Background: GLUT4 glucose transporters are trapped and sequestered intracellularly in adipocytes by TUG.
Results: Insulin stimulates TUG cleavage, which separates regions of TUG that bind GLUT4 and Golgi matrix proteins. Cleavage is required for highly insulin-responsive GLUT4 translocation.
Conclusion: TUG proteolysis liberates GLUT4 trapped at the Golgi matrix.
Significance: Endoproteolytic cleavage is a novel biochemical mechanism for insulin action to regulate glucose uptake.
Keywords: GLUT4, Golgi, Insulin, Membrane trafficking, Molecular cell biology, Protein translocation, ubiquitin-like modification, unconventional secretion
Abstract
To promote glucose uptake into fat and muscle cells, insulin causes the translocation of GLUT4 glucose transporters from intracellular vesicles to the cell surface. Previous data support a model in which TUG traps GLUT4-containing vesicles and tethers them intracellularly in unstimulated cells and in which insulin mobilizes this pool of vesicles by releasing this tether. Here we show that TUG undergoes site-specific endoproteolytic cleavage, which separates a GLUT4-binding, N-terminal region of TUG from a C-terminal region previously suggested to bind an intracellular anchor. Cleavage is accelerated by insulin stimulation in 3T3-L1 adipocytes and is highly dependent upon adipocyte differentiation. The N-terminal TUG cleavage product has properties of a novel 18-kDa ubiquitin-like modifier, which we call TUGUL. The C-terminal product is observed at the expected size of 42 kDa and also as a 54-kDa form that is released from membranes into the cytosol. In transfected cells, intact TUG links GLUT4 to PIST and also binds Golgin-160 through its C-terminal region. PIST is an effector of TC10α, a GTPase previously shown to transmit an insulin signal required for GLUT4 translocation, and we show using RNAi that TC10α is required for TUG proteolytic processing. Finally, we demonstrate that a cleavage-resistant form of TUG does not support highly insulin-responsive GLUT4 translocation or glucose uptake in 3T3-L1 adipocytes. Together with previous results, these data support a model whereby insulin stimulates TUG cleavage to liberate GLUT4 storage vesicles from the Golgi matrix, which promotes GLUT4 translocation to the cell surface and enhances glucose uptake.
Introduction
To increase glucose uptake into fat and muscle cells, insulin stimulates the translocation of GLUT4 glucose transporters (1–3). In unstimulated cells, GLUT4 is sequestered intracellularly in insulin-responsive “GLUT4 storage vesicles” (GSVs)7 (4, 5). GSVs are formed in a cell type-specific manner, which is differentiation-dependent in cultured 3T3-L1 adipocytes. These vesicles constitute a pool that is mobilized within minutes after insulin addition. Efficient GLUT4 sequestration in GSVs limits the number of GLUT4 transporters that are present at the surface of unstimulated cells and restricts basal glucose uptake. Insulin is then able to increase plasma membrane GLUT4 abundance severalfold and to markedly enhance the rate of glucose uptake. GSV mobilization is quantitatively the most important of the trafficking steps at which insulin acts to modulate overall GLUT4 distribution, yet how GSVs are retained within unstimulated cells and how they are translocated upon insulin stimulation are not well understood.
We previously identified TUG (tether containing a UBX domain, for GLUT4) as a regulator of GLUT4 trafficking and proposed that it regulates GSV sequestration and release (6, 7). In 3T3-L1 adipocytes, TUG is a 60-kDa protein present in the cytosol and on membranes, which binds and sequesters GLUT4 intracellularly in the absence of insulin. To translocate GLUT4, insulin stimulates the release of GLUT4 from TUG (6, 8). Release occurs prior to significant GLUT4 movement out of a light microsome (LM) fraction, which contains GSVs, and the number of released proteins determines the number of GLUT4 molecules that are translocated to the cell surface within 3–6 min after insulin addition (1, 6, 9). Depletion of TUG using RNAi mimics much of the effect of insulin to stimulate GLUT4 translocation and glucose uptake (7). Like acute insulin stimulation, TUG depletion mobilizes ∼60-nm vesicles to the cell surface; these vesicles carry GSV cargoes and fuse directly with the plasma membrane (10). This vesicle size is characteristic of GSVs and distinct from other exocytic vesicles (10, 11). During ongoing (≥15-min) insulin exposure, GSV cargoes recycle in ∼150-nm vesicles characteristic of endosomes and bypass a TUG-regulated membrane trafficking step. Thus, GLUT4 participates in two different exocytic pathways, which are active at distinct times after insulin addition in 3T3-L1 adipocytes. The data support the idea that the initially translocated vesicles are GSVs that are regulated by TUG, whereas subsequent GLUT4 exocytosis bypasses this GSV sequestration compartment and returns directly to the plasma membrane from endosomes.
The notion that insulin stimulates the release of sequestered GSVs to the cell surface and that GLUT4 then participates in an endosomal recycling pathway is consistent with previous data suggesting a “quantal release” mechanism for GLUT4 mobilization (12, 13). According to this model, discrete packets of GLUT4 molecules are liberated from a sequestered storage compartment into the recycling pool, yet the mechanism by which GLUT4 is released from TUG is not understood. Here we show that TUG is a substrate for site-specific endoproteolytic cleavage, which is required to translocate GLUT4. Our data support the concept that TUG proteolysis is a novel enzymatic activity by which insulin mobilizes GSVs to stimulate glucose uptake.
EXPERIMENTAL PROCEDURES
Materials
3T3-L1 and 293T cells were cultured as described (7). Retroviruses were used to express a TUG short hairpin RNA (shRNA) and/or various cDNAs in 3T3-L1 adipocytes as described (7). TUG C terminus and GLUT4 antisera were described previously (6, 7). Antisera to the TUG N terminus were raised in rabbits using a synthetic peptide (residues 1–26 of 60-kDa TUG) conjugated to KLH and were affinity-purified using a shorter peptide (residues 10–26). Antisera to PIST were purchased from Abcam (Cambridge, MA) and were also raised using a 13-residue peptide corresponding to the C terminus of murine PIST. Peptide syntheses were done at the W. M. Keck Biotechnology Facility at Yale. Conjugation to KLH, immunizations, and antisera collection were done by Covance, Inc. (Denver, PA) and OpenBiosystems (Huntsville, AL). Anti-Myc (9E10) and HA (HA.11) antibodies were from Roche Applied Science and Covance. Other antibodies (suppliers) were directed to the following: Hsc70 (Stressgen Bioreagents), insulin receptor β-chain (Upstate Biotechnology), phosphotyrosine (P-Tyr-100; Cell Signaling Technology), GLUT1 (Chemicon), syntaxin-6 and -16 (Synaptic Systems, GmbH), Vti1a (BD Transduction Laboratories), VAMP4 and VAMP3 (Abcam), transferrin receptor (Pharmingen and Santa Cruz Biotechnology, Inc.), GFP (Clontech and Invitrogen), and TC10α (Sigma). An antibody to Golgin-160 was kindly provided by Dr. Carolyn Machamer (Johns Hopkins University, Baltimore, MD). MG-132 and lactacystin were from Sigma and Calbiochem, respectively, and were used at 10 μm unless otherwise indicated. 293T cells were transfected using Fugene6 (Roche Applied Science) or Lipofectamine 2000 (Invitrogen).
Retroviruses containing a TUG shRNA and shRNA-resistant, wild type TUG were described previously (7). FLAG-tagged TUGUL, TUGULΔGG, and TUGUL K134R were made using PCR, cloned in pcDNA3.1-TOPO (Invitrogen), and sequenced. An HA-tagged ubiquitin plasmid was a kind gift of Drs. Hamid Band and Dirk Bohmann. Coding sequences for TUG GGAA, S165M, and K380R were made by overlap PCR using the shRNA-resistant cDNA template. GLUT4 containing seven Myc tags in the first exofacial loop was constructed by using PCR to insert a stop codon and eliminate the GFP from a previously described reporter (9). Sequence encoding PIST was amplified by PCR from murine skeletal muscle cDNA (Clontech) and cloned in pcDNA3.1 TOPO (Invitrogen). The neuronal splice variant (nPIST) was obtained (14). An HA epitope tag was added at the N terminus using PCR, and the clone was expressed using the pBICD4 retrovirus vector (6, 15). An expression plasmid encoding GFP-tagged Golgin-160 was a kind gift from Dr. Carolyn Machamer. All constructs were verified by sequencing. For expression of full-length TUG or TUG UBX-Cter containing IgG binding domains from Protein A, the pRAV-FLAG vector was used (16) and was a kind gift from Dr. Xuedong Liu (University of Colorado). These plasmids were transfected in 293T cells, and purifications were done as described (16). For retroviral expression in 3T3-L1 adipocytes, cDNAs were cloned in the pBICD2 or pBICD4 vectors (6, 15). Retrovirus packaging, transduction, and selection of infected cells were done as described (9, 15, 17).
Recombinant Proteins and Binding Experiments
GST-TUG, GST-TUG-UBX-Cter, and GST alone were cloned and expressed as described (7). In vitro translation of PIST was done in the presence of [35S]methionine, and binding experiments were carried out as described (7). For pull-down experiments using 3T3-L1 adipocyte lysates, cells were lysed in TNET buffer (1% Triton X-100, 150 mm NaCl, 20 mm Tris (pH 8.0), 2 mm EDTA) and incubated with immobilized GST-TUG or GST. Bound proteins were eluted in SDS-PAGE sample buffer, separated by SDS-PAGE, and visualized by GelCode Coomassie staining (Pierce).
RNA Interference
RNAi of TC10α employed synthetic siRNAs purchased from Thermo Scientific Dharmacon. The target sequences were as follows: TC10-1, GATAGGTGCATGCTGCTAT; TC10-2, CTATGATCGTCTGAGGCCT; luciferase, CGTACGCGGAATACTTCGA. siRNA duplexes were transfected into 3T3-L1 adipocytes by electroporation using siPORT buffer (Ambion) as described (7). Retrovirus expression of an shRNA to deplete TUG in 3T3-L1 adipocytes was described previously (7).
Immunoblotting and Immunoprecipitation
Denaturing lysis was done at >80 °C in 1% SDS, 50 mm Tris, pH 8.0, 150 mm NaCl, 2 mm EDTA, 20 mm iodoacetamide (Sigma), and Complete tablets (Roche Applied Science; 1 tablet/20 ml). Protein concentrations were assayed in triplicate using micro-BCA (Pierce) or EZQ (Invitrogen) kits and a PerkinElmer Victor3 plate reader. SDS-PAGE and immunoblotting were done as described (9).
To immunoprecipitate proteins after denaturing lysis, DNA was sheared using a needle or by sonication, debris was pelleted, and lysates were diluted 10-fold using PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate. For lysates of 293 cells transfected with FLAG-TUGUL, a FLAG M2 affinity matrix (Covance) was used overnight at 4 °C. After washing, bound proteins were eluted using sample buffer or an excess of FLAG peptide. Eluted proteins were analyzed by SDS-PAGE and immunoblotting. Sequence alignments were done using ClustalX, and were further adjusted manually (18).
Pulse-Chase Experiments
The protocol was adapted from Ref. 19. 3T3-L1 adipocytes overexpressing TUG were cultured in 10-cm dishes, starved overnight, and then placed in DMEM lacking Cys and Met for 1 h. Cells were labeled for 10 min at 37 °C using 0.7 mCi/dish of EXPRESS35S Protein Labeling Mix, a mixture of radiolabeled Cys and Met (PerkinElmer Life Sciences). Cells were washed and then chased in DMEM containing nonradioactive Cys and Met and 500 μm cycloheximide. Pairs of plates were chased with or without insulin as described (9). At intervals, cells were lysed in boiling 1% SDS as above. Lysates were passed through a 22-gauge needle and then centrifuged to pellet insoluble debris. Supernatants were diluted 10-fold using TNET (20 mm Tris, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100). Immunoprecipitations were done overnight using 75 μl of crude antisera per sample. Eluted proteins were analyzed by SDS-PAGE, and gels were dried and exposed using a Storm PhosphorImager (GE Healthcare).
Subcellular Fractionation
Plasma membrane (PM), LM, and heavy microsome (HM) fractions were isolated as described (7, 9). Equal protein amounts in each fraction were immunoblotted. To separate total membranes from cytosol, each 10-cm plate of 3T3-L1 adipocytes was homogenized in 1 ml of an ice-cold TES buffer (250 mm sucrose, 10 mm Tris, pH 7.4, 0.5 mm EDTA, and 20 mm iodoacetamide) using a Dounce-type tissue grinder. Homogenates were centrifuged for 30 min at 2 °C in a TLA120.2 rotor (Beckman) at 100,000 rpm to pellet membranes. The pellet was resuspended in SDS-PAGE sample buffer, EZQ protein assays were done on both the supernatant (cytosol) and pellet (total membranes), and equal amounts of protein were analyzed by immunoblotting.
Confocal Microscopy
3T3-L1 adipocytes were grown on coverslips, insulin-stimulated, fixed, and permeabilized as described (19). To visualize GLUT4, cells stably expressing a GFP-tagged GLUT4 reporter protein were used (9). Syntaxin-6 staining was detected using an AlexaFluor594-conjugated goat anti-rabbit IgG secondary antibody. Images were acquired on an Axiovert 100 M microscope equipped with an LSM510 scanning unit and ×63/1.3 numerical aperture plan Apochromat objective (Zeiss) as described (7). Postprocessing was done at 12-bit pixel depth and linear Gamma using ImageJ and Adobe Photoshop.
Glucose Uptake
2-Deoxyglucose uptake assays were done as described (7). To control for nonspecific uptake, 20 μm cytochalasin B or 200 μm oxybenzylcarbonyl-His-Phe-Phe-O-ethyl ester was used (20). Similar results were obtained using each of these reagents. Assays were done in triplicate, and data are plotted as mean ± S.E. Significance was assessed by a two-tailed Student's t test.
Translocation of Newly Synthesized GLUT4
The protocol was adapted from Ref. 21. Control 3T3-L1 adipocytes, cells stably expressing a TUG shRNA, or cells with the shRNA together with shRNA-resistant forms of TUG were electroporated using pB-GLUT4–7myc-GFP plasmid (6, 9). Cells were replated on coverslips after electroporation. Twenty hours after electroporation, staining of surface-exposed Myc epitope tag was done essentially as described (9, 22). Cells were washed three times with warm DMEM and incubated in DMEM for 4 h. Insulin (160 nm) was added for 15 min at 37 °C to the “insulin” cells, whereas “basal” cells were kept in DMEM. Cells were washed three times with PBS, fixed with 2% paraformaldehyde in PBS for 5 min at room temperature, washed three times with PBS, stained with mouse anti-Myc (9E10) for 1 h at room temperature, washed three times with PBS, stained with AlexaFluor 546 goat anti-mouse IgG for 35 min at room temperature, washed three times with PBS, and mounted using ProLong Gold antifade mounting reagent (Invitrogen).
Images were acquired using a Zeiss plan-neofluar ×40/1.3 numerical aperture oil objective on a Zeiss Axiovert 200 M wide field microscope driven by AxioVision imaging software (Carl Zeiss, Inc.). AlexaFluor 546 was imaged using a rhodamine filter set, and GFP was imaged using an FITC filter set. Identical image acquisition settings were used for all of the images. Image analysis was performed using MetaMorph software. Image background was determined as described previously (23). This background value was then subtracted from every pixel in the field. The images were then quantified by manually outlining each cell and by taking the mean fluorescence intensities associated with the cell. The ratio of Alexa 546 fluorescence to GFP fluorescence was calculated and averaged over many cells (at least 16 for basal cells and at least 27 for insulin-stimulated cells). This ratio is a measure of surface Myc-GLUT4-GFP normalized for the total construct expressed (9, 22).
Flow Cytometry
Cells infected with retroviruses carrying CD2 or GFP markers were identified using BD Biosciences FACScan and FACSCalibur analyzers (6, 7, 9). Measurement of GLUT4 trafficking was performed as described (6, 9). Stable populations of infected cells were purified by flow sorting on a FACSVantage system at the Yale Cell Sorter Facility.
RESULTS
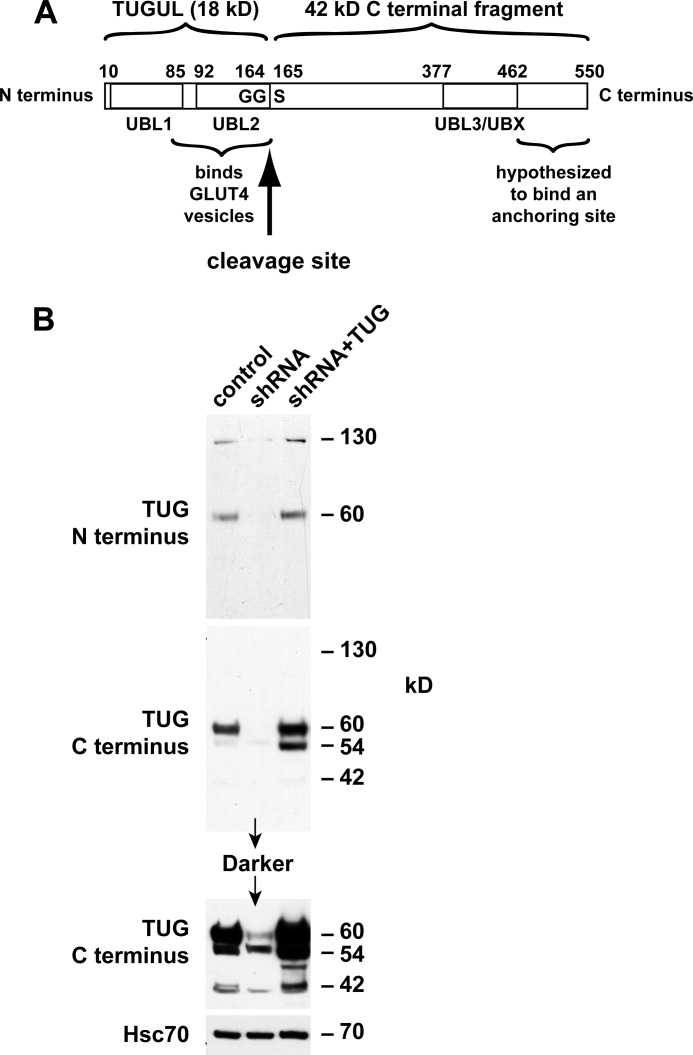
Ubiquitin and ubiquitin-like modifiers (collectively termed Ubls) are produced by endoproteolytic cleavage of precursor proteins (24). We hypothesized that TUG is a novel such precursor protein. Previous work had identified two ubiquitin-like regions in TUG, including N-terminal (residues 10–83) and UBX (residues 377–462) domains (6, 25). Based on an initial immunoblot suggesting that TUG is cleaved, we considered that TUG contains a third ubiquitin-like domain (residues 92–164) and that it may be cleaved at the 164–165 bond (Fig. 1A). Such a cleavage event would separate an N-terminal region that binds GLUT4 from a C-terminal region that was previously proposed to bind an intracellular anchoring site (6, 7).
FIGURE 1.
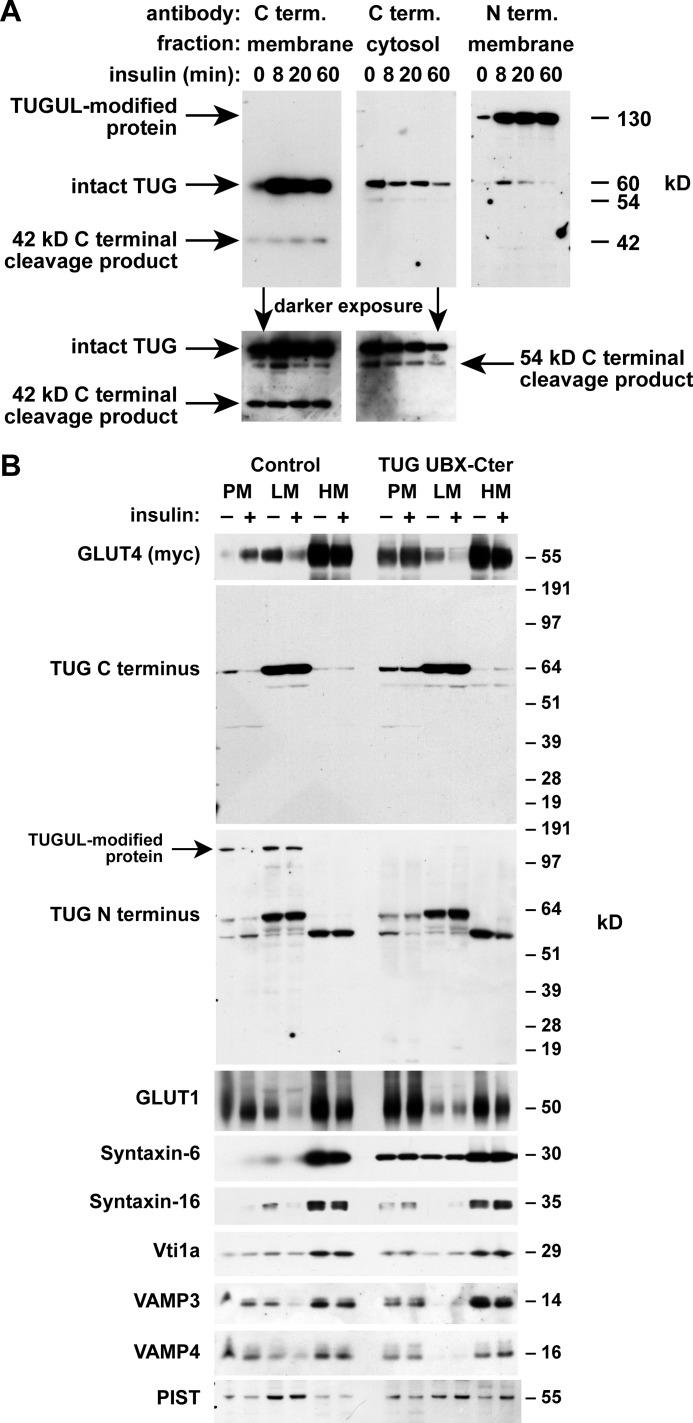
Distinct N- and C-terminal TUG derivatives in 3T3-L1 adipocytes. A, TUG is a 550-residue protein containing three ubiquitin-like domains, denoted UBL1, UBL2, and UBL3/UBX. UBL2 ends with a diglycine sequence (GG), which defines a site for endoproteolytic cleavage to produce TUGUL, which may potentially act as a novel ubiquitin-like modifier. B, control 3T3-L1 adipocytes, cells containing a TUG shRNA, and “rescued” cells containing the shRNA and shRNA-resistant TUG (shRNA + TUG cells) were lysed using denaturing conditions. Lysates were immunoblotted using antibodies to the TUG N and C termini or to Hsc70 as a control, as indicated. The experiment was repeated twice with similar results.
Ubl precursors are typically cleaved after a diglycine motif, which is present in TUG (see below). The mature Ubl (i.e. the N-terminal product) is then covalently attached through its C-terminal glycine residue to protein or lipid substrates. We hypothesized that TUG endoproteolytic cleavage gives rise to a new Ubl, which we call TUGUL (for TUG ubiquitin-like). TUGUL corresponds to residues 1–164 of intact TUG and is predicted to contain tandem β-grasp folds, similar to two known Ubls, ISG-15 and FAT10 (24). We refer to these domains as “UBL1” and “UBL2” in Fig. 1A. The UBX domain of TUG can then also be called “UBL3.” Based on previous evidence supporting the idea that TUG regulates GSVs, we hypothesized that TUG cleavage may be required to translocate these vesicles.
To test if TUGUL is produced from endogenous TUG in 3T3-L1 adipocytes, we raised an antibody to the TUG N terminus. This antibody detected a 130-kDa protein, as well as intact, 60-kDa TUG on immunoblots of whole cell lysates (Fig. 1B). Both proteins were depleted by TUG shRNA and rescued by shRNA-resistant TUG, supporting the concept that the 130-kDa protein is a TUGUL-modified (i.e. “tugulated”) substrate as well as supporting the specificity of the antibody. A previously characterized antibody to the TUG C terminus detected intact, 60-kDa TUG but not the 130 kDa band. The C terminus antibody also detected bands at 54 and 42 kDa, which were not detected by the N terminus antibody. The predicted mass of the proposed TUG C-terminal product (containing residues 165–550) is 42 kDa. As can be seen in Fig. 1B, this band was also depleted by the TUG shRNA and rescued by shRNA-resistant, wild type TUG (see the darker exposure of the C terminus immunoblot). As described below, the 54 kDa band may also be derived from intact TUG, and in Fig. 1B, its abundance was slightly reduced by the shRNA and markedly increased by shRNA-resistant TUG. These observations support the idea that endogenous TUG is cleaved in 3T3-L1 adipocytes and that cleavage produces TUGUL, which is then used for covalent, ubiquitin-like protein modification.
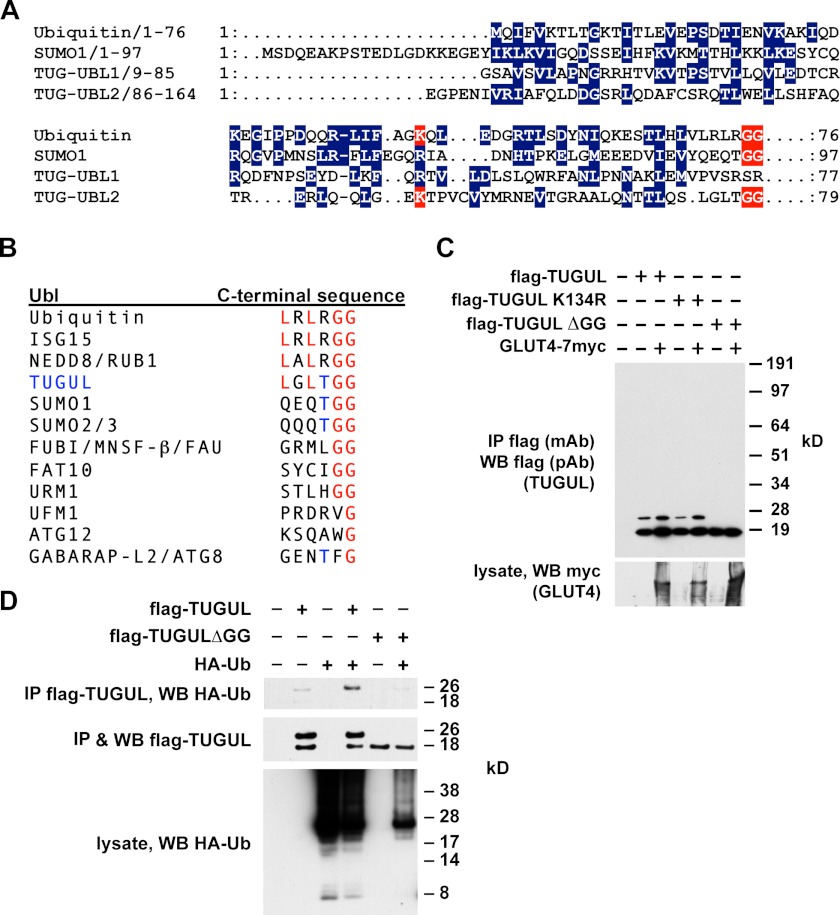
As shown in Fig. 2A, the primary sequence of TUG UBL2 has limited similarity with ubiquitin, SUMO, and other Ubls, which is not unusual for ubiquitin-like family members (24). This region was nonetheless suggested to comprise a UBX or UBX-like domain (26, 27). Importantly, TUG UBL2 terminates with a diglycine motif, defining a potential site for proteolytic cleavage by a deubiquitinating enzyme family member. Additionally, a lysine residue (Lys-134) that may be analogous to ubiquitin Lys-48, a prototypical site of polyubiquitin chain formation, is present in UBL2. As shown in Fig. 2B, the C-terminal sequence of TUGUL is most closely related to those of ubiquitin, ISG15, and NEDD8, but it shares with SUMO1 and SUMO2/3 a threonine residue immediately preceding the terminal diglycine sequence.
FIGURE 2.
Mature TUGUL has characteristics of a ubiquitin-like protein modifier. A, sequence alignments of ubiquitin, SUMO-1, and the two ubiquitin-like regions within the TUG N terminus, UBL1, and UBL2. Conserved residues are in blue. Terminal diglycine sequences of ubiquitin, SUMO-1, and TUGUL are highlighted in red. Additionally, TUG Lys-134 and ubiquitin Lys-48 are also highlighted in red. B, alignments of the C-terminal residues from several ubiquitin-like proteins and comparison with C-terminal residues from TUGUL (residues 159–164 of intact TUG). B is adapted from Ref. 95. C, FLAG-tagged TUGUL, TUGUL K134R, or TUGUL ΔGG (which lacks the terminal diglycine sequence) were coexpressed with GLUT4 by transient transfection of 293T cells. Cells were lysed in boiling 1% SDS, and then lysates were diluted with Triton X-100, and proteins were immunoprecipitated (IP) and immunoblotted (WB) as indicated. D, cells were transfected with FLAG-tagged TUGUL or TUGUL ΔGG and HA-tagged ubiquitin (Ub). After denaturing lysis, proteins were immunoprecipitated and immunoblotted as indicated. Experiments were repeated at least twice, with similar results.
To test whether mature TUGUL, if produced, would be able to modify a target protein, we transfected 293T cells. As shown in Fig. 2C, FLAG-tagged TUGUL was present not only at 18 kDa, which corresponds to the predicted size of this protein, but also as a 26-kDa form. The 26-kDa form was resistant to heat and ionic detergent and was not affected by mutation of K134R. Rather, the 26-kDa TUGUL-containing product required the terminal diglycine sequence of TUGUL, which is characteristic of ubiquitin-like attachment (28). Additionally, the abundance of this putative tugulated product was increased by cotransfection of GLUT4, which binds both TUGUL (7) and Ubc9 (29), a SUMO-conjugating enzyme. We used mass spectrometry of tryptic peptides to identify the putative 26-kDa TUGUL-conjugated protein and observed peptides derived from ubiquitin. Therefore, we cotransfected HA-tagged ubiquitin together with FLAG-tagged TUGUL. As shown in Fig. 2D, these proteins were coimmunoprecipitated after denaturing lysis, supporting the idea that they are covalently attached to each other. The terminal diglycine of TUGUL was required for this association, suggesting that the 26 kDa band is tugulated ubiquitin rather than ubiquitylated TUGUL. Ubiquitin is probably not the physiological substrate of TUGUL attachment because the putative tugulated conjugate in 3T3-L1 adipocytes totals 130 kDa. Nonetheless, the data support the hypothesis that TUGUL is able to function as a ubiquitin-like protein modifier and that it may do so if it is produced physiologically by TUG cleavage.
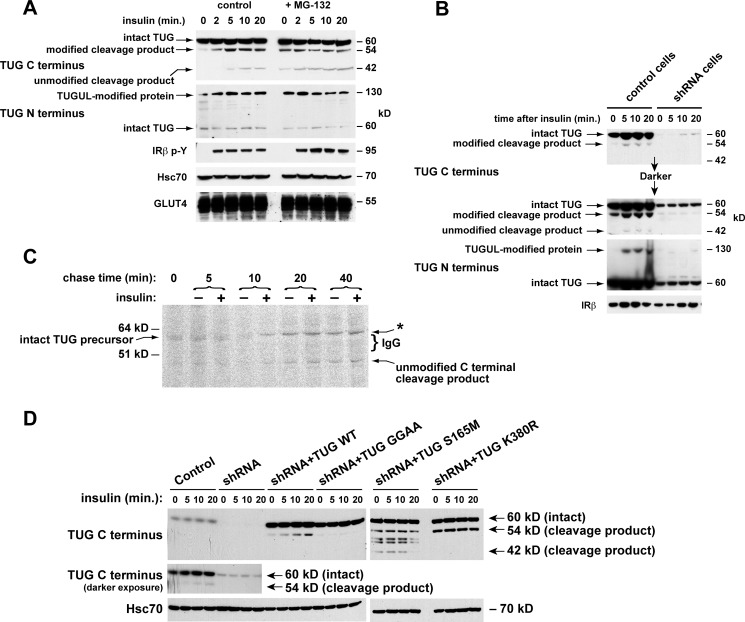
We next tested if insulin stimulates TUG cleavage in 3T3-L1 adipocytes. We predicted that cleavage generates a C-terminal product beginning with Ser-165. This residue will be unacetylated, and it is predicted to cause rapid proteasomal degradation of the product according to an N-end rule (30, 31). We therefore treated cells briefly with or without a proteasome inhibitor, MG-132, and then stimulated with insulin. Immunoblots showed that insulin stimulated the generation of the 42-kDa C-terminal product, and MG-132 increased the abundance of this protein, as predicted (Fig. 3A). The abundance of intact TUG was not markedly changed by insulin or MG-132. In control experiments, MG-132 did not affect GLUT4 translocation (not shown). These data support the notion that insulin accelerates TUG cleavage and further imply that only ∼10% of total cellular TUG is cleaved in 3T3-L1 adipocytes. As noted below, this is consistent with a model in which cleavage of one or a few TUG proteins serves to liberate each GSV.
FIGURE 3.
Production of TUG derivatives by site-specific endoproteolytic cleavage. A, cells were treated or not with 10 μm MG-132 for 40 min and then with insulin, as indicated. After denaturing lysis, immunoblots were done as indicated. Controls show phosphorylation of the insulin receptor (IRβ p-Y) and equal loading (Hsc70 and GLUT4). B, control and shRNA cells were treated with insulin, lysed in denaturing conditions, and immunoblotted as indicated. IRβ, insulin receptor β-chain. C, cells were pulse-labeled with 35S-labeled Cys and Met, chased in non-radioactive amino acids and cycloheximide and in the presence or absence of insulin, and lysed at the indicated times using denaturing conditions. Immunoprecipitations were done with the TUG C terminus antibody, and eluted material was analyzed by SDS-PAGE and phosphorimaging. IgG was visualized by Coomassie staining (bracket) and probably altered migration of the 54-kDa C-terminal product observed in other experiments (asterisk). D, control cells, shRNA cells, and shRNA cells containing shRNA-resistant wild type or mutated TUG were treated with insulin as indicated. After denaturing lysis, immunoblots were done as indicated. Experiments were repeated at least twice with similar results.
Unexpectedly, the initial band that appeared after insulin addition, using the TUG C terminus antibody, was 54 kDa, not 42 kDa (Fig. 3A). As described below, we believe that this 54-kDa protein is a modified form of the 42-kDa product. The 54 and 130 kDa bands were produced simultaneously and were detected exclusively by the TUG C and N terminus antibodies, respectively. This finding suggests that the 130- and 54-kDa proteins are generated as two products of a single biochemical process, which is accelerated by insulin and includes TUG cleavage.
To further test if the 130-, 54-, and 42-kDa proteins all derive from TUG, we used control and shRNA-containing 3T3-L1 adipocytes. Fig. 3B shows that these proteins appeared after insulin treatment of control cells and not shRNA cells. As was shown in Fig. 1B, shRNA-resistant TUG rescued the 130-kDa protein and increased the abundance of the 54 and 42 kDa bands. Densitometry of several immunoblots revealed that >97% depletion of intact, 60-kDa TUG was required to deplete the 130-kDa protein. Together, the data support the idea that insulin stimulates the production of a single, 130-kDa TUGUL-modified protein in addition to 54- and 42-kDa proteins containing a TUG C-terminal fragment.
To show directly that intact TUG is converted to a C-terminal cleavage product, we performed pulse-chase experiments. Newly synthesized proteins in 3T3-L1 adipocytes overexpressing intact TUG were pulse-labeled for 10 min using 35S-labeled Cys and Met. Cells were chased using non-radioactive amino acids and cycloheximide, and pairs of samples chased in the absence or presence of insulin were lysed at intervals using denaturing conditions. Immunoprecipitations were done using an antibody to the TUG C terminus, and eluates were analyzed by SDS-PAGE and phosphorimaging. Fig. 3C shows that intact 60-kDa TUG was isolated from cells chased for 0 or 5 min and in cells chased for 10 min in the absence of insulin. At later times, intact TUG was not present, and the 42-kDa C-terminal product was observed. The effect of insulin to accelerate the conversion of 60-kDa TUG to the 42-kDa product was particularly evident at 10 min into the chase period. At later times, the 42-kDa product was always more abundant in insulin-treated cells than in unstimulated cells from the same time point. A ∼62 kDa band also appeared in cells chased for 10 min in the presence of insulin, and this band also was more abundant in cells chased in insulin than in unstimulated cells from the same time point (Fig. 3C, asterisk). Coomassie staining revealed abundant IgG, used for the immunoprecipitation, immediately below this band (Fig. 3C, bracket). Therefore, this band may be the 54-kDa product noted in Figs. 1B and 3 (A and B), which migrates above the IgG in Fig. 3C. Importantly, intact 60-kDa TUG was converted to the predicted 42-kDa C-terminal product, and this conversion was accelerated, at least to some degree, by insulin.
To define the scissile bond as that joining residues 164 and 165, we used forms of TUG containing specific mutations. First, we changed the diglycine sequence to dialanine to create TUG GGAA, a form predicted to be cleavage-resistant (32). Second, we changed Ser-165 to Met to create TUG S165M, which is predicted to produce a more stable C-terminal cleavage product. Finally, we mutated Lys-380 to Arg to create TUG K380R, which may disrupt modification or degradation of the C-terminal product. These proteins were expressed as shRNA-resistant forms in 3T3-L1 adipocytes containing an shRNA to deplete native TUG. Cells were treated with insulin, lysed in denaturing conditions, and immunoblotted to detect C-terminal products.
Fig. 3D shows that insulin stimulated the generation of the 54-kDa modified product in control 3T3-L1 adipocytes, as best observed on a darker exposure. This 54-kDa product was depleted by TUG shRNA and rescued by shRNA-resistant, wild type TUG (in shRNA + TUG WT cells). In shRNA + TUG GGAA cells, the 54 kDa band was not produced, supporting the prediction that TUG GGAA is cleavage-resistant. In shRNA + TUG S165M cells, the 42-kDa unmodified C-terminal product accumulated as predicted and was observed even in unstimulated cells. The S165M mutation also stabilized the 54-kDa modified product as well as other forms that may reflect alternative degradation pathways. In shRNA + TUG K380R cells, the 54-kDa product accumulated, suggesting that Lys-380 is a site that is used for ubiquitin-mediated degradation. These data support the idea that insulin stimulates TUG cleavage between residues 164 and 165.
The degree to which insulin stimulated the production of TUG cleavage products was variable in 3T3-L1 adipocytes. In some experiments, the effect of insulin was minimal or absent, because TUG derivatives were observed in unstimulated 3T3-L1 cells. In other experiments, the effect of insulin to increase the abundance of TUG products was more marked. Together with data below identifying an insulin signal required for TUG proteolytic processing, we conclude that insulin stimulates cleavage. The variability in insulin's effect may result from post-lysis proteolysis, which is typical of Ubls and could increase the abundance of products observed in unstimulated cells (33). We used heat and ionic detergent as well as iodoacetamide to try to prevent such an artifact. It is also likely that TUG is cleaved at some rate in unstimulated 3T3-L1 adipocytes and that insulin accelerates this rate rather than acting as a binary switch, as described further below. Finally, insulin's ability to stimulate GLUT4 translocation is cell type-specific and, in 3T3-L1 cells, differentiation-dependent. Thus, we considered that the degree to which insulin can stimulate TUG cleavage may also be affected by the degree of adipocyte differentiation.
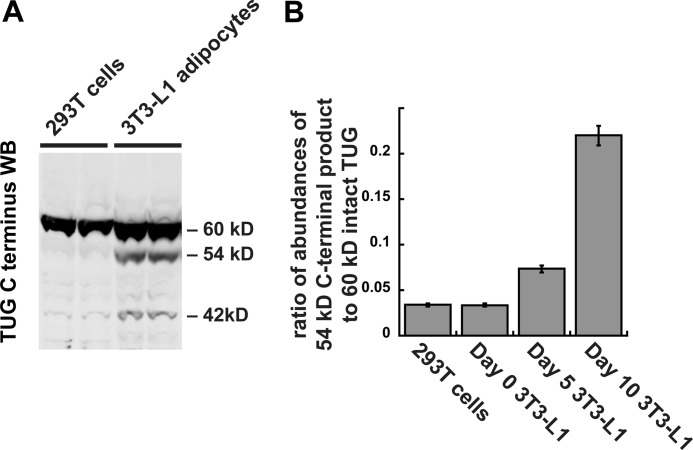
To test directly the idea that TUG proteolysis occurs in a differentiation-dependent manner, we performed immunoblots of non-adipose 293T cells and of 3T3-L1 cells at various stages of differentiation. As shown in Fig. 4A, the TUG C terminus antibody readily detected the 42- and 54-kDa products in 3T3-L1 adipocytes, but these products were barely discernable in 293T cells. Fig. 4B quantifies the abundance of the major 54-kDa product, relative to the intact 60-kDa TUG protein, in various cells. The 54-kDa product was present at only ∼3% of the abundance of intact TUG in 293T cells and in confluent, undifferentiated 3T3-L1 preadipocytes. The abundance of this product was only ∼7% of that of intact TUG in partially differentiated 3T3-L1 cells. At this stage of differentiation (day 5), GLUT4 is just beginning to be expressed, and the translocation mechanism is present but perhaps not fully developed. In fully differentiated 3T3-L1 adipocytes (day 10), the 54-kDa product was present at >20% of the abundance of intact TUG. As shown below, the number of TUG molecules that are cleaved corresponds well with the number of GSVs that are present in each cell. The results support the idea that TUG proteolytic processing is cell type-specific and that in 3T3-L1 cells it is highly dependent upon adipocyte differentiation.
FIGURE 4.
TUG processing occurs in 3T3-L1 adipocytes but not in non-adipocyte cells. A, 293T cells and 3T3-L1 adipocytes were lysed using denaturing conditions, and immunoblots (WB) were done using the TUG C terminus antibody. Duplicate samples are shown. B, 293T cells and 3T3-L1 cells at the indicated stages of adipocyte differentiation were lysed using denaturing conditions and immunoblotted using the TUG C terminus antibody. Immunoblots were quantified, and the ratio of band intensities at 54 and 60 kDa is plotted. Error bars, S.E.; n = 2 for 293T cells and day 0 3T3-L1 cells; n = 6 for day 5 and day 10 3T3-L1 cells.
Processing of TUG on Membranes Containing GSVs
To study where TUG processing occurs, we isolated membrane and cytosol fractions from 3T3-L1 adipocytes. As previously noted, TUG is present in both of these fractions, and insulin often causes no change or a slight increase in the abundance of membrane-associated TUG, which is in an LM fraction (6, 7). In Fig. 5A, insulin caused an increase in the abundance of intact TUG on membranes. The 42-kDa unmodified C-terminal product was generated exclusively on membranes. The 54-kDa modified C-terminal product was observed in both fractions and was more abundant in cytosol in several experiments. Like the 42-kDa product, the 130-kDa putative TUGUL-modified protein was produced on membranes. The 130-kDa product was more abundant than intact TUG, which, together with other data, suggests that it may be more stable. These data support the idea that TUG proteolytic processing and TUGUL modification occurs on membranes.
FIGURE 5.
TUG processing occurs on membranes containing GLUT4 storage vesicles. A, 3T3-L1 adipocytes were treated with insulin as indicated, and then membrane and cytosol fractions were isolated and immunoblotted to detect the TUG C and N termini. B, PM, LM, and HM fractions were isolated from basal and insulin-treated control 3T3-L1 adipocytes and from cells expressing dominant negative TUG UBX-Cter. All cells contained a Myc-tagged GLUT4 reporter. Fractions were immunoblotted as indicated. Experiments were repeated at least twice with similar results.
Conversion of the 42-kDa C-terminal product to the 54-kDa form probably occurs after cleavage of intact TUG. This is because 1) the 54-kDa protein is present in a different location than the 42- and 130-kDa products, and 2) cleavage-resistant TUG does not accumulate as a ∼72-kDa form, which would be predicted if modification preceded cleavage. Because the SUMO-conjugating enzyme, Ubc9, binds and regulates GLUT4 (29, 34), we tested if the 54-kDa form is a sumoylated form of the 42-kDa product. We were not able to obtain evidence that the 54-kDa protein contains SUMO, and identification of the modification will require further work. Even so, one possibility is that the modification may be involved in removing the unmodified C-terminal cleavage product from an intracellular anchoring site, as discussed below.
To further examine where TUG processing takes place, we isolated PM, LM, and HM fractions. We used control 3T3-L1 adipocytes and cells containing a truncated form of TUG, UBX-Cter (residues 377–550), which acts in a dominant negative manner to block intracellular GLUT4 sequestration in cells not treated with insulin (6, 7). In control cells, insulin redistributed GLUT4 out of the LM fraction, which contains GSVs, and to the PM (Fig. 5B). TUG UBX-Cter disrupted this response by inhibiting the basal, intracellular retention of GLUT4, as observed previously (7). Intact TUG was detected in the LM fraction by both the N and C terminus antibodies and was redistributed slightly to the PM by UBX-Cter. The 130-kDa TUGUL-modified protein was detected by the N terminus antibody in the LM and PM fractions of control cells, and this protein was not present in cells containing TUG UBX-Cter. This band was also not detected by the C terminus antibody, consistent with data shown above and with the idea that it contains only the N-terminal cleavage product. Because ubiquitin-like processing may occur in non-denaturing conditions, such as cell homogenates, the data in Fig. 5B do not address whether insulin affects the location or abundance of the TUGUL-modified protein. The UBX-Cter fragment itself was not visible on the TUG C terminus immunoblot. This fragment is present at low abundance relative to intact TUG, and it may also be degraded in cell homogenates. As discussed further below, if intact TUG is a substrate and not an enzyme, then UBX-Cter may act irreversibly (in modified form) to inhibit GLUT4 retention. In summary, the TUGUL-modified protein is produced on membranes that contain GSVs. TUG UBX-Cter not only inhibits GLUT4 retention but also blocks the generation of the TUGUL-modified protein on these membranes.
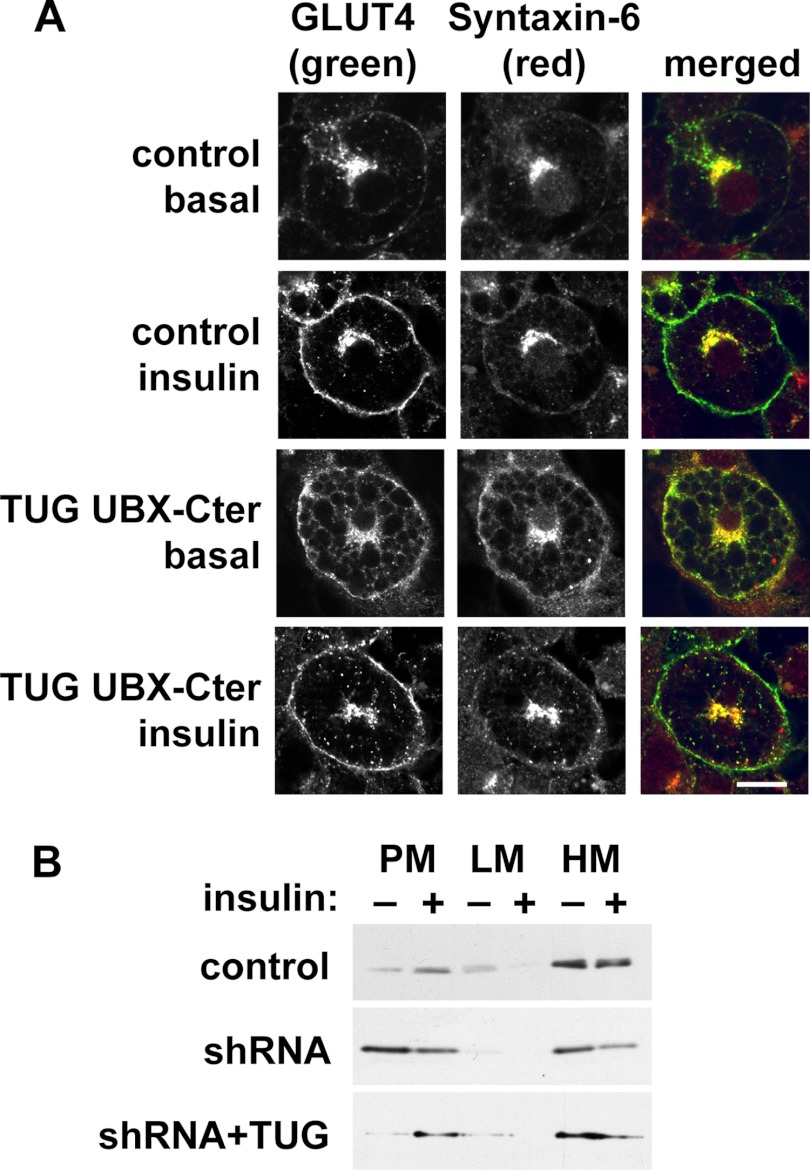
Previous data implicate a SNARE complex containing syntaxin-6, syntaxin-16, Vti1a, and VAMP4 (or VAMP3) in trafficking of GSVs at the trans-Golgi network (TGN) (35–39). Among these proteins, syntaxin-6 was dramatically redistributed to PM by dominant negative TUG UBX-Cter (Fig. 5B). The syntaxin-6-interacting protein, PIST, which is discussed further below, was also present in the LM fraction and was slightly dispersed by TUG UBX-Cter. Of note, syntaxin-6 was identified in GSV-enriched membranes by mass spectrometry and is abundant in the TGN and in endosomes (40–42). Confocal microscopy confirmed that syntaxin-6 is redistributed to the cell surface by TUG UBX-Cter (Fig. 6A). Similar results were obtained by subcellular fractionation of shRNA cells, in which syntaxin-6 was redistributed to PM in unstimulated cells (Fig. 6B). The data support the idea that TUG regulates GSVs that arise from syntaxin-6-containing membranes, possibly the TGN or endosomes.
FIGURE 6.
TUG disruption mobilizes syntaxin-6 to the plasma membrane. A, basal and insulin-treated control and UBX-Cter-containing 3T3-L1 adipocytes were imaged using confocal microscopy. GLUT4 was detected by a GFP tag, and syntaxin-6 was detected by immunostaining. Scale bar, 10 μm. B, control, shRNA, and shRNA + TUG 3T3-L1 adipocytes were treated with insulin, subjected to subcellular fractionation, and immunoblotted as indicated to detect syntaxin-6. Experiments were repeated at least twice with similar results.
TUG Cleavage Liberates GLUT4 from the Golgi Matrix and Requires TC10α
The observation that TUG disruption redistributed syntaxin-6 prompted us to test if proteins that associate with syntaxin-6 may bind and regulate TUG and GLUT4. An excellent candidate was PIST (also called FIG, GOPC, and CAL), which binds syntaxin-6 and controls the PM targeting of CFTR proteins, frizzled isoforms, ClC3-B, AMPA-type glutamate receptors, and other receptors (1, 14, 43–48). PIST is an effector of the TC10α GTPase (49), which mediates insulin-stimulated GLUT4 translocation (50) and also regulates the cell surface targeting of CFTR proteins and AMPA receptors (51–53).
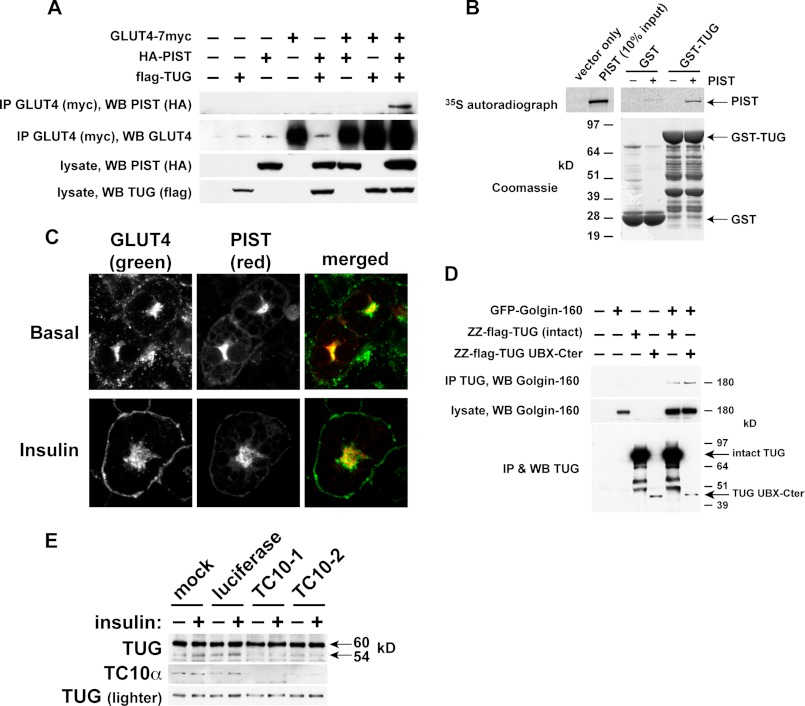
Fig. 7A shows that PIST can be coimmunoprecipitated in a complex together with TUG and GLUT4. PIST was purified with GLUT4 only when TUG was coexpressed, supporting the concept that these proteins participate in a linear arrangement, with TUG interposed between GLUT4 and PIST. We used recombinant proteins to show that TUG and PIST can bind each other directly (Fig. 7B). Endogenous PIST colocalized with TUG and with insulin-responsive GLUT4 in LM fractions and was not translocated by insulin, as noted above (Fig. 5B). By confocal microscopy, HA-tagged PIST colocalized with perinuclear GLUT4 in 3T3-L1 adipocytes and was not translocated to the cell surface by insulin (Fig. 7C). These data support the idea that GLUT4, TUG, and PIST can form a complex and that this complex may regulate GSVs.
FIGURE 7.
Evidence that TUG cleavage separates GLUT4 from the Golgi matrix and requires TC10α. A, tagged forms of GLUT4, PIST, and TUG were transfected in 293T cells as indicated. GLUT4 was immunoprecipitated, and bound proteins were analyzed by SDS-PAGE and immunoblotting as indicated. B, PIST was translated in vitro in the presence of 35S-labeled Cys and Met and was incubated with recombinant GST-TUG or with GST alone. Bound proteins were analyzed by SDS-PAGE and autoradiography. C, basal and insulin-stimulated 3T3-L1 adipocytes stably expressing HA-PIST and GLUT4-GFP were imaged by confocal microscopy. Scale bar, 10 μm. D, GFP-tagged Golgin-160 and protein A (ZZ)-tagged TUG were transfected in 293T cells, and TUG was purified by binding to immobilized IgG. Eluates and lysates were immunoblotted as indicated. E, 3T3-L1 adipocytes were electroporated with synthetic siRNAs directed to TC10α or luciferase (as a control) or mock-electroporated. Cells were treated with or without insulin and then lysed using denaturing conditions and immunoblotted as indicated. All experiments were repeated at least twice with similar results.
PIST binds the Golgi matrix protein Golgin-160, which like TUG is required for the intracellular retention of GLUT4 in unstimulated cells (54, 55). Accordingly, we tested if TUG binds Golgin-160. Fig. 7D shows that TUG and Golgin-160 were coprecipitated from transfected cells. Moreover, a truncated fragment of TUG was able to purify Golgin-160, which maps the interaction to the TUG C-terminal region (residues 377–550). This UBX-Cter fragment functions as a dominant negative protein, as noted above, and it was hypothesized to bind an anchoring site but not GLUT4 or other GSV components. By excluding endogenous TUG from this site, it blocks the ability of intact TUG to retain GLUT4 within unstimulated cells. Therefore, the data implicate Golgin-160 and PIST as components of this intracellular anchoring site. Together, the data support a model whereby cleavage separates N- and C- terminal regions of TUG that bind GSVs and the Golgi matrix, respectively.
As noted above, TC10α is required to transmit an insulin signal for GLUT4 translocation (50), and we considered that TC10α may signal through PIST to promote TUG cleavage. To test the hypothesis that TUG proteolytic processing requires TC10α, we transfected 3T3-L1 adipocytes with siRNAs. Fig. 7E shows that in cells in which TC10α was depleted, basal and insulin-stimulated production of the 54-kDa TUG C-terminal product was markedly reduced. By contrast, in control 3T3-L1 adipocytes that were mock-transfected or transfected with an irrelevant siRNA, the 54-kDa product was observed and was increased after insulin stimulation. These data indicate that TC10α is required for basal as well as insulin-stimulated TUG processing. Additionally, the data highlight the physiologic significance in 3T3-L1 adipocytes of interactions of TUG with PIST and Golgin-160. Considering this together with previous results, we conclude that insulin signaling through TC10α is required to accelerate TUG proteolytic processing.
If TUG traps GSVs at the Golgi matrix and if TUG cleavage liberates GSVs for exocytic translocation, then the number of TUG proteins that are cleaved should relate stoichiometrically to the number of insulin-responsive GSVs in each cell. Previous data show that there are about 300,000 copies of GLUT4 per 3T3-L1 adipocyte (56). Of this number, about 30–40% are in GSVs in unstimulated cells, and the remainder are in endosomes, GSV donor membranes, and the plasma membrane (1). Thus, in a typical 3T3-L1 adipocyte, about 100,000 molecules of GLUT4 are present in GSVs in the absence of insulin. Other work has shown that there are about 5–6 molecules of GLUT4 in each GSV (57). Therefore, there are ∼20,000 GSVs in each unstimulated 3T3-L1 adipocyte.
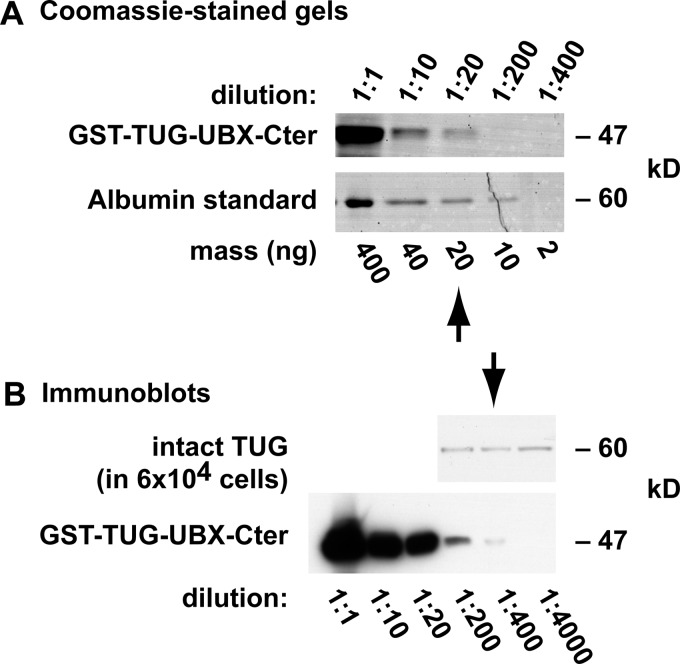
To determine the number of TUG molecules that are present in each 3T3-L1 adipocyte, we used a recombinant TUG protein. Fig. 8A compares Coomassie-stained GST-TUG-UBX-Cter and an albumin standard. Ten microliters of a 1:20 dilution of the recombinant TUG stock contained 20 ng of this protein, or about 2.5 × 1011 molecules (based on the molecular mass of GST-TUG-UBX-Cter, which is 47 kDa). Fig. 8B presents immunoblots of recombinant GST-TUG-UBX-Cter and 3T3-L1 adipocyte whole cell lysates, which were done using the antibody directed to the TUG C terminus. The lysates contained material from 6 × 104 cells, and the number of intact, 60-kDa TUG proteins that were present was similar to that in 1 ng of the recombinant GST-TUG-UBX-Cter sample, or about 1.25 × 1010 molecules. Therefore, each 3T3-L1 adipocyte contains ∼200,000 TUG proteins.
FIGURE 8.
Quantification of TUG abundance in 3T3-L1 adipocytes. A, serial dilutions of recombinant GST-TUG-UBX-Cter and a bovine serum albumin standard were subjected to SDS-PAGE and staining with GelCode Coomassie. Densitometry showed that 10 μl of a 1:20 dilution of the recombinant TUG protein contained 20 ng, or about 2.5 × 1011 molecules (based on the molecular mass of GST-TUG-UBX-Cter, 47 kDa). B, immunoblots of a whole cell lysate from 3T3-L1 adipocytes and of recombinant GST-TUG-UBX-Cter were performed using an antibody directed to the C terminus of TUG. Densitometry showed that ∼1.25 × 1010 molecules of intact TUG (60 kDa) were present in 6 × 104 cells, so that ∼200,000 molecules of TUG are present in each 3T3-L1 adipocyte.
If ∼10% of the intact TUG molecules are cleaved in response to insulin (Fig. 3), then about 20,000 TUG molecules undergo insulin-stimulated proteolysis in each 3T3-L1 adipocyte. This number corresponds very well to the number of GSVs that are present in each 3T3-L1 adipocyte, which was also calculated at ∼20,000, as noted above. These calculations are approximate, but they nonetheless support the idea that only one molecule of TUG (or at most a few molecules) is involved in “tethering” each insulin-responsive GSV at the Golgi matrix in unstimulated 3T3-L1 adipocytes. The data further support the idea that the amount of TUG cleavage we observe could reasonably be expected to liberate the insulin-responsive pool of GSVs for translocation to the cell surface.
Highly Insulin-responsive GLUT4 Translocation Requires TUG Cleavage
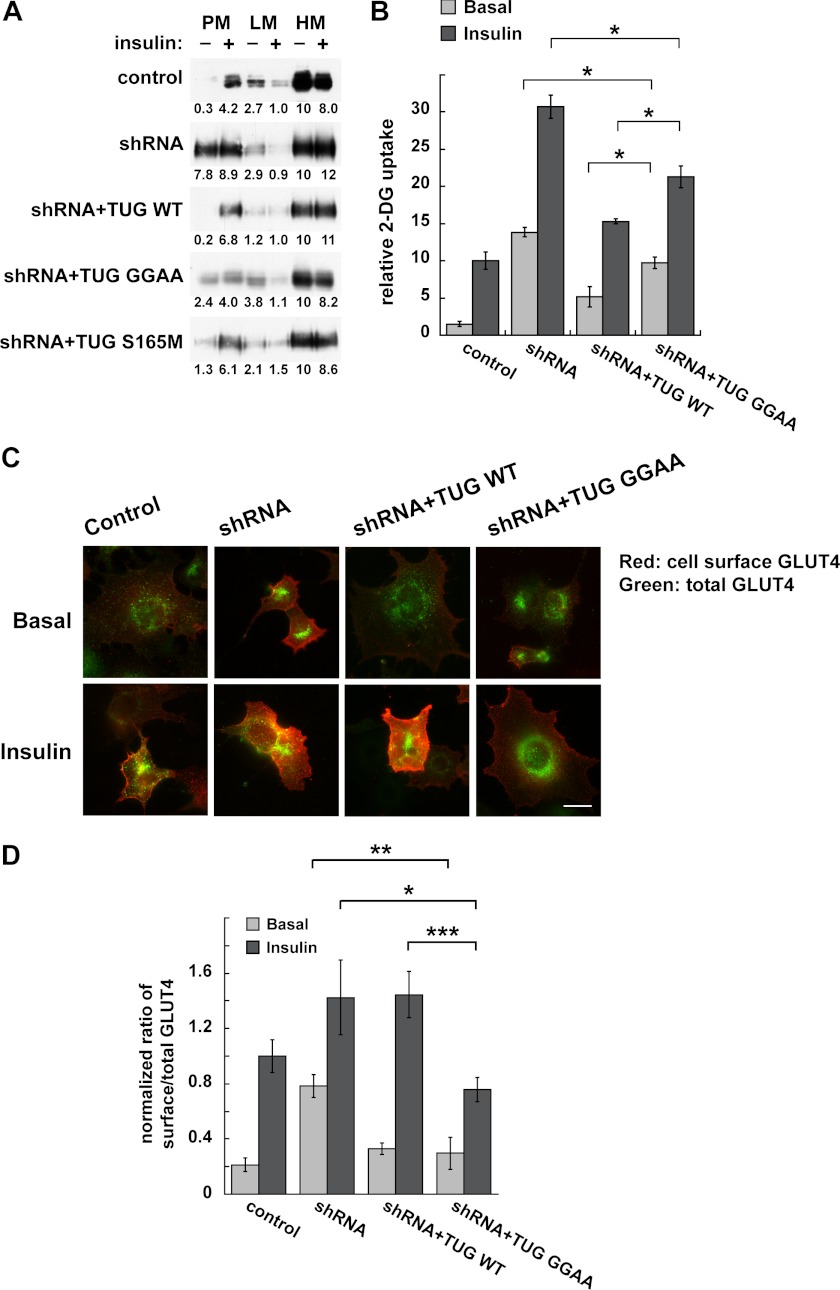
To test directly if TUG cleavage is required to translocate GLUT4, we used several approaches. Initially, we used differential centrifugation to isolate PM, LM, and HM fractions, as previously, and we immunoblotted endogenous GLUT4 in these fractions (7). In control 3T3-L1 adipocytes, insulin stimulated the translocation of GLUT4 out of the LM fraction and to the PM fraction (Fig. 9A). This effect was disrupted in shRNA cells and rescued in shRNA + TUG WT cells. In shRNA + TUG GGAA cells, containing cleavage-resistant TUG, GLUT4 responded poorly to insulin. GLUT4 was more abundant in the basal PM fraction, and insulin caused only slight additional movement out of the LM and to the PM fraction. In shRNA + TUG S165M cells, GLUT4 movement was similar to that in shRNA + TUG WT cells, suggesting that rapid degradation of the C-terminal product is not required for acute insulin action. We conclude that TUG cleavage is required for highly insulin-responsive GLUT4 targeting.
FIGURE 9.
TUG cleavage is required for regulated GLUT4 targeting in 3T3-L1 adipocytes. A, PM, LM, and HM fractions were isolated from basal and insulin-treated cells containing TUG shRNA and shRNA-resistant proteins, as indicated, and were immunoblotted for GLUT4. Band intensities are indicated and were normalized to GLUT4 in the HM fraction of unstimulated cells (set to 10) in each case. Similar results were obtained in at least two independent experiments for each cell line. B, basal and insulin-stimulated 2-deoxyglucose (2-DG) uptake was measured in the indicated cells and is plotted relative to insulin-stimulated control cells. Results shown are mean ± S.E. (error bars); n = 3; *, p < 0.05. C and D, targeting of newly synthesized GLUT4 in the indicated cells was measured 18–20 h after electroporation of a dual Myc- and GFP-tagged GLUT4 reporter. C, epifluorescence microscopy was used to image cell surface and total amounts of the reporter. D, images were quantified, and ratios were calculated and plotted relative to insulin-stimulated control cells. Results shown are mean ± S.E.; n = 16 basal and 27–30 insulin-stimulated cells in each group; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We next tested if TUG cleavage is required for insulin to regulate glucose uptake. Fig. 9B shows that, similar to previous data, shRNA-mediated TUG depletion enhanced basal glucose transport (7). Insulin caused a 2-fold further increase, which may be due to TUG-independent fusion of vesicles containing GLUT4 at the plasma membrane or to effects of insulin on GLUT4 and GLUT1 in endosomes (10). Effects of the shRNA were rescued by shRNA-resistant, wild type TUG. Cleavage-resistant TUG GGAA did not fully rescue the effect of the shRNA in either basal or insulin-stimulated conditions. Basal glucose uptake remained somewhat increased but not to the level observed in the shRNA cells, consistent with the fractionation data (Fig. 9A). Insulin-stimulated glucose uptake was also increased and was also less than that observed in the shRNA cells. Because of the increased basal uptake, insulin stimulated only a 2-fold further increase, which is not typical of highly insulin-responsive glucose uptake. This is similar to the ∼2-fold increase in PM GLUT4 that we observed by fractionation of shRNA + TUG GGAA cells. Interpretation of both the fractionation and glucose uptake data is informed by our previous observation that GLUT4 participates in two distinct exocytic pathways, only one of which is regulated by TUG (10). When flux through the TUG-regulated pathway is inhibited, by cleavage-resistant, mutated TUG, GLUT4 is still able to cycle in endosomes. Thus, impaired trapping of endocytosed TUG in GSVs may be predicted to result in increased basal glucose uptake and a diminished (but not absent) effect of insulin stimulation. The data are consistent with this scenario. We conclude that TUG cleavage is required for highly insulin-responsive glucose uptake.
To test more directly if cleavage-resistant TUG blocks GLUT4 flux through GSVs, we examined newly synthesized GLUT4. In 3T3-L1 adipocytes, transiently transfected GLUT4 enters GSVs directly from the biosynthetic pathway (21, 58). The effect on GSVs can then be examined, without interference from GLUT4 recycling through endosomes (10). Thus, we predicted that in shRNA + TUG GGAA cells, basal retention of newly synthesized GLUT4 would be fully rescued, and translocation would be selectively inhibited. Accordingly, we transfected a GLUT4 reporter in cells in which TUG proteins were stably manipulated and used microscopy to quantify the ratio of surface to total GLUT4 shortly after transfection (9, 21). As shown in Fig. 9C, we observed insulin-stimulated translocation in control cells, disrupted basal GLUT4 retention in shRNA cells, and rescue of insulin-stimulated translocation in shRNA + TUG WT cells. In shRNA + TUG GGAA cells, the basal intracellular retention of GLUT4 was fully rescued, and there was a dramatic defect in insulin-stimulated translocation, as predicted. The data were quantified in Fig. 9D, which shows that both wild type and cleavage-resistant TUG fully rescued the effect of the shRNA to increase basal targeting of GLUT4 to the cell surface. In shRNA + TUG WT cells, insulin-stimulated translocation was slightly greater than that in control cells, consistent with previous data implying that TUG overexpression enlarges the pool of GSVs (6, 7). In shRNA + TUG GGAA cells, which contain similarly overexpressed, cleavage-resistant TUG, insulin-stimulated translocation was impaired. We conclude that TUG proteolytic processing is required to mobilize GSVs to the cell surface in 3T3-L1 adipocytes.
DISCUSSION
Glucose uptake is tightly regulated by insulin, which controls the number of GLUT4 glucose transporters present in plasma membranes of fat and muscle cells. GLUT4 is very efficiently sequestered within unstimulated cells. Insulin can then act within minutes to stimulate a large increase in plasma membrane GLUT4 abundance and does so by causing the acute mobilization of GLUT4 out of intracellular membranes. Insulin acts at multiple sites in GLUT4 trafficking to modulate its overall distribution within cells. The site of regulation that is quantitatively most important is the pool of insulin-responsive storage vesicles, termed GSVs (4). These vesicles are formed in a cell type-specific, differentiation-dependent manner and are thought to exist as a preformed population within unstimulated cells (59). GLUT4 and only a few other proteins accumulate selectively in GSVs in the absence of insulin (5). These vesicles are mobilized acutely after insulin addition, which accounts for the bulk of insulin action to insert GLUT4 at the cell surface. How GSVs are efficiently sequestered in unstimulated cells and how they are mobilized by insulin stimulation are not well understood.
The data presented here support a model in which insulin stimulates TUG cleavage to mobilize GSVs to the cell surface and to augment glucose uptake. The simplest interpretation is that cleavage releases a tethering mechanism that retains the vesicles at the Golgi matrix in unstimulated cells. It is also possible that one or both TUG cleavage products functions as a signal. In particular, TUGUL liberation and covalent attachment to a target protein may activate vesicle translocation to the cell surface. The ideas that cleavage releases a tether and that TUGUL acts as a signal to promote translocation are not mutually exclusive. For example, one can envision a model in which TUG cleavage releases GSVs from the Golgi matrix, and TUGUL modification somehow activates kinesin or myosin motor proteins to promote movement of the released GSVs to the cell surface (60–64). Alternatively, the liberated TUGUL might activate proteins that participate in vesicle tethering, docking, or fusion at the plasma membrane (10). Regardless, TUG cleavage is here shown to be a novel biochemical mechanism that is required for fully insulin-responsive GLUT4 translocation and glucose uptake in 3T3-L1 adipocytes.
We previously proposed that TUG is an essential component of a functional tether that binds and sequesters GLUT4 within unstimulated cells (6, 7). Data presented here expand upon this model and implicate site-specific endoproteolytic cleavage of TUG in the release of this sequestration mechanism. Like GLUT4 translocation, TUG cleavage occurs in a differentiation-dependent manner in 3T3-L1 adipocytes. We show that intact TUG can link GLUT4 to Golgi matrix proteins, PIST and Golgin-160. Cleavage then separates an N-terminal region that binds GLUT4 from a C-terminal region that binds Golgin-160. Cleavage may thus “untether” GSVs from the Golgi matrix. We further show that TC10α is required for the production of TUG cleavage products, consistent with the idea that insulin signaling through TC10α is required to accelerate TUG cleavage and to mobilize GSVs. The number of TUG molecules that are cleaved corresponds approximately to the number of GSVs present in each cell. Finally, we show that cleavage is required for highly insulin-responsive GLUT4 translocation and glucose uptake.
TUG endoproteolysis is the first step in a complicated biochemical processing mechanism that has yet to be fully elucidated. We do not know the identity of the enzyme that mediates TUG cleavage. Our data support the idea that TUG is precursor to a novel ubiquitin-like modifier, TUGUL, and the cleaving enzyme may therefore be similar to known deubiquitinating enzymes. Mature TUGUL is covalently attached to ubiquitin in transfected 293T cells, suggesting that it would be able to modify a target protein if produced by cleavage of endogenous TUG. Our data show that endoproteolysis occurs in 3T3-L1 adipocytes and that the TUG N terminus is incorporated into a single, ∼130-kDa protein. This putative tugulated protein remains unidentified at present despite our efforts to precipitate it using the N terminus antibody or an epitope tag (which disrupted the localization of intact TUG expressed using a retrovirus in 3T3-L1 adipocytes). If the ∼130-kDa protein contains an 18-kDa TUGUL molecule, then the target of TUGUL modification in 3T3-L1 adipocytes is ∼110 kDa. Several proteins in this size range have been implicated in GLUT4 trafficking (60–63, 65, 66). The observation that TUGUL binds GLUT4 (7), together with the finding that the 130-kDa protein is present in a PM fraction (Fig. 5B), suggests that the tugulated protein may move together with GSVs to the cell surface. Further work will be required to determine if this is the case and to identify the tugulated substrate.
By analogy to known Ubls, our data suggest that activating (E1), conjugating (E2), and ligating (E3) enzymes may mediate TUGUL attachment in 3T3-L1 adipocytes. Ubc9 is well described as a SUMO E2, which binds GLUT4 and controls its targeting and, probably secondarily, its stability (1, 29, 34, 65, 67). It is formally possible that Ubc9 could mediate TUGUL conjugation. GLUT4 itself may function as an E3 because it binds TUGUL and may form a scaffold for other enzymes involved in the reaction. Daxx is another protein that binds GLUT4, Ubc9, and SUMO, which has been proposed to scaffold these proteins together with other molecules (63, 65). Other GSV cargoes, notably IRAP, sortilin, and LRP1, may be more important than GLUT4 itself for the formation and regulation of GSVs (4, 5). Understanding whether these proteins may contribute to enzymatic activities for TUG proteolysis and TUGUL modification will require further research.
The TUG C-terminal product is present as a 54-kDa protein as well as at the predicted mass of 42 kDa. The 54-kDa form appears to be a covalently modified form of the 42-kDa product because it is observed after cell lysis in denaturing conditions. Although the molecular characterization of this modification will require further work, present data show that the modified form can be stabilized by mutations (e.g. K380R and S165M) within the future TUG C-terminal product. The modification probably occurs after cleavage of the intact TUG protein. The 54-kDa form is present in the cytosol, whereas the 130- and 42-kDa products are both present exclusively in a membrane fraction, suggesting that they are the initial products. In addition, cleavage-resistant TUG does not accumulate as a ∼72-kDa form, which might be predicted if the modification preceded cleavage. Thus, one possibility is that the modification is somehow involved in removing the C-terminal cleavage product from the anchoring site.
Whether the TUG C-terminal product has an additional function is not known. We recently reported that TUG disassembles the hexameric p97/VCP ATPase into its monomeric subunits (68). The C-terminal half of TUG is highly conserved evolutionarily, and TUG is widely expressed and regulates Golgi dynamics in HeLa cells, possibly through its action on p97. We hypothesize that its function is adapted in a cell type-specific manner to control GLUT4 exocytosis. A TUG fragment corresponding to the 42-kDa C-terminal product was fully capable of disassembling p97 hexamers in vitro. Although we did not observe a role of the TUG N terminus to modulate this activity, one possibility is that TUG cleavage and p97 oligomeric status are coordinately regulated in adipocytes. Because p97 interacts with clathrin, SNAREs, and other membrane trafficking proteins, its regulation could well control GLUT4 (69–72). For example, the role of TUG to trap GLUT4 intracellularly may require its action to disassemble p97 hexamers and, presumably, to inhibit p97 ATPase activity.
After GLUT4 mobilization, a TUG C-terminal product must be removed from the Golgi matrix. p97 is well known to extract proteins from the endoplasmic reticulum (ER) for ER-associated degradation, and the putative yeast ortholog of TUG, Ubx4p, has been implicated in this process (73). Possibly, p97 is involved in TUG proteolytic processing and/or in extracting the TUG C-terminal product, given its roles to modulate proteolysis in other instances and to extract cleavage products of the membrane-bound transcription factors, Spt23p/Mga2p and SREBP (74–78). Remarkably, in transfected HeLa cells, the 42-kDa TUG C-terminal product was targeted to the nucleus and caused nuclear accumulation of endogenous p97 (68). Thus, it is possible that a C-terminal fragment produced by TUG cleavage in adipocytes is also targeted to the nucleus. Of note, Ubx4p has been implicated in controlling RNA polymerase II turnover (79). Whether TUG functions similarly and whether TUG cleavage coordinates glucose uptake and gene expression remain to be seen.
Our data show that TUG interacts with PIST and Golgin-160, implying that it may link GLUT4 to these proteins to sequester GSVs at the Golgi matrix. Other proteins may also bind the TUG C terminus to mediate intracellular sequestration (6). Golgin-160 was previously shown to be required for full intracellular retention of GLUT4 in unstimulated 3T3-L1 adipocytes (55). A similar function has been described for p115, which binds IRAP and LRP1 and is present in a similar location at the cis-Golgi, at the ER-Golgi intermediate compartment, and at ER exit sites (66, 80–83). These data fit well with our observation that TUG is present primarily at the ER-Golgi intermediate compartment, ER exit sites, and cis-Golgi in HeLa cells (68). Together with data supporting the idea that GSVs fuse directly with the plasma membrane (10), we conclude that GSVs may follow an unconventional secretion pathway (3, 84).
Our data show that TC10α is required for the production of TUG cleavage products and indicate that TC10α is coupled through its effector, PIST, directly to TUG and GLUT4. TC10α was previously shown to be required for insulin-stimulated GLUT4 translocation in 3T3-L1 adipocytes (50) and signals through its effectors, CIP4 and Exo70, to regulate GLUT4 trafficking (85–87). PIST is yet another effector, which may couple TC10α activation directly to the release of GSVs. Of note, TC10α also acts through PIST to regulate CFTR, which follows an unconventional secretion pathway to reach the cell surface (44, 51, 52, 84, 88, 89). Insulin signaling through TC10α is probably coordinated with signaling through Akt2 to AS160/Tbc1D4 and downstream Rab GTPases (2). By activating the appropriate Rab isoform(s), signaling through AS160 may cause the released GSVs to traffic to the plasma membrane and not to some other compartment. Conversely, in unstimulated cells, GSVs that are captured by TUG and Golgi matrix proteins may not be entirely static but may participate in an intracellular cycle into and out of Golgi membranes. Such a cycle could efficiently sequester GLUT4 and would fit well with the function of Golgi matrix proteins to capture vesicles and to direct their fusion at an appropriate cisterna (90). Thus, similar to activated Rab proteins, intact TUG may serve as an adaptor to link particular vesicles, GSVs, to Golgi matrix proteins and thus to mediate their capture and retention.
We conclude that site-specific proteolysis is required for insulin to regulate glucose uptake in 3T3-L1 adipocytes. Whether this mechanism pertains in muscle is not known. GLUT4 and TUG abundances are highly correlated in skeletal muscles, and insulin stimulates the dissociation of TUG from GLUT4 (8, 91). Thus, it seems likely that a similar process is involved in insulin action in muscle. More broadly, our data imply that GLUT4 trafficking shares features with other molecular systems that employ protein degradation to confer tight control and vectorial directionality. We are not aware of other instances in which insulin acts through site-specific proteolysis. The nearest example may be SREBP1c, which is mobilized into COPII vesicles by insulin and encounters a protease at the Golgi complex (92, 93). As well, mechanisms similar to the one we describe may control other protein-exposing, nonsecretory exocytoses and may regulate a wide range of physiology (94). Finally, our results may contribute to understanding of the pathogenesis of insulin resistance and type 2 diabetes.
Acknowledgments
We thank Mark Hochstrasser, Thomas Gniadek, Derek Toomre, Emily Stoops, Whitney Harris, and Hongjie Li for assistance, advice, and comments on the manuscript. We thank Xuedong Liu, Dirk Bohmann, Hamid Band, and Carolyn Machamer for reagents. This work used the Cell Biology Core of the Yale Diabetes Endocrinology Research Center (supported by National Institutes of Health Grant P30DK45735) as well as services provided by the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK075772 (to J. S. B.). This work was also supported by American Diabetes Association Grants 1-05-RA-17 and 1-12-BS-16, by a pilot grant from the Yale O'Brien Kidney Center P30DK079310, and by a Distinguished Young Scholar in Medical Research Award from the W. M. Keck Foundation (to J. S. B.).
- GSV
- GLUT4 storage vesicle
- CFTR
- cystic fibrosis transmembrane conductance regulator
- ER
- endoplasmic reticulum
- LM
- light microsome
- HM
- heavy microsome
- PM
- plasma membrane
- SUMO
- small ubiquitin-like modifier
- TGN
- trans-Golgi network
- UBL
- ubiquitin-like
- UBX
- ubiquitin regulatory X.
REFERENCES
- 1. Rubin B. R., Bogan J. S. (2009) Intracellular retention and insulin-stimulated mobilization of GLUT4 glucose transporters. Vitam. Horm. 80, 155–192 [DOI] [PubMed] [Google Scholar]
- 2. Huang S., Czech M. P. (2007) The GLUT4 glucose transporter. Cell Metab. 5, 237–252 [DOI] [PubMed] [Google Scholar]
- 3. Bogan J. S. (2012) Regulation of glucose transporter translocation in health and diabetes. Annu. Rev. Biochem. 81, 507–532 [DOI] [PubMed] [Google Scholar]
- 4. Bogan J. S., Kandror K. V. (2010) Biogenesis and regulation of insulin-responsive vesicles containing GLUT4. Curr. Opin. Cell Biol. 22, 506–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kandror K. V., Pilch P. F. (2011) The sugar is sIRVed. Sorting Glut4 and its fellow travelers. Traffic 12, 665–671 [DOI] [PubMed] [Google Scholar]
- 6. Bogan J. S., Hendon N., McKee A. E., Tsao T. S., Lodish H. F. (2003) Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature 425, 727–733 [DOI] [PubMed] [Google Scholar]
- 7. Yu C., Cresswell J., Löffler M. G., Bogan J. S. (2007) The glucose transporter 4-regulating protein TUG is essential for highly insulin-responsive glucose uptake in 3T3-L1 adipocytes. J. Biol. Chem. 282, 7710–7722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schertzer J. D., Antonescu C. N., Bilan P. J., Jain S., Huang X., Liu Z., Bonen A., Klip A. (2009) A transgenic mouse model to study glucose transporter 4myc regulation in skeletal muscle. Endocrinology 150, 1935–1940 [DOI] [PubMed] [Google Scholar]
- 9. Bogan J. S., McKee A. E., Lodish H. F. (2001) Insulin-responsive compartments containing GLUT4 in 3T3-L1 and CHO cells. Regulation by amino acid concentrations. Mol. Cell. Biol. 21, 4785–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu Y., Rubin B. R., Orme C. M., Karpikov A., Yu C., Bogan J. S., Toomre D. K. (2011) Dual-mode of insulin action controls GLUT4 vesicle exocytosis. J. Cell Biol. 193, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kandror K. V., Coderre L., Pushkin A. V., Pilch P. F. (1995) Biochem. J. 307, 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Govers R., Coster A. C., James D. E. (2004) Insulin increases cell surface GLUT4 levels by dose-dependently discharging GLUT4 into a cell surface recycling pathway. Mol. Cell. Biol. 24, 6456–6466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muretta J. M., Romenskaia I., Mastick C. C. (2008) Insulin releases Glut4 from static storage compartments into cycling endosomes and increases the rate constant for Glut4 exocytosis. J. Biol. Chem. 283, 311–323 [DOI] [PubMed] [Google Scholar]
- 14. Yue Z., Horton A., Bravin M., DeJager P. L., Selimi F., Heintz N. (2002) A novel protein complex linking the delta 2 glutamate receptor and autophagy. Implications for neurodegeneration in lurcher mice. Neuron 35, 921–933 [DOI] [PubMed] [Google Scholar]
- 15. Liu X., Constantinescu S. N., Sun Y., Bogan J. S., Hirsch D., Weinberg R. A., Lodish H. F. (2000) Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal. Biochem. 280, 20–28 [DOI] [PubMed] [Google Scholar]
- 16. Knuesel M., Wan Y., Xiao Z., Holinger E., Lowe N., Wang W., Liu X. (2003) Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol. Cell Proteomics 2, 1225–1233 [DOI] [PubMed] [Google Scholar]
- 17. Naviaux R. K., Costanzi E., Haas M., Verma I. M. (1996) The pCL vector system. Rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70, 5701–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) The CLUSTAL_X windows interface. Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bogan J. S., Lodish H. F. (1999) Two compartments for insulin-stimulated exocytosis in 3T3-L1 adipocytes defined by endogenous ACRP30 and GLUT4. J. Cell Biol. 146, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hertel J., Struthers H., Horj C. B., Hruz P. W. (2004) A structural basis for the acute effects of HIV protease inhibitors on GLUT4 intrinsic activity. J. Biol. Chem. 279, 55147–55152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khan A. H., Capilla E., Hou J. C., Watson R. T., Smith J. R., Pessin J. E. (2004) Entry of newly synthesized GLUT4 into the insulin-responsive storage compartment is dependent upon both the amino terminus and the large cytoplasmic loop. J. Biol. Chem. 279, 37505–37511 [DOI] [PubMed] [Google Scholar]
- 22. Lampson M. A., Racz A., Cushman S. W., McGraw T. E. (2000) Demonstration of insulin-responsive trafficking of GLUT4 and vpTR in fibroblasts. J. Cell Sci. 113, 4065–4076 [DOI] [PubMed] [Google Scholar]
- 23. Hao M., Maxfield F. R. (2000) Characterization of rapid membrane internalization and recycling. J. Biol. Chem. 275, 15279–15286 [DOI] [PubMed] [Google Scholar]
- 24. Hochstrasser M. (2009) Origin and function of ubiquitin-like proteins. Nature 458, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tettamanzi M. C., Yu C., Bogan J. S., Hodsdon M. E. (2006) Solution structure and backbone dynamics of an N-terminal ubiquitin-like domain in the GLUT4-regulating protein, TUG. Protein Sci. 15, 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alexandru G., Graumann J., Smith G. T., Kolawa N. J., Fang R., Deshaies R. J. (2008) UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1α turnover. Cell 134, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kloppsteck P., Ewens C. A., Förster A., Zhang X., Freemont P. S. (2012) Regulation of p97 in the ubiquitin-proteasome system by the UBX protein-family. Biochim. Biophys. Acta 1823, 125–129 [DOI] [PubMed] [Google Scholar]
- 28. Wilkinson K. D., Audhya T. K. (1981) Stimulation of ATP-dependent proteolysis requires ubiquitin with the COOH-terminal sequence Arg-Gly-Gly. J. Biol. Chem. 256, 9235–9241 [PubMed] [Google Scholar]
- 29. Giorgino F., de Robertis O., Laviola L., Montrone C., Perrini S., McCowen K. C., Smith R. J. (2000) The sentrin-conjugating enzyme mUbc9 interacts with GLUT4 and GLUT1 glucose transporters and regulates transporter levels in skeletal muscle cells. Proc. Natl. Acad. Sci. U.S.A. 97, 1125–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Varshavsky A. (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sriram S. M., Kim B. Y., Kwon Y. T. (2011) The N-end rule pathway. Emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 12, 735–747 [DOI] [PubMed] [Google Scholar]
- 32. Butt T. R., Khan M. I., Marsh J., Ecker D. J., Crooke S. T. (1988) Ubiquitin-metallothionein fusion protein expression in yeast. A genetic approach for analysis of ubiquitin functions. J. Biol. Chem. 263, 16364–16371 [PubMed] [Google Scholar]
- 33. Laney J. D., Hochstrasser M. (2011) Curr. Protoc. Protein Sci., Chapter 14, Unit 14.15 [DOI] [PubMed] [Google Scholar]
- 34. Liu L. B., Omata W., Kojima I., Shibata H. (2007) The SUMO conjugating enzyme Ubc9 is a regulator of GLUT4 turnover and targeting to the insulin-responsive storage compartment in 3T3-L1 adipocytes. Diabetes 56, 1977–1985 [DOI] [PubMed] [Google Scholar]
- 35. Perera H. K., Clarke M., Morris N. J., Hong W., Chamberlain L. H., Gould G. W. (2003) Syntaxin 6 regulates Glut4 trafficking in 3T3-L1 adipocytes. Mol. Biol. Cell 14, 2946–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shewan A. M., van Dam E. M., Martin S., Luen T. B., Hong W., Bryant N. J., James D. E. (2003) GLUT4 recycles via a trans-Golgi network (TGN) subdomain enriched in Syntaxins 6 and 16 but not TGN38. Involvement of an acidic targeting motif. Mol. Biol. Cell 14, 973–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Proctor K. M., Miller S. C., Bryant N. J., Gould G. W. (2006) Syntaxin 16 controls the intracellular sequestration of GLUT4 in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 347, 433–438 [DOI] [PubMed] [Google Scholar]
- 38. Bose A., Guilherme A., Huang S., Hubbard A. C., Lane C. R., Soriano N. A., Czech M. P. (2005) The v-SNARE Vti1a regulates insulin-stimulated glucose transport and Acrp30 secretion in 3T3-L1 adipocytes. J. Biol. Chem. 280, 36946–36951 [DOI] [PubMed] [Google Scholar]
- 39. Watson R. T., Hou J. C., Pessin J. E. (2008) Recycling of IRAP from the plasma membrane back to the insulin-responsive compartment requires the Q-SNARE syntaxin 6 but not the GGA clathrin adaptors. J. Cell Sci. 121, 1243–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Larance M., Ramm G., Stöckli J., van Dam E. M., Winata S., Wasinger V., Simpson F., Graham M., Junutula J. R., Guilhaus M., James D. E. (2005) Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J. Biol. Chem. 280, 37803–37813 [DOI] [PubMed] [Google Scholar]
- 41. Foster L. J., Rudich A., Talior I., Patel N., Huang X., Furtado L. M., Bilan P. J., Mann M., Klip A. (2006) Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 5, 64–75 [DOI] [PubMed] [Google Scholar]
- 42. Wendler F., Tooze S. (2001) Syntaxin 6. The promiscuous behavior of a SNARE protein. Traffic 2, 606–611 [DOI] [PubMed] [Google Scholar]
- 43. Charest A., Lane K., McMahon K., Housman D. E. (2001) Association of a novel PDZ domain-containing peripheral Golgi protein with the Q-SNARE (Q-soluble N-ethylmaleimide-sensitive fusion protein (NSF) attachment protein receptor) protein syntaxin 6. J. Biol. Chem. 276, 29456–29465 [DOI] [PubMed] [Google Scholar]
- 44. Cheng J., Moyer B. D., Milewski M., Loffing J., Ikeda M., Mickle J. E., Cutting G. R., Li M., Stanton B. A., Guggino W. B. (2002) A Golgi-associated PDZ domain protein modulates cystic fibrosis transmembrane regulator plasma membrane expression. J. Biol. Chem. 277, 3520–3529 [DOI] [PubMed] [Google Scholar]
- 45. Yao R., Maeda T., Takada S., Noda T. (2001) Identification of a PDZ domain-containing Golgi protein, GOPC, as an interaction partner of frizzled. Biochem. Biophys. Res. Commun. 286, 771–778 [DOI] [PubMed] [Google Scholar]
- 46. Gentzsch M., Cui L., Mengos A., Chang X. B., Chen J. H., Riordan J. R. (2003) The PDZ-binding chloride channel ClC-3B localizes to the Golgi and associates with cystic fibrosis transmembrane conductance regulator-interacting PDZ proteins. J. Biol. Chem. 278, 6440–6449 [DOI] [PubMed] [Google Scholar]
- 47. He J., Bellini M., Xu J., Castleberry A. M., Hall R. A. (2004) Interaction with cystic fibrosis transmembrane conductance regulator-associated ligand (CAL) inhibits β1-adrenergic receptor surface expression. J. Biol. Chem. 279, 50190–50196 [DOI] [PubMed] [Google Scholar]
- 48. Wente W., Efanov A. M., Treinies I., Zitzer H., Gromada J., Richter D., Kreienkamp H. J. (2005) The PDZ/coiled-coil domain containing protein PIST modulates insulin secretion in MIN6 insulinoma cells by interacting with somatostatin receptor subtype 5. FEBS Lett. 579, 6305–6310 [DOI] [PubMed] [Google Scholar]
- 49. Neudauer C. L., Joberty G., Macara I. G. (2001) PIST. A novel PDZ/coiled-coil domain binding partner for the Rho-family GTPase TC10. Biochem. Biophys. Res. Commun. 280, 541–547 [DOI] [PubMed] [Google Scholar]
- 50. Chang L., Chiang S. H., Saltiel A. R. (2007) TC10α is required for insulin-stimulated glucose uptake in adipocytes. Endocrinology 148, 27–33 [DOI] [PubMed] [Google Scholar]
- 51. Cheng J., Wang H., Guggino W. B. (2004) Modulation of mature cystic fibrosis transmembrane regulator protein by the PDZ domain protein CAL. J. Biol. Chem. 279, 1892–1898 [DOI] [PubMed] [Google Scholar]
- 52. Cheng J., Wang H., Guggino W. B. (2005) Regulation of cystic fibrosis transmembrane regulator trafficking and protein expression by a Rho family small GTPase TC10. J. Biol. Chem. 280, 3731–3739 [DOI] [PubMed] [Google Scholar]
- 53. Cuadra A. E., Kuo S. H., Kawasaki Y., Bredt D. S., Chetkovich D. M. (2004) AMPA receptor synaptic targeting regulated by stargazin interactions with the Golgi-resident PDZ protein nPIST. J. Neurosci. 24, 7491–7502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hicks S. W., Machamer C. E. (2005) Isoform-specific interaction of Golgin-160 with the Golgi-associated protein PIST. J. Biol. Chem. 280, 28944–28951 [DOI] [PubMed] [Google Scholar]
- 55. Williams D., Hicks S. W., Machamer C. E., Pessin J. E. (2006) Golgin-160 is required for the Golgi membrane sorting of the insulin-responsive glucose transporter GLUT4 in adipocytes. Mol. Biol. Cell 17, 5346–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Calderhead D. M., Kitagawa K., Tanner L. I., Holman G. D., Lienhard G. E. (1990) Insulin regulation of the two glucose transporters in 3T3-L1 adipocytes. J. Biol. Chem. 265, 13801–13808 [PubMed] [Google Scholar]
- 57. Kupriyanova T. A., Kandror V., Kandror K. V. (2002) Isolation and characterization of the two major intracellular Glut4 storage compartments. J. Biol. Chem. 277, 9133–9138 [DOI] [PubMed] [Google Scholar]
- 58. Watson R. T., Khan A. H., Furukawa M., Hou J. C., Li L., Kanzaki M., Okada S., Kandror K. V., Pessin J. E. (2004) Entry of newly synthesized GLUT4 into the insulin-responsive storage compartment is GGA-dependent. EMBO J. 23, 2059–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu Z., Kandror K. V. (2002) Translocation of small preformed vesicles is responsible for the insulin activation of glucose transport in adipose cells. Evidence from the in vitro reconstitution assay. J. Biol. Chem. 277, 47972–47975 [DOI] [PubMed] [Google Scholar]
- 60. Semiz S., Park J. G., Nicoloro S. M., Furcinitti P., Zhang C., Chawla A., Leszyk J., Czech M. P. (2003) Conventional kinesin KIF5B mediates insulin-stimulated GLUT4 movements on microtubules. EMBO J. 22, 2387–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yip M. F., Ramm G., Larance M., Hoehn K. L., Wagner M. C., Guilhaus M., James D. E. (2008) CaMKII-mediated phosphorylation of the myosin motor Myo1c is required for insulin-stimulated GLUT4 translocation in adipocytes. Cell Metab. 8, 384–398 [DOI] [PubMed] [Google Scholar]
- 62. Bose A., Guilherme A., Robida S. I., Nicoloro S. M., Zhou Q. L., Jiang Z. Y., Pomerleau D. P., Czech M. P. (2002) Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature 420, 821–824 [DOI] [PubMed] [Google Scholar]
- 63. Lalioti V. S., Vergarajauregui S., Tsuchiya Y., Hernandez-Tiedra S., Sandoval I. V. (2009) Daxx functions as a scaffold of a protein assembly constituted by GLUT4, JNK1, and KIF5B. J. Cell. Physiol. 218, 416–426 [DOI] [PubMed] [Google Scholar]
- 64. Guo H. L., Zhang C., Liu Q., Li Q., Lian G., Wu D., Li X., Zhang W., Shen Y., Ye Z., Lin S. Y., Lin S. C. (2012) The Axin/TNKS complex interacts with KIF3A and is required for insulin-stimulated GLUT4 translocation. Cell Res. 10.1038/cr.2012.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lalioti V. S., Vergarajauregui S., Pulido D., Sandoval I. V. (2002) The insulin-sensitive glucose transporter, GLUT4, interacts physically with Daxx. Two proteins with capacity to bind Ubc9 and conjugated to SUMO1. J. Biol. Chem. 277, 19783–19791 [DOI] [PubMed] [Google Scholar]
- 66. Hosaka T., Brooks C. C., Presman E., Kim S. K., Zhang Z., Breen M., Gross D. N., Sztul E., Pilch P. F. (2005) p115 interacts with the GLUT4 vesicle protein, IRAP, and plays a critical role in insulin-stimulated GLUT4 translocation. Mol. Biol. Cell 16, 2882–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kampmann U., Christensen B., Nielsen T. S., Pedersen S. B., Ørskov L., Lund S., Møller N., Jessen N. (2011) GLUT4 and UBC9 protein expression is reduced in muscle from type 2 diabetic patients with severe insulin resistance. PLoS One 6, e27854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Orme C. M., Bogan J. S. (2012) The ubiquitin regulatory X (UBX) domain-containing protein TUG regulates the p97 ATPase and resides at the endoplasmic reticulum-Golgi intermediate compartment. J. Biol. Chem. 287, 6679–6692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pleasure I. T., Black M. M., Keen J. H. (1993) Valosin-containing protein, VCP, is a ubiquitous clathrin-binding protein. Nature 365, 459–462 [DOI] [PubMed] [Google Scholar]
- 70. Ramanathan H. N., Ye Y. (2012) The p97 ATPase associates with EEA1 to regulate the size of early endosomes. Cell Res. 22, 346–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kondo H., Rabouille C., Newman R., Levine T. P., Pappin D., Freemont P., Warren G. (1997) p47 is a cofactor for p97-mediated membrane fusion. Nature 388, 75–78 [DOI] [PubMed] [Google Scholar]
- 72. Uchiyama K., Totsukawa G., Puhka M., Kaneko Y., Jokitalo E., Dreveny I., Beuron F., Zhang X., Freemont P., Kondo H. (2006) p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Dev. Cell 11, 803–816 [DOI] [PubMed] [Google Scholar]
- 73. Alberts S. M., Sonntag C., Schäfer A., Wolf D. H. (2009) Ubx4 modulates cdc48 activity and influences degradation of misfolded proteins of the endoplasmic reticulum. J. Biol. Chem. 284, 16082–16089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48 (UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107, 667–677 [DOI] [PubMed] [Google Scholar]
- 75. Hitchcock A. L., Krebber H., Frietze S., Lin A., Latterich M., Silver P. A. (2001) The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell 12, 3226–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rape M., Jentsch S. (2004) Productive RUPture. Activation of transcription factors by proteasomal processing. Biochim. Biophys. Acta 1695, 209–213 [DOI] [PubMed] [Google Scholar]
- 77. Baek G. H., Kim I., Rao H. (2011) The Cdc48 ATPase modulates the interaction between two proteolytic factors Ufd2 and Rad23. Proc. Natl. Acad. Sci. U.S.A. 108, 13558–13563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Burg J. S., Espenshade P. J. (2011) Regulation of HMG-CoA reductase in mammals and yeast. Prog. Lipid Res. 50, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Verma R., Oania R., Fang R., Smith G. T., Deshaies R. J. (2011) Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol. Cell 41, 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nelson D. S., Alvarez C., Gao Y. S., García-Mata R., Fialkowski E., Sztul E. (1998) The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J. Cell Biol. 143, 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Brandon E., Gao Y., Garcia-Mata R., Alvarez C., Sztul E. (2003) Membrane targeting of p115 phosphorylation mutants and their effects on Golgi integrity and secretory traffic. Eur. J. Cell Biol. 82, 411–420 [DOI] [PubMed] [Google Scholar]
- 82. Kim D. W. (2003) Characterization of Grp1p, a novel cis-Golgi matrix protein. Biochem. Biophys. Res. Commun. 303, 370–378 [DOI] [PubMed] [Google Scholar]
- 83. Hicks S. W., Horn T. A., McCaffery J. M., Zuckerman D. M., Machamer C. E. (2006) Golgin-160 promotes cell surface expression of the β-1 adrenergic receptor. Traffic 7, 1666–1677 [DOI] [PubMed] [Google Scholar]
- 84. Grieve A. G., Rabouille C. (2011) Golgi bypass. Skirting around the heart of classical secretion. Cold Spring Harb. Perspect. Biol. 3, a005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chang L., Adams R. D., Saltiel A. R. (2002) The TC10-interacting protein CIP4/2 is required for insulin-stimulated Glut4 translocation in 3T3L1 adipocytes. Proc. Natl. Acad. Sci. U.S.A. 99, 12835–12840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lodhi I. J., Chiang S. H., Chang L., Vollenweider D., Watson R. T., Inoue M., Pessin J. E., Saltiel A. R. (2007) Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab. 5, 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Inoue M., Chiang S. H., Chang L., Chen X. W., Saltiel A. R. (2006) Compartmentalization of the exocyst complex in lipid rafts controls Glut4 vesicle tethering. Mol. Biol. Cell 17, 2303–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Marie M., Dale H. A., Sannerud R., Saraste J. (2009) The function of the intermediate compartment in pre-Golgi trafficking involves its stable connection with the centrosome. Mol. Biol. Cell 20, 4458–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gee H. Y., Noh S. H., Tang B. L., Kim K. H., Lee M. G. (2011) Rescue of ΔF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell 146, 746–760 [DOI] [PubMed] [Google Scholar]
- 90. Munro S. (2011) The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb. Perspect. Biol. 3, a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Castorena C. M., Mackrell J. G., Bogan J. S., Kanzaki M., Cartee G. D. (2011) Clustering of GLUT4, TUG, and RUVBL2 protein levels correlates with myosin heavy chain isoform pattern in skeletal muscles, but AS160 and TBC1D1 levels do not. J. Appl. Physiol. 111, 1106–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yellaturu C. R., Deng X., Cagen L. M., Wilcox H. G., Mansbach C. M., 2nd, Siddiqi S. A., Park E. A., Raghow R., Elam M. B. (2009) Insulin enhances post-translational processing of nascent SREBP-1c by promoting its phosphorylation and association with COPII vesicles. J. Biol. Chem. 284, 7518–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Osborne T. F., Espenshade P. J. (2009) Evolutionary conservation and adaptation in the mechanism that regulates SREBP action. What a long, strange tRIP it's been. Genes Dev. 23, 2578–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chieregatti E., Meldolesi J. (2005) Regulated exocytosis. New organelles for non-secretory purposes. Nat. Rev. Mol. Cell Biol. 6, 181–187 [DOI] [PubMed] [Google Scholar]
- 95. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains. Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]