Background: The MtrB sensor kinase is a component of the essential MtrAB signal transduction system.
Results: MtrB localizes to septa independent of phosphorylation; MtrB septal association and phosphorylation are necessary for cell division and expression of MtrA targets.
Conclusion: MtrB septal localization and MtrA-regulon expression are linked.
Significance: MtrB septal assembly triggers the activation of the MtrAB signal transduction pathway.
Keywords: Bacterial Signal Transduction, Cell Division, Gene Expression, Mutant, Mycobacterium, Mycobacterium tuberculosis, Protein Phosphorylation, Response Regulator, Protein Localization, Sensor Kinase
Abstract
The mechanisms responsible for activation of the MtrAB two-component regulatory signal transduction system, which includes sensor kinase MtrB and response regulator MtrA, are unknown. Here, we show that an MtrB-GFP fusion protein localized to the cell membrane, the septa, and the poles in Mycobacterium tuberculosis and Mycobacterium smegmatis. This localization was independent of MtrB phosphorylation status but dependent upon the assembly of FtsZ, the initiator of cell division. The M. smegmatis mtrB mutant was filamentous, defective for cell division, and contained lysozyme-sensitive cell walls. The mtrB phenotype was complemented by either production of MtrB protein competent for phosphorylation or overproduction of MtrAY102C and MtrAD13A mutant proteins exhibiting altered phosphorylation potential, indicating that either MtrB phosphorylation or MtrB independent expression of MtrA regulon genes, including those involved in cell wall processing, are necessary for regulated cell division. In partial support of this observation, we found that the essential cell wall hydrolase ripA is an MtrA target and that the expression of bona fide MtrA targets ripA, fbpB, and dnaA were compromised in the mtrB mutant and partially rescued upon MtrAY102C and MtrAD13A overproduction. MtrB septal assembly was compromised upon FtsZ depletion and exposure of cells to mitomycin C, a DNA damaging agent, which interferes with FtsZ ring assembly. Expression of MtrA targets was also compromised under the above conditions, indicating that MtrB septal localization and MtrA regulon expression are linked. We propose that MtrB septal association is a necessary feature of MtrB activation that promotes MtrA phosphorylation and MtrA regulon expression.
Introduction
Mycobacterium tuberculosis, the causative agent of tuberculosis, grows slowly with an average cell duplication time of 22 h in nutrient broth. One characteristic feature of tuberculosis is persistence, which is a state in which the bacterium is believed to maintain a metabolically active but quiescent state with limited bacterial turnover (1). In liquid culture M. tuberculosis has been shown to shift between an active replicative state and a metabolically active but nonreplicative persistent state depending upon the growth conditions employed (1). It is believed that the pathogen exhibits both of these growth stages during infection. The mining of M. tuberculosis genomic sequence data indicates that the pathogen operates a host of regulatory networks and pathways that aid in its survival under various stressful growth conditions (2). One such regulatory network is the paired histidine-aspartate two-component regulatory signal transduction (2CRS)6 system that includes a membrane bound sensor kinase and a cytosolic response regulator (RR). In response to environmental stimuli, the membrane-bound sensor kinase undergoes autophosphorylation at a conserved histidine residue and then transfers the high energy phosphate to a unique aspartic acid residue on the RR that then promotes or represses select gene transcription (for review, see Refs. 3 and 4).
The M. tuberculosis genome contains 11 paired 2CRSs and several orphan sensor kinases and RRs (2). MtrAB is a system wherein MtrA is the RR and MtrB is the sensor kinase, and collectively, MtrAB is one of the two 2CRSs that have been shown to be essential for M. tuberculosis survival (5–7). Deretic and co-workers (5, 6) showed that mtrA expression was up-regulated after macrophage infection with Mycobacterium bovis BCG, a nonpathogenic vaccine strain, but was relatively unaffected in M. tuberculosis infection, indicating that the expression and possibly the activity of MtrA could influence M. tuberculosis virulence. Earlier, we showed that the overproduction of a phosphorylation-competent wild type (WT) MtrA (MtrAWT) compromised the ability of M. tuberculosis to replicate after infection, whereas the simultaneous overproduction of MtrA and the sensor kinase MtrB reversed the growth defect, suggesting that one role of the MtrB sensor kinase is to help regulate the phosphorylation status of MtrA (8). More recent data have identified oriC and the promoters (P) for dnaA and fbpB as MtrA targets (9). Other recent findings implicate MtrB in cell wall homeostasis in part through its association with the lipoprotein LpqB (10). Although the characterization of an Mycobacterium avium mtrB transposon mutant and a Corynebacterium glutamicum mtrB deletion mutant implicated MtrB in cell wall metabolism (11, 12), the role, if any, of MtrB in M. tuberculosis cell division and cell wall synthesis remains elusive.
MtrB activation is critical for MtrA phosphorylation and subsequent expression of MtrA targets, which are referred to as the MtrA regulon. However, how the MtrB kinase is activated has not been determined, and the features that are important for the activation process are still largely unknown. The present study was designed to address the latter important issue. Given the involvement of the MtrAB system in maintaining cell wall integrity, we considered the possibility that MtrB activation is connected to its association with the septa and possibly its synthesis. Consistent with this assumption, we show that MtrB associates with the septa and poles as well as the membranes and that this septal association is an important feature of its activation process (i.e. via phosphorylation) and optimal MtrA regulon expression.
MATERIALS AND METHODS
Strains and Bacterial Growth Conditions
Escherichia coli strain Top10 used for cloning purposes was propagated in Luria-Bertani (LB) broth or LB agar supplemented with ampicillin (50 mg/ml), kanamycin (50 mg/ml), or hygromycin (200 mg/ml). Mycobacterium smegmatis mc2155 and M. tuberculosis strains were grown in Middlebrook 7H9 broth supplemented with oleic acid-albumin-dextrose-catalase and sodium chloride and appropriate antibiotics as needed.
Molecular Techniques
Plasmids and oligonucleotide primers used in this study are listed in supplemental Tables S1 and S2, respectively. mtrB related constructs: Full-length mtrB or its truncated derivative lacking the first 233 codons (mtrBsol) were amplified using primer combinations MVM883-MVM814 and MR162-MVM814, respectively, and cloned as a PacI-XbaI fragment in pRD3 under the inducible tetracycline promoter (Ptet) by replacing the ftsZ coding region (13) to create plasmids pKS4 and pSVM25, respectively. The recombinant plasmids were integrated at the attB locus in M. tuberculosis and M. smegmatis to create Mtb-Ptet::mtrB-gfp/Mtb-Ptet::mtrBsol-gfp and Msmeg-Ptet::mtrB-gfp/Msmeg-Ptet::mtrBsol-gfp, respectively (see Table 1). To express gfp-mtrB, PCR amplified gfp (primers gfp-F and gfp-R) and mtrB (primers mtrB-F and mtrB-R) coding regions were cloned as a fusion in replicating plasmid pLR52 under the inducible Ptet to create pNM28 (supplemental Tables S1 and S2). In some experiments the mtrB coding region was amplified using primers MtrBstartNde and MVM814 and cloned downstream of the amidase promoter (Pami) in integration proficient vector pJFr19 (8) to create pRD102. MVM892F-MVM892R and MVM891-MVM891R were used along with the QuikChange site-directed mutagenesis kit (Stratagene Corp., La Jolla, CA) to mutate mtrB at codon 305 to create plasmid constructs pKS4H305D and pKS4H305Y expressing Ptet::mtrBH305D-gfp and Ptet::mtrBH305Y-gfp, respectively (Table 1). MVM877-MVM878 primers were used to amplify the mtrBsol coding regions of mtrB, mtrBH305D, and mtrBH305Y, and these were cloned into the pMALc4E vector to express the corresponding maltose-binding protein (MBP) fusions (see pDS21, pSVM16, and pSVM17 in supplemental Table S1). MtrAY102C was created by site-specific mutagenesis using Y102C-F and Y102C-R primers essentially as described for MtrAD13A (14). The coding regions of mtrAD13A and mtrAY102C were cloned into pJFR19 and pMALc4E (14).
BACTH Vectors
The mtrB and ftsZ coding regions were cloned into the low copy vectors pKT25 or pKNT25 and the high copy vectors pUT18 or pUT18C (see supplemental Tables S1 and S2). All cloned fragments were confirmed by sequencing.
Disruption of the mtrBMsmeg Gene at Its Native Chromosomal Locus
A two-step recombination protocol was used for disruption of the M. smegmatis mtrB gene (15). A suicidal recombination delivery vector with a 90% deletion of the mtrB coding region was prepared in three steps. First, a 835-bp region upstream of the mtrB gene was amplified using the primers MR91 and MR92 and cloned as a ScaI-HindIII fragment in the p2NIL vector to create pRD89 ( supplemental Table S2). Next, a 1007-bp fragment containing the 82-bp mtrB 3′ end to ∼900 bp of the downstream gene lpqB was amplified using primers MR89 and MR90 and cloned as a HindIII-NotI fragment into pRD89 to create pRD90. Finally, a 6-kb PacI cassette carrying the lacZ and sacB genes was cloned into pRD90 to create the suicidal recombination vector pRD91. This plasmid was electroporated into M. smegmatis, and single crossover (SCO) recombinants were selected as described (16). SCOs were confirmed by PCR and Southern before processing for double crossovers (DCOs). In some experiments pRD102 (supplemental Table S1) was integrated in an SCO strain at the attB locus and processed for DCOs. MVM883-MVM814 and MVM233-MVM814 primer combinations were used to confirm the sizes of mtrB gene at native and attB loci. MVM223 binds within the Pami. Next, the integrated pRD102 plasmid in the DCO strain was swapped with the pMV306K (Kmr, empty vector) to create the mtrB deletion strain as previously described (16).
Purification of FtsZ, MtrB, and MtrA Proteins
His-FtsZTB, MBP fusion derivatives of MtrAWT, MtrAD13A, MtrAY102C, soluble MtrB, MtrBH305D, and MtrBH305Y, and His- MtrAD13A were purified following the protocols as previously described (14, 17). Preliminary experiments indicated that MBP-MtrAD13A was not active, whereas His-MtrAD13A was active; hence, His-MtrAD13A was used to evaluate the promoter DNA binding experiments.
Phosphorylation Assay
Phosphorylation of WT and mutant MtrB proteins (5 μm) with [γ-32P]ATP was performed essentially as described (14).
Electrophoretic Mobility Shift Assay (EMSA)
Interactions of MtrA and phosphorylated MtrA (MtrA∼P) with 5′-6-carboxyfluorescein-labeled PripA or oriC were assessed by EMSA as described (9). A 200-bp ripA upstream region was amplified using 6-carboxyfluorescein-labeled primers MVM782 and MVM783. MtrA or MtrA∼P was incubated at 0.1, 0.25, 0.5, or 1.0 μm with 200 fmol of PripA in 50 mm Tris-HCl, pH 7.5, buffer containing 50 mm NaCl, 10 mm MgCl2, 10 mm CaCl2, and 5% glycerol, 20 pmol of poly(dI/dC), 1 μg of sheared salmon sperm DNA, and 1 μg of BSA. Samples were incubated for 15 min at 37 °C, resolved on polyacrylamide gels, and protein-DNA complexes were visualized using a Bio-Rad Molecular Imager Fx. For controls, PripA was incubated with EnvZ kinase, and the complexes were processed as described above.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP was carried with formaldehyde-cross-linked M. tuberculosis lysates and anti-MtrA and mock antibodies as described (9).
RNA Extraction, Reverse Transcription, and Quantitative Real-time (qRT) PCR
Total RNA was extracted from 7H9 broth-grown cultures of M. smegmatis as previously described (8, 9, 16, 18, 19). RNA was reverse-transcribed using random hexamers (Invitrogen) and 200 units of Moloney murine leukemia virus reverse transcriptase (Promega), and the cDNA obtained was used to perform qRT-PCR in a Bio-Rad iCycler iQ5TM Real-Time PCR detection system using 2× iQ SYBR Supermix (Bio-Rad). The primers used for qRT-PCR are listed in supplemental Table S3. The threshold cycle (Ct) value of each gene of interest was normalized to the Ct value of sigA, and the -fold expression was calculated using the formula, -fold change = 2−Δ(ΔCT). Final expression data were determined from an average of three independent RNA preparations that were reverse-transcribed and quantified by real-time PCR; each gene of interest was investigated in triplicate.
Microscopy
Visualization of M. smegmatis and M. tuberculosis cells and quantification of fluorescent structures were essentially as described (13, 20). M. tuberculosis samples were always fixed in 4% paraformaldehyde before visualization. This led to quenching of fluorescence signals in some cases. Therefore, where applicable quantification of localization patterns were performed in appropriate M. smegmatis strains.
Western Blotting
MtrB protein was detected in the lysates by immunoblotting after electrophoresis on 12% SDS-PA gels as previously described (13). Parallel blots were probed with affinity-purified α-MtrB and α-SigA antibodies. SigA was used as a loading control.
Cell Wall Hydrolysis Assays
Preparations of crude cell walls containing mycolyl arabinogalactan from M. smegmatis strains, fluorescein labeling, and determination of cell wall hydrolysis activity were performed essentially as described in Chauhan et al. (16). Percent hydrolysis was calculated relative to the input fluorescent counts.
RESULTS
MtrB Is an Abundant Protein That Localizes to Cell Poles, Septal Sites, and Membranes
To begin evaluating the roles of the MtrB sensor kinase in M. tuberculosis physiology and cell wall metabolism, we determined the intracellular levels of MtrB in actively growing cells using the recombinant MtrB as an internal standard. When calculated, ∼12,000 MtrB molecules/M. tuberculosis cell were found. Slightly lower levels (∼7500 molecules) of MtrB were found in M. smegmatis, which grows more rapidly, with an average doubling time of 3 h. We reported earlier that the intracellular levels of FtsZ, MtrA, and CrgA proteins in M. tuberculosis correspond to 30,000, 4,000, and 20,000 molecules/actively growing cell, respectively (8, 20, 21). Thus, MtrB appears to be a relatively abundant protein in M. tuberculosis.
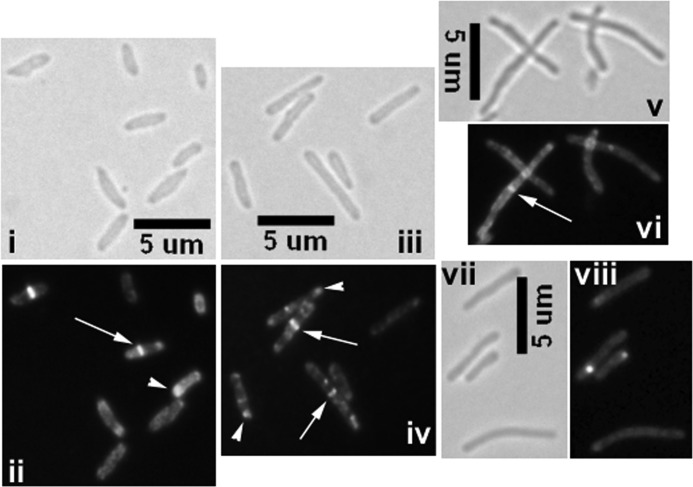
Next, we visualized MtrB-GFP localization patterns in Mtb-Ptet::mtrB-gfp carrying pKS4 integrated at the attB locus. An increase of ∼2.5-fold in MtrB levels was noted upon the addition of inducer anhydrotetracycline (supplemental Fig. S1). Under these growth conditions, bright septal, polar, and membrane localization of MtrB-GFP was evident (Fig. 1). When quantified, ∼16 and 21% of cells showed midcell and polar localizations, respectively (Fig. 1, panel ii; see arrows and arrowheads). Interestingly, many cells exhibited a wide and bulging cell phenotype (Fig. 1, panel i). The septal localization pattern of MtrB suggests its association with the septasome and/or nascent growth zones.
FIGURE 1.
Visualization of MtrB-GFP structures in M. tuberculosis and M. smegmatis. Mtb-Ptet::mtrB-gfp (i and ii), Msmeg-Ptet::mtrB-gfp (iii and iv), or Msmeg-Ptet::mtrBsol-gfp (v and vi) strains were examined by brightfield and fluorescence microscopy. Inducer anhydrotetracycline was added at a final concentration of 10 ng/ml. As controls, M. smegmatis dosS-gfp expressed from Ptet was tested (panels vii and viii). In panels ii, iv, and vi fluorescent structures are marked: arrow, midcell localization; arrowhead, polar localization. Note that Msmeg-Ptet::mtrBsol-gfp cells show diffuse fluorescence (panel vi), whereas Ptet::dosS-gfp cells do not show any distinct septal localization (panel viii).
The MtrB proteins of M. tuberculosis and M. smegmatis show significant sequence identity and 94% amino acid similarity, indicating that these proteins might be functionally conserved. Accordingly, we visualized the MtrB-GFP localization pattern in Msmeg-Ptet::mtrB-gfp. A localization pattern similar to Mtb-Ptet::mtrB-gfp was evident; quantification of the MtrB structures revealed that 20 and 39% of the cells showed midcell and the polar localizations, respectively (Fig. 1, panels iii and iv). The frequency of MtrB-GFP septal localization observed was comparable with that reported for FtsZ-GFP (13, 17). Production of MtrB-GFP in M. smegmatis did not affect cell length (data not shown).
As a control, the localization pattern of the DosS (also referred to as DevS)-GFP sensor kinase was visualized. DosS is a component of the DosRS 2CRS (2). DosS-GFP localized to membranes like the MtrB but not to the septa, which is in contrast to the observations associated with MtrB (Fig. 1, panels vii and viii). These results indicate that not every sensor kinase localizes to the septa. Next, we visualized GFP-MtrB structures and found localization patterns similar to MtrB-GFP (supplemental Fig. S2, panel ii) except that the GFP-MtrB structures at the septa appeared to be less stable and tended to bleach fast, presumably reflecting a partially active protein (see below). Nonetheless, these results suggest that the observed structures are not due to artifacts associated with the localization of the fusion protein (also see below).
The Cytosolic Domain of MtrB Is Sufficient for Septal Localization
MtrB is a 567-residue membrane-bound sensor kinase with two potential transmembrane domains (residues 42–62 and 213–233), an HAMP linker (residues 215–284), and a His-kinase A phosphoacceptor domain (residues 302–519). We recently reported that the MtrB fragment containing the C-terminal 334 residues (amino acid 234 to the end, designated as MtrBsol) is soluble and competent for exhibiting autophosphorylation and transphosphorylation activities (14). We found that the MtrBsol-GFP protein localized to midcell sites in M. smegmatis (Msmeg-Ptet::mtrBsol-gfp) but also showed an overall diffuse fluorescence throughout the cell (Fig. 1, panels v–vi). These results revealed that although the cytosolic domain is sufficient for septal localization, an intact membrane domain appears to be required for stable MtrB septal localization.
MtrB Septal Localization Requires FtsZ Assembly
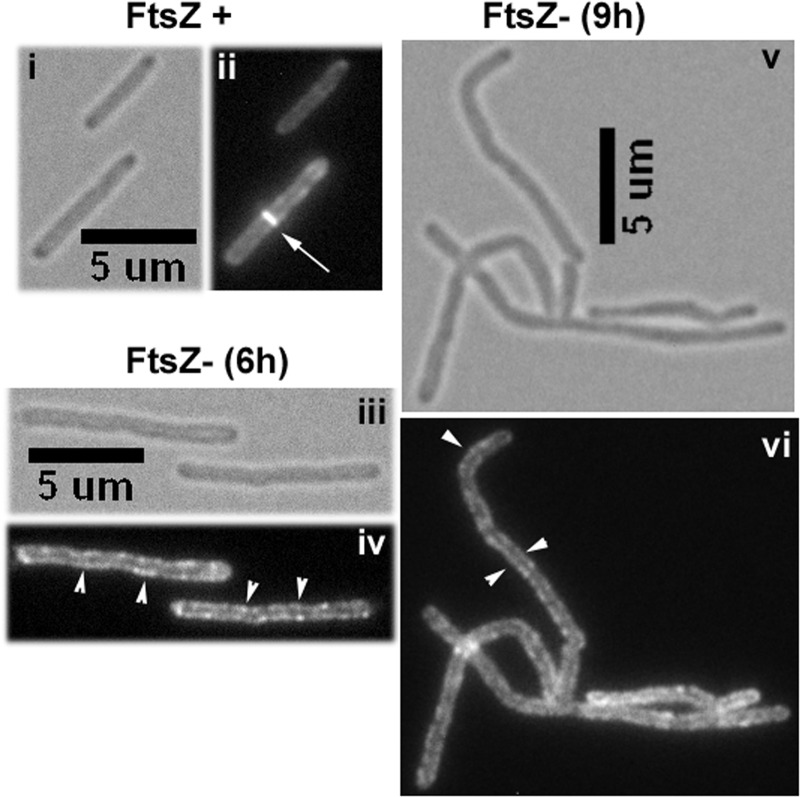
FtsZ, a homolog of tubulin, assembles as a ring at the midcell site to initiate the cell division process. Studies with other bacteria revealed that the assembly of other proteins at the septa and FtsZ interactions, either direct or indirect, with some of the assembled proteins leads to productive cell division (for a recent review, see Ref. 22). To evaluate whether MtrB-GFP septal localization was dependent on FtsZ assembly, the pRD73 plasmid was electroporated into an M. smegmatis ftsZ conditional expression strain, FZ3, where ftsZ expression is under the control of Pami (23), to create FZ3-Ptet::mtrB-gfp. In the FZ3 strain, the removal of acetamide results in the depletion of intracellular FtsZ levels and the production of filamentous cells that are sometimes branched (23). When grown in the presence of acetamide, ∼25% of cells had distinct MtrB-GFP structures at midcell sites in FZ3-Ptet::mtrB-gfp (Fig. 2, panel ii). However, when acetamide was removed to deplete FtsZ, ∼3% of the cells retained MtrB structures at septa after 6 h, which corresponds to ∼2 doubling times; nevertheless, the ability of MtrB to localize to membranes was not compromised (Fig. 2, panels iii and iv). Similar results were found 9 h after the removal of acetamide except that the cells were more elongated (Fig. 2, panels v and vi). Together, these data indicate that MtrB is recruited to midcell sites after the midcell FtsZ-ring assembly and that MtrB septal structures are compromised in the absence of FtsZ.
FIGURE 2.
MtrB localization under FtsZ depletion conditions. FZ3, M. smegmatis ΔftsZ, Pami::ftsZ strain (23) expressing Ptet::mtrB-gfp was grown with 10 ng/ml anhydrotetracycline but in the presence (i and ii) or in the absence (iii to vi) of acetamide for 6 h (iii and iv) and 9 h (v and vi) and examined by brightfield (panels i, iii, and v) and fluorescence (panels ii, iv, and vi) microscopy. Bars denote the length in microns. Note distinct Z-rings (marked with arrows) present in the panel ii are absent in panels iv and vi. Arrowhead, membrane localization.
Bacterial two-hybrid experiments (24, 25) did not show interactions between the MtrB and FtsZ proteins (supplemental Fig. S3). In these experiments constructs producing full-length FtsZ and MtrB proteins as C-terminal fusions to either the T18 or T25 domains of the Bordetella pertussis adenylate cyclase protein were cotransformed into the E. coli BTH101 strain deficient for adenylate-cyclase and spotted on minimal media agar indicator plates containing β-galactosidase substrate as described (26). No blue color was evident in the transformants carrying mtrB/ftsZ constructs (supplemental Fig. S3). In contrast, cotransformants containing ftsZ/ftsZ and the positive control gcn4/gcn4 exhibited an intense blue color. Pulldown experiments with E. coli lysates containing MBP-MtrB and His-FtsZ proteins and nickel-nitrilotriacetic acid resin showed that the FtsZ protein fractions eluted from the nickel affinity columns did not contain MtrB (supplemental Fig. S4, panel ii). Parallel experiments with His-FtsZ and MBP-ClpX, an FtsZ interacting protein (13), showed co-purification of ClpX with His-FtsZ on nickel-nitrilotriacetic acid resin (supplemental Fig. S4, panel i). Independently, co-immunoprecipitation experiments with M. smegmatis lysates containing MtrB and FtsZ proteins and anti-FtsZ antibodies showed that the FtsZ immunoprecipitated samples did not contain MtrB (see supplemental Fig. S4, panel iii, lane E1). In contrast, M. smegmatis lysates containing FtsZ and CrgA, a FtsZ interacting protein (20), recovered CrgA along with FtsZ in the FtsZ immunoprecipitated samples (supplemental Fig. S4, panel iii, lane E2). Together, these results confirm that FtsZ and MtrB proteins do not interact.
mtrB Contributes to, but Is Not Essential, for Cell Division
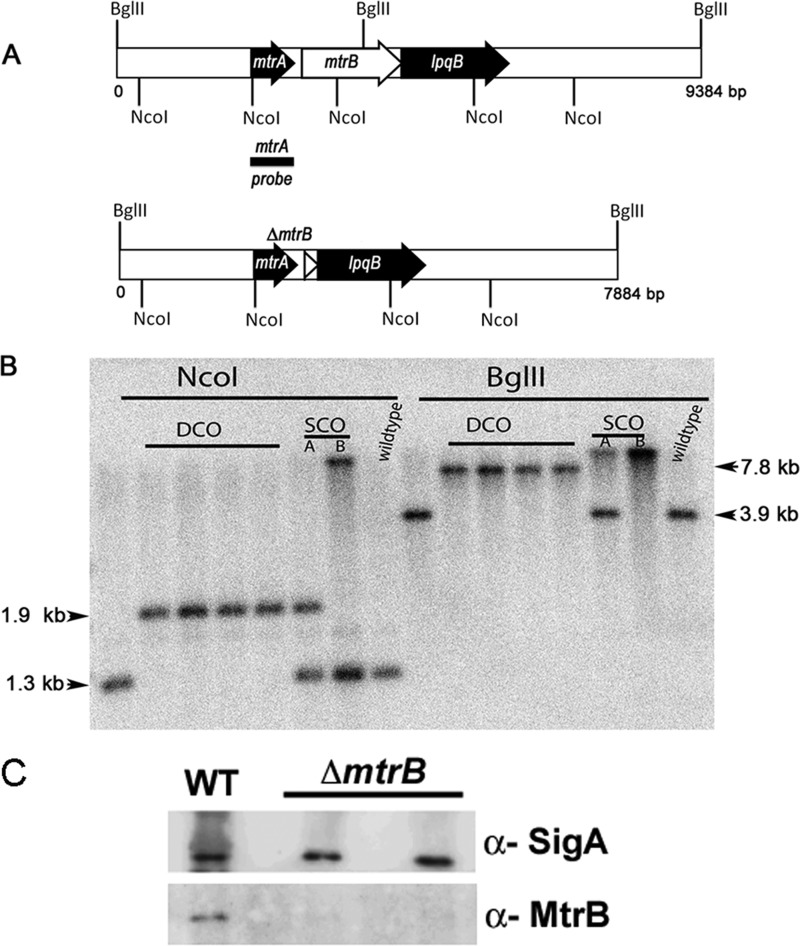
Next, we created an M. smegmatis mtrB deletion strain to evaluate its roles (Fig. 3). Our initial attempts to disrupt the mtrB gene in the WT M. smegmatis background were unsuccessful, as PCR analysis revealed that 80 of 80 DCOs contained the intact mtrB gene at the native locus (data not shown). However, when DCOs were processed from an mtrB SCO containing pRD102 expressing Pami::mtrB integrated at the attB locus, 1 of 8 DCO recombinants exhibited a mutant pattern (data not shown), and Southern blot analysis confirmed the mtrB deletion at its native locus (Fig. 3B). The pRD102 was later swapped with an empty plasmid to create an M. smegmatis ΔmtrB strain. The absence of the MtrB protein was confirmed by immunoblotting (Fig. 3C). One pure colony was propagated and used in all subsequent experiments.
FIGURE 3.
Construction of M. smegmatis mtrB mutant. A, shown is a schematic depicting the 9.3-kb mtrB region of M. smegmatis (top panel) and the mutant mtrB region (bottom panel). The region is not drawn to scale. The locations of NcoI and BglII sites are shown. B, shown is a Southern blot confirming the deletion of mtrB. Genomic DNA was isolated from WT M. smegmatis, two SCO (A and B under SCO), and four DCO strains, digested with BglII or NcoI enzymes, transferred to nitrocellulose membrane, and probed with 32P-labeled mtrA fragment (black bar shown in the top panel). Using this probe, we detected bands corresponding to 7.8 kb for mutant and 3.9 kb for wild type with BglII digestion but 1.98 kb for mutant and 1.38 kb for WT with NcoI. Positions of SCO, DCO and chromosomal copy mtrB bands are marked. C, shown is immunoblotting of M. smegmatis mtrB strains. For confirmation of the loss of MtrB protein, protein lysates from select DCOs along with WT strain were prepared, 5 μg of protein was resolved by SDS-PAGE in 12% gels, and immunoblotting was performed with affinity purified α-MtrB antibodies. Parallel blots were processed and probed with α-SigA antibodies.
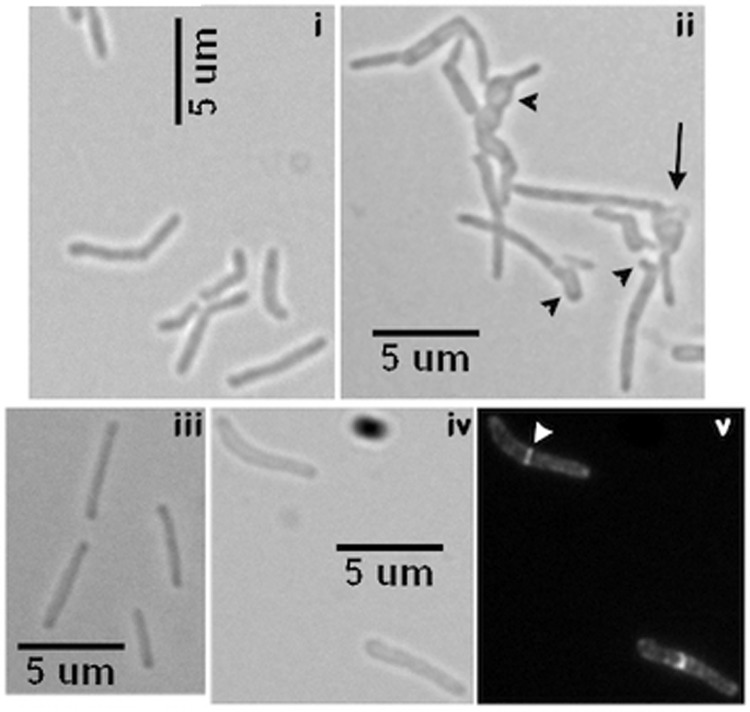
M. smegmatis ΔmtrB grew poorly in broth, was clumpy, and exhibited filamentous cells, which often had bulbous regions and exhibited a chain-like phenotype indicating possible defects in septum splitting and cell wall metabolism. Consequently, precise cell length measurements could not be made (Fig. 4, compare panels i and ii). In contrast, actively dividing WT M. smegmatis cells were short and ∼3–4 μm in length (Fig. 4, panel i). The ΔmtrB complement strain expressing either M. tuberculosis Pami::mtrB (pRD102) or Ptet::mtrB-gfp (pKS4) grew like the WT with similar cell morphology (Fig. 4, panels iii–v) and showed distinct MtrB-GFP septal, polar, and membrane localization (Fig. 4, panel v; data not shown). These latter results indicated that the mtrBTB or mtrBTB-gfp gene product can functionally replace the M. smegmatis counterpart and that MtrB activity is required for some aspect of cell division. We also found that ΔmtrB transformed with pNM28 expressing Ptet::gfp-mtrB remained filamentous and showed very few septal structures (supplemental Fig. S2, panels iii and iv), indicating a lack of complementation. Instead, fluorescent aggregates were found randomly located along the length of the cells (supplemental Fig. S2, panel iv). It is likely that either the GFP-MtrB fusion protein is partially active and/or is defective for interactions with proteins that otherwise promote stable association of MtrB with the septa.
FIGURE 4.
Phenotypic characterization of mtrB mutant. Visualization of M. smegmatis (panel i) WT and (panel ii) ΔmtrB strains by brightfield microscopy is shown. Actively growing cultures in liquid were examined. Note: the black arrow in panel ii indicates cell lysis. Black arrowheads, buds, branches, and swollen cell morphology. Complementation of ΔmtrB phenotype: panel iii, Pami::mtrBTB (pRD102); panels iv-v, Ptet::mtrB-gfp (pKS4). Panel iv represents a brightfield image, whereas v represents a fluorescence image. White arrowhead, midcell MtrB-GFP band.
MtrB Phosphorylation Activity Is Necessary for Productive Cell Division
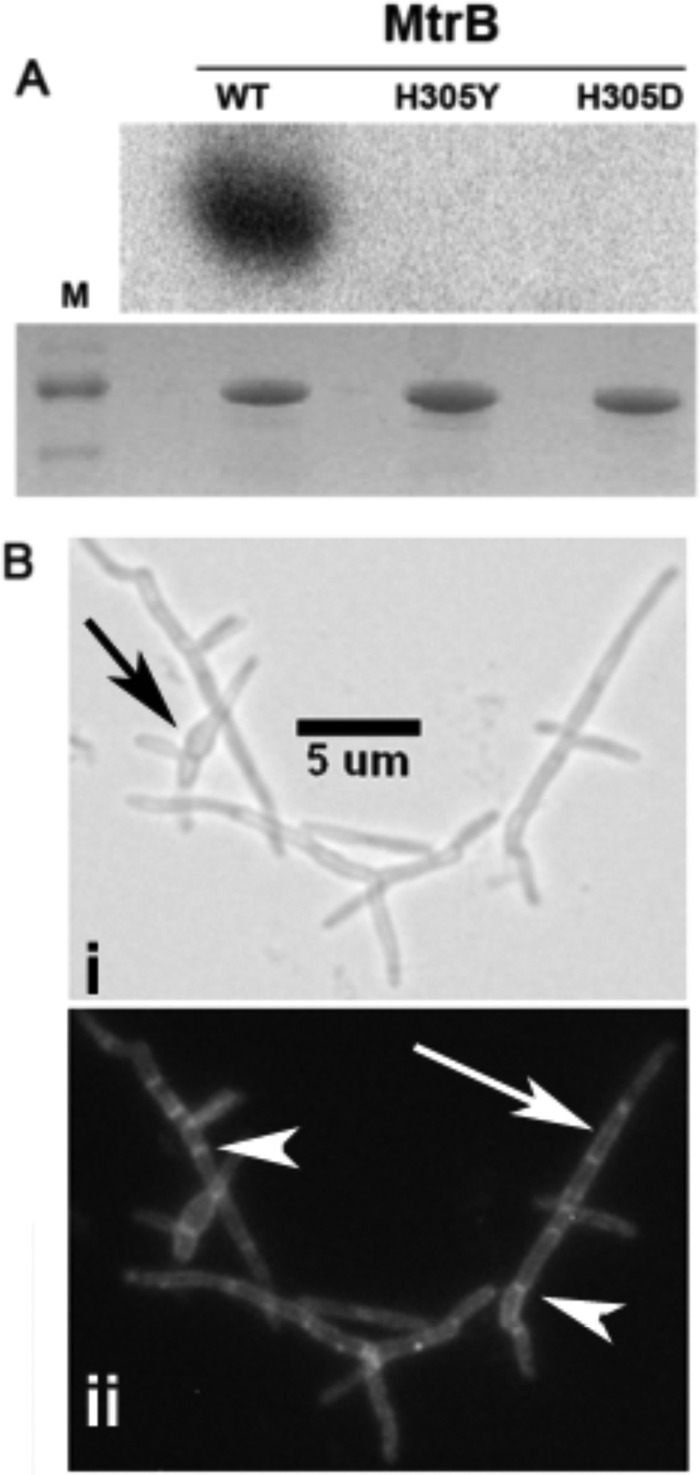
To test whether MtrB phosphorylation activity is necessary for its septal localization, we replaced the conserved histidine residue of MtrB (27) at position 305 with either tyrosine (MtrBH305Y) or aspartic acid (MtrBH305D) and characterized the mutant proteins. Incubation of MBP-MtrB proteins with [γ-32P]ATP revealed a distinctly labeled protein with WT MtrB protein confirming autophosphorylation activity (Fig. 5A, top panel). In contrast, neither MtrBH305D nor MtrBH305Y produced a labeled band (Fig. 5A, top panel). Because equal amounts of proteins were used to evaluate MtrB autophosphorylation activity (Fig. 5A, bottom panel), we concluded that the mutant MtrB proteins were defective for autophosphorylation.
FIGURE 5.
In vitro phosphorylation and in vivo localization of mutant MtrB-GFP structures. A, recombinant MtrB, MtrBH305Y, and MtrBH305D proteins were tested for autophosphorylation activity in buffer containing Ca2+ and Mg2+ and [γ-32P]ATP as described in the Materials and Methods section. Samples were incubated for 30 min and resolved by SDS-PAGE, and the radioactivity in the protein bands was visualized in a Bio-Rad Molecular Imager Fx (top panel). A parallel gel was stained with Coomassie Blue and visualized (bottom panel). M, protein size markers. Note that distinct phosphorylation was seen with wild-type MtrB but not with mutant proteins. B, visualization of MtrBH305Y-GFP structures in M. smegmatis ΔmtrB strain is shown. Panels i shows the bright-field image, whereas panel ii shows the fluorescent image. Note: these cells remained filamentous but contained distinct bands at septa. Black arrow, bulged morphology; white arrow, membrane localization; white arrowhead, septal localization.
Next, we transformed the ΔmtrB strain with Ptet::mtrBH305Y-gfp and Ptet::mtrBH305D-gfp expressing plasmids and visualized the phenotype and MtrB localization patterns in the corresponding strains (Fig. 5B, panels i and ii). As observed, these cells remained filamentous with bulgy regions (compare Fig. 5B, panel i, with Fig. 4, panels iii–v) but contained distinct membrane and septal localizations (Fig. 5B, see the white arrow and arrowheads in panel ii; only data with Ptet::mtrBH305Y-gfp are shown). We also found that ΔmtrB transformed with plasmids expressing either MtrBH305Y or MtrBH305D had a similar phenotype (data not shown). These results indicate that not only the MtrB septal localization but also the subsequent downstream events are necessary for optimal cell division.
We found that M. tuberculosis strains producing MtrBH305Y-GFP and MtrBH305D-GFP showed diffuse and less intense fluorescence in less than 2% of cells (supplemental Fig. S5). It is known that sensor kinase phosphorylation promotes its oligomerization activity (3, 4, 28). Thus, the diffuse fluorescence of MtrBH305Y-GFP and MtrBH305D-GFP could be due to defective oligomerization activity of these proteins. Alternatively, the diffuse fluorescence could also be due to the paraformaldehyde fixation step, which quenches fluorescence.
Increased Lysozyme Susceptibility of mtrB Mutant Cell Walls
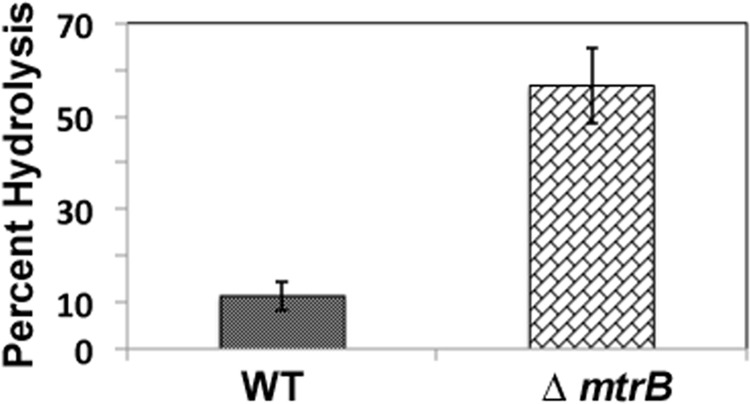
Elongated cells with the bulbous structures of the ΔmtrB strain suggested defects in cell wall peptidoglycan (PG) metabolism, which could in turn modulate access to PG of enzymes that act on glycan strands and murein sacculus (29, 30). Mycobacterial PG is generally known to be resistant to lysozyme, a muramidase that cleaves the glycosidic linkages between the sugar residues of the PG (31, 32). To test this assumption, the sensitivity of WT and ΔmtrB cell walls to lysozyme was tested. Consistent with our assumption, cell walls from ΔmtrB were relatively more sensitive to lysozyme compared with those from WT (Fig. 6).
FIGURE 6.
Cell wall hydrolysis activity of MtrB proteins. Cell wall preparations from M. smegmatis WT and ΔmtrB strains were prepared, labeled with fluorescein, and incubated with 1 mg/ml lysozyme in 50 mm Tris-HCl 8.0 and 1 mm EDTA for 4 h. Reactions were arrested by adding 4 m LiCl, and released labeled products in the supernatant relative to the total labeled input cell wall were measured in a Jasco FP 6500 fluorimeter. Average percent hydrolysis from three independent experiments is shown.
Overproduction of Mutant MtrA Proteins Reverses ΔmtrB Phenotype
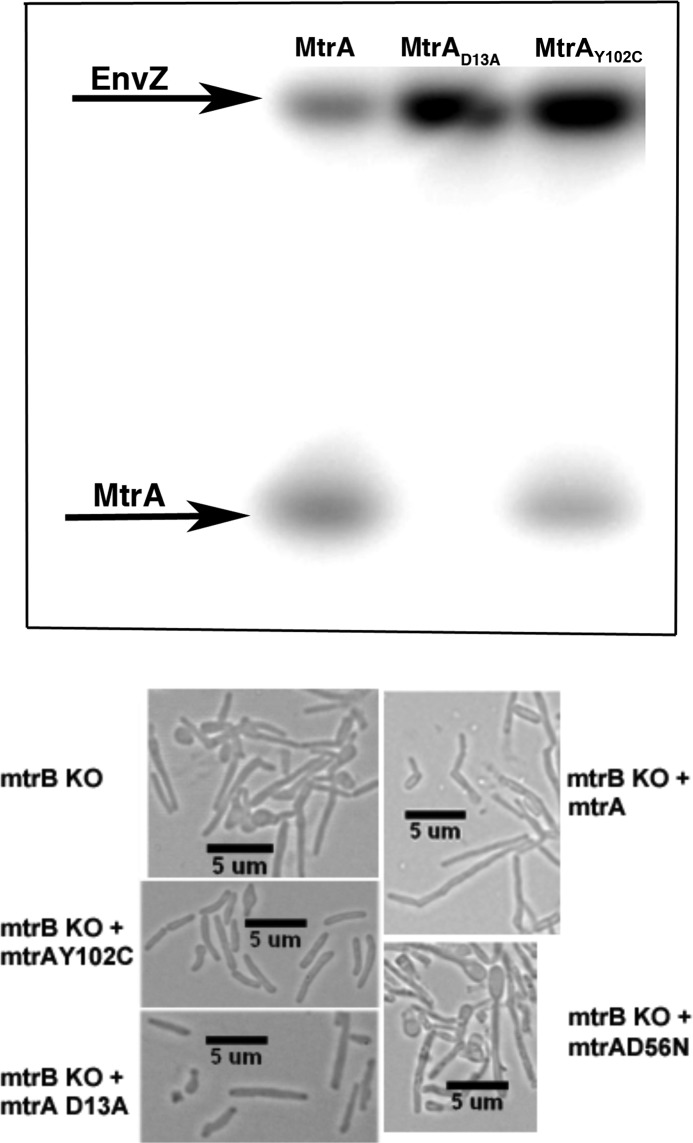
What might be the consequence of MtrB septal association? We considered the possibility that MtrB septal association promotes its autophosphorylation activity, which could in turn lead to MtrA phosphorylation and regulated expression of MtrA regulon targets including those important for cell wall metabolism. To address the issue, we first searched for MtrA proteins that no longer require MtrB for phosphorylation and yet promote the reversal of the ΔmtrB phenotype. In this regard, a candidate of interest is MtrAD13A, which has a mutation in the signal receiver domain and is phosphorylation-defective but is proficient at binding to its target PfbpB and modulating its expression (14). We further confirmed this assumption by evaluating MtrAD13A binding to the oriC target by EMSA (9). Again, MtrAD13A bound oriC proficiently in the absence of ATP (supplemental Fig. S6). Although these studies are limited in nature, we hypothesized that MtrAD13A is a constitutively active protein. MtrA crystal data indicate that the signal-receiving and DNA binding domains are locked and that tyrosine at position 102 is at the interdomain interface (33). It was proposed that mutations at this locus modulate interdomain orientation and possibly favor MtrA phosphorylation (33). Thus, MtrAY102C could be another candidate of interest. To further characterize MtrAY102C, we evaluated its phosphorylation ability by incubating with heterologous kinase EnvZ (9). It was shown earlier that MtrA is poorly phosphorylated by small phosphoryl donors but can be phosphorylated by EnvZ (8, 33). MtrAY102C, unlike MtrAD13A, was found to be phosphorylation-competent (Fig. 7, upper panel). Next, we transformed the ΔmtrB strain with integrating plasmids pMZ3 and pDS4 producing MtrAD13A and MtrAY102C, respectively, and evaluated the ΔmtrB phenotype. As seen, much of the ΔmtrB filamentous phenotype was reversed in the presence of the acetamide inducer (Fig. 7, lower panel; compare mtrAD13A and mtrAY102C panels with control ΔmtrB). However, some cells remained elongated and showed bulbous regions indicating that reversal of the phenotype was not complete. Overproduction of either MtrAWT or MtrAD56N, which is defective for phosphorylation (8), did not reverse the ΔmtrB phenotype (Fig. 7, lower panel). The MtrAD56N,unlike MtrAD13A, is not proficient at binding to its target, fbpB (14). Together, these results emphasize that activities of the MtrAY102C and MtrAD13A proteins are important for reversing the ΔmtrB phenotype.
FIGURE 7.
Characterization of the effects of mutant MtrA proteins on mtrB phenotype. Upper panel, shown is an autoradiogram evaluating the phosphorylation potential of MtrA proteins. MtrAWT, MtrAD13A, and MtrAY102C were incubated with EnvZ and [γ-32P]ATP, and products were resolved by SDS-PAGE and visualized by phosphorimaging. Lower panel, shown is morphology of ΔmtrB-producing mutant MtrA proteins. ΔmtrB transformed with plasmids producing MtrAWT (ΔmtrB + mtrAWT), phosphorylation-defective MtrA (ΔmtrB + mtrAD13A, ΔmtrB + mtrAD56N), or phosphorylation-proficient MtrA (ΔmtrB + mtrAY102C) were grown with acetamide for 16 h, and cells were visualized by brightfield microscopy. Note near restoration of WT phenotype with plasmids producing either mtrAY102C or mtrAD13A.
Essential Cell Wall Hydrolase RipA Is an MtrA Target
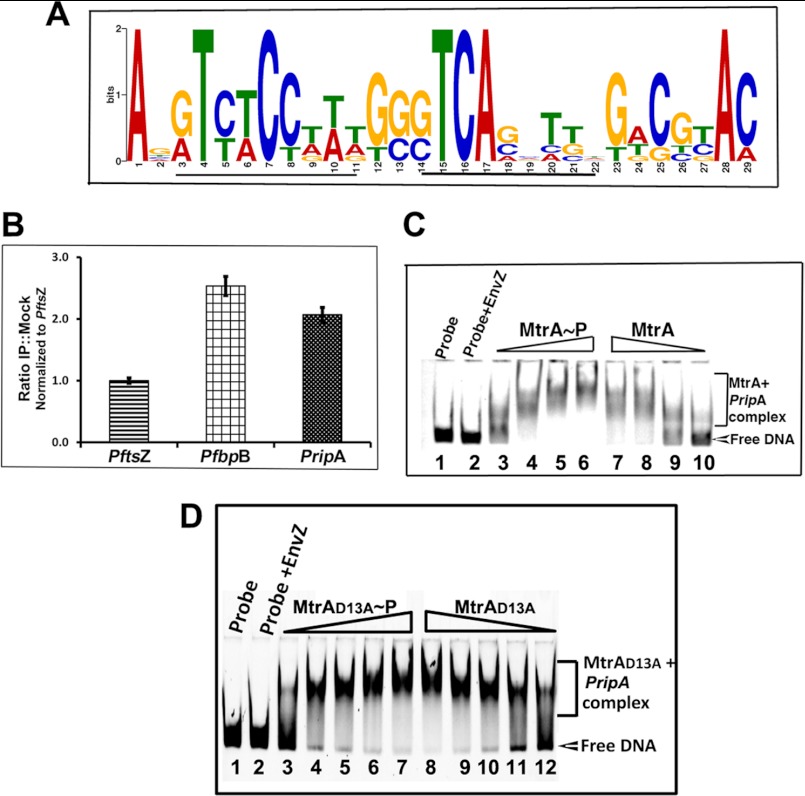
Cell wall hydrolases, which are members of the cell wall-processing enzymes, act at late stages of cell division and are critical for hydrolyzing PG layers connecting daughter cells and, hence, for daughter cell separation. Thus, defects in cell wall hydrolase activities often lead to filamentation, chain-phenotype, and possibly altered cell shapes (16, 34–36). The M. tuberculosis genome is annotated to contain several hydrolases; however, to date, RipA, ChiZ, and CwlM have been shown to possess cell wall hydrolysis activities (16, 36, 37). Furthermore, modulated levels of RipA and ChiZ result in filamentation and cell shape changes (16, 36). Given the phenotype similarities of ΔmtrB with chiZ and ripA strains, we considered the possibility that some hydrolases are members of MtrA regulon; hence, their expression levels may have been compromised in ΔmtrB. Accordingly, we first searched for the MtrA motif-like sequence in the 5′ upstream sequence of the ripA, chiZ, and cwlM coding regions (data not shown) and found that only the 5′ upstream regions of ripA of M. tuberculosis and M. smegmatis contained well conserved MtrA motif-like sequences (Fig. 8A). Next, we performed ChIP experiments and showed that the PripA, similar to the positive control PfbpB (Fig. 8B), was enriched with anti-MtrA antibodies. We further validated the ChIP data by EMSA (Fig. 8C) and identified MtrAWT -RipA complexes. Although modest, MtrAWT∼P appears to retard PripA better than MtrAWT (Fig. 8C, compare lanes 3 and 4 with 9 and 10). We also found that MtrAD13A protein retarded PripA (Fig. 8D). Interestingly, MtrAD13A bound to PripA efficiently in the absence of ATP, indicating phosphorylation-independent binding. We conclude from these experiments that ripA is an MtrA target.
FIGURE 8.
MtrA binds to ripA promoter. A, Logo analysis shows the MtrA motif in the 5′-upstream regions of the ripA gene. The 250-bp upstream sequences (also called the promoter region) of the ripA-coding region of M. smegmatis and M. tuberculosis were analyzed by MEME analysis (50). MtrA-motif like sequences were identified and used to generate MtrA-motif logo. Although not shown, the M. tuberculosis PripA region has two motifs starting at positions 90 and 142 in the + strand and at 214 in the − strand, whereas M. smegmatis has two motifs at positions 62 and 115 in the + strand and 214 in the − strand. Potential 9-base motif is underlined. B, ChIP experiments are shown. M. tuberculosis cell lysates prepared after formaldehyde cross-linking were used for immunoprecipitation with α-MtrA or mock antibodies. Protein-DNA complexes obtained were processed for removal of cross-links and subjected to PCR using primers specific to PftsZ, PripA, and PfbpB. PCR reactions were done in duplicate for different dilutions of template, products were resolved by agarose gel electrophoresis, gels were visualized by staining with SYBR Green dye in a Bio-Rad Molecular Imager Fx, and bands were quantified using Quantity One software (Bio-Rad). Results are presented as ratios of immunoprecipitates (IP) to mock and normalized against FtsZ values, and data shown are the average of at least three independent experiments. C, shown is polyacrylamide gel analysis of PripA- MtrAWT complexes. MtrAWT protein was phosphorylated by EnvZ protein in the presence of ATP. MtrAWT and MtrAWT∼P were incubated individually with 200 fmol of 6-carboxyfluorescein-labeled PripA for 15 min at 37 °C, and samples were resolved by polyacrylamide gel electrophoresis and visualized in a Bio-Rad Molecular Imager Fx. The positions of ripA probe and MtrAWT-PripA complexes are marked. Note MtrAWT∼P bound PripA better than MtrA. MtrAWT or MtrAWT∼P was used at 0.1, 0.25, 0.5, and 1.0 μm. D, shown is a polyacrylamide gel analysis of PripA-MtrA/MtrAD13A complexes. All details are as in panel C, except that MtrAD13A was used at 5, 10, 15, 20, and 25 μm.
MtrB Septal Assembly Is Necessary for Its Activation
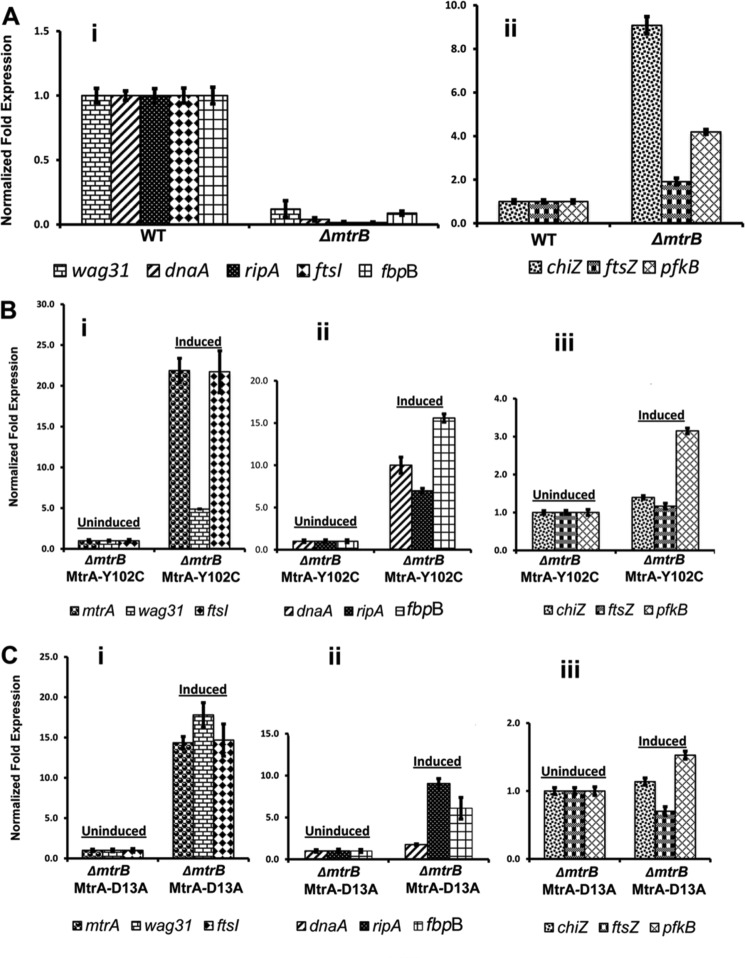
Next, to understand the correlation between MtrB septal localization and MtrA regulon expression, we evaluated the expression levels of MtrA targets dnaA (8), fbpB (9), and ripA (this study) by qRT-PCR. As controls, we measured the expression levels of ftsZ and chiZ (non-MtrA targets (9)) and pfkB, a gene linked to the dosR regulon (38). We also evaluated the expression levels of two other cell division genes, ftsI and wag31. We noted a significant reduction in the expression levels of dnaA, fbpB, and ripA in ΔmtrB cells (Fig. 9A, panel i, p values ≤ 0. 0001); however, expression levels of ftsZ, pfkB, and chiZ were not decreased (Fig. 9A, panel ii, p value ≤ 0.1271) but instead increased for chiZ and pfkB. These latter results are consistent with the reported findings that conditions that induce stress often lead to an increase in the expression levels of chiZ and pfkB (Refs. 16 and 39);see Fig. 9A, panel ii). Expression levels of ftsI and wag31 were also reduced in ΔmtrB (Fig. 9A, panel i). However, overexpression of ftsI or wag31 in the ΔmtrB background did not reverse its phenotype (data not shown). We do not know whether ftsI and wag31 are MtrA targets, but their expression could be an indirect consequence of the absence of MtrB or its septal association.
FIGURE 9.
qRT-PCR analysis of MtrA targets. Total RNA from WT and ΔmtrB cells was extracted and reverse-transcribed, and qRT-PCR was performed as described under “Materials and Methods.” Expression levels of select genes relative to housekeeping gene sigA were compared, and the values are presented as -fold difference in the expression of mutant relative to WT cells. A, shown are qRT-PCR expression levels of select targets in ΔmtrB background. Panel i includes the -fold expression levels of wag31, dnaA, ripA, ftsI, and fbpB, whereas panel ii includes the expression levels of chiZ, ftsZ, and pfkB. B, qRT-PCR expression levels of select targets in ΔmtrB background-overproducing mutant MtrAY102C. Uninduced refers to cultures grown in the absence of acetamide, whereas induced indicates cultures grown in the presence of 0.2% acetamide for 12 h. Panel i shows the expression levels of mtrA, wag31, and ftsI; panel ii shows the data for dnaA, ripA, and fbpB, and panel iii shows the qRT-PCR data for chiZ, ftsZ, and pfkB. Note the y axis scale in panel iii is different from panels i and ii. C, shown are qRT-PCR expression levels of select targets in ΔmtrB background overproducing mutant MtrAD13A. Other details are as described for panels i–iii under B.
Consistent with the reversal of the ΔmtrB phenotype, MtrAY102C and MtrAD13A overproduction increased the expression levels of dnaA, ripA, and fbpB (Fig. 9, B and C, panel ii, p values ≤ 0.01) and ftsI and wag31 (Fig. 9, B and C, panel i, p values ≤ 0.002). The expression of chiZ and ftsZ was not significantly affected (see panel iii of Fig. 9, B and C), whereas that of pfkB was modestly affected in ΔmtrB overproducing MtrAY102C (see Fig. 9B, panel iii).
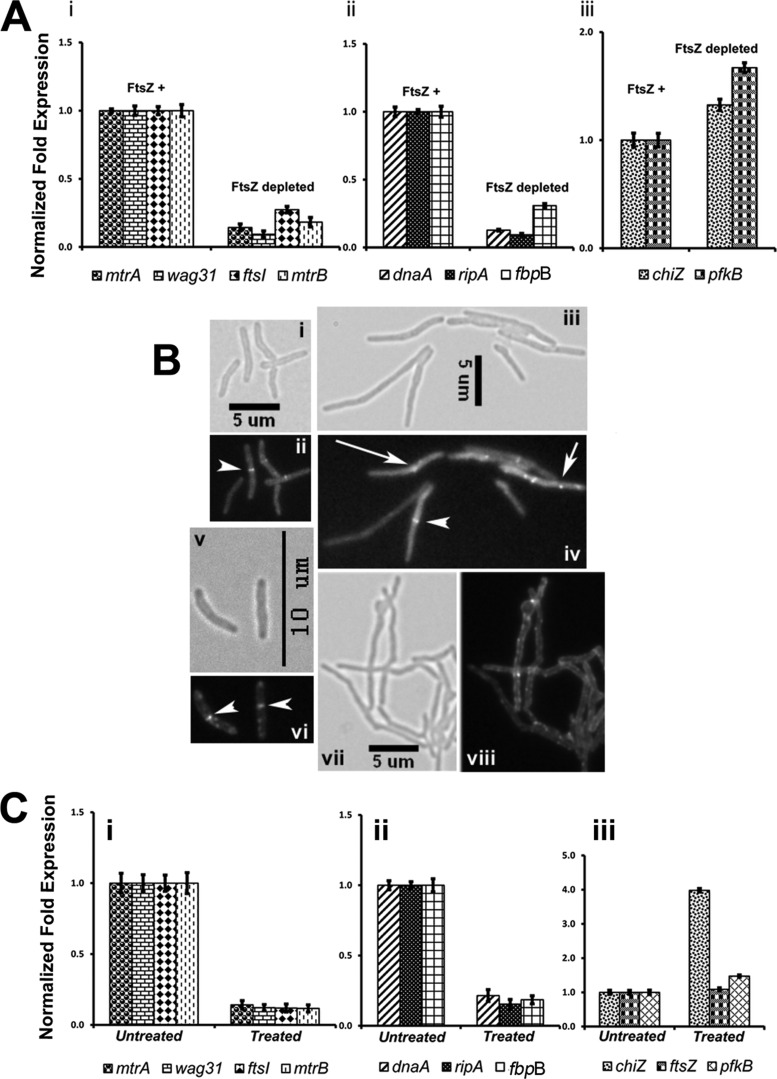
Next, we evaluated MtrA regulon expression in M. smegmatis under conditions that interfered with MtrB septal association, e.g. FtsZ depletion (see Fig. 2B). A reduction in the expression levels of dnaA, fbpB, and ripA (Fig. 10A, panel ii, p value ≤ 0.002) and ftsI, wag31, mtrA, and mtrB (Fig. 10A, panel i, p value = 0.0001) was noted under FtsZ depletion conditions, i.e. 9 h after the removal of acetamide. Similar results were also obtained 6 h after the removal of acetamide (data not shown). In contrast, the expression levels of pfkB and chiZ, which are not MtrA targets, were not significantly affected (Fig. 10A, panel iii, p values ≤ 0.1).
FIGURE 10.
Evaluation of select mtrA targets under conditions affecting MtrB-septal assembly. A, qRT-PCR expression levels of select mtrA targets under conditions affecting FtsZ-septal assembly. Actively growing M. smegmatis FZ-3, ftsZ conditional expression strain, with acetamide (referred to as FtsZ+) was harvested, washed with acetamide-free medium, and grown without acetamide (FtsZ-depleted) or with acetamide (FtsZ+) for 9 h. Total RNA from FtsZ+ and FtsZ-depleted cultures was prepared, and qRT-PCR was performed as described under Fig. 9. Panel i shows expression profiles of mtrA, mtrB, wag31, and ftsI; panel ii shows data for dnaA, ripA, and fbpB; panel iii shows data for chiZ and pfkB. Note: expression levels of chiZ and pfkB in panel iii were modestly affected. B, shown is the effect of mitomycin C exposure on MtrB-GFP and FtsZ-GFP structures. M. smegmatis expressing ftsZ-gfp (panels i–iv) or mtrB-gfp (panels (v–viii) were exposed to 0.5 μg/ml mitomycin C (panels iii, iv, vii, and viii) or grown untreated (panels i, ii, v, and vi) for 6 h, and cells were visualized by microscopy. Panels i, iii, v, and vii show brightfield images, whereas panels ii, iv, vi, and viii show fluorescent images. Septal localizations of FtsZ-GFP (panel ii and iv) and MtrB-GFP (panel vi) are marked with arrowheads. Note that majority of mitomycin C-treated cultures did not contain predominant FtsZ-GFP (panel iv) or MtrB-GFP (panel viii) septal localizations. Aberrant FtsZ-GFP localizations are marked with arrows (panel iv). C, qRT-PCR expression profiles of select MtrA-targets upon exposure to mitomycin C are shown. Total RNA from untreated and 6-h mitomycin C-treated cultures of M. smegmatis was prepared, and qRT-PCR analysis of select targets was performed as described above. Panel i shows the expression profiles of mtrA, wag31, ftsI, and mtrB; panel ii shows data for dnaA, ripA, and fbpB, whereas panel iii shows data for chiZ, ftsZ, and pfkB.
As another measure for the validation of the above concept, we exposed M. smegmatis cells to mitomycin C, a compound that damages DNA and inhibits cell division (16). Mitomycin C-exposed cells were elongated (Fig. 10B; compare panel iii with i) and defective for FtsZ-ring assembly (Fig. 10B; compare panel iv with ii). The majority of the filamentous cells either lacked Z-rings or showed aberrant FtsZ-GFP localizations. Mitomycin C exposure also compromised MtrB-GFP septal assembly (Fig. 10B, compare panels vii and viii for mitomycin C-treated samples with panels v and vi for untreated samples). These results indicated that MtrB associates with septa after the stable FtsZ-ring assembly (Fig. 2). Mitomycin C exposure also reduced the expression levels of dnaA, fbpB, and ripA (Fig. 10C, panel ii, p values ≤ 0. 0001) and mtrA, mtrB, ftsI, and wag31 (Fig. 10C, panel i, p values ≤ 0. 0001). The expression levels of ftsZ and pfkB were not affected, whereas that of chiZ was elevated (Fig. 10C, panel iii). The latter result was consistent with the published report that mitomycin C exposure leads to elevated expression of chiZ (16). Together, these results indicate that a defect in MtrB septal assembly leads to defective MtrA regulon expression and that the observed changes in the MtrA-target expression are not due to any pleiotropic effects associated with FtsZ depletion or mitomycin C exposure (Fig. 10).
DISCUSSION
The principal conclusion of our study is that MtrB septal association and expression of the MtrA regulon are linked. Our conclusion was inferred from the following observations. First, MtrB, a relatively abundant protein, associates with cell poles, membranes, and the septa; the last (Fig. 1) is independent of its phosphorylation status (Fig. 5). Second, MtrA regulon expression is compromised under conditions that interfere with MtrB septal localization (Figs. 9 and 10). Finally, the MtrB requirement for MtrA regulon expression is bypassed, albeit partially, in cells overproducing MtrAY102C and MtrAD13A, which are proteins that show altered phosphorylation potential (Fig. 9). Together, these results lead to a proposal that MtrB sensor kinase activation is promoted and/or occurs at the septa and that this process in turn leads to autophosphorylation of MtrB and the associated signal transduction process.
MtrB septal association implies that it is a septasomal complex component and has a role in cell division, and the reported mtrB phenotypes are consistent with this assumption (Figs. 4–6). Because overproduction of MtrAY102C and MtrAD13A proteins partially reversed the ΔmtrB phenotype (Fig. 7, lower panel), we reason that MtrA regulon expression is compromised in the ΔmtrB background. Because the phenotype reversal is not complete, we think that MtrB activity also directly contributes to some as yet unidentified steps of cell division. Although not directly pertinent to this study, the ΔmtrB phenotype reversal by the phosphorylation-competent MtrAY102C protein suggests that either other kinases and/or small molecule donors, e.g. acetyl∼P, promote phosphorylation of MtrAY102C in vivo.
MtrB, like other membrane-bound sensor kinases, is exposed to both extracellular and intracellular environments. Thus, MtrB association with membranes is expected; however, the observed septal association is rather unique and is yet not reported for other members of the histidine-aspartate family of sensor kinases in mycobacteria. In partial support of this statement, we showed that the DosS kinase does not localize to midcell sites (Fig. 1, panel viii). However, we note that the YycFG 2CRS in other bacteria is shown to be involved in cell division and cell wall metabolism. Like the MtrAB system, YycFG is essential in Bacillus subtilis and Staphylococcus aureus (40–42), and the YycG sensor kinase associates with midcell sites and colocalizes with FtsZ (43). There are, however, some differences between the MtrB and YycG systems. For example, YycG localization appears to be confined to septal sites, and the YycG kinase, unlike MtrB, contains a PAS domain, that is essential for sensing cellular redox status (for review, see Ref. 42). Other studies showed that the YycFG system is involved in the regulation of ftsZ expression (43) and PG metabolism (44–47). Although these data suggest that the MtrAB system in mycobacteria is functionally similar to the YycFG system of B. subtilis and other low guanine + cytosine bacteria, considering the pathogenic life cycle of M. tuberculosis and the absence of a PAS domain, there will likely be differences in the gene targets between the two systems.
Considering the MtrB membrane localization, one might assume that its septal localization could be a consequence of the joining of two flat membranes. We argue against this possibility because we showed that MtrBsol protein, which lacks the membrane domains, localized to septal sites, albeit with low efficiency (Fig. 1, panels v–vi). Additionally, the full-length MtrB was localized either as sharp foci or intense bands at septal sites. Finally, a larger percentage of M. tuberculosis and M. smegmatis cells had localizations at the cell poles compared with midcell sites. Accordingly, we suggest that MtrB localization is dynamic and that MtrB shifts from the cell membrane to the midcell and poles as cell division progresses. This hypothesis raises the possibility that a hitherto undiscovered protein(s) could guide MtrB from the membrane to septal sites and poles as cell division proceeds. Because GFP-MtrB is not proficient at complementing the mtrB mutant phenotype (supplemental Fig. S2), unlike the MtrB-GFP fusion protein (Fig. 4), we propose that the N terminus of MtrB could be crucial for productive interactions with other proteins.
Our studies led to a discovery that the ripA promoter is an MtrA target. The essential cell wall hydrolase is believed to work in concert with its interacting partner, RpfB, and promote the reactivation process wherein the M. tuberculosis pathogen exits from a non-replicating state to resume active multiplication (36, 48). Thus, the ability of MtrB to affect ripA expression via MtrA could be one way of achieving regulated cell wall metabolism. Our results also showed that the expression of chiZ, although not an MtrA target, was modulated rather indirectly (see Fig. 9). Presumably, the expression levels and, therefore, the activities of the cell wall hydrolases such ripA and chiZ need to be tightly regulated for optimized cell division.
The histidine-aspartate sensor kinase family proteins, because of the presence of an extracellular sensor domain, are implicated in sensing external signals, such as changes in nutrients, reactive oxygen, nitric oxide stress, pH, and osmolarity, and engage in the signal transduction process for modulating expression of RR regulons (4). It is known that multiple signals often activate a single 2CRS, and the same signal activates multiple 2CRSs (for review, see Refs. 4 and 49). The signals that activate the essential MtrAB system are unknown. Although our studies do not focus on the identification of MtrAB system activating signals per se, they nonetheless emphasize the importance of the steps subsequent to and/or along with the signal sensing process that is essential for the activation of the MtrAB system. We propose that the activation of the MtrAB system includes at least two steps. The first step is MtrB sensing the external signals, and the signal sensing process is currently undefined, and the second step is MtrB association with septa for which we provided evidence. We hypothesize that both of these steps should go hand-in-hand. Our results, shown in Figs. 9 and 10, support this conclusion. Thus, we envision that even though the same signal or stimuli are sensed by other histidine-aspartate sensor kinases, the ability of the MtrB protein to associate with the septa provides the much needed distinction and specificity for activation of the MtrA regulon. Future studies will focus on identifying the components, if any, in the septa that prime MtrB for activation. It is possible that MtrB associates with other septasomal complex components or that it could directly interact with the nascent PG products during the activation process. A detailed understanding of the events necessary for MtrAB activation will help us evaluate how this essential 2CRS contributes to mycobacterial growth and proliferation.
Acknowledgments
We thank members of Madiraju and Rajagopalan laboratories for stimulating discussions, Drs. D. Stankowska for help with EMSA experiments, and Sabine Ehrt for Ptet plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants AI48417 (to M. R.) and AI084734 (to M. M.).

This article contains supplemental Tables S1–S3 and Figs. S1–S6.
- 2CRS
- two-component regulatory signal transduction
- RR
- response regulator
- MtrAWT
- wild type MtrA
- SCO
- single crossover
- DCO
- double crossovers
- qRT
- quantitative real-time
- PG
- peptidoglycan.
REFERENCES
- 1. Smith I. (2003) Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 16, 463–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 3. Hoch J. A. (2000) Two component and phosphorelay signal transduction. Curr. opin. Microbiol. 3, 165–170 [DOI] [PubMed] [Google Scholar]
- 4. Stock A. M., Robinson V. L., Goudreau P. N. (2000) Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 [DOI] [PubMed] [Google Scholar]
- 5. Via L. E., Curcic R., Mudd M. H., Dhandayuthapani S., Ulmer R. J., Deretic V. (1996) Elements of signal transduction in Mycobacterium tuberculosis. In vitro phosphorylation and in vivo expression of the response regulator MtrA. J. Bacteriol. 178, 3314–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zahrt T. C., Deretic V. (2000) An essential two-component signal transduction system in. Mycobacterium tuberculosis. J. Bacteriol. 182, 3832–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haydel S. E., Malhotra V., Cornelison G. L., Clark-Curtiss J. E. (2012) The prrAB two-component system is essential for Mycobacterium tuberculosis viability and is induced under nitrogen-limiting conditions. J. Bacteriol. 194, 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fol M., Chauhan A., Nair N. K., Maloney E., Moomey M., Jagannath C., Madiraju M. V., Rajagopalan M. (2006) Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol. Microbiol. 60, 643–657 [DOI] [PubMed] [Google Scholar]
- 9. Rajagopalan M., Dziedzic R., Al Zayer M., Stankowska D., Ouimet M. C., Bastedo D. P., Marczynski G. T., Madiraju M. V. (2010) Mycobacterium tuberculosis origin of replication and the promoter for immunodominant secreted antigen 85B are the targets of MtrA, the essential response regulator. J. Biol. Chem. 285, 15816–15827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nguyen H. T., Wolff K. A., Cartabuke R. H., Ogwang S., Nguyen L. (2010) A lipoprotein modulates activity of the MtrAB two-component system to provide intrinsic multidrug resistance, cytokinetic control, and cell wall homeostasis in Mycobacterium. Mol. Microbiol. 76, 348–364 [DOI] [PubMed] [Google Scholar]
- 11. Cangelosi G. A., Do J. S., Freeman R., Bennett J. G., Semret M., Behr M. A. (2006) The two-component regulatory system mtrAB is required for morphotypic multidrug resistance in Mycobacterium avium. Antimicrob. Agents Chemother. 50, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Möker N., Brocker M., Schaffer S., Krämer R., Morbach S., Bott M. (2004) Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility, and expression of genes involved in osmoprotection. Mol. Microbiol. 54, 420–438 [DOI] [PubMed] [Google Scholar]
- 13. Dziedzic R., Kiran M., Plocinski P., Ziolkiewicz M., Brzostek A., Moomey M., Vadrevu I. S., Dziadek J., Madiraju M., Rajagopalan M. (2010) Mycobacterium tuberculosis ClpX interacts with FtsZ and interferes with FtsZ assembly. PLoS ONE 5(7):e11058, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Al Zayer M., Stankowska D., Dziedzic R., Sarva K., Madiraju M. V., Rajagopalan M. (2011) Mycobacterium tuberculosis mtrA merodiploid strains with point mutations in the signal-receiving domain of MtrA exhibit growth defects in nutrient broth. Plasmid 65, 210–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parish T., Stoker N. G. (2000) Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146, 1969–1975 [DOI] [PubMed] [Google Scholar]
- 16. Chauhan A., Lofton H., Maloney E., Moore J., Fol M., Madiraju M. V., Rajagopalan M. (2006) Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 62, 132–147 [DOI] [PubMed] [Google Scholar]
- 17. Rajagopalan M., Maloney E., Dziadek J., Poplawska M., Lofton H., Chauhan A., Madiraju M. V. (2005) Genetic evidence that mycobacterial FtsZ and FtsW proteins interact and colocalize to the division site in Mycobacterium smegmatis. FEMS Microbiol. Lett. 250, 9–17 [DOI] [PubMed] [Google Scholar]
- 18. Chauhan A., Madiraju M. V., Fol M., Lofton H., Maloney E., Reynolds R., Rajagopalan M. (2006) Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J. Bacteriol. 188, 1856–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maloney E., Stankowska D., Zhang J., Fol M., Cheng Q. J., Lun S., Bishai W. R., Rajagopalan M., Chatterjee D., Madiraju M. V. (2009) The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog. 5, e1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plocinski P., Ziolkiewicz M., Kiran M., Vadrevu S. I., Nguyen H. B., Hugonnet J., Veckerle C., Arthur M., Dziadek J., Cross T. A., Madiraju M., Rajagopalan M. (2011) Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J. Bacteriol. 193, 3246–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dziadek J., Madiraju M. V., Rutherford S. A., Atkinson M. A., Rajagopalan M. (2002) Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology 148, 961–971 [DOI] [PubMed] [Google Scholar]
- 22. Adams D. W., Errington J. (2009) Bacterial cell division. Assembly, maintenance, and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642–653 [DOI] [PubMed] [Google Scholar]
- 23. Dziadek J., Rutherford S. A., Madiraju M. V., Atkinson M. A., Rajagopalan M. (2003) Conditional expression of Mycobacterium smegmatis ftsZ, an essential cell division gene. Microbiology 149, 1593–1603 [DOI] [PubMed] [Google Scholar]
- 24. Karimova G., Dautin N., Ladant D. (2005) Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187, 2233–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Daniel R. A., Noirot-Gros M. F., Noirot P., Errington J. (2006) Multiple interactions between the transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. J. Bacteriol. 188, 7396–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hsing W., Russo F. D., Bernd K. K., Silhavy T. J. (1998) Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J. Bacteriol. 180, 4538–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bijlsma J. J., Groisman E. A. (2003) Making informed decisions. Regulatory interactions between two-component systems. Trends Microbiol. 11, 359–366 [DOI] [PubMed] [Google Scholar]
- 29. Vollmer W. (2006) The prokaryotic cytoskeleton. A putative target for inhibitors and antibiotics? Appl. Microbiol. Biotechnol. 73, 37–47 [DOI] [PubMed] [Google Scholar]
- 30. Vollmer W. (2008) Targeting the bacterial Z-ring. Chem. Biol. 15, 93–94 [DOI] [PubMed] [Google Scholar]
- 31. Dhople A. M., Hanks J. H. (1980) Effect of cholesterol, lecithin, and nucleotides on the in vitro growth of. Mycobacterium lepraemurium. Jpn. J. Exp. Med. 50, 469–471 [PubMed] [Google Scholar]
- 32. Mizuguchi Y., Tokunaga T. (1975) Genetics of Mycobacterium. Nippon Saikingaku Zasshi. 30, 297–313 [PubMed] [Google Scholar]
- 33. Friedland N., Mack T. R., Yu M., Hung L. W., Terwilliger T. C., Waldo G. S., Stock A. M. (2007) Domain orientation in the inactive response regulator Mycobacterium tuberculosis MtrA provides a barrier to activation. Biochemistry 46, 6733–6743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bernhardt T. G., de Boer P. (2003) The Escherichia coli amidase AmiC is a periplasmid septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48, 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bernhardt T. G., de Boer P. (2004) Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor murein hydrolase activity. Mol. Microbiol. 52, 1255–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hett E. C., Chao M. C., Steyn A. J., Fortune S. M., Deng L. L., Rubin E. J. (2007) A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol. Microbiol. 66, 658–668 [DOI] [PubMed] [Google Scholar]
- 37. Deng L. L., Humphries D. E., Arbeit R. D., Carlton L. E., Smole S. C., Carroll J. D. (2005) Identification of a novel peptidoglycan hydrolase CwlM in Mycobacterium tuberculosis. Biochim. Biophys. Acta 1747, 57–66 [DOI] [PubMed] [Google Scholar]
- 38. Sherman D. R., Voskuil M., Schnappinger D., Liao R., Harrell M. I., Schoolnik G. K. (2001) Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. U.S.A. 98, 7534–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ryndak M., Wang S., Smith I. (2008) PhoP, a key player in Mycobacterium tuberculosis virulence. Trends Microbiol. 16, 528–534 [DOI] [PubMed] [Google Scholar]
- 40. Dubrac S., Bisicchia P., Devine K. M., Msadek T. (2008) A matter of life and death. Cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70, 1307–1322 [DOI] [PubMed] [Google Scholar]
- 41. Fabret C., Hoch J. A. (1998) A two-component signal transduction system essential for growth of Bacillus subtilis. Implications for anti-infective therapy. J. Bacteriol. 180, 6375–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Winkler M. E., Hoch J. A. (2008) Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 190, 2645–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fukushima T., Szurmant H., Kim E. J., Perego M., Hoch J. A. (2008) A sensor histidine kinase co-ordinates cell wall architecture with cell division in Bacillus subtilis. Mol. Microbiol. 69, 621–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barendt S. M., Sham L. T., Winkler M. E. (2011) Characterization of mutants deficient in the l,d-carboxypeptidase (DacB) and WalRK (VicRK) regulon, involved in peptidoglycan maturation of Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 193, 2290–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bisicchia P., Lioliou E., Noone D., Salzberg L. I., Botella E., Hübner S., Devine K. M. (2010) Peptidoglycan metabolism is controlled by the WalRK (YycFG) and PhoPR two-component systems in phosphate-limited Bacillus subtilis cells. Mol. Microbiol. 75, 972–989 [DOI] [PubMed] [Google Scholar]
- 46. Bisicchia P., Noone D., Lioliou E., Howell A., Quigley S., Jensen T., Jarmer H., Devine K. M. (2007) The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65, 180–200 [DOI] [PubMed] [Google Scholar]
- 47. Fukushima T., Furihata I., Emmins R., Daniel R. A., Hoch J. A., Szurmant H. (2011) A role for the essential YycG sensor histidine kinase in sensing cell division. Mol. Microbiol. 79, 503–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hett E. C., Chao M. C., Deng L. L., Rubin E. J. (2008) A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog. 4, e1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zahrt T. C., Wozniak C., Jones D., Trevett A. (2003) Functional analysis of the Mycobacterium tuberculosis MprAB two-component signal transduction system. Infect. Immun. 71, 6962–6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bailey T. L., Bodén M., Buske F. A., Frith F., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S. (2009) MEME SUITE: tools for motif discovery and searching, Nucleic Acids Res. 37, W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]