Background: The bacterial phosphoryl transfer system (PTS) couples phosphoryl transfer to sugar transport.
Results: The structure of the IIAchitobiose-HPr complex completes the structure elucidation of representative cytoplasmic complexes for all four sugar branches of the PTS.
Conclusion: Phosphoryl transfer occurs without any significant backbone conformational changes.
Significance: Recognition of multiple, structurally diverse partners is facilitated by complementary interaction surfaces and side chain conformational plasticity.
Keywords: Protein Conformation, Protein Phosphorylation, Protein Structure, Protein-Protein Interactions, Signal Transduction, NMR Spectroscopy, Phosphoenolpyruvate:Sugar Phosphotransferase Pathway, Phosphoryl Transfer
Abstract
The solution structure of the complex of enzyme IIA of the N,N′-diacetylchitobiose (Chb) transporter with the histidine phosphocarrier protein HPr has been solved by NMR. The IIAChb-HPr complex completes the structure elucidation of representative cytoplasmic complexes for all four sugar branches of the bacterial phosphoryl transfer system (PTS). The active site His-89 of IIAChb was mutated to Glu to mimic the phosphorylated state. IIAChb(H89E) and HPr form a weak complex with a KD of ∼0.7 mm. The interacting binding surfaces, concave for IIAChb and convex for HPr, complement each other in terms of shape, residue type, and charge distribution, with predominantly hydrophobic residues, interspersed by some uncharged polar residues, located centrally, and polar and charged residues at the periphery. The active site histidine of HPr, His-15, is buried within the active site cleft of IIAChb formed at the interface of two adjacent subunits of the IIAChb trimer, thereby coming into close proximity with the active site residue, H89E, of IIAChb. A His89-P-His-15 pentacoordinate phosphoryl transition state can readily be modeled without necessitating any significant conformational changes, thereby facilitating rapid phosphoryl transfer. Comparison of the IIAChb-HPr complex with the IIAChb-IIBChb complex, as well as with other cytoplasmic complexes of the PTS, highlights a unifying mechanism for recognition of structurally diverse partners. This involves generating similar binding surfaces from entirely different underlying structural elements, large interaction surfaces coupled with extensive redundancy, and side chain conformational plasticity to optimize diverse sets of intermolecular interactions.
Introduction
The phosphoenolpyruvate:sugar phosphotransferase system (PTS)4 is a central bacterial signal transduction pathway in which phosphoryl transfer, via a series of bimolecular protein-protein complexes, is coupled to both sugar transport across the membrane and the regulation of many cellular processes, including catabolite repression (1–6). The first component of the PTS, enzyme I, is autophoshorylated by phosphoenolpyruvate and subsequently transfers the phosphoryl group to the histidine phosphocarrier protein (HPr). HPr then transfers the phosphoryl group to the A domain of the sugar-specific enzymes II, which are divided into four structurally distinct families corresponding to the glucose, mannose, mannitol, and lactose/chitobiose branches of the PTS. All enzymes II have similar organizations comprising A and B cytoplasmic domains, and a membrane bound sugar transporter comprising the C domain, and sometimes a D domain as well. In some instances the domains are expressed as a contiguous protein, in others as separate proteins. From IIA, the phosphoryl group is transferred to IIB, and finally onto the incoming sugar molecule bound to the transmembrane IIC domain. Despite the similar domain organization of the enzymes II, the A and B cytoplasmic domains from the different branches of the PTS bear no sequence similarity to one another, and with the exception of IIBMtl (7, 8) and IIBChb (9–11), no similarity in either ternary or quaternary structures either.
Structures of the individual cytoplasmic components of the PTS have been solved by either NMR (7, 8, 10–20) or crystallography (9, 21–35). Structures of the cytoplasmic protein-protein complexes of the PTS, however, have been intractable to crystallography, presumably due to their weak affinity making successful co-crystallization difficult. Weak binding, however, is not an impediment to NMR, and we have solved the solution structures of all the cytoplasmic binary protein complexes of the PTS (15, 16, 18, 36–43) with the exception of the IIAChb-HPr complex. These complexes provide a wealth of information for understanding the unifying mechanism whereby a common interface, coupled with side chain conformational plasticity, can be used to recognize multiple, structurally dissimilar partners, and in addition, have yielded the first direct experimental evidence for the existence of highly transient, sparsely populated encounter complexes (44–46).
In this paper we present the solution structure of the IIAChb-HPr complex, the remaining outstanding cytoplasmic complex of the PTS, thereby completing our long term goal of solving all the cytoplasmic complexes of the PTS.
EXPERIMENTAL PROCEDURES
Protein Expression and Mutagenesis
Genes encoding IIAChb* (corresponding to a NΔ13/D92L mutant of wild-type IIAChb) (20) and HPr (39, 47) were cloned into the pET-11 vector. H89E and H15D mutations of the active site histidines of IIAChb* and HPr, respectively, were introduced using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). (Residues of HPr are denoted in italics throughout.) Both mutations were designed to mimic the charge effect of phosphorylation of the active site histidines.
The IIAChb*, IIAChb*(H89E), HPr, and HPr(H15D) plasmids were introduced into Escherichia coli BL21(DE3) (Novagen) cells for protein expression and induced at an A600 ∼ 0.8 with 1 mm isopropyl β-d-thiogalactopyranoside at 37 °C. Cells were grown in either Luria-Bertani medium or minimal medium (in either H2O or D2O) with 15NH4Cl or 14NH4Cl as the sole nitrogen source, and U-[13C/1H]-, U-[12C/1H]-, U-[13C/2H]-, or U-[12C/2H]glucose as the main carbon source. Because Leu, Val, Ile, Met, Gly, Try, Ser, Phe, and Ala residues are involved in the IIAChb*-HPr binding interface, selective amino acid labeling was also employed in the preparation of NMR samples. For 2H/13C/15N-(Ile/Leu/Val)-methyl-protonated (but otherwise fully deuterated) protein samples, 100 mg of α-[13C5,3-2H1]ketoisovalerate and 50 mg of α-[13C4,3,3-2H2]ketobutyrate (Cambridge Isotopes) were added to 1 liter of D2O medium 1 h prior to induction (48). 2H/12C/14N-(Ile/Gly/Phe-protonated)-IIAChb*(H89E), 2H/12C/14N-(Leu/Met/Tyr-protonated)-IIAChb*(H89E), and 2H/12C/14N-(Val/Ala/His-protonated)-IIAChb*(H89E) samples were prepared by supplementing 1 liter of D2O medium with 300 mg of Ile/Gly/Phe/Leu/Met/Tyr/Val/Ala/His (Sigma) at natural isotopic abundance 1 h prior to induction. Cells were grown an additional 7 h after induction. Cells expressing IIAChb*(H89E) or HPr were harvested by centrifugation at 15,900 × g for 25 min. IIAChb* and IIAChb*(H89E), and HPr and HPr(H15D) were purified as described previously in Refs. 43 and 39, respectively.
NMR Data Collection and Analysis
All NMR samples were prepared in a buffer of 20 mm sodium phosphate, pH 7.4, 0.2 mm sodium azide, and either 90% H2O/10% D2O or 99.99% D2O. IIAChb* is a symmetric trimer with 3 eq binding sites for HPr. As in the case of the IIAChb*(H89E)-IIBChb(C10S) complex (43), a 1:1 mixture of IIAChb*(H89E) trimer to HPr monomer was employed to achieve optimal line widths for NMR spectroscopy. NMR spectra were recorded at 20 and 35 °C on Bruker DMX500, DMX600, DRX600, DRX800, and DRX900 spectrometers equipped with z-shielded gradient triple resonance cryoprobes. Spectra were processed with the NMRPipe package (49) and analyzed using the programs PIPP (50) and XIPP.5 Sequential and side chain assignments of IIAChb*(H89E) and HPr were derived from the following three-dimensional double and triple resonance through-bond correlation experiments (51, 52): HNCA, HN(CO)CA, HNCACB, CBCA-(CO)HN, HAHN, HNCA-TROSY, HN(CO)CA-TROSY, HNCB-TROSY, HN(CO)CB-TROSY, C(CCO)NH, H(CCO)NH, and HCCH-TOCSY. Three-dimensional 15N-separated, 13C-separated, and 13C/13C-separated nuclear Overhauser enhancement (NOE) experiments were used to facilitate side chain assignments (51).
Intermolecular NOEs were observed on the IIAChb*(H89E)-HPr complex in D2O buffer using three-dimensional 12C-filtered(F1)/13C-separated(F2) or 13C-separated(F2)/12C-filtered(F3) NOE experiments, and in H2O buffer using two-dimensional 15N-separated/13C-edited and 13C-separated/15N-edited NOE experiments (53, 54). Nine different combinations of isotope-labeled complexes were used for analysis of intermolecular NOEs (Table 1).
TABLE 1.
Labeling schemes for samples used for intermolecular NOE measurements on the IIAChb*(H89E)-HPr complex
| Isotope labeling | ||
|---|---|---|
| Sample | IIAChb*(H89E) | HPr |
| 1 | [13CH3-ILV]/[2H/13C/15N] | [1H-Ile, Gly, Phe]/[2H/12C/14N] |
| 2 | [13CH3-ILV]/[2H/13C/15N] | [1H-Leu, Met, Tyr]/[2H/12C/14N] |
| 3 | [13CH3-ILV]/[2H/13C/15N] | [1H-Val, Ala, His]/[2H/12C/14N] |
| 4 | [1H-Ile, Gly, Phe]/[2H/12C/14N] | [13CH3-ILV]/[2H/13C/15N] |
| 5 | [1H-Leu, Met, Tyr]/[2H/12C/14N] | [13CH3-ILV]/[2H/13C/15N] |
| 6 | [1H-Val, Ala, His]/[2H/12C/14N] | [13CH3-ILV]/[2H/13C/15N] |
| 7 | [1H-Ile, Gly, Phe]/[2H/12C/14N] | U-[1H/13C/15N] |
| 8 | [1H-Leu, Met, Tyr]/[2H/12C/14N] | U-[1H/13C/15N] |
| 9 | U-[2H/13C/15N] | U-[1H/12C/14N] |
Structure Calculations
NOE-derived interproton distance restraints were classified into loose approximate distance ranges of 1.8–2.7, 1.8–3.5, 1.8–5.0, and 1.8–6.0 Å corresponding to strong, medium, weak, and very weak NOE cross-peak intensities, respectively (55). An empirical correction of 0.5 Å was added to the upper distance bounds of distance restraints involving methyl groups to account for the higher apparent intensity of methyl resonances (56), and NOEs involving nonstereospecifically assigned methyl, methylene, and aromatic protons were represented by a (Σr−6)−1/6 sum (57). Backbone torsion angle restraints for the active site region (residues 13–17) of HPr were derived from backbone 1H/15N/13C chemical shifts using the program TALOS+ (58) and used in the calculations of the phosphoryl transition state. The current experiments yielded interproton distance restraints and interfacial side chain torsion angle restraints.
Structures were calculated using conjoined rigid body/torsion angle-simulated annealing (59, 60) with the program Xplor-NIH (61). The target function that is minimized comprises NOE-derived interproton distance restraints, torsion angle restraints, residual dipolar coupling restraints, 13Cα/13Cβ chemical shift restraints, a quartic van der Waals repulsion term for the nonbonded contacts, a multidimensional torsion angle data base potential of mean force (62), and a gyration volume potential to ensure optimal packing (63). Structure figures were generated using the programs VMD-XPLOR (64) and GRASP (65). Reweighted atomic probability density maps were calculated as described previously (66). The atomic coordinates and NMR experimental restraints (accession codes 2lrk and 2lrl for the unphosphorylated and phosphoryl transition state complexes, respectively) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ.
RESULTS AND DISCUSSION
Equilibrium Binding of IIAChb*(H89E) and HPr
At concentrations used in NMR experiments, wild type IIAChb is highly prone to nonspecific aggregation promoted by the presence of a disordered 13-residue N-terminal tail and divalent cations required to neutralize and coordinate three symmetry-related, buried aspartate side chains (Asp-92) located at the center of the trimer interface (20). As in previous work (20, 43), we therefore made use of the IIAChb* construct throughout the current study. IIAChb* forms a stable monodisperse trimer, and comprises a deletion of the disordered 13-residue N-terminal tail and mutation of the buried Asp-92 to Leu (20). Leu and Asp have similarly branched side chains, and the methyl groups of the three Leu-92 side chains, one from each subunit, substitute well packed hydrophobic methyl-methyl interactions at the trimer interface in place of the role fulfilled by the metal cation. These mutations do not affect phosphoryl transfer activity.
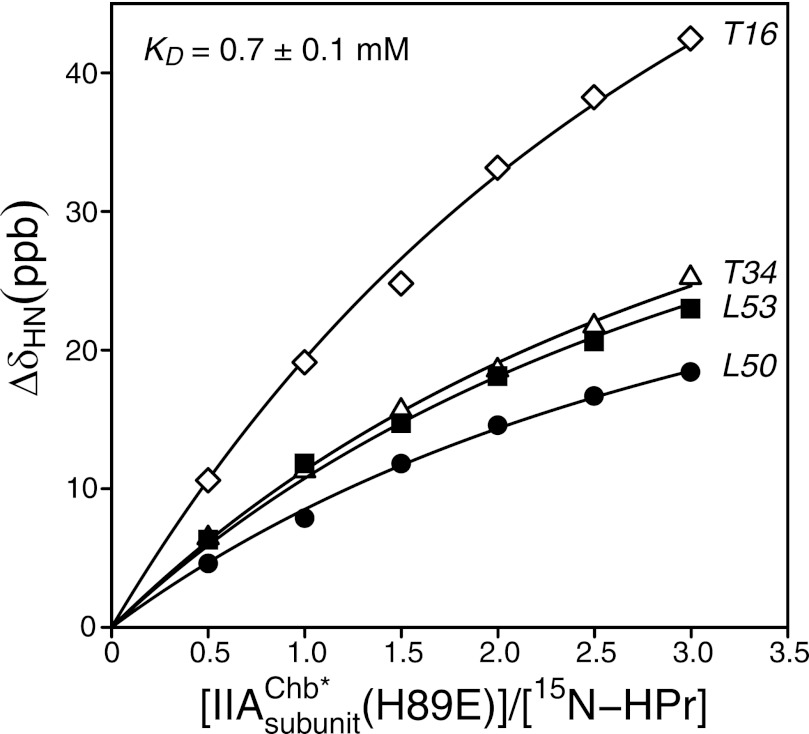
The interaction between IIAChb* and HPr was assessed by monitoring 1HN/15N chemical shift perturbations of 15N-labeled HPr upon addition of unlabeled IIAChb* (Fig. 1). Studies were carried out with HPr, IIAChb*, IIAChb*(H89E), and HPr(H15D). (Note that throughout the text, residues of HPr are printed in italics to distinguish them from residues of IIAChb*.) The latter two mutations are designed to mimic the charge effects of phosphorylation of the active site histidines at the Nϵ2 (H89E) and Nδ1 (H15D) positions. At pH 7.4 and 20 °C we could not detect any significant chemical shift perturbations upon addition of IIAChb* to either HPr or HPr(H15D) at the concentrations employed (up to ∼1.2 mm in subunits of IIAChb* with ∼0.4 mm HPr). However, measurable chemical shift perturbations were obtained upon addition of IIAChb*(H89E) to HPr yielding a KD of 0.7 ± 0.1 mm (Fig. 1). Therefore all structural studies were carried out with the IIAChb*(H89E) phosphomimetic mutant.
FIGURE 1.
Binding of IIAChb*(H89E) to 15N-labeled HPr. Backbone amide chemical shift perturbations upon titrating unlabeled IIAChb*(H89E) into a solution of 15N-labeled HPr at 20 °C. The chemical shifts were monitored using 1H-15N HSQC spectroscopy at a spectrometer 1H frequency of 600 MHz. ΔδH/N = [(Δδ15N)2/25 + (Δδ1H)2)/2]1/2 in parts per billion (ppb) (69). The IIAChb*(H89E):HPr molar ratios, expressed in terms of subunit concentration of IIAChb*(H89E), are 0, 0.5, 1.0, 1.5, 2.0, 2.5, and 3.0, with corresponding concentrations of 0.0 0.24, 0.47, 0.68, 0.87, 1.05, and 1.23 mm in subunits for IIAChb*(H89E), and 0.50, 0.48, 0.47, 0.45, 0.44, 0.42, and 0.41 mm for HPr. The solid lines represent the results of a global nonlinear least squares best fit to all the titration data simultaneously, using a simple equilibrium binding model. The optimized KD value is 0.7 ± 0.1 mm.
Structure Determination
The IIAChb*-HPr complex is in fast exchange on the chemical shift time scale (i.e. only a single set of population weighted average resonances are observed). The 1HN/15N chemical shift perturbations upon complex formation are small indicative of no significant change in backbone conformation (within the limits of the NMR method). The 1HN/15N chemical shift perturbations span residues 18–34 and 53–102 of IIAChb*, and residues 8–24 52–53, 61–62, 80, and 85 of HPr, thereby providing an approximate delineation of the interaction surfaces.
Given that three molecules of HPr can bind to the IIAChb* symmetric trimer and binding is weak, all NMR experiments were carried on samples comprising 1 mm IIAChb* (in trimer) and 1 mm HPr. Under these conditions, there is 24% free HPr and 42, 29, and 5%, singly, doubly and triply bound HPr; and 42% free IIAChb*, and 42, 14, and 2% IIAChb* with one, two, and three HPr molecules bound. Given molecular masses of ∼34 and 9.5 kDa for free IIAChb* and HPr, respectively, the population weighted average masses of IIAChb* and HPr, which determine the line widths in the NMR experiments, are ∼40 kDa each. Note that the existence of multiple bound states, as well as the presence of a significant fraction of free proteins, precludes the use of residual dipolar couplings for determining the relative orientation of the two proteins in the complex, because the apparent alignment tensor can no longer be deconvoluted into individual alignment tensors for each component in the system (43).
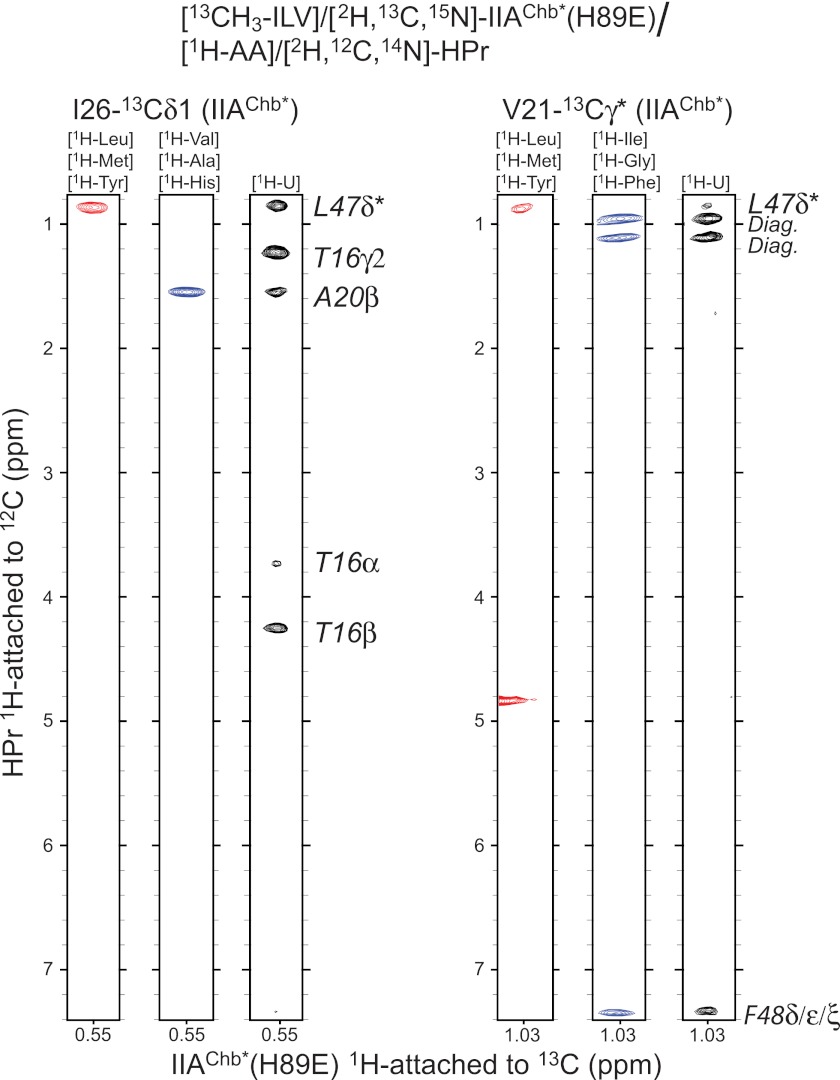
The structure of the IIAChb*-HPr complex was largely based on intermolecular NOE data derived from three-dimensional 12C-filtered/13C-separated three-dimensional NOE experiments in which NOEs are exclusively observed between protons attached to 12C and protons attached to 13C. An array of different combinations of isotopically labeled samples, comprising both uniform and residue-specific labeling (Table 1), was employed to remove any ambiguities in assignment of intermolecular NOEs. An example of the quality of the intermolecular data is provided in Fig. 2.
FIGURE 2.
Intermolecular NOEs in the IIAChb*(H89E)-HPr complex. NOEs in a three-dimensional 12C-filtered/13C-separated NOE experiment recorded in D2O are specifically observed from protons attached to 12C (in the F1 dimension) to methyl protons attached to 13C (in the F3 dimension). Strips are shown for NOEs involving the 13Cδ1 methyl group of Ile-72 (at 12.48 ppm) and one of the 13Cγ methyl groups of Val-21 (at 23.63 ppm) of IIAChb*(H89E). The amino acid (AA) specific labeling schemes used for [1H-AA]/[2H,12C,14N]HPr are shown above each strip.
The calculation strategy used to determine the structure of the complex made use of conjoined rigid body/torsion angle dynamics simulated annealing (60). In this instance, the backbone and noninterfacial side chains of the 2.0-Å resolution x-ray coordinates of free HPr (30) were treated as a rigid body with rotational and translational degrees of freedom, whereas interfacial side chains were given torsional degrees of freedom. The only coordinates of free IIAChb* available are NMR coordinates (20), which are inherently less accurate than x-ray coordinates (especially in terms of translation and packing). Thus for IIAChb* full torsional, rotational, and translational degrees of freedom were allowed with the coordinates restrained by the experimental NMR restraints (NOEs, torsion angles, dipolar couplings) obtained for free IIAChb* (20). This approach, rather than using the restrained regularized mean coordinates of free IIAChb* (20) as a rigid body, was employed for the following reasons: the interface of both partners is largely helical and structurally rigid; the active site residue (H89E) is located within a deep cleft at the interface of adjacent subunits; and therefore small errors in the backbone coordinates of the free NMR structure of IIAChb* can readily propagate and distort the docking of HPr onto IIAChb*. The backbone coordinate shifts relative to the free IIAChb* coordinates, however, are small (<1 Å) and well within the uncertainties of the NMR coordinates. In the case of the IIAChb*-IIBChb complex, on the other hand, the IIBChb interaction site comprises a loop so that uncertainties in the IIAChb* coordinates could be assimilated by simply giving the backbone of the active site loop of IIBChb torsional degrees of freedom, while treating the remaining backbone of IIBChb as well as the backbone of IIAChb* (excluding the disordered loop from residues 75–84) as rigid bodies (43).
As in the case of the weak IIAChb*-IIBChb complex, a heuristic approach was employed for interfacial side chains since the samples comprised a mixture of free and bound states (43). Thus, the interfacial side chains were given torsion angle degrees of freedom within the χ1 and where appropriate χ2 rotamers of the free structures, unless contradicted by the intermolecular NOE data. A summary of the structural statistics is provided in Table 2, a best fit superposition of the final ensemble of 100 simulated annealing structures of the complex is displayed in Fig. 3A, and a reweighted atomic probability density map for some interfacial side chains is shown in Fig. 3C.
TABLE 2.
Structural statistics
The notation of the NMR structures is as follows: <SA> are the final 100 simulated annealing structures for the IIAChb* (H89E)-HPr complex and <SAphos> are the final 70 simulated annealing structures for the corresponding phosphoryl transition state.
| <SA> | <SAphos> | ||
|---|---|---|---|
| Number of experimental NMR restraints | |||
| Intermolecular interproton distance restraints | 42 | ||
| IIAChb* intramolecular interproton distance restraintsa | 276 × 3 | ||
| IIAChb* torsion angle restraintsa | 245 × 3 | ||
| HPr torsion angle restraintsb | 36 | ||
| 1DNH RDCs for IIAChb*a | 84 × 3 | ||
| 1DNC′ RDCs for IIAChb*a | 85 × 3 | ||
| 1DCαC′ RDCs for IIAChb*a | 83 × 3 | ||
| 13Cα/13Cβ chemical shift restraints for IIAChb*a | 195 × 3 | ||
| Experimental restraints | |||
| R.m.s. deviation from interproton distance restraints (Å)c | 0.010 ± 0.002 | 0.011 ± 0.001 | |
| R.m.s. deviation from torsion angle restraints (°)c | 0.25 ± 0.04 | 0.25 ± 0.04 | |
| R.m.s. deviation from 13Cα/13Cβ shift restraints (ppm) | 1.12 ± 0.02/0.66 ± 0.02 | 1.12 ± 0.02/0.63 ± 0.01 | |
| 1DNH RDC R-factor (%)d | 7.4 ± 0.1 | 7.4 ± 0.08 | |
| 1DNC′ RDC R-factor (%)d | 17.1 ± 1.3 | 16.8 ± 0.97 | |
| 1DCαC′ RDC R-factor (%)d | 16.2 ± 0.9 | 15.9 ± 0.52 | |
| Deviations from idealized covalent geometrye | |||
| Bonds (Å) | 0.006 ± 0 | 0.006 ± 0 | |
| Angles (degree) | 0.63 ± 0 | 0.63 ± 0 | |
| Impropers (degree) | 0.63 ± 0 | 0.62 ± 0 | |
| Measures of structural qualityf | |||
| Intermolecular repulsion energy (kcal mol−1) | 1.3 ± 0.4 | 1.4 ± 0.4 | |
| Intermolecular Lennard-Jones energy (kcal mol−1) | −16.2 ± 5.2 | −16.7 ± 4.5 | |
| Number of bad contacts per 100 residues | 3.6 ± 1.1 | 3.1 ± 1.1 | |
| % Residues in most favorable region of Ramachandran map | 94.5 ± 1.0 | 94.3 ± 1.3 | |
| Coordinate precision of the complex (Å)g | |||
| Backbone (N, Cα, C′, O) atoms | 0.56 | 0.49 | |
| Interfacial sidechain heavy atoms of IIAChb* and HPrg | 1.25 | 1.07 | |
| Side chain heavy atoms of IIAChb* | 1.32 | 1.24 | |
a The intramolecular experimental restraints (NOE-derived interproton distances, torsion angles, 13Cα/13Cβ chemical shifts and residual dipolar couplings) for IIAChb* in the complex are taken from the structure determination of free IIAChb*. The interproton distance restraints comprise 82 × 3 sequential (|i − j| = 1), 86 × 3 medium range (1 <|i − j| ≤ 5), and 30 × 3 long range (|i − j| > 5) intrasubunit restraints, 78 × 3 intersubunit distances and 154 × 3 distance restraints for 77 × 3 backbone hydrogen bonds within the three helices. The torsion angle restraints comprise 87 × 3 φ, 82 × 3 ψ, 76 × 3 χ angles.
b The torsion angle restraints for HPr comprise 26 interfacial side chain torsion angles, supplemented, in the case of the calculations of the phosphoryl transition state, by 5 φ and 5 ψ backbone torsion angles (derived from chemical shifts in the complex using TALOS+ (58)) for residues 13–17 encompassing the active site His-15. The side chains of HPr given torsional degrees of freedom comprise residues 11–17, 20–21, 23–24, 27, 45–48, 51–57, and 85.
c None of the structures exhibit interproton distance violations >0.3 Å or torsion angle violations >5°.
d The RDC R-factor (70) is defined as the ratio of the r.m.s. deviation between observed and calculated values and the expected r.m.s. deviation for a random distribution of vectors. The latter is given by [2Da2 (4+3η2)/5]1/2, where Da and η are the magnitude of the alignment tensor and the rhombicity, respectively. The values of Da and η for the free IIAChb* trimer are −12.1 Hz and 0, respectively (note the rhombicity for a symmetric trimer is always 0). The R-factor scales between 0 and 100%.
e The His-P-His phosphoryl transition state formed between the Nϵ2 atom of His-89 of subunit A of IIAChb* and the Nδ1 atom of His-15 of HPr is calculated using the same experimental restraints as those used for the unphosphorylated complex with the addition of covalent geometry restraints to describe the pentacoordinate phosphoryl group in a trigonal bypiramidal geometry (37): rNϵ2(His89)-P, rNδ1(His15)-P ≤ 3.5 Å, rP-O = 1.48 Å; Nδ1(His-15)-P-Nϵ2(His-89) = 180°, Nδ1(His-15)-P-O = 90°, Nϵ2(His-89)-P-O = 90°, Cγ(His-15)-Nδ1(His-15)-P = 127.35°, Cϵ1(His-15)-Nδ1(His-15)-P = 127.35°, Cδ2(His-89)-Nϵ2(His-89)-P = 126.35°, Cϵ1(His-89)-Nϵ2(His-89)-P = 126.35°. In addition improper torsion angle restraints are used to ensure that the phosphorus atom lies in the same plane as the imidazole rings of both His-89A and His-15.
f The intermolecular repulsion energy is given by the value of the quartic van der Waals repulsion term calculated with a force constant of 4 kcal mol−1·Å−4 and a van der Waals radius scale factor of 0.78 (61). The intermolecular Lennard-Jones van der Waals interaction energy is calculated using the CHARMM19/20 parameters and is not included in the target function used to calculate the structures. The number of bad contacts per 100 residues and the percentage of residues in the most favorable region of the Ramachandran plot are calculated using PROCHECK (71). The φ/ψ, χ1/χ2, χ1 and χ3/χ4 PROCHECK g-factors are 0.76 ± 0.04, 0.60 ± 0.06, 0.21 ± 0.10, and 0.42 ± 0.07, respectively.
g Defined as the average r.m.s. difference between the final ensemble of simulated annealing structures and the mean coordinates positions.
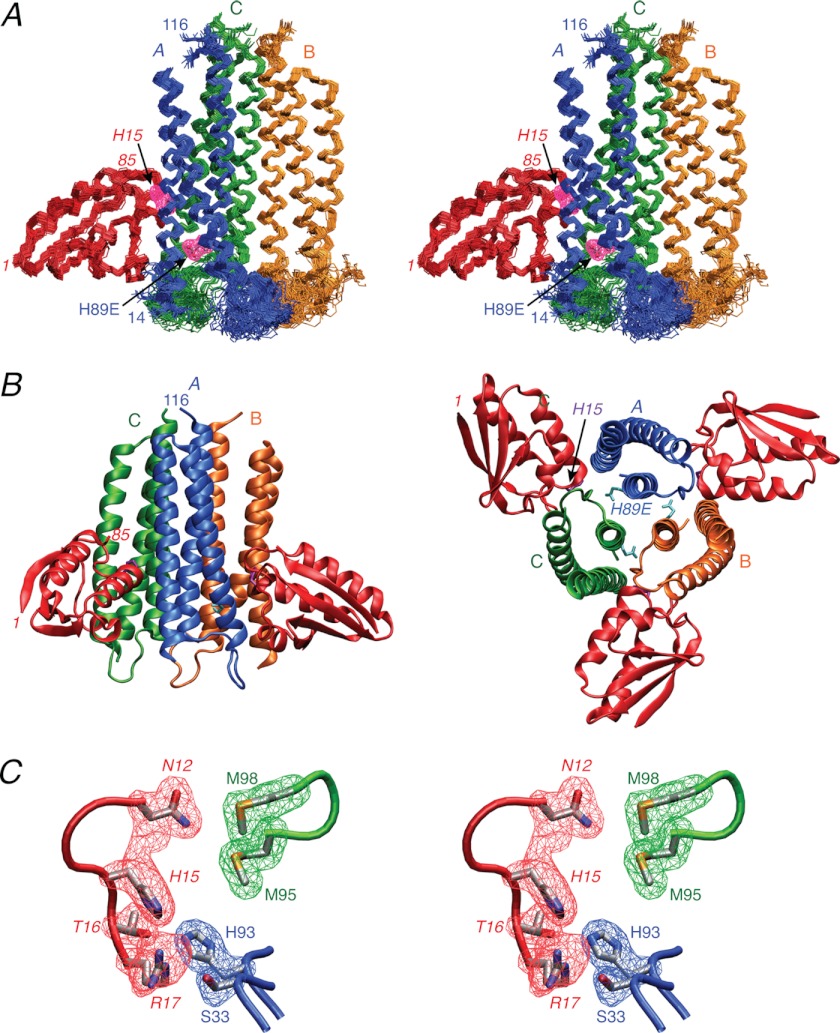
FIGURE 3.
Solution structure of the IIAChb*(H89E)-HPr complex. A, stereoview of a superposition of the backbone (N, Cα, and C) atoms of the final 100 simulated annealing structures with the A, B, and C subunits of the IIAChb*(H89E) symmetric trimer in blue, gold, and green, respectively, and HPr in red. HPr is shown interacting with the A and C subunits of IIBChb*. The purple meshes represent the atomic density probability maps (66) for the two active site residues, H89E of subunit A of IIAChb*(H89E) and His-15 of HPr. (The probability maps are drawn at a value of 20% maximum.) Note that because IIAChb*(H89E) is a symmetric trimer there are three identical binding sites formed at the interfaces between the A and C subunits, the C and B subunits, and the B and A subunits. B, ribbon diagrams of the complex showing two HPr molecules bound to the IIAChb*(H89E) trimer (left panel) and an orthogonal view depicting three molecules of HPr bound to the IIAChb*(H89E) trimer (right panel). The color coding is the same as in A. C, stereoview showing a reweighted atomic probability density map (drawn at a value of 20% maximum and calculated from the final 100 simulated annealing structures) for some of the interfacial side chains displayed as blue and green meshes for the A and C subunits for IIAChb*(H89E) and as a red mesh for HPr. The backbones are shown as tubes color coded as in A; the side chains of the restrained regularized mean structure are color coded according to atom type (carbon, gray; oxygen, red; nitrogen, blue; sulfur, yellow). Residues of HPr are labeled in italics.
The Overall Structure of the IIAChb*-HPr Complex
A ribbon diagram of the overall complex showing two and three molecules of HPr bound per trimer is displayed in Fig. 3B. Each HPr molecule interacts with two adjacent subunits of IIAChb*: specifically subunits A and C, C and B, and B and A, where the first subunit in each pair contributes the active site residue at position 89. For the purposes of describing intermolecular contacts between HPr and IIAChb*, we will restrict ourselves to the interaction surface formed at the interface of the A and C subunits of IIAChb*.
Each subunit of IIAChb* comprises 3 helices in an up, down, up topology comprising residues 17–43 (helix 1), 47–74 (helix 2), and 85–114 (helix 3) (20). HPr has three helices formed by residues 16–28 (helix 1), 47–52 (helix 2), and 70–83 (helix 3), as well as a four-stranded antiparallel β-sheet (30). The active site histidine at position 89, as well as His-93, of the A subunit of IIAChb* are located deep within a cleft formed at the interface of subunits A and C (Figs. 3B and 4A), whereas the active site His-15 of HPr is exposed at the tip of a convex protrusion on the surface of HPr (Figs. 3B and 4B), The predominant intermolecular contacts between HPr and IIAChb* involve helices. The N-terminal halves of helices 1 (residues 16–27) and 2 (residues 347–348) of HPr interact with the N-terminal halves of helices 1 (residues 18–33) and 3 (residues 89 and 93) of subunit A of IIAChb*; while the loop preceding helix 1 (residues 11–15), helix 2 (residues 47–53), and a stretch of extended strand (residues 55–57) of HPr interact with the C-terminal half of helix 2 (residues 58–73), the loop connecting helices 2 and 3 (residues 76–82), and the middle half of helix 3 (residues 91–98) of the C subunit of IIAChb*.
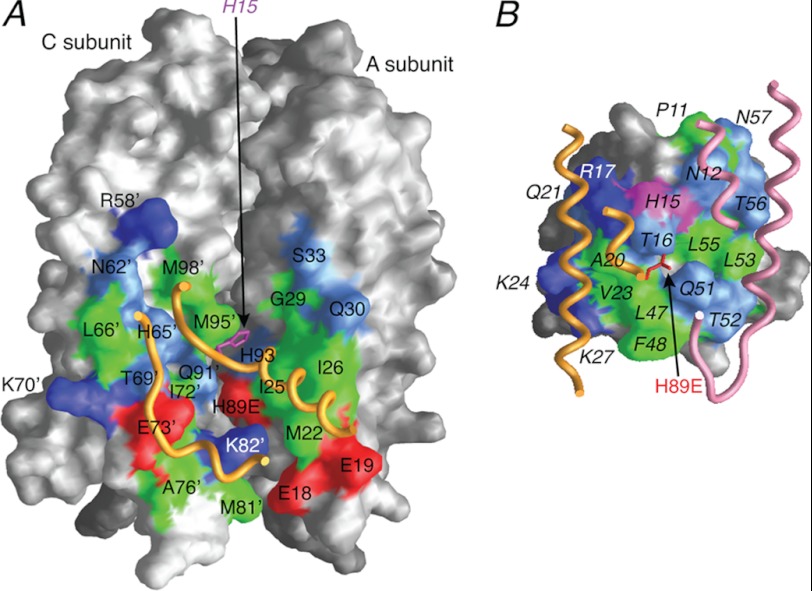
FIGURE 4.
Interaction surfaces for the IIAChb*(H89E)-HPr complex. A, interaction surface (formed by the A and C subunits) on IIAChb*(H89E) for HPr. B, interaction surface on HPr for IIAChb*(H89E). The surfaces are color coded as follows: hydrophobic residues, green; uncharged residues bearing a polar functional group, cyan; negatively charged residues, red; positively charged residues, blue. Relevant portions of the backbone and the active site residue of the interacting partner are displayed as tubes and bonds, respectively. Residues of HPr are labeled in italics. Residues from the C subunit of IIBChb*(H89E) are indicated by an apostrophe after the residue number; in addition, the surfaces of the A and C subunits of IIAChb* that do not constitute the interaction surface are colored in dark gray and light gray, respectively.
The total accessible surface area buried upon complex formation is ∼1580 Å2, comprising ∼350 Å2 and ∼450 Å2 for subunits A and C, respectively, of IIAChb*, and ∼780 Å2 for HPr (subdivided into ∼350 and ∼430 Å2 for contacts with the A and C subunits of IIAChb*, respectively). The binding site on IIAChb* for both subunits A and C comprises ∼45% hydrophobic residues, with the remainder equally divided between polar and charged residues (Fig. 4A); for HPr, the portion of the binding surface that interacts with the A subunit of IIAChb* is ∼40% hydrophobic, with the remainder equally divided between polar and charged residues (Fig. 4B, left half), while the portion of the HPr binding surface that interacts with the C subunit of IIAChb* is composed of ∼55% hydrophobic and ∼45% uncharged polar residues (Fig. 4B, right half). As in the other complexes of the PTS (15, 16, 36–40, 43), the central portions of the interaction surfaces are largely, but not exclusively, hydrophobic, interspersed by uncharged polar residues, whereas the outer edges are predominantly charged or polar (Fig. 4).
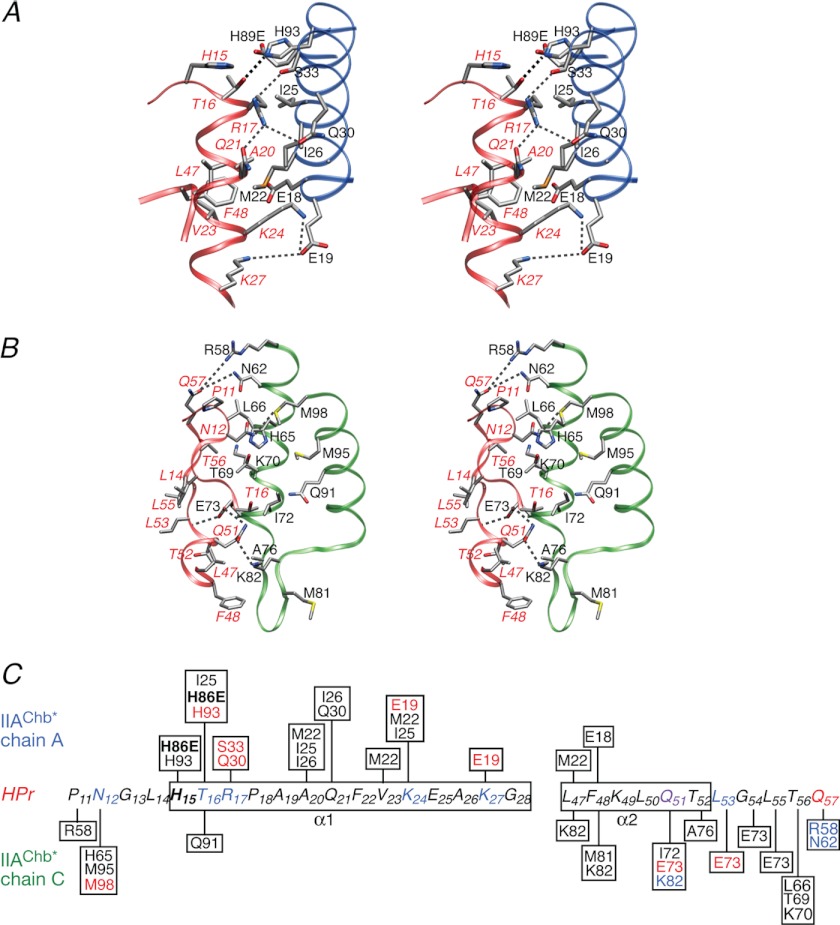
Detailed views of the side chain interactions between HPr and the A and C subunits of IIAChb* are shown in Fig. 5, A and B, respectively, together with a schematic summary of the intermolecular contacts in Fig. 5C. In contrast, to EIN, IIAGlc, IIAMtl, and IIAMan, where the charged residues in the binding site for HPr are largely negative, with very few positively charged residues (36–39), the binding surface on IIAChb* (Fig. 4A) contains an equal number of positively and negatively charged residues (three of each), of which two negative (Glu-19 and Glu-73) and two positive (Arg-58 and Lys-62) residues participate in intermolecular electrostatic interactions (Fig. 5). Indeed there are a quite number of potential hydrogen bonding, salt bridge, and longer range electrostatic interactions that serve to orient HPr and IIAChb* correctly.
FIGURE 5.
The IIAChb*(H89E)-HPr interface. A, stereoview of the interface between the A subunit of IIAChb*(H89E) and HPr with the respective backbones shown as blue and red ribbons, respectively. B, stereoview of the interface between the C subunit of IIAChb*(H89E) and HPr with the respective backbones shown as green and red ribbons, respectively. The dashed lines indicate potential intermolecular hydrogen bonds or salt bridges. The side chain atoms are colored according to atom type (carbon, gray; oxygen, red; nitrogen, blue; sulfur, yellow). Residues of HPr are labeled in italics. C, diagrammatic representation of the intermolecular contacts between the A and C subunits of IIAChb*(H89E) and HPr. Residues involved in side chain-side chain electrostatic interactions are colored in blue (donor) or red (acceptor). The active site residues, H89E of IIAChb*(H89E), and His-15 of HPr are shown in bold letters.
Thus, at the interface between HPr and the A subunit of IIAChb*, the hydroxyl group of Thr-16 forms a hydrogen bond with the Nϵ2 atom of His-93A; the guanidino group of Arg-17 forms potential salt bridges with the hydroxyl group of Ser-33A and the side chain carbonyl of Gln-30A, with the orientation of the side chain of Arg-17 further stabilized by an intramolecular interaction between its guanidino group and the side chain carbonyl of Gln-21; and Lys-24 and Lys-27 form potential salt bridge and longer range electrostatic interactions with the carboxylate of Glu-19A (Fig. 5A). In addition, the carboxylate of H89EA is sufficiently close (<5 Å) to the hydroxyl group of Thr-16 to allow for an electrostatic interaction that may explain why the IIAChb*(H89E)-HPr complex is of higher affinity than either the IIAChb*-HPr or IIAChb*-HPr(H15D) complexes.
At the interface of HPr and the C subunit of IIAChb*, the side chain carbonyl of Gln-57 has electrostatic interactions with the guanidino group of Arg-58C and the side chain amide group of Asn-62C; the side chain amide group of Asn-12 forms a potential hydrogen bond with the Sδ atom of Met-98C; the backbone amide of Leu-53 donates a potential hydrogen bond to the carboxylate of Glu-73C; and the carboxyamide group of Gln-51 forms potential hydrogen bonds with the carboxylate of Glu-73C and the side chain amino group of Lys-82C (Fig. 5B). Given that the interaction surfaces of HPr and IIAChb* are complementary both in terms of shape and distribution of residue type, it is likely that many of the above intermolecular electrostatic interactions are rather weak and transient, thereby accounting for the high equilibrium dissociation constant (KD ∼ 0.7 mm; cf. Fig. 1) for the complex.
The Phosphoryl Transition State
It is known from isotope labeling experiments that the phosphoryl transition state in complexes of the PTS comprises a pentacoordinate phosphoryl group in a trigonal bipyramidal geometry, with donor and acceptor atoms at apical positions and the oxygen atoms of the phosphoryl group lying in an equatorial plane (67, 68). The His-89A(Nϵ2)-P and His-15(Nδ1)-P distances can lie anywhere between 1.8 and 3.5 Å corresponding to pure associative and pure dissociative transition states, respectively, and the phosphorus atom lies in the plane of the imidazole group of both active site histidines.
To model the transition state, we therefore carried out conjoined rigid body/torsion angle-simulated annealing calculations using exactly the same protocol and experimental restraints as those used for the unphosphorylated complex but with the addition of covalent geometry restraints for the pentacoordinate phosphoryl group and the introduction of backbone torsional degrees of freedom for residues 13–17 of HPr encompassing the active site. The overall backbone r.m.s. shift between the restrained regularized mean structures of the transition state and unphosphorylated complexes is 0.5 Å overall, and 0.3 Å for the interface (Fig. 6B), which is well within the errors of the NMR coordinates. In addition, there are only minor perturbations in side chain positions (Fig. 6B). Thus, one can conclude that the transition state can be readily accommodated without any significant perturbation in backbone conformation. Furthermore, agreement with the experimental restraints and indicators of structural quality are unaffected by the introduction of the phosphoryl transition state (Table 1).
FIGURE 6.
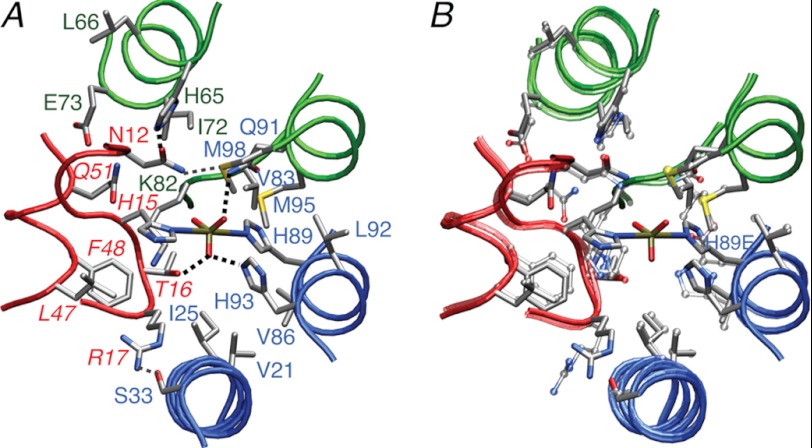
The phosphoryl transition state of the IIAChb*-HPr complex. A, environment surrounding the His-89A-P-His-15 pentacoordinate phosphoryl transition state. The backbone is displayed as transparent tubes with HPr in red, and the A and C subunits of IIAChb* in blue and green, respectively. B, identical view to A showing a superposition of the structure of the IIAChb*(H89E)-HPr complex (transparent tubes and bonds) with the structure of the IIAChb*-P-HPr transition state (opaque tubes and bonds). Exactly the same experimental restraints are used to calculate the two structures, but, in addition, the calculations for the transition state include geometric restraints specifying the geometry of the phosphoryl transition state and backbone torsion angle degrees of freedom for residues 13–17 of HPr encompassing the active side His-15. Color coding: red, HPr; blue, A subunit of IIAChb*; green, C subunit of IIAChb*. Side chains are displayed as stick diagrams with the atoms color coded according to type (carbon, gray; nitrogen, blue; oxygen, red; phosphorus, gold; sulfur, yellow). Residues of HPr are labeled in italics. Dashed black lines indicate hydrogen bonds to the phosphoryl group in the transition state, and the dashed gray line indicates a potential intermolecular hydrogen bond between the carboxyamide group of Asn-12 of HPr and the Met-98(Sδ) atom of the C subunit of IIAChb*.
The phosphoryl group in the transition state is hydrogen bonded to the hydroxyl group of Thr-16 of HPr, the Hϵ2 atom of His-93A of subunit A of IIAChb*, and the carboxyamide group of Gln-91C of subunit C of IIAChb* (Fig. 6A). As in other PTS complexes, the phosphoryl group is surrounded by a cluster of hydrophobic groups: Leu-47 and Phe-48 of HPr; Val-21A, Ile-25A, Val-86A, and Leu-92A of the A subunit of IIAChb*; and Ile-72C, Val-83C, and Met-95C, as well as the aliphatic portion of the side chain of Lys-82C, of the C subunit of IIAChb*.
Comparison with the IIAChb*-IIBChb Complex
HPr (30) and IIBChb (9–11) share no similarity in either overall structure or local structure surrounding the active site residue, His-15 in the case of HPr, and Cys-11 for IIBChb. Yet both proteins bind to highly overlapping binding sites on IIAChb* (this paper and Ref. 43). The interaction surfaces share 10 residues in common for subunit A and 9 for subunit C. The residues that are not shared by the two interaction surfaces are located at the peripheries of the binding sites. In the view shown in Fig. 4, the binding surface for HPr extends slightly upwards to include Ser-33A of subunit A and Arg-58C, Asn-62C, and Met-98C of subunit C, whereas the binding surface for IIBChb extends slightly downwards to include Glu-15A of subunit A, and Gly-74C and Gly-77C of subunit C (43). These small differences can be readily appreciated by the superposition of the two complexes shown in Fig. 7, and probably reflect two factors: first, the slightly larger size of the binding site on IIBChb, which comprises 29 residues versus 19 for HPr; and second the slightly more peripheral location of His-15 relative to the interaction surface compared with Cys-11 of IIBChb.
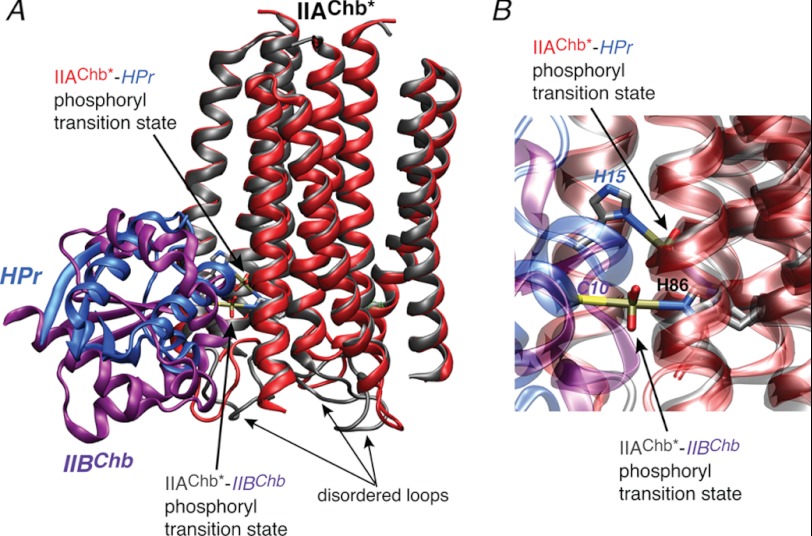
FIGURE 7.
Comparison of the IIAChb*-HPr and IIAChb*-IIBChb complexes. A, overall stereoview with IIAChb* from the two complexes best-fitted to one another, and, B, close up of the His-P-His and His-P-Cys phosphoryl transition states for the IIAChb*-HPr and IIAChb*-IIBChb complexes, respectively. The backbone is displayed as a ribbon diagram and the His-P-His and His-P-Cys transition states as stick diagrams with the atoms color coded according to type (carbon, gray; nitrogen, blue; oxygen, red; phosphorus, gold; sulfur, yellow). For the IIAChb*-HPr complex, IIAChb* and HPr are shown in red and blue, respectively; for the IIAChb*-IIBChb complex, IIAChb* and IIBChb are shown in gray and purple, respectively. The coordinates of the IIAChb*-IIBChb complex are taken from Ref. 43 (PDB code 2WWV). The small differences in the IIAChb* coordinates from the two structures is within coordinate error. Also note that the region that displays the largest apparent differences is the loop from residues 77 to 84 of IIAChb*, which is disordered in solution.
Although small, the above differences nicely illustrate the concept of redundancy in a system in which one partner, IIAChb*, recognizes multiple partners, while making use of the same active site residue (His-89A) to effect phosphoryl transfer. Thus, the four additional residues at the top edge of the IIAChb* binding surface for HPr that are not used in the interaction with IIBChb, namely Ser-33A, Arg-58C, Asn-62C, and Met-98C (Fig. 4A), are all involved in potential hydrogen bonding and electrostatic interactions with HPr (Fig. 5, A and B) that contribute to correctly orienting HPr relative to IIAChb*. The same is true of Glu-15A, located at the bottom edge of the IIAChb* binding surface for IIBChb but absent from the interaction with HPr, which forms a salt bridge with Lys-86 of IIBChb (43).
At the same time, side chain conformational plasticity allows side chains to participate in similar interactions (cf. Fig. 5 of this paper and Fig. 4 of Ref. 43). For example, Gln-30A forms a hydrogen bond with Arg-17 of HPr and Tyr-62 of IIBChb, both located in rather similar positions relative to their respective active site residues. Likewise, Glu-19A is involved in a potential salt bridge with Lys-27 of HPr and a potential electrostatic interaction with the hydroxyl group of Ser-33 of IIBChb. Finally, Glu-73C is hydrogen bonded to both the backbone amide of Leu-53 and the side chain amide of Gln-51 of HPr, and to the side chain guanidino group of Arg-24 of IIBChb. As a final example, the interaction of Met-22A with Phe-48 of HPr (Fig. 5A) is very similar to that with Tyr-84 of IIBChb, except that the hydrophobic contacts between these two pairs of residues is supplemented by a potential hydrogen bond between the Sδ atom of Met-22A and the hydroxyl group of Tyr-84.
Concluding Remarks
The structure of the IIAChb*-HPr complex in the present paper completes the structure elucidation of representative soluble complexes for all four sugar branches of the PTS (15, 16, 18, 36–40, 43). This collection of structures provides a paradigm of protein recognition in signal transduction pathways that allows for multiple recognition partners, transient interactions, and specificity.
Although the structures of the IIA components of the four sugar branches bear no sequence or structural similarity to one another, their recognition surfaces for HPr are remarkably similar in shape and residue composition. Moreover, each enzyme IIA makes use of highly overlapping surfaces to recognize both its upstream partner HPr and its downstream partner, enzyme IIB (this paper and Refs. 15, 16, 37–40, and 43).
The ability to recognize multiple different partners relies on a number of design features. First, similar surfaces are constructed from completely different underlying structural elements. Thus, the shape of the binding surfaces on HPr and the four classes of enzymes IIB are convex in shape and similar in size. Likewise, all four classes of enzymes IIA have a concave binding surface of similar size. Second, all the surfaces generally share similar features comprising predominantly hydrophobic residues, interspersed by uncharged polar residues, at the center of the interface surrounded by polar and charged residues at the periphery. Third, the interactions surfaces are all large (600–1000 Å2), thereby allowing considerable redundancy in the intermolecular interactions that have to be formed to achieve appropriate docking and orientation of the phosphoryl transfer complexes. A corollary to large surfaces and redundancy of specific intermolecular interactions is that all the complexes are transient and weak ranging from KD values of ∼10 μm to the millimolar range (this paper and Refs. 15, 16, 36–40, 42, and 43)). Fourth, conformational plasticity of amino acids with long side chains (such as Arg, Lys, and Glu) permit similar types of intermolecular interactions to occur across complexes involving one shared partner. Finally, although HPr uses the same binding surface to recognize enzyme I and all four classes of enzyme IIA, and the binding surfaces on enzymes IIA used to interact with HPr and the corresponding enzymes IIB are highly overlapping, the absence of any detectable interaction between enzyme I and any of the enzymes IIB arises through electrostatic selection. The binding surface on HPr contains no negative charges, and the charged residues on the binding surface of enzyme I are predominantly negative. In contrast, the binding surfaces on enzymes IIA and IIB comprise a mixture of positively and negatively charged residues that largely complement one another. Thus these charged residues are either involved in intermolecular salt bridges, hydrogen-bonding interactions, or participate in van der Waals contacts. Intermolecular electrostatic repulsion, however, between like-charged residues is avoided. The positively charged residues located in the binding surface of the enzymes IIA are accommodated by the binding surface of HPr, either by making use of their long side chains in hydrophobic contacts, or by electrostatic interactions with polar groups (e.g. in the case of the IIAChb*-HPr complex, Arg-58C and Lys-82C of subunit C of IIAChb* interact with the side chain carbonyls of Gln-57 and Gln-51 of HPr, respectively).
Acknowledgments
We thank Dan Garrett and Dusty Baber for software and instrumental support, respectively.
This work was supported, in whole or in part, by the intramural program of the NIDDK, National Institutes of Health and the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to G. M. C.).
This paper is dedicated to the memory of Saul Roseman who discovered the bacterial phosphotransfer system.
The atomic coordinates and experimental NMR restraints (codes 2lrk and 2lrl) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
G. S. Garrett and G. M. Clore, unpublished data.
- PTS
- phosphoenolpyruvate:sugar phosphotransferase system
- enzyme I
- EI
- HPr
- histidine-containing phosphocarrier protein
- Chb
- N,N′-diacetylchitobiose
- IIAChb
- IIBChb, and IICChb, A, B, and C domains, respectively, of the N,N′-diacetylchitobiose transporter IIChb
- IIAChb*
- double mutant of IIAChb comprising a 13-residue deletion at the N terminus and an Asp to Leu mutation at position 92 (wild type numbering)
- HSQC
- heteronuclear single quantum coherence
- TROSY
- transverse relaxation optimized spectroscopy
- NOE
- nuclear overhauser enhancement
- r.m.s.
- root mean square.
REFERENCES
- 1. Kundig W., Ghosh S., Roseman S. (1964) Phosphate bound to histidine in a protein as an intermediate in a novel phosphotransferase system. Proc. Natl. Acad. Sci. U.S.A. 52, 1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meadow N. D., Fox D. K., Roseman S. (1990) The bacterial phosphoenolpyruvate. Glycose phosphotransferase system. Annu. Rev. Biochem. 59, 497–542 [DOI] [PubMed] [Google Scholar]
- 3. Postma P. W., Lengeler J. W., Jacobson G. R. (1993) Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robillard G. T., Broos J. (1999) Structure/function studies on the bacterial carbohydrate transporters, enzymes II, of the phosphoenolpyruvate-dependent phosphotransferase system. Biochim. Biophys. Acta 1422, 73–104 [DOI] [PubMed] [Google Scholar]
- 5. Siebold C., Flükiger K., Beutler R., Erni B. (2001) Carbohydrate transporters of the bacterial phosphoenolpyruvate:sugar phosphotransferase system (PTS). FEBS Lett. 504, 104–111 [DOI] [PubMed] [Google Scholar]
- 6. Deutscher J., Francke C., Postma P. W. (2006) How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Legler P. M., Cai M., Peterkofsky A., Clore G. M. (2004) Three-dimensional solution structure of the cytoplasmic B domain of the mannitol transporter IImannitol of the Escherichia coli phosphotransferase system. J. Biol. Chem. 279, 39115–39121 [DOI] [PubMed] [Google Scholar]
- 8. Suh J. Y., Tang C., Cai M., Clore G. M. (2005) Visualization of the phosphorylated active site loop of the cytoplasmic B domain of the mannitol transporter IIMannitol of the Escherichia coli phosphotransferase system by NMR spectroscopy and residual dipolar couplings. J. Mol. Biol. 353, 1129–1136 [DOI] [PubMed] [Google Scholar]
- 9. van Montfort R. L., Pijning T., Kalk K. H., Reizer J., Saier M. H., Jr., Thunnissen M. M., Robillard G. T., Dijkstra B. W. (1997) The structure of an energy-coupling protein from bacteria, IIBcellobiose, reveals similarity to eukaryotic protein-tyrosine phosphatases. Structure 5, 217–225 [DOI] [PubMed] [Google Scholar]
- 10. Ab E., Schuurman-Wolters G., Reizer J., Saier M. H., Dijkstra K., Scheek R. M., Robillard G. T. (1997) The NMR side chain assignments and solution structure of enzyme IIBcellobiose of the phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli. Protein Sci. 6, 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ab E., Schuurman-Wolters G. K., Nijlant D., Dijkstra K., Saier M. H., Robillard G. T., Scheek R. M. (2001) NMR structure of cysteinyl-phosphorylated enzyme IIB of the N,N′-diacetylchitobiose-specific phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli. J. Mol. Biol. 308, 993–1009 [DOI] [PubMed] [Google Scholar]
- 12. Kalbitzer H. R., Hengstenberg W. (1993) The solution structure of the histidine-containing protein (HPr) from Staphylococcus aureus as determined by two-dimensional 1H-NMR spectroscopy. Eur. J. Biochem. 216, 205–214 [DOI] [PubMed] [Google Scholar]
- 13. Wittekind M., Rajagopal P., Branchini B. R., Reizer J., Saier M. H., Jr., Klevit R. E. (1992) Solution structure of the phosphocarrier protein HPr from Bacillus subtilis by two-dimensional NMR spectroscopy. Protein Sci. 1, 1363–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eberstadt M., Grdadolnik S. G., Gemmecker G., Kessler H., Buhr A., Erni B. (1996) Solution structure of the IIB domain of the glucose transporter of Escherichia coli. Biochemistry 35, 11286–11292 [DOI] [PubMed] [Google Scholar]
- 15. Hu J., Hu K., Williams D. C., Jr., Komlosh M. E., Cai M., Clore G. M. (2008) Solution NMR structures of productive and nonproductive complexes between the A and B domains of the cytoplasmic subunit of the mannose transporter of the Escherichia coli phosphotransferase system. J. Biol. Chem. 283, 11024–11037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cai M., Williams D. C., Jr., Wang G., Lee B. R., Peterkofsky A., Clore G. M. (2003) Solution structure of the phosphoryl transfer complex between the signal-transducing protein IIAGlucose and the cytoplasmic domain of the glucose transporter IICBGlucose of the Escherichia coli glucose phosphotransferase system. J. Biol. Chem. 278, 25191–25206 [DOI] [PubMed] [Google Scholar]
- 17. Garrett D. S., Seok Y. J., Liao D. I., Peterkofsky A., Gronenborn A. M., Clore G. M. (1997) Solution structure of the 30-kDa N-terminal domain of enzyme I of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system by multidimensional NMR. Biochemistry 36, 2517–2530 [DOI] [PubMed] [Google Scholar]
- 18. Schwieters C. D., Suh J. Y., Grishaev A., Ghirlando R., Takayama Y., Clore G. M. (2010) Solution structure of the 128-kDa enzyme I dimer from Escherichia coli and its 146-kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering. J. Am. Chem. Soc. 132, 13026–13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takayama Y., Schwieters C. D., Grishaev A., Ghirlando R., Clore G. M. (2010) Combined use of residual dipolar couplings and solution x-ray scattering to rapidly probe rigid-body conformational transitions in a nonphosphorylatable active-site mutant of the 128-kDa enzyme I dimer. J. Am. Chem. Soc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang C., Williams D. C., Jr., Ghirlando R., Clore G. M. (2005) Solution structure of enzyme IIAChitobiose from the N,N′-diacetylchitobiose branch of the Escherichia coli phosphotransferase system. J. Biol. Chem. 280, 11770–11780 [DOI] [PubMed] [Google Scholar]
- 21. van Montfort R. L., Pijning T., Kalk K. H., Hangyi I., Kouwijzer M. L., Robillard G. T., Dijkstra B. W. (1998) The structure of the Escherichia coli phosphotransferase IIAmannitol reveals a novel fold with two conformations of the active site. Structure 6, 377–388 [DOI] [PubMed] [Google Scholar]
- 22. Teplyakov A., Lim K., Zhu P. P., Kapadia G., Chen C. C., Schwartz J., Howard A., Reddy P. T., Peterkofsky A., Herzberg O. (2006) Structure of phosphorylated enzyme I, the phosphoenolpyruvate:sugar phosphotransferase system sugar translocation signal protein. Proc. Natl. Acad. Sci. U.S.A. 103, 16218–16223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oberholzer A. E., Bumann M., Schneider P., Bächler C., Siebold C., Baumann U., Erni B. (2005) Crystal structure of the phosphoenolpyruvate-binding enzyme I-domain from the Thermoanaerobacter tengcongensis PEP:sugar phosphotransferase system (PTS). J. Mol. Biol. 346, 521–532 [DOI] [PubMed] [Google Scholar]
- 24. Oberholzer A. E., Schneider P., Siebold C., Baumann U., Erni B. (2009) Crystal structure of enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system in the dephosphorylated state. J. Biol. Chem. 284, 33169–33176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Orriss G. L., Erni B., Schirmer T. (2003) Crystal structure of the IIBSor domain of the sorbose permease from Klebsiella pneumoniae solved to 1.75-Å resolution. J. Mol. Biol. 327, 1111–1119 [DOI] [PubMed] [Google Scholar]
- 26. Liao D. I., Kapadia G., Reddy P., Saier M. H., Jr., Reizer J., Herzberg O. (1991) Structure of the IIA domain of the glucose permease of Bacillus subtilis at 2.2-Å resolution. Biochemistry 30, 9583–9594 [DOI] [PubMed] [Google Scholar]
- 27. Liao D. I., Silverton E., Seok Y. J., Lee B. R., Peterkofsky A., Davies D. R. (1996) The first step in sugar transport. Crystal structure of the amino terminal domain of enzyme I of the E. coli PEP:sugar phosphotransferase system and a model of the phosphotransfer complex with HPr. Structure 4, 861–872 [DOI] [PubMed] [Google Scholar]
- 28. Máarquez J., Reinelt S., Koch B., Engelmann R., Hengstenberg W., Scheffzek K. (2006) Structure of the full-length enzyme I of the phosphoenolpyruvate-dependent sugar phosphotransferase system. J. Biol. Chem. 281, 32508–32515 [DOI] [PubMed] [Google Scholar]
- 29. Herzberg O., Reddy P., Sutrina S., Saier M. H., Jr., Reizer J., Kapadia G. (1992) Structure of the histidine-containing phosphocarrier protein HPr from Bacillus subtilis at 2.0-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 89, 2499–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jia Z., Quail J. W., Waygood E. B., Delbaere L. T. (1993) The 2.0-Å resolution structure of Escherichia coli histidine-containing phosphocarrier protein HPr. A redetermination. J. Biol. Chem. 268, 22490–22501 [DOI] [PubMed] [Google Scholar]
- 31. Worthylake D., Meadow N. D., Roseman S., Liao D. I., Herzberg O., Remington S. J. (1991) Three-dimensional structure of the Escherichia coli phosphocarrier protein IIIGlc. Proc. Natl. Acad. Sci. U.S.A. 88, 10382–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sliz P., Engelmann R., Hengstenberg W., Pai E. F. (1997) The structure of enzyme IIAlactose from Lactococcus lactis reveals a new fold and points to possible interactions of a multicomponent system. Structure 5, 775–788 [DOI] [PubMed] [Google Scholar]
- 33. Nunn R. S., Marković-Housley Z., Génovésio-Taverne J. C., Flükiger K., Rizkallah P. J., Jansonius J. N., Schirmer T., Erni B. (1996) Structure of the IIA domain of the mannose transporter from Escherichia coli at 1.7-Å resolution. J. Mol. Biol. 259, 502–511 [DOI] [PubMed] [Google Scholar]
- 34. Schauder S., Nunn R. S., Lanz R., Erni B., Schirmer T. (1998) Crystal structure of the IIB subunit of a fructose permease (IIBLev) from Bacillus subtilis. J. Mol. Biol. 276, 591–602 [DOI] [PubMed] [Google Scholar]
- 35. Navdaeva V., Zurbriggen A., Waltersperger S., Schneider P., Oberholzer A. E., Bahler P., Bachler C., Grieder A., Baumann U., Erni B. (2011) Phosphoenolpyruvate:sugar phosphotransferase system from the hyperthermophilic Thermoanaerobacter tengcongensis. Biochemistry 50, 1184–1193 [DOI] [PubMed] [Google Scholar]
- 36. Garrett D. S., Seok Y. J., Peterkofsky A., Gronenborn A. M., Clore G. M. (1999) Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr. Nature Struct. Biol. 6, 166–173 [DOI] [PubMed] [Google Scholar]
- 37. Wang G., Louis J. M., Sondej M., Seok Y. J., Peterkofsky A., Clore G. M. (2000) Solution structure of the phosphoryl transfer complex between the signal transducing proteins HPr and IIAglucose of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. EMBO J. 19, 5635–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cornilescu G., Lee B. R., Cornilescu C. C., Wang G., Peterkofsky A., Clore G. M. (2002) Solution structure of the phosphoryl transfer complex between the cytoplasmic A domain of the mannitol transporter IIMannitol and HPr of the Escherichia coli phosphotransferase system. J. Biol. Chem. 277, 42289–42298 [DOI] [PubMed] [Google Scholar]
- 39. Williams D. C., Jr., Cai M., Suh J. Y., Peterkofsky A., Clore G. M. (2005) Solution NMR structure of the 48-kDa IIAMannose-HPr complex of the Escherichia coli mannose phosphotransferase system. J. Biol. Chem. 280, 20775–20784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suh J. Y., Cai M., Williams D. C., Jr., Clore G. M. (2006) Solution structure of a post-transition state analog of the phosphotransfer reaction between the A and B cytoplasmic domains of the mannitol transporter IIMannitol of the Escherichia coli phosphotransferase system. J. Biol. Chem. 281, 8939–8949 [DOI] [PubMed] [Google Scholar]
- 41. Suh J. Y., Iwahara J., Clore G. M. (2007) Intramolecular domain-domain association/dissociation and phosphoryl transfer in the mannitol transporter of Escherichia coli are not coupled. Proc. Natl. Acad. Sci. U.S.A. 104, 3153–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suh J. Y., Cai M., Clore G. M. (2008) Impact of phosphorylation on structure and thermodynamics of the interaction between the N-terminal domain of enzyme I and the histidine phosphocarrier protein of the bacterial phosphotransferase system. J. Biol. Chem. 283, 18980–18989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jung Y. S., Cai M., Clore G. M. (2010) Solution structure of the IIAChitobiose-IIBChitobiose complex of the N,N′-diacetylchitobiose branch of the Escherichia coli phosphotransferase system. J. Biol. Chem. 285, 4173–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tang C., Iwahara J., Clore G. M. (2006) Visualization of transient encounter complexes in protein-protein association. Nature 444, 383–386 [DOI] [PubMed] [Google Scholar]
- 45. Suh J. Y., Tang C., Clore G. M. (2007) Role of electrostatic interactions in transient encounter complexes in protein-protein association investigated by paramagnetic relaxation enhancement. J. Am. Chem. Soc. 129, 12954–12955 [DOI] [PubMed] [Google Scholar]
- 46. Fawzi N. L., Doucleff M., Suh J. Y., Clore G. M. (2010) Mechanistic details of a protein-protein association pathway revealed by paramagnetic relaxation enhancement titration measurements. Proc. Natl. Acad. Sci. U.S.A. 107, 1379–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garrett D. S., Seok Y. J., Peterkofsky A., Clore G. M., Gronenborn A. M. (1997) Identification by NMR of the binding surface for the histidine-containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the Escherichia coli phosphotransferase system. Biochemistry 36, 4393–4398 [DOI] [PubMed] [Google Scholar]
- 48. Goto N. K., Gardner K. H., Mueller G. A., Willis R. C., Kay L. E. (1999) A robust and cost-effective method for the production of Val, Leu, Ile(δ1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J. Biomol. NMR 13, 369–374 [DOI] [PubMed] [Google Scholar]
- 49. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 50. Garrett D. S., Powers R., Gronenborn A. M., Clore G. M. (1991) A common sense approach to picking two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 95, 214–220 [DOI] [PubMed] [Google Scholar]
- 51. Clore G. M., Gronenborn A. M. (1991) Structures of larger proteins in solution. Three- and four-dimensional heteronuclear NMR spectroscopy. Science 252, 1390–1399 [DOI] [PubMed] [Google Scholar]
- 52. Yang D., Kay L. E. (1999) Improved 1HN-detected triple resonance TROSY-based experiments. J. Biomol. NMR 13, 3–10 [DOI] [PubMed] [Google Scholar]
- 53. Clore G. M., Gronenborn A. M. (1998) Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 16, 22–34 [DOI] [PubMed] [Google Scholar]
- 54. Cai M., Huang Y., Zheng R., Wei S. Q., Ghirlando R., Lee M. S., Craigie R., Gronenborn A. M., Clore G. M. (1998) Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nature Struct. Biol. 5, 903–909 [DOI] [PubMed] [Google Scholar]
- 55. Clore G. M., Gronenborn A. M. (1998) New methods of structure refinement for macromolecular structure determination by NMR. Proc. Natl. Acad. Sci. U.S.A. 95, 5891–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Clore G. M., Gronenborn A. M., Nilges M., Ryan C. A. (1987) Three-dimensional structure of potato carboxypeptidase inhibitor in solution. A study using nuclear magnetic resonance, distance geometry, and restrained molecular dynamics. Biochemistry 26, 8012–8023 [DOI] [PubMed] [Google Scholar]
- 57. Nilges M. (1993) A calculation strategy for the structure determination of symmetric dimers by 1H NMR. Proteins 17, 297–309 [DOI] [PubMed] [Google Scholar]
- 58. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+. A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Clore G. M. (2000) Accurate and rapid docking of protein-protein complexes on the basis of intermolecular nuclear overhauser enhancement data and dipolar couplings by rigid body minimization. Proc. Natl. Acad. Sci. U.S.A. 97, 9021–9025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schwieters C. D., Clore G. M. (2001) Internal coordinates for molecular dynamics and minimization in structure determination and refinement. J. Magn. Reson. 152, 288–302 [DOI] [PubMed] [Google Scholar]
- 61. Schwieters C. D., Kuszewski J., Clore G. M. (2006) Using Xplor-NIH for NMR molecular structure determination. Progr. NMR Spectros. 48, 47–62 [Google Scholar]
- 62. Clore G. M., Kuszewski J. (2002) χ1 rotamer populations and angles of mobile surface side chains are accurately predicted by a torsion angle database potential of mean force. J. Am. Chem. Soc. 124, 2866–2867 [DOI] [PubMed] [Google Scholar]
- 63. Schwieters C. D., Clore G. M. (2008) A pseudopotential for improving the packing of ellipsoidal protein structures determined from NMR data. J. Phys. Chem. B 112, 6070–6073 [DOI] [PubMed] [Google Scholar]
- 64. Schwieters C. D., Clore G. M. (2001) The VMD-XPLOR visualization package for NMR structure refinement. J. Magn. Reson. 149, 239–244 [DOI] [PubMed] [Google Scholar]
- 65. Nicholls A., Sharp K. A., Honig B. (1991) Protein folding and association. Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 [DOI] [PubMed] [Google Scholar]
- 66. Schwieters C. D., Clore G. M. (2002) Reweighted atomic densities to represent ensembles of NMR structures. J. Biomol. NMR 23, 221–225 [DOI] [PubMed] [Google Scholar]
- 67. Begley G. S., Hansen D. E., Jacobson G. R., Knowles J. R. (1982) Stereochemical course of the reactions catalyzed by the bacterial phosphoenolpyruvate:glucose phosphotransferase system. Biochemistry 21, 5552–5556 [DOI] [PubMed] [Google Scholar]
- 68. Herzberg O. (1992) An atomic model for protein-protein phosphoryl group transfer. J. Biol. Chem. 267, 24819–24823 [PubMed] [Google Scholar]
- 69. Grzesiek S., Stahl S. J., Wingfield P. T., Bax A. (1996) The CD4 determinant for down-regulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry 35, 10256–10261 [DOI] [PubMed] [Google Scholar]
- 70. Clore G. M., Garrett D. S. (1999) R-factor, free R, and complete coss-validation for dipolar coupling refinement of NMR structures. J. Am. Chem. Soc. 121, 9008–9012 [Google Scholar]
- 71. Laskowski R. A., McArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK. A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]