Background: Lipids play an important role in membrane protein function (lipid allostery).
Results: The co-crystal structure of lipid-free Pla protease and a substrate peptide shows that the substrate displaces the active site nucleophilic water molecule.
Conclusion: Removal of LPS from Pla causes subtle conformational changes that inactivate the enzyme.
Significance: The structure provides the first explanation for lipid allostery of an outer membrane protein.
Keywords: Bacterial Pathogenesis, Lipopolysaccharide (LPS), Membrane, Plasminogen, Protease, Lipid Allostery, Outer Membrane Protease, Plasminogen Activator, Yersinia pestis Pla
Abstract
Omptins constitute a unique family of outer membrane proteases that are widespread in Enterobacteriaceae. The plasminogen activator (Pla) of Yersinia pestis is an omptin family member that is very important for development of both bubonic and pneumonic plague. The physiological function of Pla is to cleave (activate) human plasminogen to form the plasma protease plasmin. Uniquely, lipopolysaccharide (LPS) is essential for the catalytic activity of all omptins, including Pla. Why omptins require LPS for enzymatic activity is unknown. Here, we report the co-crystal structure of LPS-free Pla in complex with the activation loop peptide of human plasminogen, its natural substrate. The structure shows that in the absence of LPS, the peptide substrate binds deep within the active site groove and displaces the nucleophilic water molecule, providing an explanation for the dependence of omptins on LPS for enzymatic activity.
Introduction
It is becoming increasingly clear that membrane lipids do not just provide a suitable but inert environment for membrane proteins, but that they can also directly modulate membrane protein function, a phenomenon dubbed “lipid allostery” (1). The biological function of membrane proteins can be affected by membrane lipid composition as well as by specific protein-lipid interactions. Examples of the latter class are the interaction of cholesterol with G protein-coupled receptors and the activation of the inward rectified K+ channel Kir2.2 by phosphatidylinositol 4,5-bisphosphate (2, 3). To date, however, no explanation for the modulation of bacterial outer membrane (OM)2 protein function by membrane lipids has been reported.
Yersinia pestis, a highly pathogenic bacterium, is the causative agent of plague in humans and has been classified by the Centers for Disease Control and Prevention as a Category A microbe. An important virulence factor of Y. pestis is the 292-amino acid long OM protease Pla (plasminogen activator), the physiological function of which is to activate (cleave) human plasminogen to generate the circulating serine protease plasmin. Plasmin degrades fibrin clots (fibrinolysis), noncollagenous proteins of base membranes and the extracellular matrix, to facilitate cell migration (4). Pla has been shown to be essential for development of bubonic plague by facilitating rapid dissemination of bacteria from the initial site of infection (5). Pla is also important for the progression of pneumonic plague, because it allows Y. pestis to replicate rapidly in lungs (6).
Pla belongs to a unique family of OM proteases known as omptins, which are widely distributed in Gram-negative bacterial pathogens that infect humans and plants (7–9). Omptins share high sequence identities (40–74%). The solved structures of two members of the family, Escherichia coli OmpT (10) and Y. pestis Pla (11), show that they have very similar structures with a barrel composed of 10 antiparallel transmembrane β-strands connected by five long extracellular loops. Despite their high identity and structural similarity, omptins differ greatly in their target specificity for proteolysis, invasive/adhesive properties, and virulence association, probably due to adaptations to the life style of the host bacterium (7–9).
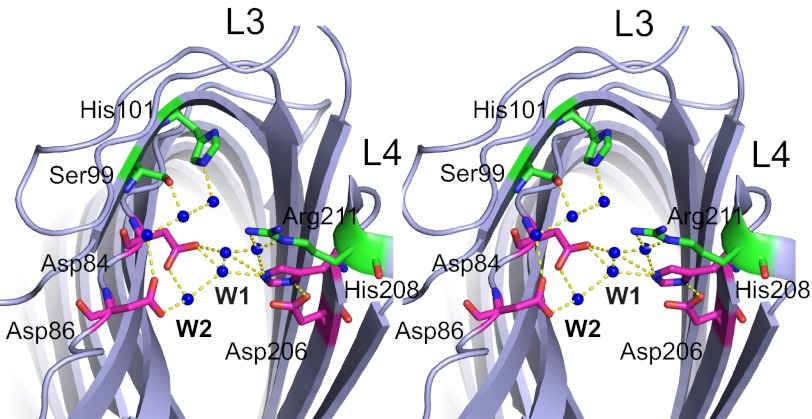
Omptins appear to constitute a unique class of proteases because their conserved active site residues (Asp84, Asp86, Asp206, and His208 in Pla, Fig. 1) differ from the active site consensus sequences found in serine, aspartic or metalloproteases. Crystal structures, mutagenesis studies, and molecular dynamics simulations for OmpT suggest a unique catalytic mechanism for proteases (10, 12, 13). According to this proposed mechanism, the catalytic Asp206-His208 dyad activates a nucleophilic water molecule that attacks the scissile carbonyl carbon atom. The Asp84-Asp86 dyad likely coordinates the nucleophilic water molecule either directly or indirectly via another water molecule (Fig. 1). In addition to the active site residues, both Ser99 and His101 located close to the active site have been shown to be important for omptin activity (14) because they coordinate the Asp84-Asp86 dyad via bridging water molecules (12) (Fig. 1).
FIGURE 1.
Stereoview of the Pla active site viewed from the extracellular side (PDB ID 2X55). The active site residues (Asn84, Asp86, Asp206, and His208) are shown in magenta; the other residues that are important for activity are shown in green (Ser99, His101, and Arg211). Active site water molecules are shown as blue spheres. The nucleophilic water is labeled W1, and the water molecule between the Asp84-Asp86 dyad is labeled W2. Hydrogen bonding of water molecules and amino acids are shown as yellow dashes.
Another unique feature of omptins is that catalytic activity requires the presence of bound lipopolysaccharide (LPS), a complex glycolipid that makes up the outer leaflet of the OM. LPS binds at a defined position on the outside of the barrel (14–16). Although crystal structures of apo-Pla in the absence and presence of bound LPS showed a subtle narrowing of the active site groove in the presence of LPS (13), it is unclear how this conformational change could activate the enzyme.
EXPERIMENTAL PROCEDURES
Purification of Pla for Crystallization
Cloning, expression, and site-directed mutagenesis of Pla (D84N) were carried out as described previously (11). BL21(DE3) T1 phage-resistant cells (New England Biolabs) were transformed with the pB22-Pla D84N construct. The cells were grown to A600 ∼ 0.6 at 37 °C and then induced with 0.1% arabinose at 20 °C overnight. Cells were harvested by centrifugation at 4500 rpm for 30 min (J6-MC; Beckman Coulter). Cell pellets were suspended in TSB (20 mm Tris, 300 mm NaCl, 10% glycerol, pH 8.0), and cells were lysed by sonication (3 × 40-s intervals) (Branson Digital Sonifier). Total membranes were obtained by centrifugation at 40,000 rpm for 40 min (Ti-45 rotor, Beckman L8-70M ultracentrifuge). Membranes were homogenized in TSB and solubilized in 1% n-dodecyl-N,N-dimethylamine-N-oxide (LDAO; Anatrace) for 1 h at 4 °C followed by centrifugation at 40,000 rpm for 30 min to remove precipitates and unsolubilized membranes. The membrane extract was applied to a 10-ml nickel column. The column was washed with 10 column volumes of TSB containing 0.2% LDAO and 15 mm imidazole. The proteins were eluted with 3 column volumes of TSB containing 0.2% LDAO and 200 mm imidazole. The proteins were further purified by gel filtration chromatography using 10 mm Tris, 50 mm NaCl, and 0.05% LDAO, pH 8.0. For final polishing and detergent exchange, another gel filtration chromatography column was performed in 10 mm Tris, 100 mm LiCl, 0.35% C8E4, 0.05% decyl-β-maltoside, pH 8.0. The purified protein was concentrated to 10–15 mg/ml using 50-kDa molecular mass cutoff filters (Amicon), dialyzed overnight against gel filtration buffer, and flash-frozen in liquid nitrogen. For the preparation of active Pla used for cleavage of the activation loop peptide, protein was extracted in 1.5% β-octylglucoside to prevent loss of bound LPS (11). The nickel column and the first gel filtration column were also done in β-octylglucoside (0.8%).
Crystallization of Pla-Alp11 and Structure Determination
An 11-residue activation loop peptide of human plasminogen (Alp11) with the sequence 557KCPGRVVGGCK567 was synthesized (Biomatik, Wilmington, DE) with the substitution V567K (numbering for intact human plasminogen) to increase solubility. The peptide was dissolved in protein buffer and added to Pla D84N at a final concentration of ∼10 mm. Hanging-drop crystallization trials were set up using several commercial screens (MemGold and Structure I/II). Several initial hits were obtained, and one of those (Structure II/#8) was optimized to give thin plates that grew to sufficient size after several weeks of incubation. The crystals were obtained by adding 1 μl of protein-peptide solution to 1 μl of mother liquor containing 45% (v/v) 2-methyl-2,4-pentanediol, 0.2 m NH4KH2PO4, and 0.1 m NaOAc, pH 5.5. The crystals of the complex belonged to space group I222 and contained one molecule/asymmetric unit. Crystals were flash-frozen in liquid nitrogen directly from the mother liquor.
Diffraction data were collected at 100 K at the National Synchrotron Light Source (Brookhaven National Laboratory) at beamline X6A and processed with HKL2000 (17). The crystals had to be annealed by blocking the cryostream once for ∼ 2 s to improve diffraction and decrease the mosaicity dramatically. The structure of Pla-Alp11 was solved at 2.0 Å resolution by molecular replacement in Phaser (18) using the high resolution apo-Pla structure (Protein Data Bank (PDB) ID 2X55) as the search model. Model (re)building was performed manually within COOT (19), and the structure was refined using Phenix (20). Structure validation was performed within Phenix. 94.3% of the residues are present in favored regions, 5.3% in allowed regions, and 0.4% in disallowed regions of the Ramachandran plot. Final refinement statistics and analysis are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parameters | Pla-Alp11 |
|---|---|
| Data collection | |
| Space group | I222 |
| Cell dimensions | |
| a, b, c (Å) | 52.4, 63.1, 201.2 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50-2.03 |
| Rsym or Rmerge | 10.1 (42.7)a |
| I/σI | 23.4 (5.2) |
| Completeness (%) | 99.8 (99.7) |
| Redundancy | 5.5 (5.1) |
| Refinement | |
| Resolution (Å) | 19.8-2.03 |
| No. reflections | 20,964 (1,917) |
| Rwork/Rfree | 18.04/23.19 |
| No. atoms | |
| Protein | 2,203 |
| Peptide | 64 |
| Detergent/ion | 21/5 |
| Water | 106 |
| B-factors | |
| Protein | 33.1 |
| Peptide | 57.4 |
| Detergent/ion | 46.8 |
| Water | 38.5 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.156 |
a Values in parentheses are for the highest resolution shell.
Cleavage of Alp11 by Pla D84N
Pla-mediated proteolysis of Alp11 was carried out overnight by incubating Pla D84N purified in β-octylglucoside and Alp11 in the crystallization buffer (45% 2-methyl-2,4-pentanediol, 0.2 m NH4KH2PO4, and 0.1 m NaOAc, pH 5.5) at 25 °C. The reaction products were analyzed using electrospray ionization MS (Proteomics and Mass Spectrometry Facility, University of Massachusetts Medical School, Shrewsbury, MA) under nonreducing and reducing (10 mm DTT, 30 min, 60 °C) conditions. The sequences of the resulting peptides were confirmed by tandem mass spectrometry.
RESULTS
Structure of the Pla-Alp11 Complex
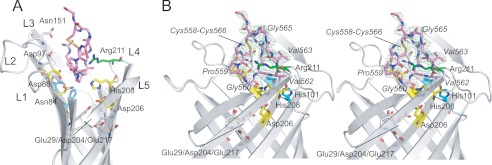
Wild-type Pla crystallized only in its LPS-bound active form, even when the protein was extracted and purified in LDAO (11). Moreover, soaking and co-crystallization attempts of peptides with wild-type Pla proved unsuccessful, presumably due to cleavage of the peptide substrate. We therefore employed the low activity (∼15% relative to WT) Pla mutant D84N to obtain a 2.0 Å co-crystal structure of Pla in complex with the Alp11 of human plasminogen (Table 1). As is the case for the apo-protein, Pla within the complex forms an ∼70 Å long monomeric vase-like barrel that consists of 10 antiparallel β-strands (S1–S10) connected by 5 long extracellular loops (L1–L5). The active site is located in a deep groove formed by the extracellular loops (Fig. 2A) and consists of two residue couples: Asn84/Asp86 in S3 on one side of the barrel and Asp206/His208 in S7 on the other side. The Alp11 peptide is bound within the active site groove, between both active site couples (Fig. 2A). In the following discussion, we will use P1 and P1′ for the peptide residues immediately prior to and following the scissile bond, i.e. Arg561 is the P1 residue and Val562 is the P1′ residue (21). The Alp11 peptide is present in the native, oxidized form, with clear density visible for the Cys558-Cys566 disulfide bond (Fig. 2B) and for most of the peptide. The termini of the peptide are disordered, with Lys557 and Lys567 modeled as glycine residues due to the absence of side chain density. The density for the scissile peptide bond is clearly continuous, indicating that the peptide substrate has not been cleaved. Given the fact that the D84N Pla mutant has (low) catalytic activity, this result can be explained by the absence of bound LPS in the protein preparation, rendering the enzyme inactive (10). The lack of LPS is due to the fact that the protein was initially purified in LDAO (see “Experimental Procedures”), which removes LPS from its binding site on the outside of the barrel (11). Indeed, the crystal packing of the complex is such that the residues on the outside of the barrel that are normally involved in LPS binding now mediate crystal contacts, making LPS binding physically impossible within the Pla-Alp11 crystal.
FIGURE 2.
Overview of the Pla-Alp11 structure. A, side view of the extracellular part of the complex shown schematically. Extracellular loops are labeled (L1–L5). Alp11 residues (Lys557-Lys567) are shown as pink stick models. The Cys558-Cys566 disulfide bond is colored yellow. The active site residues (Asn84, Asp86, Asp206, and His208) are shown in yellow; the other residues that are important for activity are shown in cyan (Ser99 and His101) and green (Arg211). Residues in the two acidic pockets are shown in gray (Asp97, Asn151, Glu29, Asp204, and Glu217). B, stereoview of the Pla-Alp11 active site from the side, 90° rotated relative to A. Residues of the peptide are labeled in italics. 2 Fo − Fc map density is shown as a green mesh, contoured at 0.7 σ.
LPS-mediated Conformational Changes
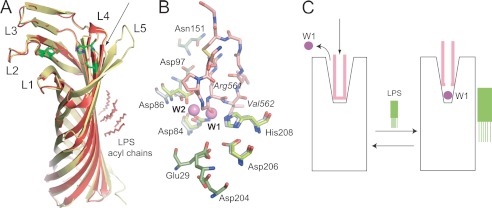
What is the effect of LPS removal on the Pla structure? The previously determined structures of apo-Pla in the presence and absence of LPS are very similar, especially in the membrane-embedded regions, with an overall root mean square deviation of 0.42 Å for the C-α atoms (Fig. 3A). The biggest structural differences between LPS-free and LPS-containing Pla are found in loops L4 and L5, both of which are present on the side of the barrel where LPS binds (Fig. 3). Most of loop L5 is not visible in the Pla-LPS structure, most likely due to autocatalytic activity at residue Lys262 prior to extraction with LDAO (14). Potentially more relevant is the conformation of loop L4 containing the catalytic residues Asp206 and His208, as well as the important residue Arg211 (13). In the LPS-free structure, the tip of this loop is shifted (∼1 Å) outward relative to the structure that contains LPS (Fig. 3A). Although this conformational shift would potentially widen the active site groove, the positions of the catalytic residues at the bottom of the groove are essentially unchanged in both structures (differences <0.5 Å). Thus, the structures of apo-Pla in the absence or presence of LPS do not give unambiguous clues as to why LPS removal inactivates the enzyme.
FIGURE 3.
Structure-based model for Pla activation by LPS. A, structural comparison of LPS-free apo-Pla (yellow; PDB ID 2X4M) and LPS-bound apo-Pla (red; PDB ID 2X55). The active site residues Asp84, Asp86, Asp206, and His208 are shown as green stick models for both structures. The conformational changes in the tip of loop L4 that arise from LPS binding are indicated with an arrow. B, close-up of the active site groove of LPS-bound apo-Pla and of the Pla-Alp11 complex. The peptide is colored salmon, with oxygens red and nitrogens blue. The active site residues are shown as sticks in gray (apo-Pla) and in green (Pla-Alp11). The nucleophilic water molecule (W1) and the water molecule W2 between Asp84 and Asp86 that are present in apo-Pla but absent in Pla-Alp11 are shown as purple spheres. Residues within the two acidic binding pockets are shown in green for Pla-Alp11 only. C, schematic model for Pla activation by LPS. In the absence of LPS (left), the activation loop of the substrate (pink) binds deep into the active site groove, displacing the nucleophilic water molecule and rendering the enzyme inactive. Upon binding of LPS, the active site groove narrows (right). When substrate binds, the catalytic water molecule remains in place to cleave the substrate.
Pla contains an active site water network, linking the catalytic residues and other residues important for enzymatic activity (Fig. 1). One of those water molecules is the putative nucleophilic water, located between Asp84 and His208 (11). Strikingly, in the Pla-Alp11 structure the peptide has displaced most of the active site waters, including the nucleophilic one (Fig. 3B). In fact, the position of the nucleophilic water molecule in the apo-structure is very close to the carbonyl oxygen atom of the P2 residue Gly560 within Pla-Alp11 (Fig. 3B). Within the complex, no potential other nucleophile is located close enough to the scissile Arg561-Val562 peptide bond to mediate catalysis. We propose that in the absence of LPS, Pla undergoes subtle conformational changes that widen the active site groove. As a result, peptide substrates can diffuse further down into the active site, replacing essential active site water molecules and rendering the enzyme inactive (Fig. 3C).
In the Pla-Alp11 complex, the Gly560-Arg561 (P2-P1) peptide bond is located between the catalytic residues Asn84 and His208, closest to the position where the nucleophilic water would be in the active enzyme. This contrasts with molecular dynamics simulations of E. coli OmpT with a short substrate peptide, which placed the scissile (P1-P1′) Arg-Arg peptide bond between the catalytic residues Asp83 and His208 (OmpT numbering) (12). Extrapolating to Pla, this would mean that the Arg561-Val562 peptide bond should be located between Asn84 and His208. Moreover, the negatively charged pocket formed by Glu29/Asp204/Glu217 and which was proposed to bind the P1 arginine guanidinium group (10, 13), is empty in the structure of the complex (Fig. 3B). Instead, the P1 arginine side chain is located at the position of the P1′ side chain in the molecular dynamics simulations, close to Asp97 and Asn151. In other words, there is a register shift of one residue between the predictions of the molecular dynamics simulations and our structural data. It is not clear whether the register shift is a consequence of the lack of LPS or whether this is the actual substrate binding mode within the active enzyme. In this context it should be noted that Pla is a very poor enzyme for plasminogen activation, with a kcat of 0.21 min−1 (Km ∼120 nm) (22), suggesting that the active site may have an unusual geometry. Structural studies of active enzyme-substrate complexes as well as biochemical studies based on the current structure will be required to shed light on this issue.
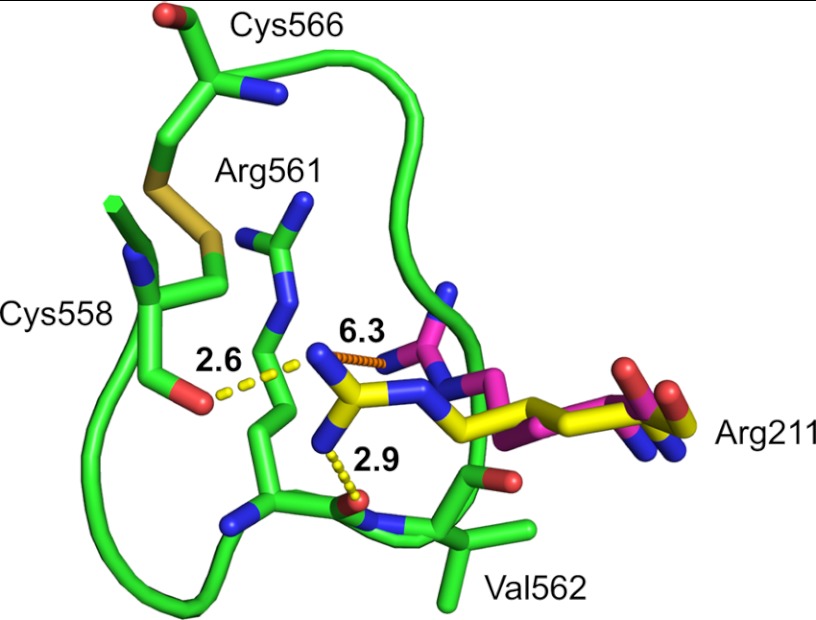
Possible Implications for the Role of Arg211
In the Pla-Alp11 structure the guanidinium group of Arg211 moves by 5–6 Å to interact with the peptide substrate (Fig. 4). The arginine side chain forms strong hydrogen bonds with the carbonyl oxygen of Cys558. In addition, a strong hydrogen bond (2.9 Å) is formed with the oxygen atom of the scissile carbonyl group (Fig. 4). Although care must be taken in the interpretation of these structural details because Pla is inactive, the observed interactions might explain the importance of Arg211 for catalytic activity. Mutations of this residue to alanine or lysine drastically lower Pla-mediated cleavage of plasminogen (∼1% compared with WT) (11, 13). The observed conformational change of Arg211 in the Pla-Alp11 structure may support our previous hypothesis that the positive charge on the arginine side chain could play a role in the stabilization of the oxyanion intermediate during catalysis, in a similar manner to the calcium ion in phospholipase A2 (23).
FIGURE 4.
Substrate-induced conformational change of Arg211. Positions of Arg211 in apo-Pla (magenta; PDB ID 2X55) and in Pla-Alp11 (yellow) are shown in aligned structures. The distance between the arginine side chains in both structures is shown as an orange dashed line. Hydrogen bonding between Pla-Alp11 Arg211 and the scissile carbonyl as well as the Cys558 carbonyl are shown as yellow dashes. For simplicity only Arg561, Val562, Cys558, and Cys566 in Alp11 are shown as green stick models. Numbers in bold show distances in Å.
Alp11 Proteolysis by Active Pla D84N
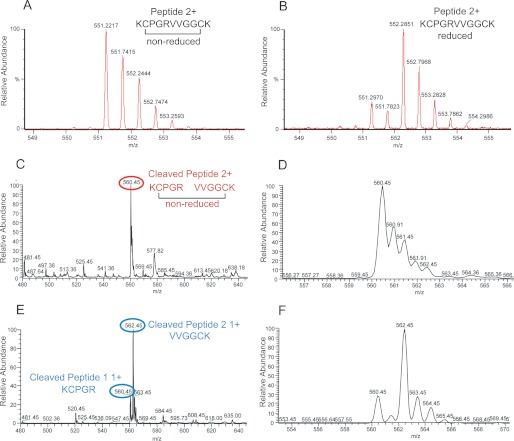
To rule out the possibility that the observed position of the Alp11 peptide within the complex is an artifact due to the small size of the peptide relative to full-length plasminogen (∼90 kDa), we purified the Pla D84N mutant in β-octylglucoside, ensuring that LPS remains bound on the outside of the barrel and preserving enzymatic activity (11). After incubating the protease with the Alp11 peptide in crystallization buffer (see “Experimental Procedures”), the reaction products were analyzed by mass spectrometry (Fig. 5). The results show that (i) the Alp11 peptide is present in the oxidized form, (ii) Pla D84N is active and cleaves Alp11 in the crystallization buffer, and (iii) the Arg561-Val562 peptide bond is cleaved, identical to the cleavage site in full-length human plasminogen (22). We conclude that the Alp11 peptide is a genuine substrate of Pla that is cleaved in the same way as its physiological macromolecular substrate, provided that LPS is bound to the protein.
FIGURE 5.
Analysis of Pla-mediated proteolysis of Alp11. A, mass spectrum obtained by infusion of 100 μg of intact Alp11 (expected M2H+ 551.29). B, mass spectrum obtained by infusion of 100 μg of reduced Alp11 (expected M2H+ 552.29). C, mass spectrum obtained by infusion of 100 ng/μl nonreduced Pla-treated Alp11. Cleaved, disulfide-linked peptide peak (+2) at 560.45 m/z is labeled with a red circle. (expected M2H+ 560.29). D, close-up view of the +2 peak in C. E, mass spectrum obtained by infusion of 100 ng/μl reduced Pla-treated Alp11. Cleaved peptide 1 (KCPGR; +1) and peptide 2 (VVGGCK; +1) peaks at 560.45 and 562.45 m/z are labeled with blue circles (peptide 1 (KCPGR) MH+ 559.29; peptide 2 (VVGGCK) MH+ 561.29). F, close-up view of the peaks in E. The sequences of the resulting peptides were confirmed by tandem mass spectrometry. The values in parentheses for A–F are the expected (calculated) m/z values for the peptides.
DISCUSSION
The structural data presented here provide an explanation for the LPS dependence of omptins for enzymatic activity. An interesting question is why omptins require LPS to be active. By virtue of the fact that the outer leaflet of the OM consists almost exclusively of LPS, omptin activity will always be turned on. Together with the fact that a number of different forms of LPS can activate omptins (15), it is therefore unlikely that LPS serves as a regulator of Pla activity within the OM. An alternative, intriguing possibility is that the requirement for LPS is a consequence of a need to keep the protease inactive within the periplasmic space during OM targeting and integration. This hypothesis would extend the known roles of lipid allostery in membrane protein function and would imply that native-like structure in OM proteins could be formed prior to their insertion into the OM.
Acknowledgments
We thank the staff of NSLS beamline X6A for beam time and for assistance during data collection and Stephanie Maniatis for mass spectrometry analysis.
The atomic coordinates and structure factors (code 4DCB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- OM
- outer membrane
- Alp11
- 11-residue activation loop peptide of human plasminogen
- LDAO
- n-dodecyl-N,N-dimethylamine-N-oxide
- PDB
- Protein Data Bank
- Pla
- plasminogen activator.
REFERENCES
- 1. Coskun U., Simons K. (2011) Cell membranes: the lipid perspective. Structure 19, 1543–1548 [DOI] [PubMed] [Google Scholar]
- 2. Hansen S. B., Tao X., MacKinnon R. (2011) Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oates J., Watts A. (2011) Uncovering the intimate relationship between lipids, cholesterol, and GPCR activation. Curr. Opin. Struct. Biol. 21, 802–807 [DOI] [PubMed] [Google Scholar]
- 4. Plow E. F., Ploplis V. A., Busuttil S., Carmeliet P., Collen D. (1999) A role of plasminogen in atherosclerosis and restenosis models in mice. Thromb. Haemost. 82, 4–7 [PubMed] [Google Scholar]
- 5. Sebbane F., Jarrett C. O., Gardner D., Long D., Hinnebusch B. J. (2006) Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. U.S.A. 103, 5526–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lathem W. W., Price P. A., Miller V. L., Goldman W. E. (2007) A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science 315, 509–513 [DOI] [PubMed] [Google Scholar]
- 7. Haiko J., Suomalainen M., Ojala T., Lähteenmäki K., Korhonen T. K. (2009) Invited review. Breaking barriers: attack on innate immune defences by omptin surface proteases of enterobacterial pathogen. Innate Immun. 15, 67–80 [DOI] [PubMed] [Google Scholar]
- 8. Hritonenko V., Stathopoulos C. (2007) Omptin proteins: an expanding family of outer membrane proteases in Gram-negative Enterobacteriaceae. Mol. Membr. Biol. 24, 395–406 [DOI] [PubMed] [Google Scholar]
- 9. Kukkonen M., Korhonen T. K. (2004) The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int. J. Med. Microbiol. 294, 7–14 [DOI] [PubMed] [Google Scholar]
- 10. Vandeputte-Rutten L., Kramer R. A., Kroon J., Dekker N., Egmond M. R., Gros P. (2001) Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J. 20, 5033–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eren E., Murphy M., Goguen J., van den Berg B. (2010) An active site water network in the plasminogen activator Pla from Yersinia pestis. Structure 18, 809–818 [DOI] [PubMed] [Google Scholar]
- 12. Baaden M., Sansom M. S. (2004) OmpT: molecular dynamics simulations of an outer membrane enzyme. Biophys J. 87, 2942–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kramer R. A., Vandeputte-Rutten L., de Roon G. J., Gros P., Dekker N., Egmond M. R. (2001) Identification of essential acidic residues of outer membrane protease OmpT supports a novel active site. FEBS Lett. 505, 426–430 [DOI] [PubMed] [Google Scholar]
- 14. Kukkonen M., Lähteenmäki K., Suomalainen M., Kalkkinen N., Emödy L., Lång H., Korhonen T. K. (2001) Protein regions important for plasminogen activation and inactivation of α2-antiplasmin in the surface protease Pla of Yersinia pestis. Mol. Microbiol. 40, 1097–1111 [DOI] [PubMed] [Google Scholar]
- 15. Kramer R. A., Brandenburg K., Vandeputte-Rutten L., Werkhoven M., Gros P., Dekker N., Egmond M. R. (2002) Lipopolysaccharide regions involved in the activation of Escherichia coli outer membrane protease OmpT. Eur. J. Biochem. 269, 1746–1752 [DOI] [PubMed] [Google Scholar]
- 16. Kukkonen M., Suomalainen M., Kyllönen P., Lähteenmäki K., Lång H., Virkola R., Helander I. M., Holst O., Korhonen T. K. (2004) Lack of O-antigen is essential for plasminogen activation by Yersinia pestis and Salmonella enterica. Mol. Microbiol. 51, 215–225 [DOI] [PubMed] [Google Scholar]
- 17. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18. Storoni L. C., McCoy A. J., Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 19. Emsley P., Cowtan K. (2004) COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 21. Schechter I., Berger A. (1967) On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 22. Sodeinde O. A., Subrahmanyam Y. V., Stark K., Quan T., Bao Y., Goguen J. D. (1992) A surface protease and the invasive character of plague. Science 258, 1004–1007 [DOI] [PubMed] [Google Scholar]
- 23. Scott D. L., White S. P., Otwinowski Z., Yuan W., Gelb M. H., Sigler P. B. (1990) Interfacial catalysis: the mechanism of phospholipase A2. Science 250, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]