Background: The enzymology of aminophospholipid-processing enzymes is not well understood.
Results: Structures of HRAS-like tumor suppressors resemble those of thiol proteases with Cys-His-His catalytic triad.
Conclusion: An aminophospholipid acyl group first modifies the Cys residue of HRAS-like tumor suppressor enzymes before it is transferred onto a second substrate.
Significance: This study provides a structural basis for the enzymatic mechanism of HRAS-like tumor suppressors.
Keywords: Enzyme Catalysis, Lipase, Lipids, Phosphatidylcholine, Structural Biology
Abstract
Lecithin:retinol acyltransferase-like proteins, also referred to as HRAS-like tumor suppressors, comprise a vertebrate subfamily of papain-like or NlpC/P60 thiol proteases that function as phospholipid-metabolizing enzymes. HRAS-like tumor suppressor 3, a representative member of this group, plays a key role in regulating triglyceride accumulation and energy expenditure in adipocytes and therefore constitutes a novel pharmacological target for treatment of metabolic disorders causing obesity. Here, we delineate a catalytic mechanism common to lecithin:retinol acyltransferase-like proteins and provide evidence for their alternative robust lipid-dependent acyltransferase enzymatic activity. We also determined high resolution crystal structures of HRAS-like tumor suppressor 2 and 3 to gain insight into their active site architecture. Based on this structural analysis, two conformational states of the catalytic Cys-113 were identified that differ in reactivity and thus could define the catalytic properties of these two proteins. Finally, these structures provide a model for the topology of these enzymes and allow identification of the protein-lipid bilayer interface. This study contributes to the enzymatic and structural understanding of HRAS-like tumor suppressor enzymes.
Introduction
Glycerophospholipids are major cellular constituents that provide the structural and physicochemical basis for many cell membrane functions. Phospholipids also act as mediators in many vital processes, including membrane trafficking, cytoskeletal rearrangements, and signal transduction, where they serve as molecular signaling precursors for a variety of lipid mediators such as fatty acids, sphingolipids, inositols, lysophospholipids, endocannabinoids, and prostaglandins (1, 2). In response to intra- and extracellular stimuli, the production and cellular location of lipid mediators are tightly controlled by numerous enzymes that employ lipids as substrates, including phospholipases and lipid-dependent acyltransferases (3).
A recently characterized group of proteins involved in phospholipid metabolism are vertebrate representatives of the papain-like or NlpC/P60 thiol protease superfamily. This class of proteins consists of five proteins known as HRAS-like tumor suppressors (HRASLS1–5)4 and lecithin:retinol acyltransferase (LRAT) (4). HRASLS enzymes are also referred to as LRAT-like proteins because of their sequence homology to LRAT, the best characterized member of this group, which catalyzes the formation of retinyl esters by transferring an acyl group from the sn-1 position of phosphatidylcholine (PC) onto retinol (vitamin A). Except for HRASLS5, these enzymes possess a single C-terminal transmembrane-spanning segment. All members of this family share a highly conserved NCEHFV motif, which contains a reactive Cys residue involved in catalytic activity (Fig. 1A) (5, 6). Despite conservation of their catalytic sites, HRASLS proteins catalyze disparate reactions, many of which remain uncharacterized. In contrast, the enzymatic activities and physiological significance of LRAT in vision and vitamin A metabolism have been well understood (5, 7, 8). Functions of HRASLS3 also known as H-rev107 or AdPLA have recently been studied in the Adpla−/− mouse model (9). HRASLS3 deficiency leads to markedly reduced adipose tissue mass and triglyceride content but normal adipogenesis accompanied by higher energy expenditure and increased fatty acid oxidation in adipocytes. This profound effect on triglyceride metabolism identifies HRASLS3 as a major regulator of adipocyte lipolysis and a crucial factor in the development of obesity. However, the molecular mechanism responsible for the Adpla−/− mouse phenotype, originally attributed to the phospholipase A2 (PLA2) activity of HRASLS3 and consequent deficiency in prostaglandin E2 in these null mice, remains obscure because of contradictory reports about the enzymatic specificity of HRASLS3 with respect to the sn-1/sn-2 position of PC (9–12). Interestingly, biochemical characterization of other members of the LRAT-like protein family, including HRASLS1, -2, and -4, indicates a common PLA1,2 activity with a predominant specificity for the sn-1 acyl position. In contrast, HRASLS5 reportedly exhibits LRAT-like acyltransferase activity by abstracting an acyl group from the sn-1 position of PC and transferring it onto the amine group of phosphatidylethanolamine (PE) to form N-acylphosphatidylethanolamine (N-acyl-PE), the precursor of the bioactive endocannabinoid, anandamide (13, 14).
FIGURE 1.
Sequence alignment and purification of LRAT-like protein members. A, sequences of human HRASLS1–5 and LRAT were aligned by using ClustalW2 available at the EMBL-EBI server. Conserved and homologous residues are shown with yellow and green backgrounds, respectively. The conserved His residues and six amino acid stretches containing the catalytic Cys residue are highlighted in red. The predicted C-terminal transmembrane segments are marked with a gray background. B, SDS-PAGE mobility of purified HRASLS proteins. Two μg of each protein were separated in a 15% gel and stained with Coomassie Blue R-250. Single protein bands indicate the high purity of protein preparations. C, schematic representations of truncated C-terminal His6-tagged HRASLS1–4 expressed in E. coli.
The physiological importance of LRAT-like proteins in key metabolic pathways encouraged us to undertake a comprehensive characterization of this class of proteins. Here, we describe and compare mechanisms of enzymatic activity for selected HRASLS proteins, including the formation and stability of their covalent acyl enzyme intermediates. We also demonstrate alternative acyltransferase enzymatic activities of HRASLS enzymes that likely reflect their physiological functions. Finally, we report crystallographic structures of HRASLS2 and HRASLS3, providing information about the active site architecture of this family of enzymes at atomic resolution. These structures also allowed identification of the protein-lipid interface as well as construction of a preferred model of the membrane topology. These findings could facilitate the development of therapeutic modulators of LRAT-like enzymes for the treatment of metabolic syndromes causing obesity.
EXPERIMENTAL PROCEDURES
Materials
cDNAs encoding human HRASLS1, -2, and -4 amino acid sequences (gi:9966859, gi:8923526, and gi:149588791, respectively) were synthesized de novo (DNA 2.0, Menlo Park, CA). Clones for human HRASLS3 and HRASLS5 (gi:189571621 and gi:21707989) were obtained from the American Type Culture Collection. Phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). Protein crystallization screens were obtained from Hampton Research (Aliso Vieja, CA).
Expression and Purification of HRASLS Proteins
Expression of HRASLS proteins in a soluble form required truncation of each protein sequence at the predicted C-terminal transmembrane helix (Fig. 1A). NdeI and XhoI restriction sites were introduced at the ends of HRASLS1–5 coding sequences by PCR with the following primers: HRASLS1, 5′-GCAGATCATATGGCGTTTAATGATTGCTTCAGTTTGAACTACC-3′ and 5′-CGTCTACTCGAGTCGGTTGGCCTGCTCTGAAACTCC-3′; HRASLS2, 5′-GCAGATCATATGGCTTTGGCCAGACCAAGACC-3′ and 5′-CGTCTACTCGAGCTGGTCACTGCGGGAGACGCC-3′; HRASLS3, 5′-GCAGATCATATGCGTGCGCCCATTCCAGAGC-3′ and 5′-CGTCTACTCGAGATCTCTGACCTGGTCACTGCGGGCG-3′; and HRASLS4, 5′-GCAGATGGATCCCACCACCACCACCACCACGCTTCGCCACACCAAGAGCCC-3′ and 5′-CGTCTAGAATTTCACTTGGCCTTTTCCACCTGTTTACAGC-3′. Amplified cDNAs were cloned into the pET30b bacterial expression vector (Invitrogen). A common purification strategy derived from the protocol published by Han et al. (15) was used for all HRASLS enzymes. Recombinant proteins were expressed in the BL21(DE3) Escherichia coli strain grown in the presence of 50 μm kanamycin. Protein expression was induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside and carried out at 22 °C for 5 h. Bacteria then were harvested by centrifugation (6000 × g, 15 min, 4 °C), treated with lysozyme (10 μg/ml), and disrupted by osmotic shock (16). The lysate was cleared by centrifugation at 36,000 × g for 30 min at 4 °C. After adjustment of pH and ionic strength to 50 mm Tris-HCl, pH 8.0, and 250 mm NaCl, the supernatant was loaded onto a 5-ml His-trap column (GE Healthcare). The column was subsequently washed with 50 ml of 50 mm Tris-HCl, pH 8.0, containing 250 mm NaCl and 10 mm imidazole, and the protein was eluted by raising imidazole to 250 mm in the same buffer. The volume of combined fractions containing protein was reduced to 5 ml in a 10-kDa molecular mass cutoff Amicon Ultra (Millipore, Billerica, MA), and the solution was loaded onto a gel filtration column (120 ml) packed with Superdex-200 (GE Healthcare) and equilibrated with 10 mm MES-NaOH, pH 6.5, and 5 mm dithiothreitol (DTT). After gel filtration, eluted protein was loaded onto a 5-ml HiTrap-SP cation exchanger column (GE Healthcare) equilibrated with 10 mm MES-NaOH, pH 6.5, and 5 mm DTT. Protein was eluted with a linear gradient of NaCl in the equilibration buffer (0–0.5 m, 30 min, 1 ml/min). Protein purity in collected fractions was examined by SDS-PAGE, and the protein identity was verified by MS. Finally, the protein solution was concentrated to 6, 2, 9, or 16 mg/ml for HRASLS1–4, respectively, in a 10-kDa molecular mass cutoff Amicon Ultra, snap-frozen in liquid nitrogen, and stored at −80 °C.
Expression and Purification of LRAT-GST Fusion Protein
The soluble form of enzymatically active LRAT was obtained by expressing a GST-fused mouse protein (residues 30–186) in E. coli and then purifying it as described in our previous report (17).
LC/MS Analysis of Intact Proteins and Peptides
All experiments were performed with LXQ and LTQ Velos linear ion trap mass spectrometers equipped with an electrospray ionization source (Thermo Scientific, Waltham, MA). An Agilent Technologies 1100 Series HPLC system (Agilent Technology, Santa Clara, CA) was interfaced with the LXQ MS, and the LTQ Velos instrument was equipped with an Accela 600 quaternary pump for in-line protein, peptide, and lipid separation. To record spectra of intact proteins, 0.5 μg of protein was loaded onto an XBridge BEH300 C4 column (3.5 μm, 2.1 × 50 mm) (Waters, Milford, MA) and eluted with a linear (5–100%) gradient of acetonitrile in water over 20 min at a flow rate of 0.25 ml/min. Both solvents contained 0.1% formic acid. Intact protein masses were deconvoluted manually or by using ProMass for Xcalibur software (Thermo Scientific). The same column and solvents were employed to separate products of pepsin digestion except the acetonitrile gradient was developed over 40 min. Pepsin digestion of the proteins in solution was performed at room temperature, pH 2.0, for 15 min followed by immediate injection onto the column. Products of the digest were analyzed with a Mass Matrix Database Search Engine and then verified manually (18).
Self-acylation Assays
Acylated forms of HRASLS proteins (5 μg) were detected after their incubation with short acyl chain PCs (0.05–2 mm) in 0.1 ml of 10 mm Tris-HCl, pH 8.0, containing 1 mm DTT at 25 °C. Molecular ratios between the proteins and lipids ranged from 1:150 to 1:6000. The reaction mixtures were directly subjected to LC/MS and analyzed as described above.
Phospholipase Enzymatic Assays
Phospholipase activity of the HRASLS enzymes was examined by incubating these enzymes (4 μg) with 0.5 mm 7:0–7:0 PC in 0.1 ml of 10 mm Tris-HCl, pH 8.0, containing 1 mm DTT at 25 °C. The reaction was quenched at various time points with 0.1 ml of methanol, and the substrate as well as lyso-PC products were separated and quantified by LC/MS as described below. 1-Hexanoyl-2-(6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl)-sn-glycero-3-phosphocholine (6:0–6:0-NBD-PC) was used as a substrate to determine the positional specificity of phospholipase activity. Because the commercially available compound contained significant amounts of hexanoyl-NBD acid, 6:0–6:0-NBD-PC was pre-purified by HPLC prior to use. This sn-2 chromophore-labeled PC was incubated in the presence of proteins for up to 3 h under conditions described above. Products of the enzymatic reaction were separated by chromatographic methods described for intact proteins and peptides and quantified with a diode array detector (Agilent) set at 460 nm. Chemical identities of eluted compounds were confirmed by MS.
Aminophospholipid Acyl Rearrangement Assays
The acyltransferase enzymatic assay was performed under essentially the same reaction conditions as described for the phospholipase assay, but the initial phospholipid substrate consisted of either an equimolar mixture of 7:0–7:0 PC and 6:0–6:0 PE or 7:0–7:0 PC and 6:0–6:0 PC, each at a final concentration of 0.25 mm. A 2 mm lipid concentration was used for acyl transfer between 7:0–7:0 PC and 16:0–16:0 PC. Enzymatic reactions were initiated by addition of 5 μg of the test protein to 0.1 ml of total reaction mixture. After incubation at 25 °C, reactions were quenched at various time points with 0.1 ml of methanol, and the phospholipid composition was determined by LC/MS.
Phospholipids were quantified based on the reference peaks of added internal standards. Because isotopically labeled short acyl chain phospholipids were not commercially available, the phospholipid internal standards used represented the same class of lipids as the quantification target but with a different acyl chain length. To compensate for slight differences in ionization, correction factors were calculated for each of the ion pairs in 1:10 to 10:1 concentration ratios. Thus, 6:0–6:0 PC was used for calculation of the correction factor for 7:0–7:0 PC (0.95) that was then used to determine amount of the 7:0–6:0 PC in the experimental samples. 8:0 Lyso-PC served as an internal standard for 6:0 lyso-PC (correction factor 1.63). Quenched reaction mixtures were supplemented with known amounts of internal standards (0.15–0.5 nmol) and directly injected onto an XBridge BEH300 C4 (3.5 μm, 2.1 × 50 mm) column. The column temperature was 25 °C. Chromatographic separation of phospholipids, including PC, PE, and the corresponding lysophospholipids, was achieved with a linear (5–100%) gradient of acetonitrile/water (0.1% formic acid) over 20 min at a flow rate of 0.25 ml/min. The eluent was directed into an LTQ Velos MS via an electrospray ionization source working in positive mode, and the mass spectrometer was tuned for 7:0–7:0 PC to ensure adequate sensitivity. The resulting ion chromatograms were used to quantify the intensities of isolated selected ions. Areas under the peaks of interest were calculated by using X-calibur 2.2 (Thermo Scientific).
N-Acyltransferase Activity of HRASLS3
The ability of purified recombinant HRASLS3 to act enzymatically as a PE N-acyltransferase was assessed with 18:1–16:0 PC as an acyl donor and 17:0–17:0 PE as an acyl acceptor (Avanti Polar Lipids). Assays were carried out in 50 mm glycine-NaOH, pH 9.0, containing 2 mm DTT and 0.1% Nonidet P-40 in a final reaction volume of 0.2 ml at 37 °C. PC (final concentration, 40 μm) and PE (final concentration, 75 μm) were added in CHCl3 to glass tubes used for these incubations; the CHCl3 was evaporated under N2, and the phospholipids were resolubilized with the detergent-containing assay buffer and vigorous mixing. The reaction was started by addition of purified HRASLS3 (1–5 μg) and later quenched by addition of 320 μl of CHCl3/CH3OH (2:1 (v/v)) containing 0.5 nmol of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-nonadecanoyl (18:1–18:1 PE-N-19:0) as an internal standard. The CHCl3 phase containing the extracted phospholipids was removed and used for LC/MS analysis of products carried out on a Xevo TQ MS ACQUITY UPLC system (Waters). Samples in autosampler vials were maintained at 4 °C in the autosampler until 5-μl aliquots were injected onto the LC/MS. A BEH C18 column (2.1 × 100 mm, 1.7-μm particle size, Waters), preceded by a 2.1 × 5-mm guard column packed with the same material, was employed to assess N-acyl-PE formation. Initial HPLC solvents were 5% solvent A (H2O) and 95% solvent B (methanol); both solvents contained 10 mm ammonium acetate. Solvent B increased linearly over 2 min to 100% B and remained at 100% for 3 min (5-min total run), at a flow rate of 0.5 ml/min. The column temperature was 40 °C. The source temperature was 150 °C, desolvation temperature 500 °C, desolvation gas 1000 liter/h, cone voltage 76 V, and collision energy 50 eV.
Crystallization of HRASLS2 and HRASLS3 and Preparation of Heavy Atom Derivatives
HRASLS2 and HRASLS3 crystals were grown by the sitting drop vapor diffusion method. HRASLS2 was crystallized by mixing 1.5 μl of protein solution (2 mg/ml) in 10 mm MES-NaOH, pH 6.5, containing 200 mm NaCl and 10 mm DTT with 1.5 μl of crystallization solution consisting of either 1.1 m dibasic ammonium tartrate, pH 7.0, or 1.5 μl of 0.2 m MgCl2 hexahydrate, 0.1 m BisTris-HCl, pH 5.5, and 25% (w/v) polyethylene glycol (PEG) 3350. The resulting drops were incubated at room temperature over 0.1 ml of the same crystallization solution. Crystals typically first appeared after 5–7 days and grew to final size in 2 weeks. Most crystals were rod shaped and reached dimensions of ∼500 × 50 × 50 μm. Crystals grown in dibasic ammonium tartrate were cryoprotected by brief soaking in synthetic mother liquor supplemented with 15% (v/v) glycerol and flash cooled in liquid nitrogen prior to x-ray exposure.
Crystals of HRASLS3 were obtained by mixing 1.5 μl of protein solution (6 mg/ml) in 10 mm MES-NaOH, pH 6.5, containing 200 mm NaCl, 10 mm DTT with 1.5 μl of 0.1 m BisTris-HCl, pH 6.5, 28% (w/v) PEG 2000 monomethyl ether and incubating the drops at room temperature. Crystals appeared in 7–14 days and grew to final size after another 2–4 days. These crystals had an elongated shape and a hexagonal cross-section with approximate dimensions of ∼300 × 20 × 20 μm. Native crystals were harvested by direct flash cooling in liquid nitrogen. Four heavy atom derivatives of HRASLS3 were prepared to enable de novo phasing. Mature crystals were soaked for 10–30 min in synthetic mother liquor containing 1 mm of either ethyl mercury thiosalicylate, potassium tetrachloroplatinate(II) (K2PtCl4), or potassium gold cyanide (KAu(CN)2) prior to flash cooling in liquid nitrogen. The iodide derivative was obtained by rapid submersion of crystals in synthetic mother liquor containing 1 m potassium iodide (KI) for ∼30 s followed by immediate flash cooling in liquid nitrogen (19).
X-ray Diffraction Data Collection, Phasing, and Structural Refinement
Diffraction data were collected at the NSLS X29 and APS NE-CAT 24-ID-C beamlines. Data obtained from the gold-, mercury-, and platinum-derivatized HRASLS3 crystals were collected at x-ray energies just above the respective LIII absorption edges to exploit the anomalous scattering of these atoms for phase determination. The iodine-derivatized crystal was collected at a remote energy, where iodine exhibits significant anomalous scattering. Data reduction was performed using XDS (20, 21), HKL2000 (22), and TRUNCATE (23). HRASLS3 crystals belonged to space group P3121 and contained one monomer per asymmetric unit. Riso values indicated good isomorphism between native and derivative HRASLS3 crystals (supplemental Table 1). The scaled data sets were input into autoSHARP (24) for experimental phase determination using both isomorphous and anomalous differences. Initial heavy atom substructures were located with SHELXC/D (25). Heavy atom substructures for each derivative were refined and completed, and initial protein phases were calculated with SHARP (26). Density modification with SOLOMON (27) resulted in a high quality electron density map that allowed automated protein model building in ARP/wARP (24). This initial model was refined against a second isomorphous native HRASLS3 data set that extended to higher resolution than the original native data set used for phase determination. The new data set displayed significant diffraction anisotropy (2.4 Å resolution in the a* and b* directions and 1.95 Å resolution in the c* direction with I/σI >3), and thus was submitted to the diffraction anisotropy server for ellipsoidal truncation and scaling (28). This procedure markedly improved the appearance of the electron density maps and also resulted in superior model refinement statistics. Structure refinement was conducted in PHENIX (29) and REFMAC (30) with TLS B-factor refinement (31), and COOT was used to manually adjust the model (32, 33). The refinement converged to an Rfree value of 21.6% with excellent stereochemistry as assessed with the MolProbity Server (supplemental Table 2) (34). Residues 1–5, 40–57, and 126–140 were not modeled because of the weak or absent electron density of these regions. Arg-18 and Cys-89 were modeled in split conformations, and 43 water molecules were included in the final model.
A high resolution data set extending to 1.25 Å resolution was collected on an HRASLS2 crystal at NE-CAT beamline ID-24C by using the continuous vector scanning option to maximize diffraction resolution and distribute the dose over the entire crystal. These crystals belonged to space group P21 and contained one monomer per asymmetric unit. The HRASLS2 structure was solved by molecular replacement with PHASER_MR (35) and the refined model of HRASLS3 as a search model. Adjustments to the protein sequence were made with COOT (33), and the initial model was refined with SHELXL (25) and REFMAC (30, 36). The resolution used for refinement was gradually extended from 1.8 to 1.25 Å. B-factors were refined anisotropically in the final stages of refinement, which reduced Rfree by about 2%. Cys-113 was found to adopt two conformations, one of which contained extra electron density near the Sγ atom. Because of the short distance between the peak maximum and the Sγ atom, we interpreted this density as originating from an oxidative modification of the Cys residue. This alternative conformation of Cys-113 was thus modeled as a sulfenic acid moiety. The refinement converged to an Rfree value of 17.8% with excellent stereochemical statistics as assessed with the Molprobity server (supplemental Table 2) (34). Residues 1, 40–51, 106–110, and 128–137 and the side chains of residues 7, 57, and 126 were omitted from the final model because of weak or absent electron densities. Pro-8, Ser-17, His-23, and His-119 in addition to Cys-113 were modeled in two conformations, and 105 water molecules were included in the final model. The coordinates and structure factors were deposited in the Protein Data Bank under accession codes 4DPZ and 4DOT for HRASLS2 and HRASLS3, respectively.
RESULTS
Expression and Purification of Recombinant Human HRASLS Proteins
Sequence alignment of proteins belonging to the LRAT-like family revealed an overall common architecture featuring variable N-terminal sequences, conserved catalytic domains, and a hydrophobic membrane-anchoring region predicted to be a single transmembrane α-helix at the C terminus (Fig. 1A). Expression of full-length HRASLS proteins in bacteria results in formation of insoluble inclusion bodies (15). Therefore, to minimize aggregation and ensure robust expression efficiency, we generated constructs lacking the hydrophobic C-terminal domain for HRASLS1–5. Conveniently, high homology between these HRASLS proteins allowed the development of a three-step purification protocol involving His tag affinity, gel filtration, and cation exchange chromatography, resulting in reproducible, highly purified homogeneous preparations (Fig. 1, B and C). The only exception was HRASLS5, which after bacterial expression in truncated or full-length forms did not produce a sufficient yield of properly folded protein for further characterization.
HRASLS Proteins Share a Common Enzymatic Strategy
In previous studies, we provided direct evidence that LRAT adopts a catalytic strategy similar to papain-like thiol proteases in which deprotonated Cys-161 exerts a nucleophilic attack on a carbonyl carbon of the PC ester bond leading to transient covalent acylation of the protein (6, 17). Thus, an MS-based assay developed to trap the LRAT thioester intermediate was used to investigate the catalytic mechanism of the HRASLS proteins. Electrospray ionization allowed analyses of the intact proteins, whereas the charge state distributions of protein ions revealed information about the polypeptide mass. Spectra of purified HRASLS3 (Fig. 2), HRASLS1, HRASLS2, and HRASLS4 (supplemental Fig. S1) recorded under acidic conditions provided well defined charged states, and the mass spectra deconvolution revealed protein masses identical to those calculated from amino acid sequences. Incubation of HRASLS2–4 with 7:0–7:0 PC caused the rapid appearance of an extra chromatographic peak in addition to signals from the intact protein and phospholipid (Fig. 2A and supplemental Fig. S1). The MS spectrum corresponding to this extra peak was closely related to that of the tested protein but indicated an increase in its molecular mass by 112 Da, a mass shift corresponding exactly to the mass of a C7 fatty acid moiety. Thus, the presence of 7:0–7:0 PC led to covalent modification of HRASLS2–4. Similar results were obtained for PE and lipid substrates with acyl chain lengths between 5 and 16 carbons. However, the efficiency of protein modification decreased with longer acyl chain lengths, especially for HRASLS2 and HRASLS3, because of reduced phospholipid aqueous solubility. Unlike lipases, the soluble form of HRASLS enzymes and LRAT can access phospholipids in monomeric rather than ordered micellar form. In this sense, the soluble form of LRAT-like proteins exhibit features characteristic of esterases (37). Consequently, the efficiency of self-acylation of HRASLS2 and HRASLS3 drops with increasing length of acyl chains and lower critical micelle concentration of lipid substrates was discussed previously for LRAT (17).
FIGURE 2.
Self-acylation of HRASLS3 in the presence of PC. Ten μg of HRASLS3 was incubated with 0.5 mm 7:0–7:0 PC in 10 mm Tris-HCl, pH 8.0, 1 mm DTT for 10 min at 25 °C. The mixture was then analyzed by LC/MS. A, HPLC separation and MS spectra of the protein and the lipid substrate. Peaks 1a and 1b represent intact protein and its acyl modified form, respectively. Peak 2 corresponds to 7:0–7:0 PC. The MS spectrum, shown in the lower panel, displays a selected group of multiply charged protein ions for the unmodified and acylated protein. Deconvolution of the HRASLS3 spectrum reveals a protein mass of 15,871 Da, which is identical to the theoretical mass of the recombinant His6-tagged protein. Incubation of this protein with 7:0–7:0 PC led to a 112-Da shift in the experimentally obtained protein mass as calculated from the additional series of ions labeled in red. This mass difference matches a heptanoic acid moiety carried by the lipid substrate. B, determination of the acylation site by tandem MS. A pepsin digest of HRASLS3 after incubation with 7:0–7:0 PC revealed a +1 peptide (m/z = 1103.5), with a fragmentation pattern that specified Cys-113 as the site of covalent acyl modification. Comparison of the native (top) and modified peptide (bottom) MS/MS spectra indicates a shift in masses corresponding to a series of b and y ions consistent with a C7 thioester modification at Cys-113. Ions that reveal the 112-Da shift are highlighted in blue and red for native and acylated peptide, respectively.
The only protein that did not undergo acylation under our experimental conditions was HRASLS1. This discrepancy could result from differences in substrate specificity, a requirement for specific assay conditions or possible incorrect folding of the protein upon expression in a bacterial system.
Acyl modifications of HRASLS proteins were stable, allowing identification of lipid-modified residues. MS examination of an HRASLS3 pepsin digest preincubated with 7:0–7:0 PC revealed the presence of an ion (m/z = 1103.5) that corresponded to the NCEHFVNE peptide with an added C7 acyl chain. The identity of this peptide was confirmed by its MS/MS fragmentation pattern. A 112-Da shift in the b-ion series observed between the modified and unmodified peptide clearly indicated the Cys-113 residue as part of a formed thioester (Fig. 2B). Analogous analysis performed on HRASLS2 revealed the very same acylation site (supplemental Fig. S2). Thus, we directly demonstrated that HRASLS proteins, similar to LRAT, were self-acylated in the presence of PC as an integral part of their enzymatic activity. Hence, members of the LRAT-like family could share a common catalytic strategy.
Stability of Acylated Forms of HRASLS Proteins and Implications for the Enzymatic Activity of These Proteins
Adoption of a thiol protease enzymatic strategy by HRASLS proteins agrees with the reported hydrolysis of phospholipid carboxyl groups by these enzymes. However, the well characterized acyltransferase activity of LRAT suggests that the same mechanism can be used by HRASLS enzymes to catalyze acyltransferase reactions in the presence of proper acyl donors and acceptors. We surmised that the ability of these enzymes to act as acyltransferases should relate to the stability of their acylated forms in aqueous solution. Thus, we investigated the stability and hydrolysis rates of the acylated Cys residues formed when HRASLS2 and -3, and LRAT were exposed to 7:0–7:0 PC. Incubation of HRASLS2, HRASLS3, and GST-tLRAT (a catalytically active, truncated, and soluble form of LRAT) (17) with this phospholipid substrate at pH 8.0, which is optimal for hydrolytic activity, did not cause rapid breakdown of the lipid substrates (Fig. 3). The lyso-PC product accumulated slowly over an extended period of incubation with a rate near the assay detection limit of 0.04 nmol/min/μg of protein for HRASLS2, which was more efficient than HRASLS3 or GST-tLRAT. Despite the slow rates of phospholipid hydrolysis, each of the tested proteins became efficiently acylated under the experimental conditions employed indicating their enzymatic competence. The acyl-modified forms of HRASLS2 and HRASLS3 persisted over the whole assay period, mimicking behavior of LRAT (17). The high stability of this lipid modification was underscored by the fact that these acylated proteins could be separated from PC by gel filtration chromatography (data not shown).
FIGURE 3.
Relationship between the lipolytic activity and stability of acylated forms of HRASLS enzymes. A, breakdown of 7:0–7:0 PC upon incubation with selected LRAT-like proteins as follows: HRASLS2 (●), HRASLS3 (▿), HRASLS4 (▾), and GST-tLRAT (□). Reaction products were separated and analyzed by LC/MS. The lipid substrate was quantified based on the ratio between the areas under peaks corresponding to intensities of m/z = 482.4 [MH]+ (7:0–7:0 PC) and the internal standard (6:0–6:0 PC, m/z = 454.4 [MH]+) ions. B, differences in stabilities of the acylated forms of HRASLS3 (top) and HRASLS4 (bottom) upon incubation with 7:0–7:0 PC. Mixtures of protein and phospholipid were subjected to LC/MS at different time points after incubation. The chromatograms show base peak intensities. The resolved peaks are labeled as follows: 1a, protein; 1b, acylated form; 2, 7:0–7:0 PC; 3, lyso-PC; arrow, disappearance of acylated form; *, disappearance of 7:0–7:0 PC. The modified form of HRASLS3 persists throughout the 2-h incubation, whereas the HRASLS4 thioester adduct is hydrolyzed within 30 min accompanied by progressive decrease of the substrate. Changes in stability of protein thioester adducts are quantified in C. All experiments were repeated three times in triplicate. Error bars, S.D.
In marked contrast to HRASLS2, HRASLS3, and GST-tLRAT, HRASLS4 exhibited a much higher 7:0–7:0 PC hydrolysis rate of 1 nmol/min/μg of protein (Fig. 3A). This robust enzymatic activity was accompanied by the rapid acyl-protein complex disappearance indicating a direct relationship between the hydrolytic enzymatic activity and decay of the thioester reaction intermediate (Fig. 3, B and C). The above results indicate that water-driven decomposition of the thioester intermediate is not preferred by either HRASLS2 or HRASLS3. Thus, their acyltransferase activity could dominate in the presence of a suitable acyl acceptor.
To determine the specificity of the observed phospholipase activity, we used 1-hexanoyl-2-(6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl)-sn-glycero-3-phosphocholine (6:0–6:0-NBD-PC), an sn-2 fluorescently labeled PC, as a substrate. Absorbance of 460 nm of light by the NBD chromophore allowed direct quantification of lipid hydrolysis products. A representative chromatogram showing separation of this substrate and the reaction products is shown in Fig. 4. The presence of both 1-lyso-2-NBD PC and free 6-NBD-hexanoic acid indicated that HRASLS enzymes possess both sn-1 and sn-2 activity. However, HRASLS2 showed a slight preference for the sn-2 position, whereas nearly 70% of HRASLS4-dependent cleavage occurred at the sn-1 position of this substrate. HRASLS3 did not reveal a significant preference for either site (Fig. 4C).
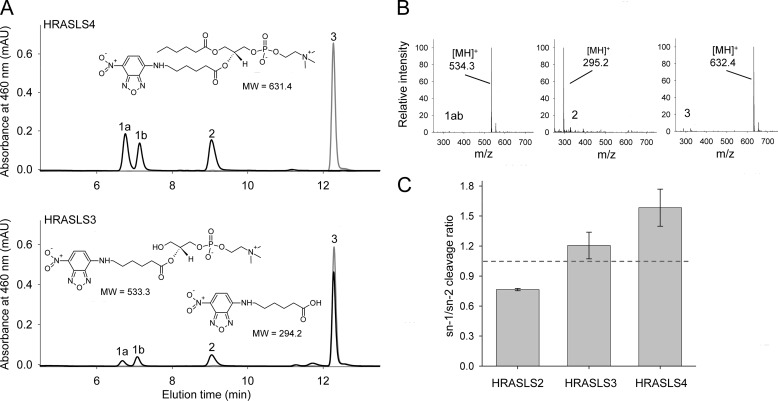
FIGURE 4.
sn-1/sn-2 specificity of HRASLS2–4 lipolytic activity. The preferential site of ester bond hydrolysis in PC was determined by using a phospholipid substrate labeled with NBD chromophore at the sn-2 position (6:0–6:0-NBP PC, structure shown in A, top). A, HRASLS4 (10 μg) incubated with 0.1 mm 6:0–6:0-NBP PC for 15 min under standard conditions used for the lipolysis assay yielded two products with characteristic absorbance for the NBD chromophore (black line) and molecular masses of 533.3 and 294.2 Da, respectively (B). These compounds were identified as 1-hydroxy-2-hexanoyl-NBD-lyso-PC (A, peaks 1a and 1b) and 6-NBD-hexanoic acid (A, peak 2). The structures of both products are shown in A, bottom. Two peaks that correspond to the lyso-PC product indicate a preferential intramolecular acyl shift from the sn-2 to the sn-1 position. Slower phospholipid hydrolytic activity was documented for HRASLS2, and HRASLS3 required a prolonged 3-h incubation to detect any reaction products (A, bottom; data for HRASLS3 shown) leaving most of the substrate intact (peak 3). Control samples, incubated without protein, did not reveal products of 6:0–6:0-NBP PC degradation (A, gray lines). C, absorbance-based quantification of the lipolysis product indicates a lack of strict preference for the cleavage site. Hydrolysis occurred at both the sn-1 and sn-2 positions with a slight preference for the sn-2 position by HRASLS2 and for sn-1 by HRASLS4. HRASLS3 did not show a significant preference toward either site. Error bars, S.D. Experiments were performed in triplicate.
HRASLS Proteins Function as Acyltransferases
The capability of HRASLS enzymes to catalyze acyl transfer was demonstrated for HRASLS5, which transfers an acyl group from the sn-1 position of PC onto an amino group of PE to form N-acyl-PE (14). A trace of similar activity was seen for HRASLS3 and other members of LRAT-like protein family as well (13, 38). To further understand this activity, we incubated HRASLS2, HRASLS3, and HRASLS4 with 18:1–16:0 PC in the presence of 17:0–17:0 PE. Purified recombinant HRASLS enzymes catalyzed the transfer of either the sn-1 or the sn-2 acyl moieties of PC to form an N-acyl-PE, the first metabolic step needed for N-acylethanolamine formation. As deduced from the LC/MS/MS profiles provided in supplemental Fig. S3, both the sn-1 and sn-2 groups of 18:1–16:0 PC can be transferred to the 17:0–17:0 PE acceptor forming either 17:0–17:0 PE-N-18:1 or 17:0–17:0 PE-N-16:0. There were significant differences in acyl transfer as well as lipolysis rates between tested proteins. All tested proteins revealed acyltransferase and lipase activity. However, HRASLS2 and HRASLS3 predominantly catalyzed acyl transfer, whereas lipase activity was dominant for HRASLS4. Apparent reaction rates are listed in supplemental Fig. S3C.
In an attempt to reproduce these data with short chain phospholipids, we incubated HRASLS2–4 with a mixture of 7:0–7:0 PC and 6:0–6:0 PE (1:1 molar ratio) and analyzed the lipid products of these reactions with LC/MS. N-Acyl-PE production was not detected in any of these experimental samples. However, we did note that the reaction mixtures contained 6:0–7:0 PC (m/z = 468.4 [MH]+) and 7:0–6:0 PE (m/z = 426.4 [MH]+), both products of fatty acid remodeling of the lipid substrates (Fig. 5). For HRASLS2 and HRASLS3, the amounts of detected phospholipids with mixed acyls were relatively small and comparable with those of lyso-PC obtained upon incubating these proteins with PC. Interestingly, in the case of HRASLS4, which effectively hydrolyzes PC and PE, both 6:0 lyso-PC and 7:0 lyso-PE were easily detectable. Similar results were obtained with a mixtures of 6:0–6:0 PC and 7:0–7:0 PC or 7:0–7:0 PC and 16:0–16:0 PC indicating that acyl remodeling occurs also between PCs and is not limited to short acyl chain substrates (Fig. 6).
FIGURE 5.
Aminophospholipid acyl chain remodeling activity of HRASLS enzymes. HRASLS proteins were incubated with an equimolar mixture of 7:0–7:0 PC and 6:0–6:0 PE. Reaction mixtures were quenched with methanol and supplement with internal standards, and their lipid composition was determined by LC/MS at various time points. A, in addition to 7:0 lyso-PC and 6:0 lyso-PE, the expected products of substrate hydrolysis, samples containing HRASLS4, evidenced the presence of 6:0 lyso-PC resulting from acyl chain remodeling prior to PC cleavage. Selected chromatographic ion peaks for m/z = 370.4 (7:0 lyso-PC) and m/z = 356.3 (6:0 lyso-PC) are outlined in gray and black, respectively. B, quantification of 6:0 lyso-PC production upon incubation with HRASLS4. 8:0-lyso-PC was used as an internal standard. C, phospholipid acyl chain remodeling in the presence of HRASLS3. Incubation of aminophospholipids with HRASLS3 led to formation of phospholipid products with a mixed acyl composition (6:0–7:0 PC and 7:0–6:0 PE). Chromatograms represent an experiment in which production of mixed acyl phospholipids was enhanced by the presence of HRASLS4 that served as an internal source of 1-hydroxyl-2-heptanoyl-PC. Selected chromatographic ion peaks for m/z = 482.4 (7:0–7:0 PC, substrate) and m/z = 468.4 (6:0–7:0 PC, product) in the top panel and m/z = 412.4 (6:0–6:0 PE, substrate) and m/z = 426.4 (7:0–6:0 PE, product) in the bottom panel are outlined in gray and black, respectively. D, averaged MS spectrum recorded between 11.5 and 13.5 min of elution from the chromatogram shown in C. Indicated [MH]+ ions represent substrates and products of the enzymatic reaction as well as the internal standard (6:0–6:0 PC) used for lipid quantification (m/z = 454.4). E, effect of HRASLS4 on HRASLS2- and HRASLS3-dependent acyl chain remodeling. Enzymatic activity of HRASLS4 or PLA1 from R. oryzae was used to generate 1-hydroxyl-2-heptanoyl-lyso-PC in situ. Open symbols correspond to data points for HRASLS2 collected in the absence of lyso-PC producing enzymes (○) and in the presence of HRASLS4 (Δ) or PLA1 (□). Black colored symbols represent HRASLS3 incubated with HRASLS4 (▾), PLA1 (■). or without any additional protein (●). Error bars, S.D. calculated from two independent experiments, each performed in triplicate.
FIGURE 6.
HRASLS2- and HRASLS3-dependent acyl chain remodeling between PCs. HRASLS proteins were incubated with an equimolar mixture of 7:0–7:0 PC and 6:0–6:0 PC or 7:0–7:0 PC and 16:0–16:0 PC (1:3 molar ratio) for 2 h at 25 °C. Reaction mixtures were quenched with methanol, and the lipid composition was examined by LC/MS (LTQ Velos or LXQ). HRASLS2 (A) and HRASLS3 (B) catalyzed formation of phospholipid products with mixed acyl chain lengths (6:0–7:0 PC). Chromatograms present selected ion signals for m/z = 482.4 (7:0–7:0 PC, substrate) and m/z = 454.4 (6:0–6:0 PC, substrate) and m/z = 468.4 (mixed acylated PC, product). C, averaged MS spectrum recorded between 11.5 and 13.5 min of elution indicates the presence of [MH]+ ions for substrates and the product of this enzymatic reaction. D, MS/MS fragmentation pattern of an ion corresponding to the mixed acylated PC product. Molecular identification of this product was achieved by recording its MS/MS fragmentation in a negative mode with 50% acetonitrile in methanol (v/v) containing 10 mm ammonium formate used as a solvent. The parent ion (m/z = 512.1) represents a typical formate adduct to PC. Its fragmentation (MS2) reveals m/z = 452.1 ions that are characteristic for PC loss of a methyl group (15 Da). Further induced decomposition of this ion (MS3) led to the appearance of two ions m/z = 340.1 and 354.1 that are products of C7 (112 Da) and C6 (98 Da) acyl loss. Similar acyl chain rearrangement occurred in the presence of long acyl chain PC (E and F). Chromatograms represent selected ion signals for m/z = 482.4 (7:0–7:0 PC, substrate) and m/z = 734.7 (16:0–16:0 PC, substrate) and m/z = 608.5 (mixed acylated PC, product). Inset in E represent averaged MS spectrum recorded between 15 and 15.5 min of elution that indicate mass of 7:0–16:0 PC (m/z = 608.5 [M + H]+ and m/z = 630.5 [M + H + Na]+).
Based on these results, we assume that all of these enzymes form an intermediary acyl-enzyme complex from which the acyl group can be transferred to either water to yield lysophospholipid and free fatty acid or to a second molecule of lysophospholipid to produce a remodeled phospholipid. In the case of HRASLS4, these remodeled phospholipids are subsequently hydrolyzed to form lysophospholipids.
Rearrangement of lysophospholipids via intramolecular I-II acyl migration represents a serious problem for accurate determination of acyl acceptor specificities of the HRASLS enzymes (39). Aqueous conditions and the alkaline pH used in enzymatic assays here promoted interconversion of 1-lyso-2-acyl-PC into 1-acyl-2-lyso-PC, which composed >95% of this lysophospholipid population at equilibrium achieved within minutes. Externally added 1-heptanoyl-2-lyso-PC or any other tested 1-acyl-2-lysophospholipids failed to increase the production of mixed acylated PCs by HRASLS proteins, suggesting that only 1-lyso-2-acyl phospholipids were used as substrates. To test this idea, we used the efficient, sn-1-preferential phospholipid hydrolytic activity of HRASLS4 or PLA1 from Rizopus oryzae to produce 1-lyso-2-acyl-phospholipid in situ during the enzymatic assay. Samples containing HRASLS, 7:0–7:0 PC, and 6:0–6:0 PE were supplemented with HRASLS4 at a concentration 1000 times lower than that of the tested enzyme or 0.5 μg of PLA1 (7 units/mg) and 1 mm CaCl2. Addition of HRASLS4 or PLA1 increased the yield of 6:0–7:0 PC up to 10-fold, indicating that predominantly 1-lyso-2-acyl phospholipids serve as fatty acid moiety acceptors (Fig. 5E). These results confirm the regiospecificity of HRASLS-mediated acyl transfer described by Shinohara et al. (38).
Crystallization of HRASLS2 and HRASLS3
To gain structural insights into the enzymatic activities of HRASLS proteins, two members of the LRAT-like protein family, human recombinant HRASLS2 and HRASLS3, were crystallized. Despite high homology (69% identities and 83% homology) and a common purification scheme, these two proteins required different processing and crystallization conditions. HRASLS2 displayed a strong tendency for nonspecific oligomerization at concentrations above 4 mg/ml by forming a white precipitate. Interestingly, this characteristic did not adversely affect its crystallization that occurred over a broad pH range (5.5–8.5) and in multiple PEG and PEG-derived precipitants (PEG 2000 to 8000). However, the only ionic precipitant that promoted HRASLS2 crystallization was dibasic ammonium tartrate. Crystals obtained under these conditions diffracted to d spacings corresponding to 1.25 Å. Unlike HRASLS2, HRASLS3 could be concentrated to 15 mg/ml but crystallized under much more stringent conditions with respect to pH and precipitant (pH 6.0–7.0; 26–30% PEG 2000 monomethyl ether at room temperature). For optimal crystal growth, the protein concentration of HRASLS3 could not exceed 3 mg/ml in the crystallization drop.
Structural Basis of Enzymatic Activities of HRASLS
The crystal structure of human recombinant HRASLS3 was solved by the multiple isomorphous replacement with anomalous scattering technique. Data from four isomorphous heavy atom derivatives were used to calculate initial phases, which were characterized by an overall figure of merit of 0.47 and 0.51 for acentric and centric reflections, respectively (supplemental Table S1). The initial electron density map displayed good connectivity and clear secondary structure features. Following density modification and phase extension to the full resolution range of the native data set, the resulting electron density map was of sufficient quality to allow automated model building of a large portion of the protein chain. The initial structural model was subsequently refined to an Rfree value of 21.6% at 1.96 Å resolution (supplemental Table S2). The HRASLS3 structure then served as a molecular replacement search model to solve the HRASLS2 structure, which was refined to an Rfree = 17.8% at 1.25 Å resolution (supplemental Table S2).
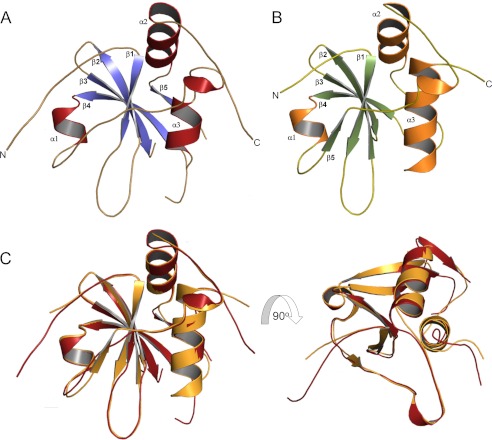
The basic structural motif of HRASLS2 and HRASLS3 was composed of a four-strand antiparallel β-sheet and three α-helices (Fig. 7, A and B). The longest α-helix (α3) was packed against the β-sheet, and the two other shorter α-helices were located on the sides. These two α-helical and β-sheet regions were well separated. Aside from slight differences in the lengths of certain β-sheets, HRASLS2 and -3 structures superimposed well with a root mean square deviation of 0.41 Å for Cα carbons (Fig. 7C). The overall architecture of these two proteins was similar to the classical α+β-fold with antiparallel β-sheets and segregated α- and β-regions. Despite some differences caused by insertions and circular permutations in the sequences of these proteins, their structures were largely reminiscent of papain-like proteases (4, 40).
FIGURE 7.
Structures of HRASLS2 and HRASLS3. A, ribbon diagram of HRASLS2 with α-helices and β-strands colored in red and blue, respectively. B, structure of HRASLS3 represented as ribbons with secondary structures colored in orange and green. C, structural comparison of HRASLS2 and HRASLS3. The structures were superimposed based on Cα positions in COOT by using the SSM superimpose tool.
Active Site Architecture
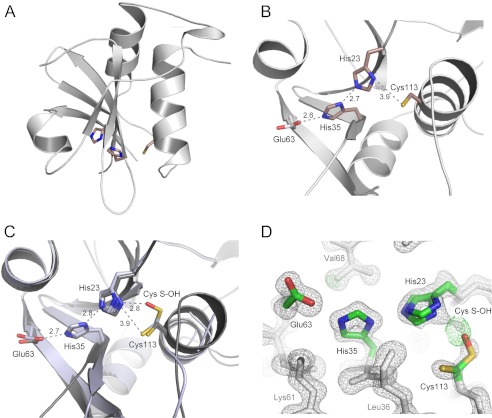
A highly conserved catalytic Cys-113, identified as the acylation site, is located near the N terminus of α3. This arrangement defines the active site location, which is embedded into a well defined grove formed by the extended loops between β1-β2, β3-β4, and the N terminus of the α3 helix (Figs. 8 and 9, A and B). The side chain of Cys-113 is packed against a β-sheet core of the protein, placing it in close proximity to the conserved His-23 from the β2 strand. The β-sheet is spread open on one end allowing formation of a hydrogen bond between His-23 and Cys-113 (Fig. 8B). There are no Asp or Glu residues present near His-23. The third polar residue in this catalytic triad is a second His residue (His-35) in the neighboring β3 strand. The His residue at this position is conserved in more than 60% of NlpC/P60s and in all HRASLS proteins with the exception of HRASLS1 (40, 41). In such an arrangement of catalytic residues, His-23 serves as a general base that deprotonates the Sγ atom of Cys-113, whereas Nδ1 of the His-35 side chain hydrogen bonds with the His-23 imidazole ring to stabilize its proper orientation.
FIGURE 8.
Architecture of the HRASLS2 and -3 active sites. A, location of key residues involved in catalysis within the structure of HRASLS3. B, orientation and local interaction of the active site residues. Distances are shown in Å. C, comparison of the active site residues positioned in HRASLS2 (gray) and HRASLS3 (light blue) structures with alternative conformations of His-23 and Cys-113 and its oxidative modification. D, 1.25 Å resolution, σA-weighted 2Fo − Fc electron density map contoured at 1.6σ for HRASLS2 is represented as a gray mesh. The green mesh represents a σA-weighted Fo − Fc omit electron density map contoured at 3.5σ. The appearance of the green mesh electron density next to the Sγ atom of Cys-113 is highly suggestive of an oxidative modification of this residue.
FIGURE 9.
HRASLS3 active site groove and proposed phospholipid membrane topology of HRASLS proteins. A, molecular surface of the protein with its distinctive aminophospholipid-binding groove. Blue and yellow colors correspond to Sγ and Nδ1 atoms of Cys-113, His-23, and His-35. B, charge distribution on the surface of HRASLS3. Negative charges are shown in red and positive charges in blue. The electrostatics calculations were performed with APBS (68). C, HRASLS3 with depicted regions absent in the crystal structure due to genetic manipulation (C-terminal helix) or inadequate electron density (residues 41–57). Secondary structure prediction for the 41–57 fragment of HRASLS3 listed in this panel was performed with the PSIPRED tool available at the University College London, Department of Computer Science server. D, proposed membrane topology for HRASLS3. Missing residues were modeled using MODELLER 9.10 (69) with secondary structure restraints introduced according to theoretical predictions. The color scheme for the molecule surface displays the relative hydrophobicity of the side chains. Blue color corresponds to polar and red indicates hydrophobic residues. E, ribbon representation of the HRASLS3 structure with modeled C-terminal α-helix and 41–57 segment in the phospholipid bilayer. Regions interacting with the membrane hydrophobic core as well as selected residues in the amphipathic α-helix are shown in red.
As expected, positions of key residues in the two structures involved in catalysis superimposed well. However, the high resolution structure of HRASLS2 provides additional information regarding the active site dynamics. Notably, Cys-113 was observed in two distinct conformations (Fig. 8, C and D). Originally, the alternative orientation of this residue was modeled in its reduced form. However, a consistent area of 2Fo − Fc and positive Fo − Fc electron density at the covalent bond distance of the Sγ atom strongly suggested a possible covalent modification of this residue. Thus, we assumed that the alternative conformer of Cys-113 represents sulfenic acid, an oxidized form of the Cys residue (Fig. 8, C and D). Consistent with this interpretation, the B-factor of the additional oxygen atom refined to a value close to the Sγ atom of this residue. Sulfenic acids are generally labile and can easily be oxidized further to their sulfinic or sulfonic forms unless they are stabilized by a network of hydrogen bonds (42). The stability of the sulfenic acid moiety results from interaction with Nδ1 of His-23 and the amide nitrogen of the Trp-24 backbone. The presence of the oxygen atom affects the position of the His-23 imidazole ring in which a slightly tilted alternative conformation can be clearly observed in the electron density map.
Protein-Membrane Interface and Membrane Topology
LRAT is an integral membrane protein that assumes a single membrane-spanning topology with a cytoplasmic N terminus and a C-terminal segment facing the endoplasmic reticulum lumen (43). The C-terminal transmembrane helix is indispensable for post-translational targeting and anchoring of LRAT to the endoplasmic reticulum. It also determines accessibility of the lipid substrates for this enzyme (44). Even though the C-terminal transmembrane domain has a variable sequence, it is predicted from hydropathy plots to be present in other members of the LRAT-like protein family. The only exception could be HRASLS5 where characteristics of this region are poorly defined (Fig. 1A). Although the most hydrophobic putative membrane-spanning portion of these proteins was truncated to obtain crystals, the resulting structures of HRASLS2 and HRASLS3 provide valuable information about the orientation of these molecules with respect to the lipid bilayer. Based on the location of the active site and the substrate binding groove that requires a proximal orientation to the lipid membrane for access to phospholipids, we postulate a likely orientation for HRASLS enzymes with respect to the membrane. The opening of this groove is surrounded by hydrophobic residues that constitute the most nonpolar segment of these proteins and thus are likely to integrate monotopically into the lipid bilayer (Fig. 9, A and B). A region that contributes to the HRASLS2 and HRASLS3 membrane interaction is the 40–57-amino acid segment between β3 and β4. Although the electron density for this region was inadequate for model building in both crystal structures, structure prediction indicated a high propensity for an α-helix. This in turn suggested an amphiphilic helix with a hydrophobic interface mainly composed of Val-50, Met-51, and Leu-54 in HRASLS3 and Val-50, Leu-51, and Leu-54 in HRASLS2 that could interact with the nonpolar core of a bilayer (Fig. 9, C–E). Thus, the reason for the disorder observed in this particular region could stem from the lack of a proper interfacial environment in the protein crystal, as lipid membrane interactions have been shown to stabilize amphiphilic helices (45). The model of HRASLS membrane topology proposed above is supported by thermodynamic calculations of rotational and translational positions of HRASLS3 in a phospholipid bilayer using the PPM 2.0 algorithm (available at Orientation of Protein in Membrane database server) (46). Analysis of the experimentally determined HRASLS3 structure as well as models of HRASLS3 containing the putative amphipathic loop (residues 40–57) and the C-terminal membrane-spanning α-helix (residues 133–155), individually or in combination, all resulted the same HRASLS3 membrane orientation consistent with our hypothesis (supplemental Fig. S4). An additional, much smaller, region for protein-lipid interaction is the hydrophobic loop between β3 and β4 with the exposed Phe-19 and Phe-20 residues in HRASLS2 and -3, respectively.
DISCUSSION
Here, we provide experimental evidence for a common enzymatic strategy shared by members of the LRAT-like protein family. Adaptation of the thiol-based protease mechanism in which a reactive Cys residue becomes transiently acylated provides a universal platform for both their hydrolase and acyltransferase activities. The ability of HRASLS2–5 to cleave PC and PE has been studied previously (13, 38). However, in each case, the rate of hydrolysis was significantly lower than found for PLA2 (15). Such modest PLA2 enzymatic activity along with a surprisingly high stability of the thioester catalytic intermediate comparable with that observed for the well characterized acyltransferase LRAT (5, 17) suggested a potential acyltransferase activity that could be fully expressed only in the presence of an appropriate acyl acceptor. In a series of experiments, we identified that 1-lyso-2-acyl-PC and 1-lyso-2-acyl-PE are acyl acceptors for HRASLS2–4. This finding suggests the involvement of HRASLS enzymes in acyl chain remodeling, a process that provides biologically active lipid mediators that continuously maintain the phospholipid composition of mammalian cells (47). Thus, this robust enzymatic activity of the HRASLS acyltransferase could explain the physiological role of these proteins. HRASLS3 is a major regulator of triglyceride metabolism in adipocytes and peroxisomal functions in culture models of other tissues, emphasizing the physiological significance of this protein (9, 12). However, the primary mechanism responsible for the physiological effects of HRASLS3 is still a matter of debate. The proposed inadequate prostaglandin signaling in Adpla−/− mice that affects triglyceride storage was linked to the PLA2 specificity of this protein, whereas a predominant PLA1 activity was observed in tissue culture models (9, 11, 48). The molecular mechanism by which HRASLS3 overexpression causes disruption of peroxisomes and ether-phospholipid production in cells is also unclear (12). Thus, we cannot exclude the existence of alternative protein- or tissue-specific acyl acceptors for HRASLS proteins within living cells. This possibility is reinforced by the action of HRASLS5 and other HRASLS proteins, which facilitate formation of N-acyl-PE (supplemental Fig. S3) (14). This is the first of two metabolic steps required for N-acylethanolamine formation. After formation, N-acylethanolamine must be acted upon by an N-acyl-PE-specific phospholipase D, forming the corresponding N-acylethanolamine and phosphatidic acid (49, 50). Biologically important N-acylethanolamines include the endocannabinoid N-arachidonoylethanolamide (anandamide), as well as N-palmitoylethanolamine, N-oleoylethanolamine, and N-stearoylethanolamine, which lack affinity for the canonical cannabinoid receptors (49–54). There is increasing evidence that endocannabinoid signaling is dysregulated in metabolic diseases like hyperglycemia and obesity (49–51, 54, 55). Moreover, it is known that N-oleoylethanolamine induces satiety and, together with N-palmitoylethanolamine and N-stearoylethanolamine, it enhances N-arachidonoylethanolamide actions in modulating metabolic processes (50–52). Given the capability of HRASLS3 to catalyze N-acyl-PE formation, it is possible that this enzyme can play a physiological role in modulating metabolic functions regulated by this poorly understood class of bioactive lipids.
The crystal structures of two human representatives of LRAT-like proteins reported in this study feature an architecture closely resembling that of the NlpC/P60 peptidase domain, an evolutionally conserved fold found in several archaeal, bacterial, and viral enzymes that catalyze related but distinct reactions such as murein degradation, amide hydrolysis, and acyl transfer reactions (4, 41). NlpC/P60 domain-containing proteins feature an unconventional Cys-His-His catalytic triad in their active sites despite being structurally related to the classical papain-like α+β-fold with four antiparallel β-strands packed against an α-helix. Such an assembly of catalytic residues is known only for the bacterial β-ketoacyl-acyl carrier (56). In this unusual arrangement, His-23 serves as a general base that lowers the pKa of the Cys-113 sulfhydryl group, whereas the second His residue of the triad (His-35) provides hydrogen bonds with the His-23 side chain stabilizing its proper orientation. The position of His-35 is also restrained by interaction with the side chain of Glu-63 (Fig. 8). His-35 is conserved in HRASLS2–4 and is substituted by a chemically similar Gln or Asn residue in HRASLS1 and LRAT, respectively. Interestingly, substitution of His-35 by Asn, Asp, Gln, or Glu residues, all of which could serve a similar hydrogen bonding role, is found in nearly 40% of NlpC/P60 proteins (41). His/Asn exchange is also observed in HRASLS1. Because bacterial expression of human recombinant HRASLS1 did not produce the active form of this protein, it is not clear how this substitution affects the enzymatic properties HRASLS1 relative to other members of this family. But the recent characterization of HRASLS1 expressed in COS-1 cells provides evidence for PLA1,2 and acyltransferase activities similar to those of other HRASLS proteins (38).
The high resolution structure of HRASLS2 provides important details about the reactivity and dynamics of the active site. Two alternative conformations of the catalytic Cys-113 side chain were clearly distinguished in the electron density map (Fig. 8D). Although the distances between Sγ of Cys-113 and Nδ1 of His-23 were similar, these two rotamers differed in orientation of the thiol group with respect to the imidazole ring. The interaction between hydrogen donor and acceptor is critical for deprotonation of the Cys sulfhydryl group and subsequent nucleophilic attack on the acyl donor. Because hydrogen bond properties depend on the angle of such bond formation, the existence of two conformations could imply differences in their reactivity (57). The more preferable geometry and presumably stronger interaction with Nδ1 characteristic of one of these Cys-113 positions suggest it is the active conformation for this residue. Oxidation of Cys-113 to sulfenic acid that occurs only in one of the two orientations seems to support this hypothesis. Susceptibility to spontaneous Cys residue oxidation strongly depends on the ionization state, as thiolates are far more nucleophilic than their protonated counterparts (58). Thus, a highly selective orientation-dependent modification of Cys-113 should specify the most reactive and catalytically competent conformation. Mapping interactions of thioester or anionic tetrahedral intermediates will require their trapping within the active site. However, the architecture of the cleft and especially the close vicinity of the sulfenic acid rotamer to the amide bonds of Glu-114 and Trp-24 backbone could indicate the location of an oxyanion hole involved in stabilizing the tetrahedral anion. The nonoxidized Cys-113 conformation faces away from any potential interactions of this type, including the side chain of Tyr-21 that previously was proposed to stabilize the oxyanion intermediate in NlpC/P60 Spr lipoprotein (41). There are no structural factors that would favor or stabilize any particular Cys-113 rotamer. Thus, the interchangeable character of these two states is probably responsible for presence of active (acylated) and inactive (not acylated) protein populations detected by MS upon incubation of HRASLS2 and other HRASLS proteins with excess phospholipids (Fig. 2 and supplemental Fig. S1). Moreover, the high propensity of Cys-113 to undergo oxidation explains the need of reducing agents such as DTT for full activity of these proteins.
Although the structural and catalytic similarities between acyltransferases and hydrolases have been recognized, the molecular basis for their different reactivity has been a long standing mystery. Hydrolysis is a common side reaction of acyltransferases, especially in the absence of a specific acceptor (59, 60). Comparison of x-ray crystal structures of acyltransferases, including those with bound substrate analogues, do not reveal a common molecular mechanism responsible for discrimination between acyltransferase and hydrolytic activity. Most structures indicate specific interactions between the alcohol and acyltransferase that favor acyl transfer over hydrolysis. Recent studies by Jiang et al. (61) shed new light on this problem by showing that orientation of the active site oxyanion loop determines the dominant type of enzymatic activity. In esterases, a main chain carbonyl oxygen that is part of the oxyanion loop acts as a base via a bridging water molecule to activate the attacking water molecule. But in acyltransferases, the NH of a main chain acts as an acid via a bridging water molecule to deactivate the attacking water molecule. Thus, discrimination between the hydrolytic and acyltransferase activity is governed at the water activation level. Furthermore, deactivated water is susceptible to displacement by larger alcohol or amine acyl acceptors, which can then be activated directly by the side chain of a His residue for the acyl transfer reaction.
HRASLS2 and HRASLS3 reveal greater selectivity toward acyl acceptor than the acyl donor. They use sn-1 lysophospholipids as well as PE but neither ethanolamine nor glycerophosphoethanolamine. Although one cannot exclude preferential binding of certain substrates, activation of a water molecule could also play a key role in the preferential acyltransferase activity of these enzymes. Direct experimental approaches to prove this theory require both crystallization of HRASLS2 or HRASLS3 with a sulfonate analog of a tetrahedral intermediate that mimics the attack of water on the thioester and identification of an oxyanion loop within the active site. Alternatively, determination of the HRASLS4 structure, an enzyme that exhibits predominant hydrolase activity, together with its direct comparison of its active with other HRASLS proteins could provide direct evidence for a role of the oxyanion loop orientation in enzymatic activity.
The crystal structures of HRASLS2 and HRASLS3 suggest a preferential orientation of these molecules with respect to the phospholipid membrane (Fig. 9 and supplemental Fig. S4). The active site location in a shallow groove at the periphery of these proteins is characteristic of many enzymes such as phospholipases A2, C, and D acting at membrane-water interface (62–64). Docking of the active site at the polar lipid phosphate region of the bilayer determines substrate accessibility without thermodynamically unfavorable extraction of lipids from the membrane. The primary structural element that allows membrane insertion of HRASLS enzymes (with the possible exception of HRASLS5) is a C-terminal transmembrane helix. However, secondary protein-lipid interactions are probably critical for proper active site membrane embedding and could affect substrate specificity. We identified two regions potentially involved in hydrophobic interactions. Residues 41–57 that constitute a disordered loop between β3 and β4 in the crystal structure have a propensity to adopt an α-helical conformation with well defined hydrophobic and polar interfaces (Fig. 9, C–E). Interaction of this region with a lipid bilayer could promote its re-folding as documented for many other amphiphilic helices (65). Analogous sequences are present in other HRASLS proteins. This region is especially interesting in LRAT where an 11-residue Leu-, Ile-, and Val-rich extension confers significantly more hydrophobicity (Fig. 1). Unlike LRAT, soluble forms of HRASLS proteins tested in this study did not spontaneously bind to asolectin liposomes (data not shown). Thus, the enlarged hydrophobic patch of LRAT could be solely responsible for binding LRAT to the membranes in the absence of its N- and C-terminal hydrophobic segments. Such an interaction is in agreement with the lipid interface model proposed for LRAT based on physicochemical measurements and could explain the low aqueous solubility of this truncated protein (66, 67).
In summary, this study provides detailed insights into the enzymology, structural features, active site architecture, and membrane topology of a physiologically important class of proteins that catalyze reactions at the phospholipid membrane-water interface. Key roles for HRASLS3 and LRAT in the regulation of triglyceride and vitamin A homeostasis position these proteins as potential drug targets for treatment of metabolic syndromes. Thus, these results should facilitate the rational design of small molecules that alter the enzymatic functions of these proteins.
Acknowledgments
We thank Dr. Xiongying Tu for initial x-ray diffraction data collection on HRASLS3 crystals, Dr. Tivadar Orban for help in modeling missing regions in HRASLS3, and Drs. Leslie Webster, Jr., and Michael Maguire for help in preparation of this manuscript. Use of the Advanced Photon Source, an Office of Science User Facility operated for the United States Department of Energy Office of Science by the Argonne National Laboratory, was supported by the United States Department of Energy under Contract DE-AC02-06CH11357. Data for this study were obtained at beamline X29 of the National Synchrotron Light Source. Its financial support is derived principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy and by National Institutes of Health Grant P41RR012408 from NCRR Grant P41GM103473 from NIGMS.
This work was supported, in whole or in part, by National Institutes of Health Grants EY008061 (to K. P., M. G., and A. E. S.), EY009339 (to K. P., M. G., P. D. K, and A. E. S.), DK079221 (to W. B.), and EY019718 (to D. T. L.). This work was based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines supported by National Institutes of Health Grants 5P41RR015301–10 from NCRR and Grant 8 P41 GM103403-10 from NIGMS.

This article contains supplemental Figs. S1–S4, Tables S1 and S2, and additional references.
The atomic coordinates and structure factors (codes 4DPZ and 4DOT) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HRASLS
- HRAS-like tumor suppressor
- LRAT
- lecithin:retinol acyltransferase
- N-acyl-PE
- N-acylphosphatidylethanolamine
- NBD
- 6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl group
- PEG
- polyethylene glycol
- PLA
- phospholipase A
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids. Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fernandis A. Z., Wenk M. R. (2007) Membrane lipids as signaling molecules. Curr. Opin. Lipidol. 18, 121–128 [DOI] [PubMed] [Google Scholar]
- 3. Hannun Y. A., Obeid L. M. (2008) Principles of bioactive lipid signaling. Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 4. Anantharaman V., Aravind L. (2003) Evolutionary history, structural features, and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4, R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruiz A., Winston A., Lim Y. H., Gilbert B. A., Rando R. R., Bok D. (1999) Molecular and biochemical characterization of lecithin-retinol acyltransferase. J. Biol. Chem. 274, 3834–3841 [DOI] [PubMed] [Google Scholar]
- 6. Kiser P. D., Golczak M., Maeda A., Palczewski K. (2012) Key enzymes of the retinoid (visual) cycle in vertebrate retina. Biochim. Biophys. Acta 1821, 137–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Batten M. L., Imanishi Y., Maeda T., Tu D. C., Moise A. R., Bronson D., Possin D., Van Gelder R. N., Baehr W., Palczewski K. (2004) Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J. Biol. Chem. 279, 10422–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Golczak M., Imanishi Y., Kuksa V., Maeda T., Kubota R., Palczewski K. (2005) Lecithin:retinol acyltransferase is responsible for amidation of retinylamine, a potent inhibitor of the retinoid cycle. J. Biol. Chem. 280, 42263–42273 [DOI] [PubMed] [Google Scholar]
- 9. Jaworski K., Ahmadian M., Duncan R. E., Sarkadi-Nagy E., Varady K. A., Hellerstein M. K., Lee H. Y., Samuel V. T., Shulman G. I., Kim K. H., de Val S., Kang C., Sul H. S. (2009) AdPLA ablation increases lipolysis and prevents obesity induced by high fat feeding or leptin deficiency. Nat. Med. 15, 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duncan R. E., Sarkadi-Nagy E., Jaworski K., Ahmadian M., Sul H. S. (2008) Identification and functional characterization of adipose-specific phospholipase A2 (AdPLA). J. Biol. Chem. 283, 25428–25436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uyama T., Morishita J., Jin X. H., Okamoto Y., Tsuboi K., Ueda N. (2009) The tumor suppressor gene H-Rev107 functions as a novel Ca2+-independent cytosolic phospholipase A1/2 of the thiol hydrolase type. J. Lipid Res. 50, 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uyama T., Ichi I., Kono N., Inoue A., Tsuboi K., Jin X. H., Araki N., Aoki J., Arai H., Ueda N. (2012) Regulation of peroxisomal lipid metabolism by catalytic activity of tumor suppressor H-rev107. J. Biol. Chem. 287, 2706–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uyama T., Jin X. H., Tsuboi K., Tonai T., Ueda N. (2009) Characterization of the human tumor suppressors TIG3 and HRASLS2 as phospholipid-metabolizing enzymes. Biochim. Biophys. Acta 1791, 1114–1124 [DOI] [PubMed] [Google Scholar]
- 14. Jin X. H., Okamoto Y., Morishita J., Tsuboi K., Tonai T., Ueda N. (2007) Discovery and characterization of a Ca2+-independent phosphatidylethanolamine N-acyltransferase generating the anandamide precursor and its congeners. J. Biol. Chem. 282, 3614–3623 [DOI] [PubMed] [Google Scholar]
- 15. Han B. G., Cho J. W., Cho Y. D., Kim S. Y., Yoon H. J., Song H. K., Cheong H. K., Jeon Y. H., Lee D. K., Lee S., Lee B. I. (2010) Expression, purification, and biochemical characterization of the N-terminal regions of human TIG3 and HRASLS3 proteins. Protein Expr. Purif. 71, 103–107 [DOI] [PubMed] [Google Scholar]
- 16. Burger A., Berendes R., Voges D., Huber R., Demange P. (1993) A rapid and efficient purification method for recombinant annexin V for biophysical studies. FEBS Lett. 329, 25–28 [DOI] [PubMed] [Google Scholar]
- 17. Golczak M., Palczewski K. (2010) An acyl-covalent enzyme intermediate of lecithin:retinol acyltransferase. J. Biol. Chem. 285, 29217–29222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu H., Freitas M. A. (2007) A mass accuracy sensitive probability-based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics 8, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dauter Z., Dauter M., Rajashankar K. R. (2000) Novel approach to phasing proteins. Derivatization by short cryo-soaking with halides. Acta Crystallogr. D Biol. Crystallogr. 56, 232–237 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kabsch W. (2010) Integration, scaling, space-group assignment, and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23. Collaborative Computational Project Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 24. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 25. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 26. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Generation, representation, and flow of phase information in structure determination. Recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 27. Abrahams J. P., Leslie A. G. (1996) Methods used in the structure determination of bovine mitochondrial F1-ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 28. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D. (2006) Toward the structural genomics of complexes. Crystal structure of a PE-PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 31. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 32. Afonine P. V., Grosse-Kunstleve R. W., Chen V. B., Headd J. J., Moriarty N. W., Richardson J. S., Richardson D. C., Urzhumtsev A., Zwart P. H., Adams P. D. (2010) phenix.model_vs_data. A high level tool for the calculation of crystallographic model and data statistics. J. Appl. Crystallogr. 43, 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 37. Chahinian H., Sarda L. (2009) Distinction between esterases and lipases. Comparative biochemical properties of sequence-related carboxylesterases. Protein Pept. Lett. 16, 1149–1161 [DOI] [PubMed] [Google Scholar]
- 38. Shinohara N., Uyama T., Jin X. H., Tsuboi K., Tonai T., Houchi H., Ueda N. (2011) Enzymological analysis of the tumor suppressor A-C1 reveals a novel group of phospholipid-metabolizing enzymes. J. Lipid Res. 52, 1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Plückthun A., Dennis E. A. (1982) Acyl and phosphoryl migration in lysophospholipids. Importance in phospholipid synthesis and phospholipase specificity. Biochemistry 21, 1743–1750 [DOI] [PubMed] [Google Scholar]
- 40. Ren X., Lin J., Jin C., Xia B. (2010) Solution structure of the N-terminal catalytic domain of human H-REV107. A novel circular permutated NlpC/P60 domain. FEBS Lett. 584, 4222–4226 [DOI] [PubMed] [Google Scholar]
- 41. Aramini J. M., Rossi P., Huang Y. J., Zhao L., Jiang M., Maglaqui M., Xiao R., Locke J., Nair R., Rost B., Acton T. B., Inouye M., Montelione G. T. (2008) Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli. Structural evidence for a novel cysteine peptidase catalytic triad. Biochemistry 47, 9715–9717 [DOI] [PubMed] [Google Scholar]
- 42. Claiborne A., Yeh J. I., Mallett T. C., Luba J., Crane E. J., 3rd, Charrier V., Parsonage D. (1999) Protein-sulfenic acids. Diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry 38, 15407–15416 [DOI] [PubMed] [Google Scholar]
- 43. Moise A. R., Golczak M., Imanishi Y., Palczewski K. (2007) Topology and membrane association of lecithin:retinol acyltransferase. J. Biol. Chem. 282, 2081–2090 [DOI] [PubMed] [Google Scholar]
- 44. Mateja A., Szlachcic A., Downing M. E., Dobosz M., Mariappan M., Hegde R. S., Keenan R. J. (2009) The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461, 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. White S. H., Wimley W. C. (1999) Membrane protein folding and stability. Physical principles. Annu. Rev. Biophys. Biomol. Struct. 28, 319–365 [DOI] [PubMed] [Google Scholar]
- 46. Lomize A. L., Pogozheva I. D., Mosberg H. I. (2011) Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in membranes. J. Chem. Inf. Model. 51, 930–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. MacDonald J. I., Sprecher H. (1991) Phospholipid fatty acid remodeling in mammalian cells. Biochim. Biophys. Acta 1084, 105–121 [DOI] [PubMed] [Google Scholar]
- 48. Wolf G. (2009) Adipose-specific phospholipase as regulator of adiposity. Nutr. Rev. 67, 551–554 [DOI] [PubMed] [Google Scholar]
- 49. Ezzili C., Otrubova K., Boger D. L. (2010) Fatty acid amide signaling molecules. Bioorg. Med. Chem. Lett. 20, 5959–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Farrell E. K., Merkler D. J. (2008) Biosynthesis, degradation, and pharmacological importance of the fatty acid amides. Drug Discov. Today 13, 558–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Matias I., Petrosino S., Racioppi A., Capasso R., Izzo A. A., Di Marzo V. (2008) Dysregulation of peripheral endocannabinoid levels in hyperglycemia and obesity. Effect of high fat diets. Mol. Cell. Endocrinol. 286, S66–78 [DOI] [PubMed] [Google Scholar]
- 52. Hoareau L., Buyse M., Festy F., Ravanan P., Gonthier M. P., Matias I., Petrosino S., Tallet F., d'Hellencourt C. L., Cesari M., Di Marzo V., Roche R. (2009) Anti-inflammatory effect of palmitoylethanolamide on human adipocytes. Obesity 17, 431–438 [DOI] [PubMed] [Google Scholar]
- 53. Fu J., Oveisi F., Gaetani S., Lin E., Piomelli D. (2005) Oleoylethanolamide, an endogenous PPAR-α agonist, lowers body weight and hyperlipidemia in obese rats. Neuropharmacology 48, 1147–1153 [DOI] [PubMed] [Google Scholar]
- 54. Verhoeckx K. C., Voortman T., Balvers M. G., Hendriks H. F., Wortelboer H. M., Witkamp R. F. (2011) Presence, formation, and putative biological activities of N-acyl serotonins, a novel class of fatty acid-derived mediators, in the intestinal tract. Biochim. Biophys. Acta 1811, 578–586 [DOI] [PubMed] [Google Scholar]
- 55. Matias I., Gonthier M. P., Petrosino S., Docimo L., Capasso R., Hoareau L., Monteleone P., Roche R., Izzo A. A., Di Marzo V. (2007) Role and regulation of acylethanolamides in energy balance. Focus on adipocytes and beta-cells. Br. J. Pharmacol. 152, 676–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang W., Jia J., Edwards P., Dehesh K., Schneider G., Lindqvist Y. (1998) Crystal structure of β-ketoacyl-acyl carrier protein synthase II from E. coli reveals the molecular architecture of condensing enzymes. EMBO J. 17, 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Torshin I. Y., Weber I. T., Harrison R. W. (2002) Geometric criteria of hydrogen bonds in proteins and identification of “bifurcated” hydrogen bonds. Protein Eng. 15, 359–363 [DOI] [PubMed] [Google Scholar]
- 58. Poole L. B., Karplus P. A., Claiborne A. (2004) Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol. 44, 325–347 [DOI] [PubMed] [Google Scholar]
- 59. Liu M., Subbaiah P. V. (1994) Hydrolysis and transesterification of platelet-activating factor by lecithin-cholesterol acyltransferase. Proc. Natl. Acad. Sci. U.S.A. 91, 6035–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fournand D., Arnaud A. (2001) Aliphatic and enantioselective amidases. From hydrolysis to acyl transfer activity. J. Appl. Microbiol. 91, 381–393 [DOI] [PubMed] [Google Scholar]
- 61. Jiang Y., Morley K. L., Schrag J. D., Kazlauskas R. J. (2011) Different active-site loop orientation in serine hydrolases versus acyltransferases. ChemBioChem 12, 768–776 [DOI] [PubMed] [Google Scholar]
- 62. Scott D. L., White S. P., Otwinowski Z., Yuan W., Gelb M. H., Sigler P. B. (1990) Interfacial catalysis. The mechanism of phospholipase A2. Science 250, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. de Giuseppe P. O., Ullah A., Silva D. T., Gremski L. H., Wille A. C., Chaves Moreira D., Ribeiro A. S., Chaim O. M., Murakami M. T., Veiga S. S., Arni R. K. (2011) Structure of a novel class II phospholipase D. Catalytic cleft is modified by a disulfide bridge. Biochem. Biophys. Res. Commun. 409, 622–627 [DOI] [PubMed] [Google Scholar]
- 64. Essen L. O., Perisic O., Katan M., Wu Y., Roberts M. F., Williams R. L. (1997) Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C. Biochemistry 36, 1704–1718 [DOI] [PubMed] [Google Scholar]
- 65. Ladokhin A. S., White S. H. (2004) Interfacial folding and membrane insertion of a designed helical peptide. Biochemistry 43, 5782–5791 [DOI] [PubMed] [Google Scholar]
- 66. Bussieres S., Cantin L., Desbat B., Salesse C. (2012) Binding of a truncated form of lecithin:retinol acyltransferase and Its N- and C-terminal peptides to lipid monolayers. Langmuir, 28, 3516–3523 [DOI] [PubMed] [Google Scholar]
- 67. Bussières S., Buffeteau T., Desbat B., Breton R., Salesse C. (2008) Secondary structure of a truncated form of lecithin-retinol acyltransferase in solution and evidence for its binding and hydrolytic action in monolayers. Biochim. Biophys. Acta 1778, 1324–1334 [DOI] [PubMed] [Google Scholar]
- 68. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of Nanosystems. Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2007) Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. Chapter 2, Unit 2.9 [DOI] [PubMed] [Google Scholar]