Background: The BRC repeat is essential for BRCA2 to bind RAD51 and promote homologous recombination.
Results: A BRC repeat variant is essential for RECQL5 to bind RAD51 and suppress homologous recombination.
Conclusion: The BRC repeat can be utilized to either promote or suppress homologous recombination.
Significance: Discovery of multiple functions of the BRC repeat is important for understanding regulation of homologous recombination.
Keywords: DNA Damage Response, DNA Recombination, DNA Repair, DNA Replication, DNA-Protein Interaction, BRCA2, BRC Repeat, Homologous Recombination, RAD51, RECQL5, Srs2
Abstract
The BRC repeat is a structural motif in the tumor suppressor BRCA2 (breast cancer type 2 susceptibility protein), which promotes homologous recombination (HR) by regulating RAD51 recombinase activity. To date, the BRC repeat has not been observed in other proteins, so that its role in HR is inferred only in the context of BRCA2. Here, we identified a BRC repeat variant, named BRCv, in the RECQL5 helicase, which possesses anti-recombinase activity in vitro and suppresses HR and promotes cellular resistance to camptothecin-induced replication stress in vivo. RECQL5-BRCv interacted with RAD51 through two conserved motifs similar to those in the BRCA2-BRC repeat. Mutations of either motif compromised functions of RECQL5, including association with RAD51, inhibition of RAD51-mediated D-loop formation, suppression of sister chromatid exchange, and resistance to camptothecin-induced replication stress. Potential BRCvs were also found in other HR regulatory proteins, including Srs2 and Sgs1, which possess anti-recombinase activities similar to that of RECQL5. A point mutation in the predicted Srs2-BRCv disrupted the ability of the protein to bind RAD51 and to inhibit D-loop formation. Thus, BRC is a common RAD51 interaction module that can be utilized by different proteins to either promote HR, as in the case of BRCA2, or to suppress HR, as in RECQL5.
Introduction
Homologous recombination (HR)3 is critical for the error-free repair of chromosomal lesions, such as DNA double strand breaks, and also for the recovery of damaged replication forks (1, 2). During meiosis, HR generates crossovers among homologous chromosomes to ensure their proper segregation at the first meiotic division (1, 3). Thus, HR is indispensable for the maintenance of genome integrity and meiotic chromosome segregation (4). In fact, defects in HR can increase cancer and failed meiosis (4). On the other hand, untimely and inappropriate HR can cause gross chromosome rearrangements, including translocations, deletions, and inversions, with potentially mutagenic or oncogenic consequences (2, 4). Therefore, HR is tightly controlled in cells by a variety of pro- and anti-recombinogenic regulatory mechanisms.
Human BRCA2 and its orthologs are an important positive regulatory element for HR (5–7). BRCA2 directly interacts with RAD51 recombinase and recruits it to double strand breaks. BRCA2 promotes the formation of the RAD51-ssDNA nucleoprotein filament that pairs with and invades a homologous DNA duplex to initiate homologous DNA repair. Moreover, BRCA2 modulates the DNA binding selectivity of RAD51 to stimulate strand exchange (8, 9). The primary RAD51 interaction domain in BRCA2 and its orthologs consists of a varying number of the BRC repeat, a module of ∼35 amino acid residues (10, 11). The BRC repeat directly associates with RAD51 or RAD51-DNA filaments, regulates the DNA binding activity of RAD51, enhances the exchange of the ssDNA-binding factor replication protein A (RPA) by RAD51 on ssDNA, and promotes RAD51-dependent homologous DNA pairing (8, 9, 12, 13). Structural and functional analyses have revealed that the BRC repeat harbors two motifs, referred to as motif 1 and motif 2, which are separated by a linker sequence (11, 14). Motif 1 binds the oligomerization interface of RAD51 to regulate RAD51 filament assembly, whereas motif 2 binds a separate region of RAD51, and a functional BRC repeat requires the two motifs working in tandem pairs (14).
To date, the BRC repeat has not been discovered in any other proteins, but we report here its presence in RECQL5, where it also regulates HR. RECQL5 is one of five RecQ-like DNA helicases in mammalian cells (15, 16). Genetic studies in mouse and chicken DT40 cells have implicated RECQL5 in the suppression of HR, including inhibition of sister chromatid exchange (SCE) (17, 18), in the preservation of genome integrity upon genotoxic stress (19) and in cancer avoidance (20). RECQL5 directly interacts with RAD51 and inhibits RAD51-mediated homologous DNA pairing by dismantling the RAD51 presynaptic filament assembled on ssDNA (20, 21). A previous study has mapped a RAD51-interacting region between residues 654 and 725 of RECQL5 and shown that several residues within this region are important for binding and regulation of RAD51 recombinase in vitro and for HR regulation in vivo (21). However, the region was not defined as a recognizable domain. Here, we show that the RAD51 interaction domain in RECQL5 consists of two motifs with a strong resemblance to the conserved motifs found in the BRCA2-BRC repeats. We have named this RECQL5 domain BRCv (BRC variant) because it also harbors notable differences from the BRC repeats in BRCA2. Importantly, we demonstrate that both motifs of BRCv are required for RECQL5 to interact with RAD51, to inhibit RAD51-mediated homologous DNA pairing, to suppress SCE, and to tolerate replication stress. Sequence-based comparisons reveal that potential BRCvs are present in other HR regulatory proteins, suggesting that the BRC repeat and its variants may be employed by many proteins in the regulation of HR.
EXPERIMENTAL PROCEDURES
Cell Culture
Chicken DT40 cell lines were maintained in RPMI medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, 1% chicken serum, 1.5% penicillin-streptomycin (Invitrogen), and 10 mm HEPES (pH 7.9). HEK-293 cells were grown in DMEM containing 10% fetal bovine serum.
Antibodies
RAD51 (H-92) and RAD51 (ab63801) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and Abcam, respectively. Anti-FLAG M2 and RNA polymerase II (Pol II) (ARNA-3) monoclonal antibodies were obtained from Sigma and Fitzgerald, respectively.
Protein Purification
Wild-type and mutant forms of human RECQL5 were expressed in Escherichia coli and purified as described previously (20). Human RAD51 K133R, RPA, and HOP2-MND1 complex were purified according to previously published protocols (22, 23). Wild type and mutant forms of Srs2 were expressed in E. coli and purified as described (24, 25). Yeast Rad51 protein was expressed in yeast cells and purified as described (26).
Immunoprecipitation
RecQL5-associated complexes were isolated from total cell lysate of HEK-293 cells transiently expressing FLAG-tagged RECQL5 (FRECQL5). Cell pellets were suspended in three volumes of lysis buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 5 mm MgCl2, and 0.5% Nonidet P-40) containing 1 mm PMSF, 1 mm DTT, and protease inhibitor mixture. FLAG immunoprecipitation (IP) was done according to the manufacturer's protocol (Sigma). In brief, cell lysates were diluted three times with lysis buffer and incubated with anti-FLAG M2-agarose affinity gel for at least 12 h at 4 °C. The resin was washed three times for 10 min each with lysis buffer and then treated with the FLAG peptide to elute proteins. The eluate was subjected to immunoblotting.
Affinity Pull-down
His9-tagged wild type or mutant Srs2 was incubated with yeast Rad51 in 30 μl of buffer (25 mm Tris-HCl, pH 7.5, 0.01% Igepal, 1 mm 2-mercaptoethanol, and 10 mm imidazole) containing 100 mm KCl for 30 min at 4 °C and then mixed with 10 μl of Ni-NTA-agarose (Qiagen) for 2 h at 4 °C to capture the tagged Srs2 and Rad51. The resin was washed three times with 50 μl of the same buffer and then treated with 20 μl of 2% SDS to elute proteins. The supernatant, last wash, and SDS eluate (10 μl each) were analyzed by SDS-PAGE.
ATPase Assay
The ATPase assay was carried out as described (20). Briefly, RECQL5 or one of the indicated RECQL5 mutants (25 nm) was incubated with φX174 ssDNA (75 μm nucleotides) and 1.5 mm [γ-32P]ATP for the indicated times. For wild type and mutant Srs2, 15 nm proteins was used. Analysis was done by thin layer chromatography (20).
D-loop Assay
This was conducted at 37 °C as described (20), by incubating the RAD51-K133R protein (1 μm) with radioactively labeled 90-mer oligonucleotide D1 (3 μm nucleotides) for 5 min, followed by the incorporation of RPA (135 nm) together with RECQL5 or the indicated RECQL5 mutant (25 nm) and a 5-min incubation. Then HOP2-MND1 (300 nm) and pBluescript replicative form I DNA (50 μm base pairs) were added to complete the reaction. After 10 min of incubation, the reaction mixtures were deproteinized and analyzed, as described (20). The D-loop assay for wild type and mutant forms of Srs2 was conducted according to Ref. 24.
Complementation Analyses
Complementation analyses were done as described previously (27) with some modifications. In brief, FLAG-tagged RECQL5 constructs were generated by using a QuikChange multisite-directed mutagenesis kit and transfected into BLM−/−/RecQL5−/− chicken DT40 cells by using nucleofection solution. 1 × 106 cells in pellets were suspended into 100 μl of nucleofection solution, and 2–5 μg of DNA was then added to the suspended solutions. Due to the low transfection efficiency, each construct was digested with XhoI, and the linearized plasmids were used for transfection. Transfected solutions were incubated for 24 h in a 6-well plate with 2.0 ml of medium. Stable clones were isolated from the primary transfectant pools using limiting dilution into 96-well plates and selection for resistance to zeocin.
SCE Assay
The SCE assay followed the published protocol (27, 28) with minor modifications. In brief, cell cultures were grown through two cell cycles in the presence of 10 μm 5-bromodeoxyuridine (BrdU). Colcemid was added to a final concentration of 0.1 μg/ml to accumulate mitotic cells 2 h prior to the harvesting of cells. Harvested cells were then treated with 75 mm KCl for 20 min at room temperature and then fixed with 3:1 (v/v) methanol-glacial acetic acid. The fixed cell suspension was dropped onto a glass slide and air-dried. The cells on the slides were incubated with 10 μg of Hoechst 33258/ml in 50 mm phosphate buffer (pH 6.8) for 20 min and rinsed with MacIlvaine solution. The cells were exposed to UV for 60 min and then incubated in 2× SSC at room temperature for 60 min. The cells were finally stained with 3% Giemsa solution for 20–40 min and examined under a light microscope.
Camptothecin (CPT) Sensitivity Assay
This assay was done as described (27).
Homology Modeling
The homology model was generated as described previously (27, 29, 30).
RESULTS
RECQL5 Interacts with RAD51 through a Conserved Region
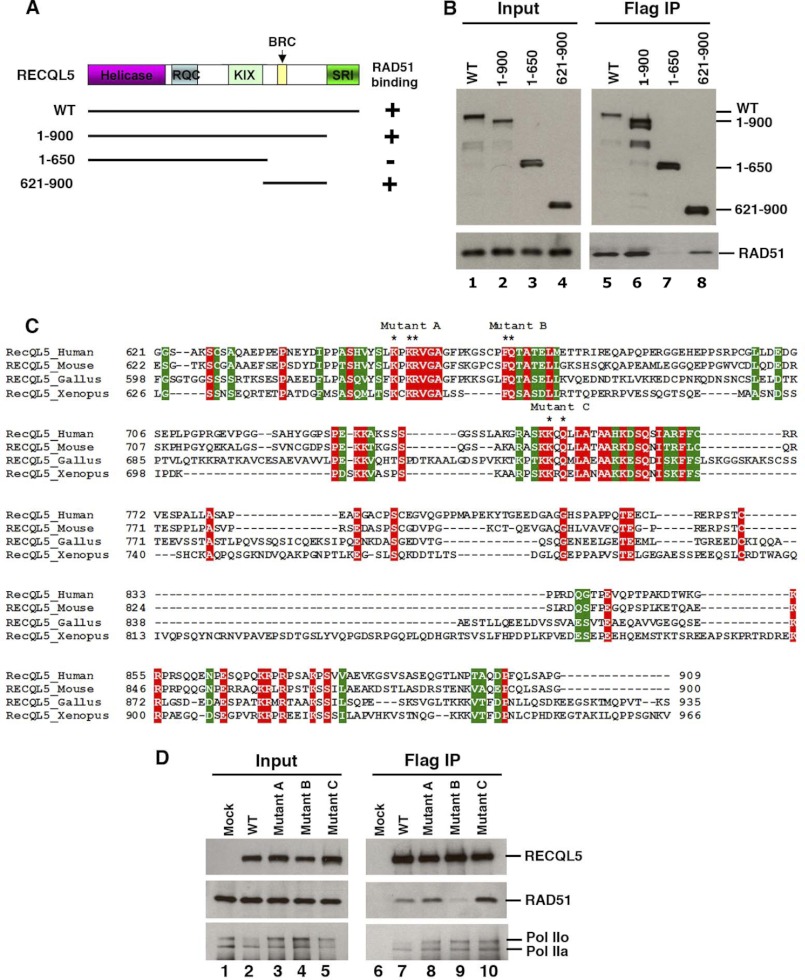
A RAD51-interacting region has been previously identified in RECQL5 by an in vitro assay using a series of RECQL5 fragments (21). We confirmed and extended the previous analysis in vivo by expressing a series of FLAG-tagged RECQL5 fragments (Fig. 1A) in HEK-293 cells and performing immunoprecipitation-coupled Western blot (IP-Western) to identify the RAD51 interaction domain. This strategy has been previously used to identify the KIX and SRI domains as the Pol II interaction domains of RECQL5 (27). We found that the region (residues 621–900) between the KIX and SRI domains is both necessary and sufficient for RAD51 association (Fig. 1, A and B, lane 8). Because RAD51 is highly conserved in evolution, it seemed likely that its binding residues in RECQL5 should also be well conserved. Therefore, we mutagenized several conserved residues within this region (Fig. 1C) and found that mutant B (F666A/Q667A) substantially reduced RAD51 association, whereas mutant A (K651A/K653A/R654A) and mutant C (K750A/Q752A) retained normal association (Fig. 1, C and D). Thus, the region containing Phe-666 and Gln-667 is required for RAD51 association, in agreement with the previous in vitro data indicating that Phe-666 and residues between positions 652 and 674 are required for RAD51 interaction (21). We noted that although mutant B has reduced association with RECQL5, it retained normal association with Pol II (Fig. 1D), indicating that RECQL5-RAD51 association is independent of RECQL5-Pol II interactions.
FIGURE 1.
RECQL5 interacts with RAD51 independently of the KIX and SRI domains. A, schematic diagram of the wild type and truncated forms of FLAG-RECQL5. The N-terminal helicase, RECQ C-terminal (RQC), KIX, and SRI domains are shown along with the newly identified BRC repeat. The RAD51 interaction data are summarized on the right. B, immunoblotting shows the associations between different RECQL5 mutants and RAD51. HEK-293 cells were transfected with expression plasmid of full-length and various RECQL5 deletion constructs. The mixtures of FLAG IP were analyzed by immunoblotting with antibodies to FLAG and RAD51. C, sequence alignment of RECQL5 from different species. The residues mutated are marked with asterisks. All of the residues were converted to alanine. D, IP-Western shows that different RECQL5 point mutants coimmunoprecipitated with different amounts of RAD51 but with comparable amounts of Pol IIo and Pol IIa. The FLAG-tagged RECQL5 and its different point mutants were transfected into HEK-293 cells. Co-immunoprecipitation was performed using the FLAG antibody.
A BRC Repeat Variant Is Present in the RAD51 Interaction Domain of RECQL5
Sequence analysis revealed that the RAD51 interaction domain of RECQL5 contains two conserved motifs that are either identical or similar to those previously described in the BRC repeat of BRCA2 (Fig. 2A) (14). For example, the consensus sequence of motif 1 in RECQL5, FX(T/S)A, is identical to the corresponding sequence of the BRC repeat (FXXA) (Fig. 2A). In addition, the motif 2 sequence of RECQL5, LLDE, is also similar to the consensus sequence of motif 2 of the BRC repeat (hhXa, where “h” represents a hydrophobic and “a” represents an acidic residue). Based on these observations, we have named the RECQL5 region that encompasses the two conserved motifs as BRCv (BRC repeat variant).
FIGURE 2.
A single BRC repeat variant in RECQL5 is essential for RAD51 interaction. A, sequence alignment showing similarities between BRCvs of RECQL5 from different species and the eight BRC repeats of human BRCA2. Motif 1 and motif 2 are shown by red and green, respectively, with their consensus sequences shown at the top. The asterisks indicate a key conserved residue in the linker region that is also important for RAD51 association, but this residue is located at different positions between BRCv and BRC. A blue divider in the middle is used to highlight the difference in length of linker sequences between BRCA2-BRC and RECQL5-BRCv. The number of linker residues is shown in brackets. The species include human (HOMSA), mouse (MUSMU), chicken (GALGA), Xenopus (XENTR), fish (FUGRI), and Drosophila (DROME1 and DROME2). Notably, Drosophila RECQL5 has two potential BRCvs, whereas that from other species has only one. B, sequence alignment of putative BRCvs identified in four helicases (Srs2, Mph1, Sgs1, and Pif1) from different yeast species. The asterisks indicate the residues mutated to alanine. C, homology model of the RECQL5-BRC repeat. RAD51-BRC4 (BRCA2) (Protein Data Bank entry 1N0W) is shown overlaid with the homology model of RECQL5-BRCv. Cyan, RAD51; orange, BRCA2-BRC4; green, RECQL5-BRCv. Residues associated with motif 1 and motif 2 are shown in stick representations and labeled accordingly. D, IP-Western shows that RECQL5 with mutations in either motif 1 or 2 co-immunoprecipitated with reduced levels of RAD51, whereas that with mutations in both motifs almost completely lost RAD51 association. The numbers below lanes 1–5 indicate relative levels of RAD51 that co-immunoprecipitated with an indicated RECQL5 mutant, with the wild type protein being set as 1.0.
A Homology Model Predicts That BRCv Binds RAD51-like BRC
Structure predictions suggested that RECQL5-BRCv, like the BRCA2-BRC repeat, is largely devoid of regular secondary structure (31). A homology model for the RECQL5-BRCv repeat was generated in the Swissmodel server, using the RAD51-BRC4 structure (Protein Data Bank entry 1N0W) as the template (29). The model predicts that the conserved Phe and Ala side chains of the BRCv FXXA motif 1 may make contact with the hydrophobic pocket on the core catalytic domain of RAD51, part of the oligomerization interface, as seen in the RAD51-BRC4 structure (Fig. 2, A and C) (11). The LLDE motif 2 of BRCv is expected to bind a region of RAD51 distant from the oligomerization surface. The first two hydrophobic residues (Leu-700 and Leu-701) are predicted to occupy the same pocket occupied in RAD51 by Leu-1545 and Phe-1546 of BRCA2-BRC4, and the acidic residue in the fourth position (Glu-703) is expected to form a salt bridge with Arg-250 of RAD51, as seen for Glu-1548 of BRCA2-BRC4 (Fig. 2C) (11, 14). We note that the sequence that intervenes between the two BRCv motifs, predicted by the model to form a helix and a loop, is twice the length (25 amino acids) of that of the BRCA2-BRC repeat (12 amino acids) and therefore unique to the RECQL5-BRCv.
Both Motifs of the BRCv Are Needed for RECQL5-RAD51 Association
A previous study has shown that both motifs of BRCA2-BRC4 are required for RAD51 binding and BRC function (14). We investigated whether the two motifs of BRCv are similarly required for RECQL5 to associate with RAD51 by structure-guided mutagenesis, using the homology model described above. In motif 1, we substituted F666E or A669E to disrupt hydrophobic interactions at the predicted BRCv-RAD51 interface (Fig. 2C). In motif 2, we substituted L700E or L701E to disrupt the predicted hydrophobic interactions and E703A to abolish the predicted salt bridge with RAD51-R250 (Fig. 2C) (11, 14). We found that each mutation substantially reduced the amount of RAD51 that co-immunoprecipitated with RECQL5 (Fig. 2D, lanes 2–5). The results support the inference that both motifs are integral parts of a BRC repeat variant and indispensable for RAD51 interaction.
We noted that RECQL5 mutants in which only one motif of BRCv is mutated retained partial association with RAD51 (about 10–40%). We therefore generated several double mutants that simultaneously mutate both motifs and found that these mutants were more severely defective in RAD51 association (Fig. 2D, lanes 6–9). The data suggest that BRCv can bind RAD51 through either motif and that only when both motifs are inactivated is RAD51 binding abolished.
A Conserved Residue outside Motif 1 of BRCv Contributes to RAD51 Association
In addition to the two motifs, conserved residues next to motif 1 in BRCA2-BRC4 have also been shown to contribute to RAD51 association (11, 14). In particular, mutation of one of the hydrophobic residues, Val-1532, disrupts BRCA2-RAD51 association (32). We found that the corresponding residue of Val-1532 in BRCv is highly variable in RECQL5 from different species (Fig. 2A), indicating that this residue in BRCv is dispensable for RAD51 association. In contrast, BRCv has two highly conserved hydrophobic residues next to motif 1, Leu-672 and Met-673. Substitution of Leu-672 with Ala substantially reduced RECQL5-RAD51 association (supplemental Fig. S1), consistent with the notion that residues outside of motif 1 also contribute to RAD51 association.
Motif 1 of BRCv Is More Important than Motif 2 for Suppression of D-loop Formation
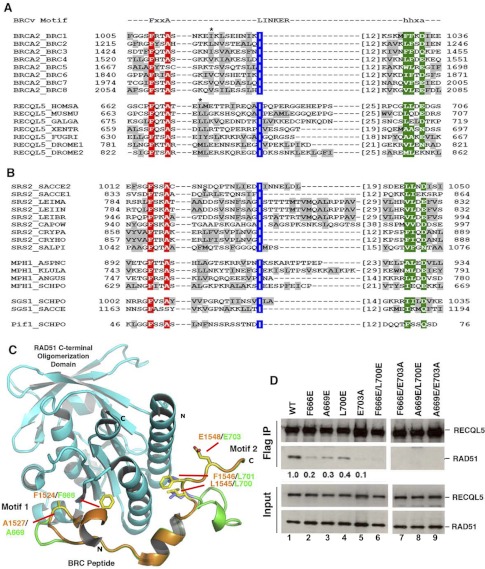
RECQL5 has been shown to possess anti-recombinase activity, evidenced by its ability to inhibit RAD51-mediated D-loop formation (20). This activity was later shown to be decreased in two mutants localized in motif 1: F666A, which substituted the conserved Phe in the FX(T/S)A consensus sequence, and Δ652–674, in which motif 1 was deleted (21). We studied whether one or both motifs of BRCv are required for the anti-recombinase activity of RECQL5, using the same D-loop assay (20). For this purpose, we expressed and purified several recombinant RECQL5 mutant proteins from E. coli that harbored mutations in either motif 1 (F666E) or 2 (L700E and E703E) or in both (F666E/L700E and F666E/E703A) (Fig. 3A). All three mutant proteins behaved like the wild type counterpart during purification and had wild-type levels of ATPase activity (Fig. 3B).
FIGURE 3.
RECQL5-RAD51 interaction mediated by BRCv motif 1 is critical for the suppression of D-loop formation. A, I, purification scheme for recombinant RECQL5 protein expressed in E. coli. II, the purified WT and mutant RECQL5 proteins were resolved by 10% SDS-PAGE and stained with Coomassie Blue. B, a graph shows ATPase activity of RECQL5 and its various point mutants. C, a schematic diagram shows the D-loop reaction scheme. D, an autoradiograph shows effects of RECQL5 and its mutants on RAD51-mediated D-loop formation. The radiolabeled single-stranded DNA and the D-loop are indicated on the left. The presence (+) or absence (−) of RECQL5 and its various mutants is shown at the top. E, quantification of the D-loop assay. Mean ± S.D. values (error bars) from at least three independent experiments are plotted.
We found that whereas the wild type RECQL5 protein inhibited D-loop formation efficiently, the motif 1 mutant was deficient in this activity (Fig. 3, C–E). The result is in agreement with published data (21). In contrast, the results showed that the two motif 2 mutants are less affected in their D-loop inhibitory activity. As expected, the two variants with mutations in both motifs were more deficient in D-loop inhibitory activity (Fig. 3, C–E). These results indicate that although both motifs of the BRCv are required for optimal association between RECQL5 and RAD51, motif 1 is more essential for anti-recombinase function, with motif 2 seeming to play only a minor role.
Both Motifs of BRCv Are Required by RECQL5 to Suppress SCE
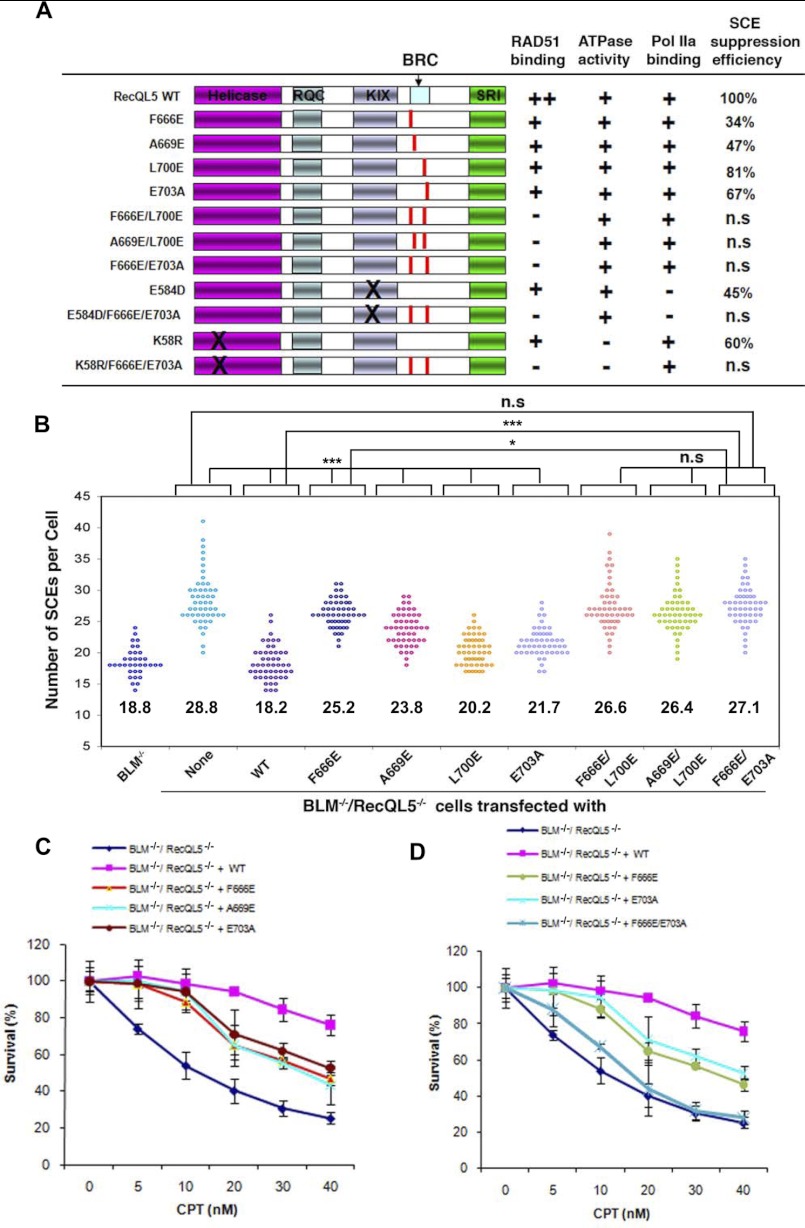
Using a trans-dominant negative assay in human HEK-293 cells, a small difference (about 15–20%) has been previously observed between a RECQL5 motif 1 mutant (F666A) and wild-type protein in the down-regulation of HR-dependent repair of double strand breaks (21). One caveat of this assay is that the exogenous protein needs to be overexpressed to a level significantly higher than that of the endogenous protein. To circumvent this requirement, we utilized RECQL5-knock-out chicken DT40 cells (18, 27) to investigate whether the conserved motifs of BRCv are needed for RECQL5 to promote genome stabilization in vivo. Because RECQL5 and BLM are functionally redundant in DT40 cells (RECQL5−/− single mutant cells lack obvious genome instability phenotypes), we studied functions of RECQL5 in RECQL5−/−/BLM−/− double mutant cells, which exhibit a higher SCE frequency and CPT sensitivity compared with BLM−/− single mutant cells (18, 27). We have previously shown that reintroduction of wild type RECQL5 protein into RECQL5−/−/BLM−/− cells restored the SCE frequency and CPT sensitivity to those of the BLM−/− cells, whereas reintroduction of RECQL5 mutants deficient in association with RNA polymerase II failed to fully rescue (27). Here we used the same RECQL5−/−/BLM−/− mutant cells to study whether various RECQL5-BRCv mutants (Fig. 4A) can correct these phenotypes.
FIGURE 4.
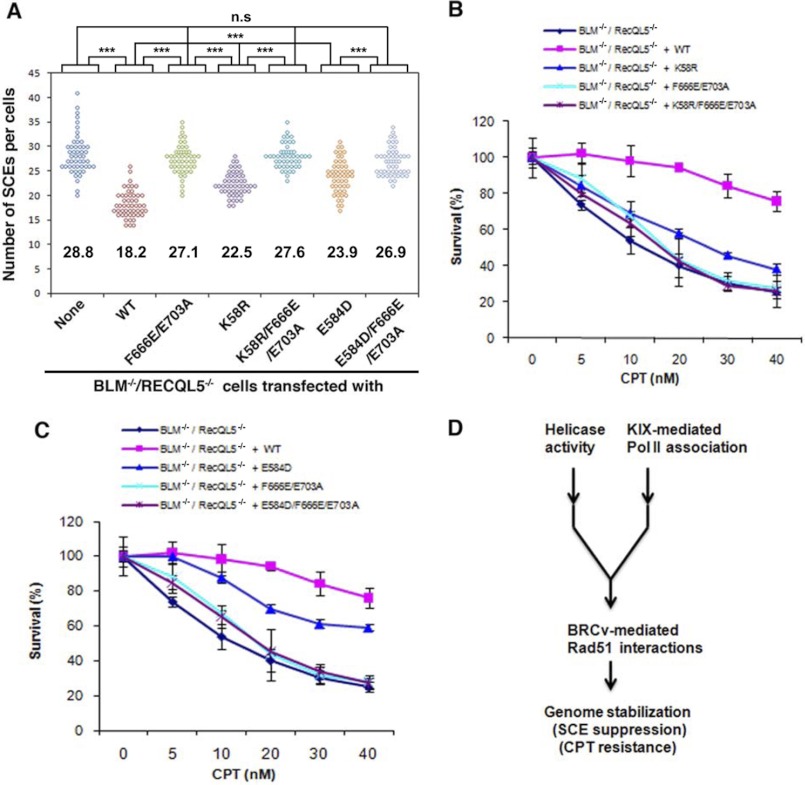
RECQL5 mutants deficient in RAD51 interaction are defective in suppression of SCE and resistance to CPT. A, summary of different human RECQL5 mutants and their corresponding activities in association with RAD51, ATPase activity, Pol IIa binding, and suppression of SCE. Different RAD51 association is indicated as follows. ++, normal; +, reduced; −, nearly completely lost. SCE values are shown as a percentage. The suppression efficiency for the wild type protein was set to 100%. Lower suppression efficiency correlates with stronger defect. n.s, statistically not significantly different from the BLM−/−/RecQL5−/− cells. B, scatter plot showing the SCE levels of BLM−/−/RecQL5−/− DT40 cells complemented by the wild type and various mutants of RECQL5 as shown in A. Each point represents the total number of SCEs in a single mitotic spread. Conditions that show significant differences of p value <0.05 or p value <0.0001 are shown as one or three asterisks, respectively. The mean number of SCEs/cell is shown for each mutant, which was determined by examining chromosome spreads of three independent groups of metaphase cells (50 cells per group). C and D, CPT sensitivity curves of BLM−/−/RecQL5−/− DT40 cells complemented with WT and different RECQL5 mutants, as indicated. Two different clones were tested for each mutant in order to ensure that the results are not due to clonal variation. Mean and S.D. (error bars) from three independent experiments are shown.
All mutants were expressed at levels comparable with that of the wild-type protein (supplemental Fig. S2). In addition, all four variants with single-motif mutations in BRCv associated with RAD51 at reduced levels by IP-Western, and those with mutations in both motifs associated with little or no RAD51 (supplemental Fig. S2, lanes 1–8). These results are in agreement with the finding that both motifs of BRCv are required for normal association with RAD51 in human cells (Fig. 2D, lanes 6–9).
We noticed that the levels of RAD51 that associate with three single mutants (F666E, A669E, and L700E) were somewhat higher than those observed in human cells (compare Supplemental Fig. S2, lanes 2–4, with Fig. 2D, lanes 2–4). This could be due to the use of human proteins in chicken cells, so that the observed RECQL5-RAD51 association could be somewhat different compared with that in human cells.
As reported before (18, 27), the SCE level of RecQL5−/−/BLM−/− cells was found to be significantly higher than that of BLM−/− cells (28.8 versus 18.8), and this enhanced level of SCE was completely suppressed by reintroduction of wild type RECQL5 protein (Fig. 4, A and B). In this assay, lower SCE suppression efficiency correlates with a stronger protein defect, with the suppression efficiency of the wild type protein being set as 100%. Reintroduction of either motif 1 or 2 mutants partially suppressed the enhanced SCE levels of RecQL5−/−/BLM−/− cells. Suppression efficiency ranged from 34–47% for the two motif 1 mutants (F666E and A669E) to 67–81% for the two motif 2 mutants (L700E and E703A). Transfection of the variants with mutations in both motifs (F666E/L700E, A669E/L700E, and F666E/E703A) resulted in SCE suppression efficiency that was lower than that seen with single mutants; the level of SCE in cells that expressed these compound mutants was statistically indistinguishable from that of RecQL5−/−/BLM−/− cells (Fig. 4, A and B). These results are in accord with the RAD51 binding data in Fig. 2, indicating that the RECQL5-RAD51 interactions mediated by both motifs of BRCv are required for optimal suppression of SCE. The observations that the two motif 1 mutants are more deficient in SCE suppression than the two motif 2 mutants correlate well with the D-loop inhibition data (Fig. 3) and suggest that motif 1 is more important than motif 2 for RECQL5 to regulate RAD51 activity during HR in vivo.
Both Motifs of BRCv Are Required by RECQL5 to Promote Cellular Resistance to CPT-induced Replication Stress
We studied whether RECQL5 mutants carrying mutations in different motifs of BRCv are defective in promoting resistance to CPT-induced replication stress. Consistent with our previous findings, introduction of human RECQL5 into RecQL5−/−/BLM−/− DT40 cells largely corrected the hypersensitivity of these cells to CPT (Fig. 4C) (27). Importantly, RECQL5 variants carrying mutations in either motif 1 (F666E and A669E) or motif 2 (L700E and E703A) were only partially active in rescuing the hypersensitivity to CPT, whereas variants carrying combined mutations in both motifs (F666E/E703A, F666E/L700E, A669E/L700E) were more deficient than single motif mutants in this assay (Fig. 4, C and D, and supplemental Fig. S3). These results correlate with the degree of RAD51 binding (Fig. 2D) by revealing that RECQL5 variants with single-motif mutations bound reduced amounts of RAD51 and partially supported CPT resistance, whereas variants with mutations in both motifs were more deficient in RAD51 binding and thus more defective in supporting CPT resistance. The results are also consistent with the SCE data and again suggest that both motifs of BRCv are required for the normal function of RECQL5 during genome stabilization.
BRCv Is Needed in Genome Maintenance Pathways Dependent on the Helicase Activity or the KIX Domain of RECQL5
We have previously shown that RECQL5 with mutations in either the helicase domain (K58R, which inactivates the helicase activity) or the KIX domain (E584D, which disrupts association with Pol IIa) were partially deficient in correcting the higher SCE level and CPT hypersensitivity of RecQL5−/−/BLM−/− cells, whereas RECQL5 with mutations in both domains was completely deficient, suggesting that these two domains are needed for the integrity of different genome maintenance pathways (27). We investigated whether the helicase- and KIX-dependent pathways require BRCv. For this purpose, we generated a mutant that compromises the BRCv and helicase function (K58R/F666E/E703A) and another that inactivates both the BRCv and KIX domain (E584D/F666E/E703A). IP-Western confirmed that these two mutants were as deficient in RAD51 association as the BRCv mutant (F666E/E703A). Importantly, neither the helicase nor the KIX domain mutation affected RAD51 binding (supplemental Fig. S2, lanes 9, 10, and 12).
We found that the SCE level and CPT sensitivity of RecQL5−/−/BLM−/− cells expressing the BRCv-helicase double mutant were higher (i.e. more functionally impaired) than those cells complemented by the helicase single mutant but were comparable with those cells complemented by the BRCv mutant (Fig. 5, A and B; also see Fig. 4A). Similarly, the SCE level and CPT sensitivity of cells expressing the BRCv-KIX double mutant were higher than those of cells complemented by the KIX single mutant but were comparable with those of cells complemented by the BRCv mutant (Fig. 5, A and C; also see Fig. 4A). These results suggest that both the helicase- and KIX domain-dependent genome maintenance pathways require BRCv to function.
FIGURE 5.
RECQL5 BRCv acts in the same pathway either with the helicase or the KIX domain to suppress SCE and resist CPT-induced replication stress. A, scatter plot showing the SCE levels of BLM−/−/RecQL5−/− DT40 cells complemented by various mutants of RECQL5, as indicated. See the legend to Fig. 4. The mean SCE numbers are indicated for each mutant. B and C, CPT sensitivity curves of BLM−/−/RecQL5−/− DT40 cells complemented with WT or different RECQL5 mutants, as indicated. D, model to illustrate how different domains of RECQL5 may work together to promote genome stabilization. Error bars, S.D.
Potential BRCvs Are Present in Other HR Regulatory Proteins
One implication of our studies is that BRCv may be utilized by proteins other than RECQL5 to regulate HR. When we searched protein databases with consensus sequences of the two BRCv motifs, we identified potential BRCv motifs in several HR regulatory proteins, including yeast helicases Sgs1, Srs2, Mph1, and Pif1 (see Fig. 2B). Interestingly, the first three helicases all have anti-recombinase activities similar to that of RECQL5, including disruption of RAD51-made D-loops and suppression of crossover recombination (20, 24, 25, 33, 34). The data imply that the predicted BRCv motifs in these helicases may play a similar role in modulating the interaction with Rad51 as does the BRCv motif of RECQL5.
A Predicted BRCv Motif in Srs2 Is Critical for RAD51 Association and Suppression of D-loop Formation
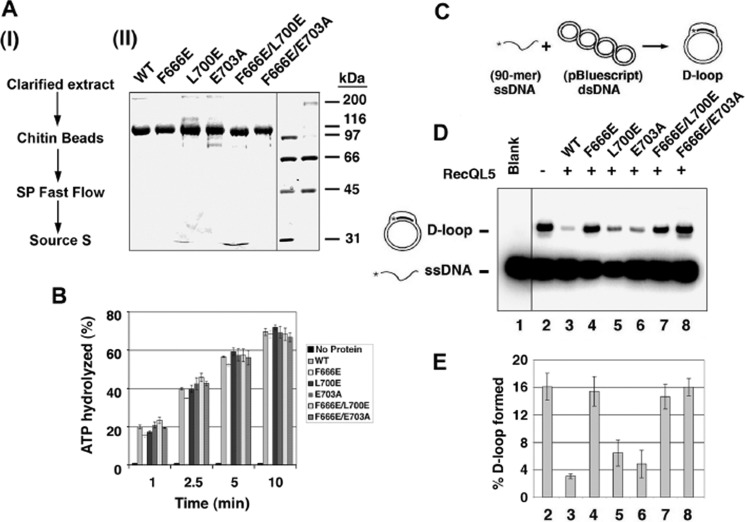
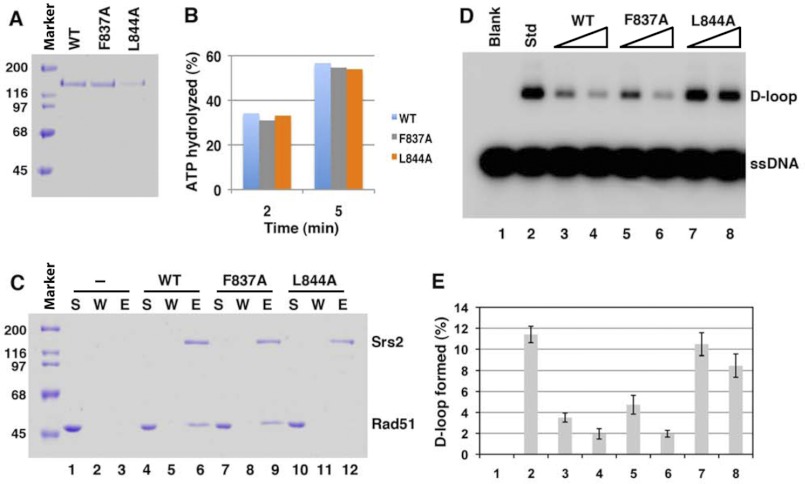
One of the putative BRCv motifs in Srs2 (see Fig. 2B) is located within the Rad51-binding region that has been previously mapped (residues 783–1038) (35). To study whether this BRCv is important for Srs2 to bind Rad51 and suppress HR, we mutated two residues that are highly conserved in RECQL5-BRCv motifs and critical for RECQL5 to bind RAD51: one within the predicted motif 1 (F837A) and the other just outside of the motif 1 (L844A) (see Fig. 2B). The Srs2 wild type and mutant proteins were expressed in E. coli, purified to near homogeneity (Fig. 6A), and analyzed for RAD51 association as described above for RECQL5. Both mutants had ATPase activity similar to that of the wild type protein (Fig. 6B), indicating that the proteins are folded properly. Importantly, the mutant L844A was strongly deficient in both Rad51 interaction (Fig. 6C) and inhibition of Rad51-mediated D-loop formation (Fig. 6, D and E), whereas the mutant F837A was largely normal in Rad51 association (Fig. 6C) but was modestly defective in suppression of D-loop formation at low protein concentrations (the suppression was about 70% for the wild type and 58% for the F837A mutant) (Fig. 6, D and E; compare lanes 5 and 3). Overall, these data support our prediction that a BRCv is present in Srs2, and this motif is critical for Srs2 to bind Rad51 and inhibit D-loop formation.
FIGURE 6.
The predicted Srs2-BRCv motif is important for Rad51 binding and for inhibition of Rad51-mediated D-loop formation. A, the purified WT and mutant (F837A and L844A) Srs2 proteins were resolved by 10% SDS-PAGE and stained with Coomassie Blue. The two mutants carry point mutations in two highly conserved residues in the predicted BRCv (see Fig. 2B). B, a bar graph showing the ATPase activity of Srs2 WT and mutant proteins. C, for affinity pull-down, WT and mutant Srs2 proteins were combined with purified Rad51 and mixed with Ni-NTA-agarose beads. The supernatant that retained unbound protein (S), the last wash (W), and the SDS eluate (E) were resolved by SDS-PAGE and stained with Coomassie Blue (lanes 4–12). As a control, Rad51 alone was incubated with Ni-NTA-agarose beads (lanes 1–3). D, autoradiograph shows inhibition of the D-loop formation by Srs2 proteins with mutations in the predicted BRCv motif. The radiolabeled D1 oligonucleotide (90-mer) was incubated with Rad51, Rad54, or RPA (lanes 2–8), with or without Srs2 proteins (WT, Srs2-F837A, or Srs2-L844A proteins) (7.5 or 15 nm); the mixture was added with pBluescript form I DNA. The reaction without Srs2 (lane 2) was designated as the standard (Std). The blank sample (lane 1) contained the DNA substrates without any proteins. E, quantification of the D-loop assay in D. The error bars represent the S.D. values from three independent experiments.
DISCUSSION
A BRC Repeat Variant in RECQL5 and Srs2 Mediates RAD51 Association
Although the BRC repeat had not been found in proteins other than BRCA2, we demonstrated that an HR regulatory protein, RECQL5, possesses a BRC repeat variant (BRCv) that resembles the BRCA2-BRC repeat in terms of structure and interaction motifs. Notably, both BRCv and BRC comprise two conserved motifs separated by a linker sequence, and the consensus sequence is identical for motif 1 and very similar for motif 2. Moreover, the two motifs in BRCv, as has been documented for BRC (14), are required for optimal RAD51 association; mutations in either motif reduce the ability of RECQL5 to bind RAD51, to inhibit RAD51-mediated D-loop formation, to suppress SCE, and to resist CPT-induced replication stress. Notably, using BRCv consensus sequences to search protein databases, we identified potential BRCv motifs in other HR regulatory proteins (Fig. 2B). We showed that a point mutant (L844A) in the predicted BRCv of Srs2 is strongly deficient in both Rad51 association and D-loop inhibition (Fig. 6). Our findings thus expand the role of the BRC repeat beyond BRCA2 and its orthologs. This strengthens the inference that the BRCv-mediated RAD51 association is important for HR regulation in general. Although the data presented herein indicate that the BRCv regulates interaction with Rad51 to mediate the anti-recombinase function of helicases, it remains an intriguing possibility that this motif may also promote Rad51 interaction in a prorecombination role under other circumstances.
We note that RECQL5 and several other vertebrate helicases (FBH1, RTEL1, and PARI) have been proposed to be functionally equivalent to yeast Srs2 (20, 36–38). Our finding that Srs2 contains a functional BRCv favors RECQL5 as the Srs2 equivalent in humans.
BRCv and BRC Have both Common and Unique Features
The structural and functional similarities between BRCv and BRC suggest that the two domains may engage the same epitope on RAD51. For example, motif 1 of BRCv is predicted to bind the same RAD51 oligomerization interface (86FXXA89) as does motif 1 of BRC, which is expected to interfere with a crucial contact between RAD51 monomers to prevent RAD51 filament formation (11). On the other hand, motif 2 in BRCv is expected to interact with a pocket on RAD51 that is distant from the oligomerization surface (11, 14). In support of this model, RECQL5 motif 1 mutants are deficient in inhibiting RAD51-mediated D-loop formation, whereas RECQL5 motif 2 mutants have reduced, albeit still significant, inhibitory activity. Moreover, motif 1 mutants of RECQL5 are more defective than motif 2 mutants in SCE suppression. These data suggest that interaction mediated by motif 1 at the RAD51 oligomerization interface is more important in regulating RAD51 activity than interaction by motif 2 at the distant pocket. This may help to rationalize the observation that motif 1 is strictly conserved in RECQL5 orthologs through evolution, whereas motif 2 is not (Fig. 2A).
BRCv and BRC nevertheless differ from one another in several ways. First, the residues near motif 1 are different (Fig. 2A) between the two. In the BRCA2-BRC repeats, hydrophobic residues scattered in the linker between motif 1 and motif 2 are required for the interaction between the linker and RAD51 as the repeat wraps around RAD51 (11, 14, 32). This hydrophobic patch is not found in RECQL5-BRCv. Likewise, a pair of hydrophobic residues conserved in the RECQL5-BRCv immediately adjacent to the FXXA motif 1 are not found in the BRCA2-BRC repeats. One of these residues (Leu-672) is also conserved in Srs2-BRCv (Leu-844), and mutation of this residue in either RECQL5 or Srs2 substantially reduces RAD51 association. Thus, interactions with RAD51 outside the two primary motifs appear to be different for BRCv versus BRC. Second, the linker sequences between the two motifs are different; the length in BRC is constant (12 residues), whereas that in BRCv is highly variable and also longer (18–25 residues) (Fig. 2A). The extra residues in BRCv are of unknown significance. Third, whereas BRC exists in multiple copies in human BRCA2, BRCv is present as a single copy in RECQL5 from most species. It was shown that BRC repeats within BRCA2 can be classified into two groups that have complementary functions and can work synergistically to activate HR (39). Speculatively, a single copy of BRCv may be sufficient for HR suppression, whereas cooperative interactions by multiple BRC repeats could be required for optimal activation of HR. Interestingly, Drosophila RECQL5 contains two putative BRCv repeats (Fig. 2A), hinting that BRCv may also function in tandem as seen in BRCA2-BRC.
We noted that the motif 1 mutant of Srs2-BRCv is only mildly defective in D-loop inhibition and is indistinguishable in Rad51 association compared with the wild type protein (Fig. 6). This differs from the motif 1 mutant of RECQL5 that is strongly defective in both assays (Fig. 2) (21). The data imply that Srs2 may be less dependent on motif 1-mediated Rad51 interactions compared with RECQL5.
Two Pathways Mediated by Helicase and KIX Domains Converge to Regulate BRCv-mediated Recombination
Previously, several functional domains were identified in RECQL5: the helicase domain that is involved in DNA translocation and the KIX and SRI domains that mediate the association with different forms of RNA Pol II (27, 40). The findings here add BRCv as a new distinct functional domain that enables RECQL5 to interact with RAD51. How do these different domains and their associated activities cooperate to accomplish RECQL5 functions? Our previously reported double mutant analyses have shown that the helicase and KIX domains act in independent pathways (27), whereas here we demonstrated that the BRCv is needed for the functional integrity of both pathways that are dependent on either the helicase or KIX domain. We suggest that BRCv-mediated RAD51 association may represent a common denominator of the two pathways that suppress SCE and confer resistance to CPT-induced replication stress (Fig. 5D). Although it seems likely that the helicase activity allows RECQL5 to translocate on DNA and dissociate RAD51 monomers from DNA in a processive fashion, it remains unclear how KIX affects BRCv-dependent RAD51 regulation. One possibility is that when the helicase/DNA translocase activity of RECQL5 is inactivated, the KIX-associated Pol II may recruit a backup helicase to power the translocation of RECQL5 on DNA in order to effect RAD51 removal.
In summary, our results suggest that the BRC repeat could be a general RAD51 regulatory module that can be used to either promote HR, as in the case of BRCA2, or repress HR, as in the case of RECQL5.
This work was supported, in whole or in part, by National Institutes of Health, NIA, Intramural Research Program Grant Z01:AG000657-09 (to W. W.) and National Institutes of Health Grants ES016532 and ES015252 (to P. S.).

This article contains supplemental Figs. S1–S3.
- HR
- homologous recombination
- RPA
- replication protein A
- SCE
- sister chromatid exchange
- Pol
- polymerase
- IP
- immunoprecipitation
- IP-Western
- immunoprecipitation-coupled Western blot
- CPT
- camptothecin
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Symington L. S. (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630–670, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu L., Hickson I. D. (2006) DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 40, 279–306 [DOI] [PubMed] [Google Scholar]
- 3. Pâques F., Haber J. E. (1999) Multiple pathways of recombination induced by double strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sung P., Klein H. (2006) Mechanism of homologous recombination. Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7, 739–750 [DOI] [PubMed] [Google Scholar]
- 5. Jensen R. B., Carreira A., Kowalczykowski S. C. (2010) Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thorslund T., McIlwraith M. J., Compton S. A., Lekomtsev S., Petronczki M., Griffith J. D., West S. C. (2010) The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1263–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu J., Doty T., Gibson B., Heyer W. D. (2010) Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carreira A., Hilario J., Amitani I., Baskin R. J., Shivji M. K., Venkitaraman A. R., Kowalczykowski S. C. (2009) The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell 136, 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shivji M. K., Mukund S. R., Rajendra E., Chen S., Short J. M., Savill J., Klenerman D., Venkitaraman A. R. (2009) The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc. Natl. Acad. Sci. U.S.A. 106, 13254–13259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davies A. A., Masson J. Y., McIlwraith M. J., Stasiak A. Z., Stasiak A., Venkitaraman A. R., West S. C. (2001) Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7, 273–282 [DOI] [PubMed] [Google Scholar]
- 11. Pellegrini L., Yu D. S., Lo T., Anand S., Lee M., Blundell T. L., Venkitaraman A. R. (2002) Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420, 287–293 [DOI] [PubMed] [Google Scholar]
- 12. Galkin V. E., Esashi F., Yu X., Yang S., West S. C., Egelman E. H. (2005) BRCA2 BRC motifs bind RAD51-DNA filaments. Proc. Natl. Acad. Sci. U.S.A. 102, 8537–8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shivji M. K., Davies O. R., Savill J. M., Bates D. L., Pellegrini L., Venkitaraman A. R. (2006) A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 34, 4000–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rajendra E., Venkitaraman A. R. (2010) Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res. 38, 82–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bachrati C. Z., Hickson I. D. (2003) RecQ helicases. Suppressors of tumorigenesis and premature aging. Biochem. J. 374, 577–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chu W. K., Hickson I. D. (2009) RecQ helicases. Multifunctional genome caretakers. Nat. Rev. Cancer 9, 644–654 [DOI] [PubMed] [Google Scholar]
- 17. Hu Y., Lu X., Barnes E., Yan M., Lou H., Luo G. (2005) Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell Biol. 25, 3431–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang W., Seki M., Narita Y., Nakagawa T., Yoshimura A., Otsuki M., Kawabe Y., Tada S., Yagi H., Ishii Y., Enomoto T. (2003) Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell Biol. 23, 3527–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu Y., Lu X., Zhou G., Barnes E. L., Luo G. (2009) Recql5 plays an important role in DNA replication and cell survival after camptothecin treatment. Mol. Biol. Cell 20, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu Y., Raynard S., Sehorn M. G., Lu X., Bussen W., Zheng L., Stark J. M., Barnes E. L., Chi P., Janscak P., Jasin M., Vogel H., Sung P., Luo G. (2007) RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21, 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwendener S., Raynard S., Paliwal S., Cheng A., Kanagaraj R., Shevelev I., Stark J. M., Sung P., Janscak P. (2010) Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 285, 15739–15745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sigurdsson S., Van Komen S., Petukhova G., Sung P. (2002) Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 277, 42790–42794 [DOI] [PubMed] [Google Scholar]
- 23. Petukhova G. V., Pezza R. J., Vanevski F., Ploquin M., Masson J. Y., Camerini-Otero R. D. (2005) The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 12, 449–453 [DOI] [PubMed] [Google Scholar]
- 24. Colavito S., Macris-Kiss M., Seong C., Gleeson O., Greene E. C., Klein H. L., Krejci L., Sung P. (2009) Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 37, 6754–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., Klein H., Ellenberger T., Sung P. (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 [DOI] [PubMed] [Google Scholar]
- 26. Sung P., Stratton S. A. (1996) Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271, 27983–27986 [DOI] [PubMed] [Google Scholar]
- 27. Islam M. N., Fox D., 3rd, Guo R., Enomoto T., Wang W. (2010) RecQL5 promotes genome stabilization through two parallel mechanisms. Interacting with RNA polymerase II and acting as a helicase. Mol. Cell Biol. 30, 2460–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perry P., Wolff S. (1974) New Giemsa method for the differential staining of sister chromatids. Nature 251, 156–158 [DOI] [PubMed] [Google Scholar]
- 29. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace. A web-based environment for protein structure homology modeling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 30. Benkert P., Künzli M., Schwede T. (2009) QMEAN server for protein model quality estimation. Nucleic Acids Res. 37, W510–W514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rost B., Liu J. (2003) The PredictProtein server. Nucleic Acids Res. 31, 3300–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ochiai K., Yoshikawa Y., Yoshimatsu K., Oonuma T., Tomioka Y., Takeda E., Arikawa J., Mominoki K., Omi T., Hashizume K., Morimatsu M. (2011) Valine 1532 of human BRC repeat 4 plays an important role in the interaction between BRCA2 and RAD51. FEBS Lett. 585, 1771–1777 [DOI] [PubMed] [Google Scholar]
- 33. Wu L., Davies S. L., Levitt N. C., Hickson I. D. (2001) Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276, 19375–19381 [DOI] [PubMed] [Google Scholar]
- 34. Prakash R., Satory D., Dray E., Papusha A., Scheller J., Kramer W., Krejci L., Klein H., Haber J. E., Sung P., Ira G. (2009) Yeast Mph1 helicase dissociates Rad51-made D-loops. Implications for crossover control in mitotic recombination. Genes Dev. 23, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S. (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 [DOI] [PubMed] [Google Scholar]
- 36. Kohzaki M., Hatanaka A., Sonoda E., Yamazoe M., Kikuchi K., Vu Trung N., Szüts D., Sale J. E., Shinagawa H., Watanabe M., Takeda S. (2007) Cooperative roles of vertebrate Fbh1 and Blm DNA helicases in avoidance of crossovers during recombination initiated by replication fork collapse. Mol. Cell Biol. 27, 2812–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barber L. J., Youds J. L., Ward J. D., McIlwraith M. J., O'Neil N. J., Petalcorin M. I., Martin J. S., Collis S. J., Cantor S. B., Auclair M., Tissenbaum H., West S. C., Rose A. M., Boulton S. J. (2008) RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135, 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moldovan G. L., Dejsuphong D., Petalcorin M. I., Hofmann K., Takeda S., Boulton S. J., D'Andrea A. D. (2012) Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 45, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carreira A., Kowalczykowski S. C. (2011) Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 108, 10448–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garcia P. L., Liu Y., Jiricny J., West S. C., Janscak P. (2004) Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 23, 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]