The role of VRK1 in DDR suggests that it may also participate in normal processes in which DNA recombination occurs, such as meiosis. Thus, VRK1 knock-out mice have defects in meiosis resulting in male and female sterility (68–70) in addition to defects in cell proliferation (71). Its D. melanogaster ortholog, NHK-1, also affects meiotic recombination (72). Recently a recessive mutation in VRK1, R385X, manifested a complex syndrome with a pontocerebellar hypoplasia, ataxia, and muscular atrophy (73). Although the pathogenic role of VRK1 in this clinical report is not clear, it is interesting to note that patients with mutations in other kinases, like ATM and ATR, also present motor coordination problems and ataxia (1). Defects in DDR pathways are also associated with spinocerebellar ataxias (7). VRK1, like NHK-1, is a nucleosomal histone kinase and thus is a likely player in all processes implicated in chromatin alterations, normal or pathological. The identification of VRK1 as a novel upstream element in DDR opens up new possibilities to understand the mechanisms regulating the specificity of cellular protection in the contexts of DNA damage and recombination.
VRK1, a late appearing nucleosomal kinase in evolution, is likely to play a coordinating role among different signaling pathways that have to be tightly organized and coordinated in time and space in response to DNA damage. Future study into these processes and their pathology will clarify how these master pathways of sensing and responding to DNA damage converge.
This work was supported in part by Ministerio de Ciencia e Innovación Grants SAF2010-14935 and CSD2007-0017, Junta de Castilla y León Grants CSI006A11-2 and GR-15, and the Kutxa-Fundación Inbiomed.
The protein interactions from this publication have been submitted to the EMBO-EBI IMEx consortium (http://www.imexconsortium.org/) through IntAct (PMID 19850723) and assigned the identifier IM-15883.

This article contains supplemental Figs. S1–S6.
- DDR
- DNA damage response
- VRK1
- vaccinia-related kinase 1
- IR
- ionizing radiation
- DSB
- double strand break
- ATM
- ataxia-telangiectasia mutated
- DNA-PK
- DNA-dependent protein kinase
- ATR
- ATM- and Rad3-related
- ATRIP
- ATR-interacting protein
- Gy
- grays
- NHK-1
- nucleosomal histone kinase-1.
REFERENCES
- 1. Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D. A., Smith S., Uziel T., Sfez S., Ashkenazi M., Pecker I., Frydman M., Harnik R., Patanjali S. R., Simmons A., Clines G. A., Sartiel A., Gatti R. A., Chessa L., Sanal O., Lavin M. F., Jaspers N. G., Taylor A. M., Arlett C. F., Miki T., Weissman S. M., Lovett M., Collins F. S., Shiloh Y. (1995) A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 268, 1749–1753 [DOI] [PubMed] [Google Scholar]
- 2. Lakin N. D., Hann B. C., Jackson S. P. (1999) The ataxia-telangiectasia related protein ATR mediates DNA-dependent phosphorylation of p53. Oncogene 18, 3989–3995 [DOI] [PubMed] [Google Scholar]
- 3. Lee S. E., Mitchell R. A., Cheng A., Hendrickson E. A. (1997) Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell. Biol. 17, 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abraham R. T. (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15, 2177–2196 [DOI] [PubMed] [Google Scholar]
- 5. Jackson S. P., Bartek J. (2009) The DNA-damage response in human biology and disease. Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoeijmakers J. H. (2009) DNA damage, aging, and cancer. N. Engl. J. Med. 361, 1475–1485 [DOI] [PubMed] [Google Scholar]
- 7. Rass U., Ahel I., West S. C. (2007) Defective DNA repair and neurodegenerative disease. Cell 130, 991–1004 [DOI] [PubMed] [Google Scholar]
- 8. Crick F. (1974) The double helix: a personal view. Nature 248, 766–769 [DOI] [PubMed] [Google Scholar]
- 9. Schultz L. B., Chehab N. H., Malikzay A., Halazonetis T. D. (2000) p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151, 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang B., Matsuoka S., Carpenter P. B., Elledge S. J. (2002) 53BP1, a mediator of the DNA damage checkpoint. Science 298, 1435–1438 [DOI] [PubMed] [Google Scholar]
- 11. Mochan T. A., Venere M., DiTullio R. A., Jr., Halazonetis T. D. (2003) 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res. 63, 8586–8591 [PubMed] [Google Scholar]
- 12. Ward I. M., Chen J. (2001) Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276, 47759–47762 [DOI] [PubMed] [Google Scholar]
- 13. Riballo E., Kühne M., Rief N., Doherty A., Smith G. C., Recio M. J., Reis C., Dahm K., Fricke A., Krempler A., Parker A. R., Jackson S. P., Gennery A., Jeggo P. A., Löbrich M. (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol. Cell 16, 715–724 [DOI] [PubMed] [Google Scholar]
- 14. Saito S., Yamaguchi H., Higashimoto Y., Chao C., Xu Y., Fornace A. J., Jr., Appella E., Anderson C. W. (2003) Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J. Biol. Chem. 278, 37536–37544 [DOI] [PubMed] [Google Scholar]
- 15. Meek D. W. (2009) Tumour suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer 9, 714–723 [DOI] [PubMed] [Google Scholar]
- 16. Lopez-Borges S., Lazo P. A. (2000) The human vaccinia-related kinase 1 (VRK1) phosphorylates threonine-18 within the mdm-2 binding site of the p53 tumour suppressor protein. Oncogene 19, 3656–3664 [DOI] [PubMed] [Google Scholar]
- 17. Vega F. M., Sevilla A., Lazo P. A. (2004) p53 Stabilization and accumulation induced by human vaccinia-related kinase 1. Mol. Cell. Biol. 24, 10366–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Valbuena A., Castro-Obregón S., Lazo P. A. (2011) Downregulation of VRK1 by p53 in response to DNA damage is mediated by the autophagic pathway. PLoS One 6, e17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schon O., Friedler A., Bycroft M., Freund S. M., Fersht A. R. (2002) Molecular mechanism of the interaction between MDM2 and p53. J. Mol. Biol. 323, 491–501 [DOI] [PubMed] [Google Scholar]
- 20. Teufel D. P., Bycroft M., Fersht A. R. (2009) Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene 28, 2112–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee C. W., Ferreon J. C., Ferreon A. C., Arai M., Wright P. E. (2010) Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 107, 19290–19295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park C. H., Choi B. H., Jeong M. W., Kim S., Kim W., Song Y. S., Kim K. T. (2011) Protein kinase Cδ regulates vaccinia-related kinase 1 in DNA damage-induced apoptosis. Mol. Biol. Cell 22, 1398–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Valbuena A., López-Sánchez I., Lazo P. A. (2008) Human VRK1 is an early response gene and its loss causes a block in cell cycle progression. PLoS One 3, e1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang T. H., Park D. Y., Choi Y. H., Kim K. J., Yoon H. S., Kim K. T. (2007) Mitotic histone H3 phosphorylation by vaccinia-related kinase 1 in mammalian cells. Mol. Cell. Biol. 27, 8533–8546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanz-García M., López-Sánchez I., Lazo P. A. (2008) Proteomics identification of nuclear Ran GTPase as an inhibitor of human VRK1 and VRK2 (vaccinia-related kinase) activities. Mol. Cell. Proteomics 7, 2199–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aihara H., Nakagawa T., Yasui K., Ohta T., Hirose S., Dhomae N., Takio K., Kaneko M., Takeshima Y., Muramatsu M., Ito T. (2004) Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes Dev. 18, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim W., Chakraborty G., Kim S., Shin J., Park C. H., Jeong M. W., Bharatham N., Yoon H. S., Kim K. T. (2012) Macro histone H2A1.2 (macroH2A1) protein suppresses mitotic kinase VRK1 during interphase. J. Biol. Chem. 287, 5278–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suzuki Y., Ogawa K., Koyanagi Y. (2010) Functional disruption of the Moloney murine leukemia virus preintegration complex by vaccinia-related kinases. J. Biol. Chem. 285, 24032–24043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Misteli T., Soutoglou E. (2009) The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 10, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pandita T. K., Richardson C. (2009) Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 37, 1363–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith E., Shilatifard A. (2010) The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol. Cell 40, 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klerkx E. P., Lazo P. A., Askjaer P. (2009) Emerging biological functions of the vaccinia-related kinase (VRK) family. Histol. Histopathol. 24, 749–759 [DOI] [PubMed] [Google Scholar]
- 33. Rappold I., Iwabuchi K., Date T., Chen J. (2001) Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 153, 613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin W. C., Lin F. T., Nevins J. R. (2001) Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15, 1833–1844 [PMC free article] [PubMed] [Google Scholar]
- 35. Tibbetts R. S., Brumbaugh K. M., Williams J. M., Sarkaria J. N., Cliby W. A., Shieh S. Y., Taya Y., Prives C., Abraham R. T. (1999) A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13, 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Valbuena A., Blanco S., Vega F. M., Lazo P. A. (2008) The C/H3 domain of p300 is required to protect VRK1 and VRK2 from their downregulation induced by p53. PLoS One 3, e2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Valbuena A., Suárez-Gauthier A., López-Rios F., López-Encuentra A., Blanco S., Fernández P. L., Sánchez-Céspedes M., Lazo P. A. (2007) Alteration of the VRK1-p53 autoregulatory loop in human lung carcinomas. Lung Cancer 58, 303–309 [DOI] [PubMed] [Google Scholar]
- 38. Barcia R., López-Borges S., Vega F. M., Lazo P. A. (2002) Kinetic properties of p53 phosphorylation by the human vaccinia-related kinase 1. Arch. Biochem. Biophys. 399, 1–5 [DOI] [PubMed] [Google Scholar]
- 39. Valbuena A., López-Sánchez I., Vega F. M., Sevilla A., Sanz-García M., Blanco S., Lazo P. A. (2007) Identification of a dominant epitope in human vaccinia-related kinase 1 (VRK1) and detection of different intracellular subpopulations. Arch. Biochem. Biophys. 465, 219–226 [DOI] [PubMed] [Google Scholar]
- 40. Buscemi G., Perego P., Carenini N., Nakanishi M., Chessa L., Chen J., Khanna K., Delia D. (2004) Activation of ATM and Chk2 kinases in relation to the amount of DNA strand breaks. Oncogene 23, 7691–7700 [DOI] [PubMed] [Google Scholar]
- 41. Sevilla A., Santos C. R., Barcia R., Vega F. M., Lazo P. A. (2004) c-Jun phosphorylation by the human vaccinia-related kinase 1 (VRK1) and its cooperation with the N-terminal kinase of c-Jun (JNK). Oncogene 23, 8950–8958 [DOI] [PubMed] [Google Scholar]
- 42. Sevilla A., Santos C. R., Vega F. M., Lazo P. A. (2004) Human vaccinia-related kinase 1 (VRK1) activates the ATF2 transcriptional activity by novel phosphorylation on Thr-73 and Ser-62 and cooperates with JNK. J. Biol. Chem. 279, 27458–27465 [DOI] [PubMed] [Google Scholar]
- 43. Bakkenist C. J., Kastan M. B. (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506 [DOI] [PubMed] [Google Scholar]
- 44. Vázquez-Cedeira M., Barcia-Sanjurjo I., Sanz-García M., Barcia R., Lazo P. A. (2011) Differential inhibitor sensitivity between human kinases VRK1 and VRK2. PLoS One 6, e23235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mochan T. A., Venere M., DiTullio R. A., Jr., Halazonetis T. D. (2004) 53BP1, an activator of ATM in response to DNA damage. DNA repair 3, 945–952 [DOI] [PubMed] [Google Scholar]
- 46. DiTullio R. A., Jr., Mochan T. A., Venere M., Bartkova J., Sehested M., Bartek J., Halazonetis T. D. (2002) 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 4, 998–1002 [DOI] [PubMed] [Google Scholar]
- 47. Gorgoulis V. G., Vassiliou L. V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R. A., Jr., Kastrinakis N. G., Levy B., Kletsas D., Yoneta A., Herlyn M., Kittas C., Halazonetis T. D. (2005) Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 [DOI] [PubMed] [Google Scholar]
- 48. Kang T. H., Park D. Y., Kim W., Kim K. T. (2008) VRK1 phosphorylates CREB and mediates CCND1 expression. J. Cell Sci. 121, 3035–3041 [DOI] [PubMed] [Google Scholar]
- 49. Smith J., Tho L. M., Xu N., Gillespie D. A. (2010) The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 108, 73–112 [DOI] [PubMed] [Google Scholar]
- 50. Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S. J. (2000) Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 97, 10389–10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ward I. M., Wu X., Chen J. (2001) Threonine 68 of Chk2 is phosphorylated at sites of DNA strand breaks. J. Biol. Chem. 276, 47755–47758 [DOI] [PubMed] [Google Scholar]
- 52. Chan D. W., Chen B. P., Prithivirajsingh S., Kurimasa A., Story M. D., Qin J., Chen D. J. (2002) Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 16, 2333–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cuadrado M., Martinez-Pastor B., Murga M., Toledo L. I., Gutierrez-Martinez P., Lopez E., Fernandez-Capetillo O. (2006) ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 203, 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fernandez-Capetillo O., Chen H. T., Celeste A., Ward I., Romanienko P. J., Morales J. C., Naka K., Xia Z., Camerini-Otero R. D., Motoyama N., Carpenter P. B., Bonner W. M., Chen J., Nussenzweig A. (2002) DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4, 993–997 [DOI] [PubMed] [Google Scholar]
- 55. Nakamura A. J., Rao V. A., Pommier Y., Bonner W. M. (2010) The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle 9, 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stiff T., O'Driscoll M., Rief N., Iwabuchi K., Löbrich M., Jeggo P. A. (2004) ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64, 2390–2396 [DOI] [PubMed] [Google Scholar]
- 57. Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., Jackson S. P. (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 58. Coster G., Goldberg M. (2010) The cellular response to DNA damage: a focus on MDC1 and its interacting proteins. Nucleus 1, 166–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tomimatsu N., Mukherjee B., Burma S. (2009) Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 10, 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mohrin M., Bourke E., Alexander D., Warr M. R., Barry-Holson K., Le Beau M. M., Morrison C. G., Passegué E. (2010) Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 7, 174–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ward I., Kim J. E., Minn K., Chini C. C., Mer G., Chen J. (2006) The tandem BRCT domain of 53BP1 is not required for its repair function. J. Biol. Chem. 281, 38472–38477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Munoz I. M., Jowsey P. A., Toth R., Rouse J. (2007) Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res. 35, 5312–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu J., Prindle M. J., Dressler G. R., Yu X. (2009) PTIP regulates 53BP1 and SMC1 at the DNA damage sites. J. Biol. Chem. 284, 18078–18084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. van Vugt M. A., Gardino A. K., Linding R., Ostheimer G. J., Reinhardt H. C., Ong S. E., Tan C. S., Miao H., Keezer S. M., Li J., Pawson T., Lewis T. A., Carr S. A., Smerdon S. J., Brummelkamp T. R., Yaffe M. B. (2010) A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G2/M DNA damage checkpoint. PLoS Biol. 8, e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Scheeff E. D., Eswaran J., Bunkoczi G., Knapp S., Manning G. (2009) Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure 17, 128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shin J., Chakraborty G., Bharatham N., Kang C., Tochio N., Koshiba S., Kigawa T., Kim W., Kim K. T., Yoon H. S. (2011) NMR solution structure of human vaccinia-related kinase 1 (VRK1) reveals the C-terminal tail essential for its structural stability and autocatalytic activity. J. Biol. Chem. 286, 22131–22138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fedorov O., Marsden B., Pogacic V., Rellos P., Müller S., Bullock A. N., Schwaller J., Sundström M., Knapp S. (2007) A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc. Natl. Acad. Sci. U.S.A. 104, 20523–20528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wiebe M. S., Nichols R. J., Molitor T. P., Lindgren J. K., Traktman P. (2010) Mice deficient in the serine/threonine protein kinase VRK1 are infertile due to a progressive loss of spermatogonia. Biol. Reprod. 82, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Choi Y. H., Park C. H., Kim W., Ling H., Kang A., Chang M. W., Im S. K., Jeong H. W., Kong Y. Y., Kim K. T. (2010) Vaccinia-related kinase 1 is required for the maintenance of undifferentiated spermatogonia in mouse male germ cells. PLoS One 5, e15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schober C. S., Aydiner F., Booth C. J., Seli E., Reinke V. (2011) The kinase VRK1 is required for normal meiotic progression in mammalian oogenesis. Mech. Dev. 128, 178–190 [DOI] [PubMed] [Google Scholar]
- 71. Valbuena A., Sanz-García M., López-Sánchez I., Vega F. M., Lazo P. A. (2011) Roles of VRK1 as a new player in the control of biological processes required for cell division. Cell. Signal. 23, 1267–1272 [DOI] [PubMed] [Google Scholar]
- 72. Lancaster O. M., Breuer M., Cullen C. F., Ito T., Ohkura H. (2010) The meiotic recombination checkpoint suppresses NHK-1 kinase to prevent reorganisation of the oocyte nucleus in Drosophila. PLoS Genet. 6, e1001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Renbaum P., Kellerman E., Jaron R., Geiger D., Segel R., Lee M., King M. C., Levy-Lahad E. (2009) Spinal muscular atrophy with pontocerebellar hypoplasia is caused by a mutation in the VRK1 gene. Am. J. Hum. Genet. 85, 281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. López-Sánchez I., Sanz-García M., Lazo P. A. (2009) Plk3 interacts with and specifically phosphorylates VRK1 in Ser342, a downstream target in a pathway that induces Golgi fragmentation. Mol. Cell. Biol. 29, 1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]