Background: ATE1 transfers Arg to protein N termini, generating the degron for the N-end rule pathway.
Results: ATE1-deficient cardiomyocytes are impaired in the PLC/PKC-MEK1-ERK axis of Gαq-mediated cardiac signaling.
Conclusion: The arginine branch of the N-end rule pathway controls G-protein signaling in cardiomyocytes in part through hypoxia-sensitive degradation of GAP proteins.
Significance: This study provides a cellular mechanism underlying cardiovascular defects observed in ATE1-deficient mice.
Keywords: Posttranslational Modification, Protease, Protein Degradation, Ubiquitin, Ubiquitylation, ATE1 R-transferase, N-recognin, Arginylation, Proteolysis
Abstract
The N-end rule pathway is a proteolytic system in which destabilizing N-terminal amino acids of short lived proteins are recognized by recognition components (N-recognins) as an essential element of degrons, called N-degrons. In eukaryotes, the major way to generate N-degrons is through arginylation by ATE1 arginyl-tRNA-protein transferases, which transfer Arg from aminoacyl-tRNA to N-terminal Asp and Glu (and Cys as well in mammals). We have shown previously that ATE1-deficient mice die during embryogenesis with defects in cardiac and vascular development. Here, we characterized the arginylation-dependent N-end rule pathway in cardiomyocytes. Our results suggest that the cardiac and vascular defects in ATE1-deficient embryos are independent from each other and cell-autonomous. ATE1-deficient myocardium and cardiomyocytes therein, but not non-cardiomyocytes, showed reduced DNA synthesis and mitotic activity ∼24 h before the onset of cardiac and vascular defects at embryonic day 12.5 associated with the impairment in the phospholipase C/PKC-MEK1-ERK axis of Gαq-mediated cardiac signaling pathways. Cardiac overexpression of Gαq rescued ATE1-deficient embryos from thin myocardium and ventricular septal defect but not from vascular defects, genetically dissecting vascular defects from cardiac defects. The misregulation in cardiovascular signaling can be attributed in part to the failure in hypoxia-sensitive degradation of RGS4, a GTPase-activating protein for Gαq. This study is the first to characterize the N-end rule pathway in cardiomyocytes and reveals the role of its arginylation branch in Gαq-mediated signaling of cardiomyocytes in part through N-degron-based, oxygen-sensitive proteolysis of G-protein regulators.
Introduction
The N-end rule pathway is a proteolytic system in which destabilizing N-terminal residues of short lived proteins function as an essential degradation determinant (1–4) (supplemental Fig. 1). Posttranslational conjugation of Arg to N-terminal Asp and Glu is a universal eukaryotic protein modification that generates the principal degron Arg (5–7). The arginylation branch of the N-end rule pathway is catalyzed by evolutionarily conserved arginyl-tRNA-protein transferase (ATE1 or R-transferase),4 which transfers Arg from Arg-tRNA to the acceptor residue Asp or Glu (6, 8). An acceptor substrate (Asp or Glu) of R-transferase can be exposed through a proteolytic cleavage of an otherwise stable polypeptide or deamidation of the pro-N-degron Asn or Gln by a specific N-terminal amidohydrolase (for reviews, see Refs. 3 and 9). In mammals, N-terminal Cys can also be converted to an arginylation-permissive pro-N-degron through a redox modification involving its oxidation into Cys sulfinic acid (CysO2(H)) or Cys sulfonic acid (CysO3(H)) (10–12) (supplemental Fig. 1). N-terminal Arg together with other primary destabilizing residues (Lys, His, Phe, Tyr, Trp, Leu, and Ile) is recognized by a family of N-recognins (UBR1, UBR2, UBR4, and UBR5 in mammals) that promote N-degron-based polyubiquitylation and subsequent proteolysis through the 26 S proteasome (13, 14) (supplemental Fig. 1). Mammalian N-recognins share the UBR box, a zinc finger domain that binds preferentially the N-terminal Arg with a dissociation constant of low μm (15). The physiological functions and mechanisms of the N-end rule pathway are reviewed in Ref. 3.
The mammalian ATE1 gene produces at least six R-transferase isoforms, including those containing either of two homologous exons, through alternative splicing of pre-mRNAs (6). Although posttranslational arginylation was reported half a century ago (5), its physiological function has remained unclear until the discovery that knock-out of ATE1 in mice resulted in embryonic death (7). ATE1-deficient embryos die at embryonic day 15.5 (E15.5)–E16.5 with defects in cardiac and vascular development. Phenotypes of ATE1−/− hearts include ventricular myocardial hypoplasia associated with disorganized ventricular trabeculation, ventricular septal defect (VSD), and an outflow tract defect called persistent truncus arteriosus (PTA; alternatively called common arterial trunk) in which the truncus arteriosus is not properly separated into the pulmonary artery and aorta. ATE1−/− embryos also exhibit frequent hemorrhages and defective remodeling and branching of small vessels. Although these results suggest that ATE1 is required for development of embryonic hearts and maturation and/or integrity of blood vessels, the cellular function of arginylation in the cardiovascular lineage remains unknown. In addition to cardiovascular development, genetic analyses in mice implicated ATE1 in spermatogenesis (16, 17), metabolic homeostasis (16), and migration of neural crest cells (18). In the plant Arabidopsis, two known R-transferases, AtATE1 and AtATE2, expressed from two separate genes are required for seed ripening and germination, shoot and leaf development, and leaf senescence (19, 20). The fly Drosophila Ate1 regulates apoptosis and viability (21). In contrast to multicellular eukaryotes, no obvious defects were observed in Saccharomyces cerevisiae cells lacking Ate1, the only R-transferase of the yeast N-end rule pathway (8). Substrates of arginylation include structurally related mammalian RGS proteins (RGS4, RGS5, and RGS16) (11, 12, 22), which act as GTPase-activating proteins for heterotrimeric G-protein α subunits of the i, q, and 12 classes. N-terminal arginylation also has been found in Drosophila inhibitor of apoptosis 1, which inhibits undesired apoptotic activities (23); the endoplasmic reticulum chaperone protein calreticulin, which assists folding of newly translocated proteins in the endoplasmic reticulum lumen (24, 25); and β-actin, one of the most abundant cellular proteins, which can be arginylated at the pro-N-degron Asp-2 or Asp-3 to control actin filament properties, actin polymerization, and lamella formation in motile cells (26). In addition, recent proteomics approaches identified a number of proteins that are isolated in an arginylated form (27, 28).
In this study, we studied the cellular function of the arginylation branch of the N-end rule pathway in embryonic hearts and primary cardiomyocytes. We show that cardiomyocytes of ATE1−/− embryonic hearts are impaired in proliferation associated with down-regulation of G-protein signaling, which can be attributed in part to the failure to mediate hypoxia-sensitive degradation of GTPase-activating proteins of G-protein signaling.
EXPERIMENTAL PROCEDURES
Experimental Animals
ATE1−/− mice were described in Ref. 7. ATE1 was inactivated by replacing exons 1 through 3 with the NLS-lacZ marker (β-galactosidase N-terminally fused with a nuclear localization signal) in CJ7 embryonic stem (ES) cells (7). ATE1−/− embryos were produced through heterozygous crosses in a 129SvImJ/C57BL/6 background. Genotyping was carried out by using polymerase chain reaction (PCR) with primers F1 (CCAGCTCATTCCTCCCACTCATGATC), R1 (GGTATTTGCTGCCGTCCTTTGGTGGTC), and R2 (CTGGAGACAAAGCCCCAGCCAGAC), which amplify 570- and 430-bp fragments for wild type and knock-out alleles, respectively. ATE1−/−;MHC-Gαq40 mice were generated by mating ATE1+/− mice with MHC-Gαq40 transgenic mice (29), which express 40 copies of Gαq transgene in the heart from α-myosin heavy chain (MHC) promoter. Animal studies were conducted according to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH Publication Number 85-23, revised in 1996) and the protocols (0812811-A1) approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh. Euthanization involved inhalant anesthetic (isoflurane) followed by intraperitoneal injection of a xylazine (10 mg/kg) and ketamine (100 mg/kg) mixture.
Primary Cardiomyocytes and Explanted Hearts
Primary cardiomyocytes from mouse embryonic hearts were isolated as described with some modifications (30). Briefly, dissected hearts at E13.5 were digested in Hanks' balanced salt solution containing 0.2% collagenase II, 0.005% trypsin, and 0.1% chicken serum for 15 min at 37 °C. The enzymes were inactivated with horse serum, and the cells were settled down by centrifugation and plated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Twenty-four hours after plating, the media were replaced by serum-free DMEM supplemented with 10 μg/ml insulin, 5.5 μg/ml transferrin, 5 μg/ml selenium, and 110 μg/ml pyruvate or with DMEM containing 10% horse serum and 5% FBS. Approximately 50% of the cells were determined to be cardiomyocytes by immunostaining with anti-sarcomeric α-actinin or anti-troponin I antibody (Santa Cruz Biotechnology). To culture embryonic hearts ex vivo, the hearts from E13.5 embryos containing outflow regions were incubated in DMEM containing 5% FBS, penicillin, and streptomycin, and the media were changed with serum-free DMEM containing supplements. The explanted hearts continued beating during incubation with 40 μm 5-bromo-2-deoxyuridine (BrdU) for 24 h in the presence or absence of agonists of G-protein-coupled receptor (GPCR). Proliferation of the hearts was examined by immunostaining BrdU on paraffin sections.
Histology and β-Galactosidase Staining
For histological analysis, embryos were fixed overnight at 4 °C in 4% paraformaldehyde (Fisher Scientific) in phosphate-buffered saline (PBS). Embryos were treated with 70% ethanol, dehydrated, embedded in paraffin wax, and sectioned transversely or sagittally with 7-μm thickness followed by staining with hematoxylin and eosin (H&E). To detect the activity of β-galactosidase on sections, embryos or tissues were fixed in 4% paraformaldehyde in PBS for 10 min, rinsed in PBS three times, and stained overnight at 37 °C in 4-chloro-5-bromo-3-indolyl β-galactoside (X-gal) solution (1.3 mg/ml potassium ferrocyanide, 1 mg/ml potassium ferricyanide, 0.3% Triton X-100, 1 mm MgCl2, 150 mm NaCl, and 1 mg/ml X-gal (Roche Applied Science) in PBS (pH 7.4)) followed by postfixation. To measure β-galactosidase activity in cultured primary cardiac cells, cells were fixed in 0.25% glutaraldehyde (Fisher Scientific) in PBS for 5 min and stained in X-gal solution for 1 h followed by immunostaining with mouse anti-sarcomeric α-actinin (Clone EA-53, Sigma) to identify cardiomyocytes.
Immunohistochemistry and Proliferation Assays
Antibodies against RGS4 and RGS16 were gifts from Susanne Mumby (University of Texas Southwestern Medical Center) and Ching Kang Chen (Caltech), respectively. Immunostaining of paraffin sections and whole-mount immunohistochemical staining of embryos were performed as described (30). For the in vivo proliferation assay, pregnant mice were intraperitoneally injected (150 mg/g) with BrdU in 250 μl of saline. 2 h postinjection, embryos were subjected to paraffin sectioning and immunostaining of BrdU (S phase marker) or phosphorylated histone H3 (M phase marker). To monitor BrdU incorporation in cultured primary cardiomyocytes, cells were incubated with 10 μm BrdU for 16 h, then fixed in 4% paraformaldehyde for 25 min at 4 °C and for 5 min at room temperature, permeabilized in 0.2% Triton X-100 for 10 min, denatured with 2 n HCl, and neutralized with 0.1 m sodium borate. Cells were coimmunostained with rat anti-BrdU antibody and rabbit anti-troponin I antibody followed by the incubation with secondary antibody (anti-rat IgG-Alexa Fluor 555 and anti-rabbit IgG-FITC, respectively) and counterstaining with 4′,6-diamidino-2-phenylindole (DAPI). For immunohistochemistry of phosphorylated histone H3 and atrial natriuretic protein in cardiomyocytes, the immunostaining was performed as was done for BrdU staining without acid treatment and neutralization. The proliferation of explanted hearts was examined by anti-BrdU immunohistochemistry on paraffin sections.
To determine the effect of extracellular ligands on the S phase index in cardiac cells, cultured cardiac cells or explanted hearts were treated every 24 h with 200 μm phenylephrine, 2 μm angiotensin II, 100 nm prostaglandin F2α, 50 ng/ml basic fibroblast growth factor, or 4 μm isoproterenol in serum-free culture media for 2 days followed by treatment with 10 μm BrdU for 16 h. To stain RGS4 on paraffin sections, samples were blocked in 10% heat-inactivated goat serum in PBS with 0.05% Tween 20 and incubated with rabbit anti-RGS4 antibody (1:50 dilution). Endogenous peroxide activity was quenched with 3% H2O2 in methanol for 30 min. Biotinylated goat anti-rabbit IgG and diaminobenzidine were used to develop the signal. Control sections and embryos were incubated with preimmune antisera instead of anti-RGS4 antibody. Whole-mount immunohistochemical staining of RGS4 on embryos was performed as described (31).
Analysis of GPCR Pathways
The enzymatic activities of signaling molecules in embryonic hearts were determined using commercial kinase assay kits (Upstate Biotechnology, Charlottesville, VA). The substrates were myelin basic protein (for extracellular signal-regulated kinases 1 and 2 (ERK1 and -2)), the peptide KKALRRQETVDAL (for Ca2+/calmodulin-dependent protein kinase II), kemptide (for protein kinase A (PKA)), and the peptide QKRPSQRSKYL (for protein kinase C (PKC)). The formation of inositol phosphate by phospholipase C (PLC) was measured using [3H]phosphatidylinositol bisphosphate as a substrate. To measure the kinetics of activation and inactivation of kinases in mitogen-activated protein kinase (MAPK) pathways, primary cardiomyocytes were starved in serum-free medium for 24 h, activated by DMEM containing 20% FBS, and subjected to immunoblotting of phosphorylated forms of kinases (11).
Pulse-Chase Analysis under Hypoxia
HEK293 cells were transiently transfected with a plasmid expressing RGS4. About 24 h after transfection, cells were grown under normoxia (20% oxygen (O2)) or hypoxia (0.1% O2) for 6 h followed by labeling with [35S]methionine/cysteine (35S Express, PerkinElmer Life Sciences). The pulse was followed by a chase for 0, 30, and 60 min in the presence of cycloheximide; preparation of extracts; immunoprecipitation; SDS-PAGE; autoradiography; and quantitation using a PhosphorImager as described previously (32). The O2 level was adjusted by mixing N2 in a hypoxic chamber (Forma Scientific).
Statistical Analysis
For experiments using embryonic hearts, at least three different litters (10 sections each) were analyzed. To determine the proliferation of cultured cardiomyocytes, more than 5,000 cells for each experimental group were counted. Samples were counted twice, and there was typically less than 10% variability per sample. To measure enzymatic activities, three hearts for each genotype were combined for a single assay, which was duplicated. The images were analyzed using ImageJ software (version 1.34s, National Institutes of Health) to count BrdU-positive cells and to measure the area of cardiomyocytes. Data are presented as mean ± S.D., and statistical analysis was performed by unpaired Student's t test or analysis of variance. A value of p <0.1 was accepted as statistically significant.
RESULTS
Cardiomyocytes of ATE1-deficient Embryos Are Impaired in Proliferation
To determine the cellular function of ATE1 in cardiac development, we observed the gross morphology of ∼1,000 embryos at E10.5–E17.5 from ATE1+/− parents in a B6/129S background (Table 1). ATE1−/− embryos normally grew until E11.5 but began to show defects in cardiac and vascular development at E12.5 apparently without other morphological defects outside the cardiovascular system. ATE1−/− embryos at E12.5 showed VSD and thin myocardium in both ventricles associated with defective circulation as evidenced by pale yolk sacs (supplemental Figs. 2 and 3). By E13.5, the mutant embryos showed additional cardiovascular phenotypes observed in the previous study (7), including atrial septal defect, poorly developed trabeculae, dilated atria, PTA, and various vascular defects (local hemorrhages, poorly branched and thinner small vessels, irregularly terminated large vessels, and pale yolk sacs) (supplemental Figs. 2 and 3). Notably, all of the ATE1−/− hearts with PTA (n = 10) also contained VSD, whereas only ∼50% of the hearts with VSD (n = 19) exhibited PTA, suggesting that VSD and PTA in ATE1−/− hearts may involve misregulation in two independent processes, for example myocardial proliferation and migration of neural crest cells, respectively. No live animals were retrieved beyond E15.5. Despite defective cardiogenesis, the expression of the following markers involved in cardiac development was comparable in E13.5 ATE1−/− hearts as determined by quantitative RT-PCR: GATA4, Nkx2.5, MEF2C, dHAND, eHAND, NPPA, β-myosin heavy chain, cardiac α-actin, skeletal α-actin, Srf, and Atp2a2 (data not shown). Thus, the cardiac defects are not mainly due to misregulation of developmental program at the transcriptional level.
TABLE 1.
Genotyping of embryos from intercrossing between ATE1+/− mice
| Age | +/+ | +/− | −/− |
|---|---|---|---|
| E10.5 | 12 | 21 | 14 |
| E11.5 | 31 | 63 | 25 |
| E12.5 | 29 | 52 | 24 (2a) |
| E13.5 | 71 | 133 | 77 (14a) |
| E14.5 | 50 | 78 | 45 (12a) |
| E15.5 | 17 | 32 | 9 (9a) |
| E16.5 | 10 | 18 | 5 (5a) |
| E17.5 | 4 | 46 | 1 (1a) |
| Postnatal | 223 | 404 | 0 |
a Found dead.
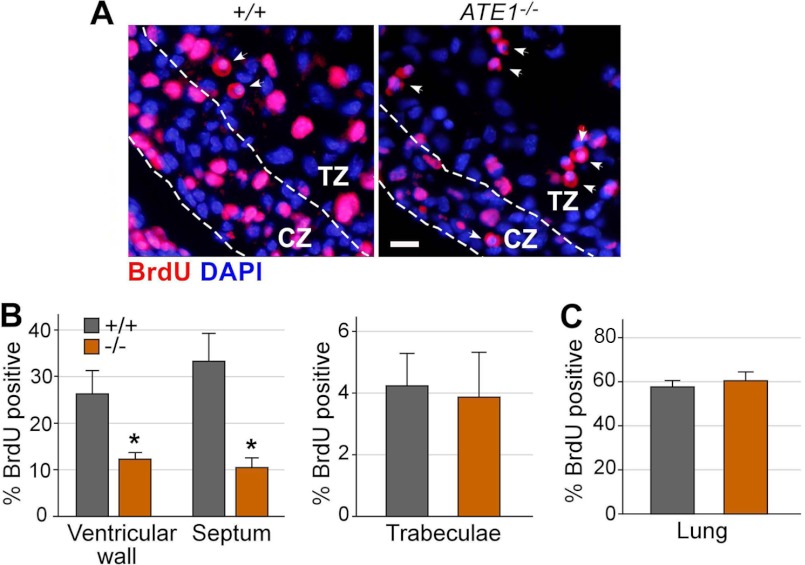
To determine the proliferation of ATE1−/− embryonic hearts, we intraperitoneally injected BrdU into pregnant females, harvested embryos at different stages, and performed immunofluorescence staining of BrdU on the cross-sections of embryos. The hearts of E12.5 ATE1−/− embryos exhibited reduced levels of S phase cells in ventricular walls (26% in +/+ versus 12% in −/−) and intraventricular septum (33% in +/+ versus 10.4% in −/−) (Fig. 1, A and B). The reduced DNA synthesis rate is unlikely due to nonspecific growth arrest as lungs of the same mutant embryos showed a normal S phase index (57% in +/+; 60.2% in −/−) (Fig. 1C). As an independent measurement, we performed an analogous assay with an antibody specific to phosphorylated histone H3, a hallmark of mitosis. The hearts of E11.5 ATE1−/− embryos exhibited reduced levels of M phase cells in ventricular walls (1.8% in +/+; 0.52% in −/−), ventricular septum (1.4% in +/+; 0.73% in −/−), and trabeculae (1.5% in +/+; 0.72% in −/−) (supplemental Fig. 4). The reduced proliferation is not because more cardiac cells are eliminated by apoptosis as TUNEL (terminal deoxynucleotidyltransferase dUTP nick end labeling) assays did not reveal a significant difference between +/+ and ATE1−/− hearts at E11.5 through E12.5 (data not shown).
FIGURE 1.
Myocardium of ATE1-deficient embryos is impaired in proliferation. A, BrdU incorporation assay of +/+ and ATE1−/− embryonic hearts at E12.5. Shown is immunohistochemistry of BrdU on cross-sections of embryonic hearts. CZ, compact zone; TZ, trabeculae zone. Arrowheads indicate red blood cells with cross-reactivity. Scale bar, 100 μm. B and C, quantitation of the BrdU incorporation assay shown in A. Data are presented as mean ± S.D.
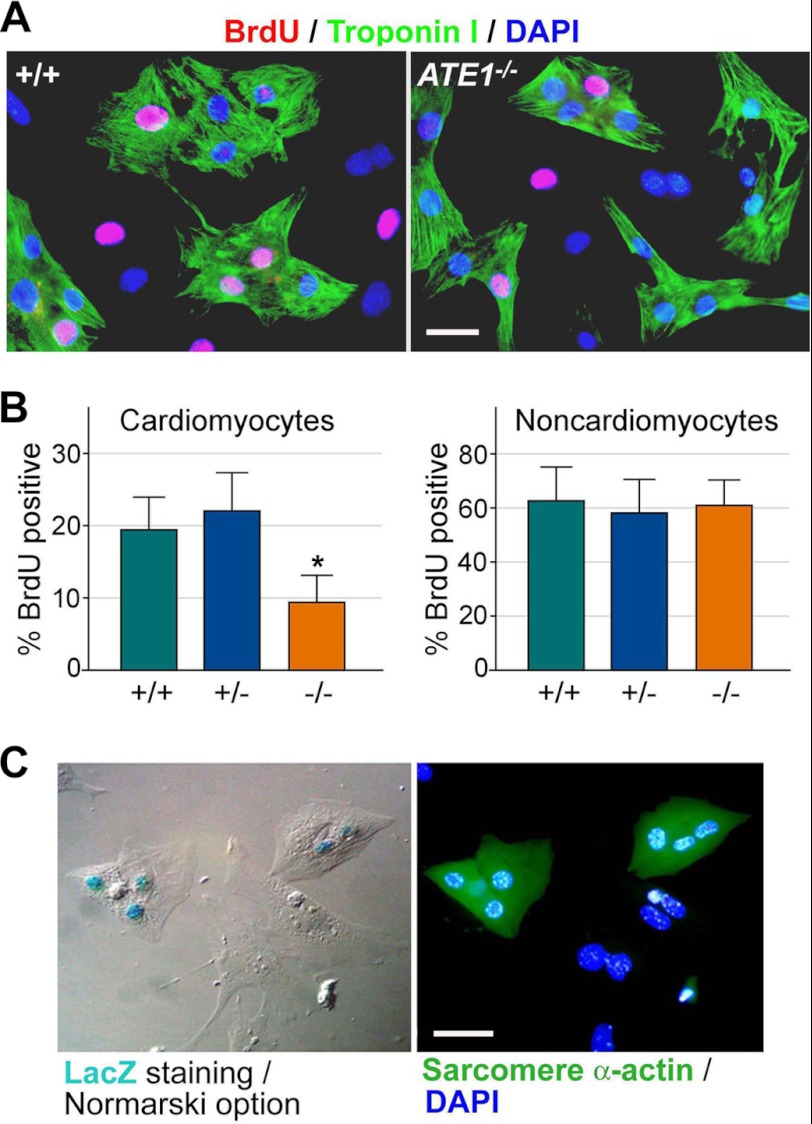
To determine whether arginylation is required for the proliferation of cultured cardiomyocytes, primary cardiac cells established from +/+ and ATE1−/− embryos at E13.5 were used for a BrdU incorporation assay. Coimmunostaining of BrdU and troponin I, a marker of cardiomyocytes, revealed a reduced proliferation in ATE1−/− cardiomyocytes (19.4% in +/+ versus 9.4% in −/−) (Fig. 2, A and B). In contrast, the difference was not obvious in troponin I-negative cardiac cells, which are mainly composed of cardiac fibroblasts with a minor contribution by endothelial cells and smooth muscle cells (62.3% in +/+; 58.1% in +/−; 60.9% in −/−) (Fig. 2B). An analogous assay with an antibody to phosphorylated histone H3 also suggested that the mitotic activity of cardiomyocytes is significantly reduced in the absence of ATE1 (supplemental Fig. 5).
FIGURE 2.
Primary cardiomyocytes from ATE1-deficient embryos at E13. 5 are impaired in proliferation. A, BrdU incorporation assay of primary cardiac cells derived from +/+ and ATE1−/− embryonic hearts at E13.5. Cardiomyocytes were identified by immunostaining of troponin I. Cardiomyocytes are distinguished from non-cardiomyocytes by immunostaining of troponin I or sarcomeric α-actinin. Scale bars, 10 μm. B, quantitation of A. C, the enzymatic staining of β-galactosidase in primary cardiac cells from ATE1+/− embryos at E13.5. Data are presented as mean ± S.D.
To determine whether ATE1 is expressed in cardiomyocytes, cultured primary cardiac cells from E13.5 ATE1+/− embryos were subjected to enzymatic staining for the reporter NLS-lacZ, which marks the ATG codon of ATE1. Immunostaining of sarcomeric α-actinin, a marker of cardiomyocytes, following lacZ staining revealed a robust expression of ATE1 in cardiomyocytes (Fig. 2C). By contrast, the expression of ATE1 was much lower or often undetectable in α-actinin-negative non-cardiomyocytes. These results suggest that ATE1 knock-out results in significantly reduced proliferation in cardiomyocytes without severe defects in the developmental program or increased apoptosis.
ATE1-deficient Embryonic Hearts Are Impaired in G-protein Signaling
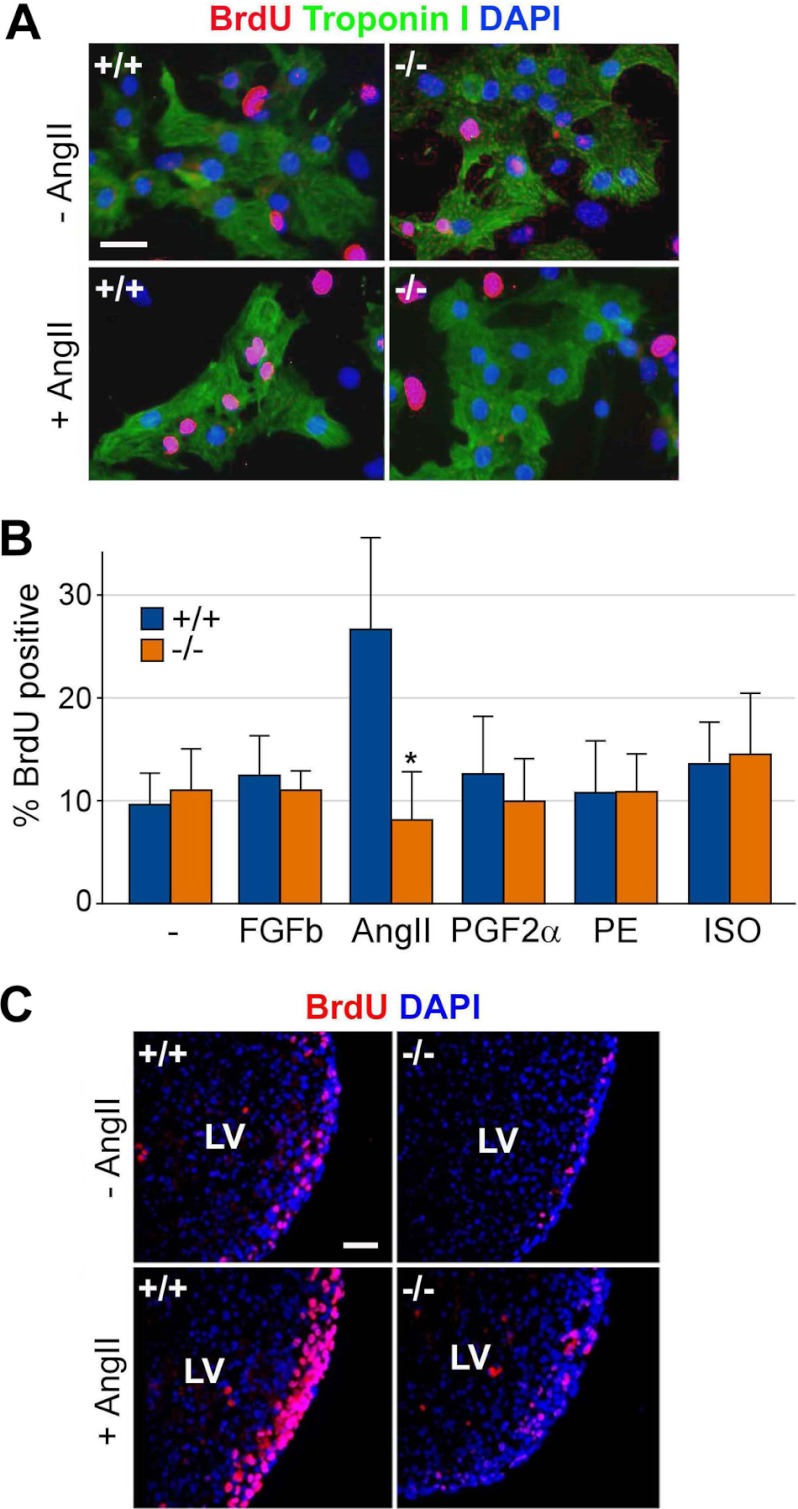
Gene mutations associated with cardiac defects have been implicated in signaling molecules, cell adhesion molecules, ion channels, and transcription factors (33). To test a potential function of arginylation in extracellular signaling pathways of cardiomyocytes, a BrdU incorporation assay was performed on primary cardiac cells from +/+ and ATE1−/− embryos at E13.5 before and after the treatment with ligands to GPCRs and receptor tyrosine kinases, including prostaglandin F2α (for prostaglandin F receptor coupled with Gq), phenylephrine (for α-adrenergic receptor coupled with Gq and Gi), basic FGF (for fibroblast growth factor basic receptor, a receptor tyrosine kinase), isoproterenol (for β-adrenergic receptor coupled with Gs), and angiotensin II (for AT1 receptor coupled with Gq and Gi). Among these, only angiotensin II significantly promoted the proliferation of troponin I-positive cardiomyocytes (Fig. 3, A and B). Notably, ATE1−/− cardiomyocytes failed to properly respond to angiotensin II compared with +/+ cells. To determine angiotensin II-induced proliferation in a more physiological condition, we used an ex vivo model for +/+ and ATE1−/− hearts from E13.5 embryos. An analogous assay on explanted hearts showed that ATE1−/− hearts are impaired in angiotensin II-induced proliferation (Fig. 3C and supplemental Fig. 6A). The mRNA level of the AT1 receptor was comparable in cultured cardiac cells from +/+ and ATE1−/− hearts as determined by quantitative RT-PCR (supplemental Fig. 6B).
FIGURE 3.
Cardiomyocytes of ATE1-deficient embryos are impaired in angiotensin II-induced G-protein signaling. A, cultured primary cardiac cells from +/+ and ATE1−/− embryos at E13.5 were treated with 2 μm angiotensin II (AngII) and subjected to a BrdU incorporation assay with coimmunostaining of troponin I to identify cardiomyocytes. Scale bar, 10 μm. B, quantitation of an analogous assay (A) in which cells were treated with various ligands to GPCR: 50 ng/ml basic fibroblast growth factor (FGFb), 2 μm angiotensin II (AngII), 100 nm prostaglandin F2α (PGF2α), 200 μm phenylephrine (PE), and 4 μm isoproterenol (ISO). C, explanted hearts from +/+ and ATE1−/− embryos at E13.5 were treated with 2 μm angiotensin II followed by a BrdU incorporation assay on cross-sections of the left ventricle (LV). Scale bar, 200 μm. Data are presented as mean ± S.D.
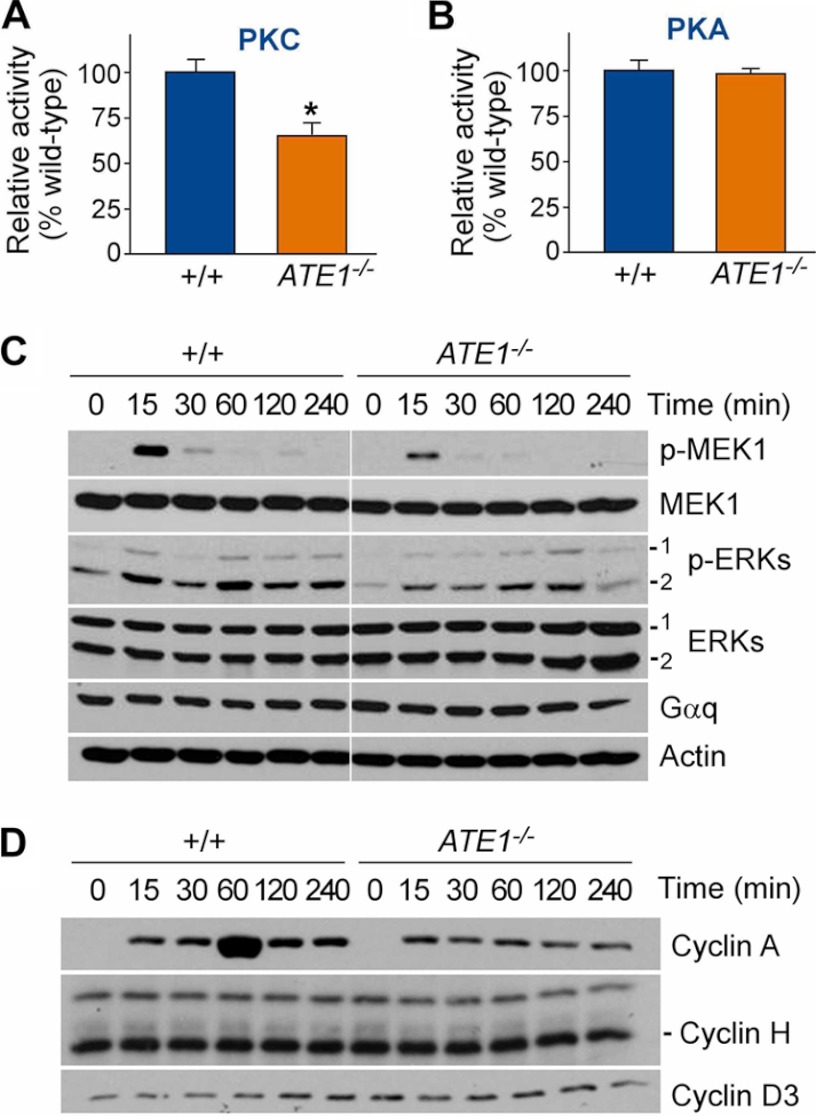
Extrinsic stimuli such as endothelin-1, angiotensin II, and phenylephrine induce cell growth in the heart through their interaction with GPCRs that activate the Gq class of GTP-binding proteins (34). Upon binding to an agonist-occupied receptor, the heterotrimeric Gq protein dissociates into individual Gαq and Gβγ subunits. GTP-bound Gαq activates PLC, which results in inositol 1,4,5-trisphosphate-mediated calcium release and diacylglycerol-mediated activation of PKC. Dissociated Gβγ has the potential to activate the small GTP-binding protein Ras and initiate a tyrosine kinase cascade, leading to activation of MAPKs. Gαq can also activate MAPKs independently from Gβγ via a PKC-dependent mechanism. To determine the function of ATE1 in Gq signaling of cardiomyocytes, we measured the enzymatic activities of signaling molecules in extracts of +/+ and ATE1−/− embryonic hearts at E13.5. ATE1−/− hearts contained reduced activities for several enzymes that mediate Gαq signaling, such as PKC (Fig. 4A) and PLC (supplemental Fig. 7A). By contrast, no difference was observed for PKA (Fig. 4B) and Ca2+/calmodulin-dependent protein kinase II (supplemental Fig. 7B), which are activated by Gs-dependent adenylyl cyclase. PLC and PKC activate G1-S progression through the MAPK pathway. To determine a specific MAPK subpathway linked to ATE1-dependent arginylation, we monitored the kinetics of activation and inactivation of candidate components in primary cardiac cells established from +/+ and ATE1−/− embryonic hearts at E13.5. Time course immunoblotting following 24-h serum starvation and subsequent serum activation identified MAPK/ERK kinase 1 (MEK1) as a component whose activity is markedly attenuated in ATE1−/− cardiomyocytes (Fig. 4C). An analogous assay for MAPKs showed that the activities of ERK1 and ERK2, which are phosphorylated by MEK1, were significantly down-regulated in ATE1−/− hearts (Fig. 4C), which was verified by the immunohistochemistry analysis (supplemental Fig. 7C) and an in vitro kinase assay (supplemental Fig. 7D).
FIGURE 4.
Characterization of G-protein signaling pathways in ATE1−/− embryonic hearts. A and B, enzymatic activities of PKC and PKA were determined using extracts from +/+ and ATE1−/− embryonic hearts at E13.5 and model substrates as described under “Experimental Procedures.” C and D, primary cardiac cells from +/+ and ATE1−/− embryos at E13.5 were subjected to 24-h serum starvation and subsequent serum activation followed by time course immunoblotting of components in MAPK pathways (C) and cyclins (D). p-ERKs, phosphorylated ERKs. Data are presented as mean ± S.D.
The MAPK pathway can induce G1-S progression through transcriptional induction of cyclin A, which binds to cyclin-dependent kinase 2. To determine the effect of ATE1 knock-out on the activation of cyclins, primary cardiac cells established from +/+ and ATE1−/− embryonic hearts at E13.5 were subjected to serum stimulation following 24-h serum starvation. Immunoblotting analysis revealed a robust induction for cyclin A in ATE1+/− cells that was markedly diminished in mutants (Fig. 4D, bottom). Under these conditions, no significant differences were observed for cyclins H and D3. As an alternative approach, we used fluorescence-activated cell sorting (FACS) analysis using cultured cardiac cells at passage number 5 (to obtain a sufficient number of cells) that were derived from +/+ and ATE1−/− embryonic hearts at E13.5. The percentage of ATE1−/− cells in S phase (16.3% in +/+ versus 5.4% in −/−), but not G0G1 phase (61.7% in +/+ versus 78.3% in −/−), was significantly lower compared with controls. These results together suggest that cardiovascular defects of ATE1−/− embryos are in part contributed by misregulation of the Gαq-PLC/PKC-MEK1-ERK1 axis of G-protein signaling in embryonic hearts.
Cardiac Overexpression of Gαq Significantly Rescues ATE1-deficient Mouse Embryos from Ventricular Septal Defects and Thin Myocardium
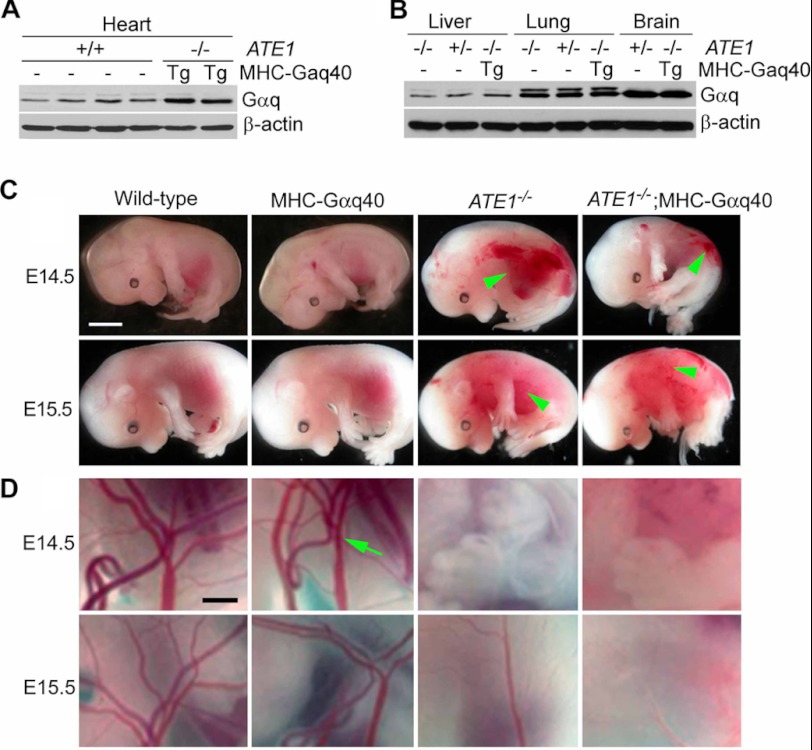
To determine whether cardiac overexpression of Gαq improves cardiac development in ATE1−/− embryos, we generated a double mutant strain (ATE1−/−;MHC-Gαq40) by crossing ATE1+/− mice in a C57BL/6J-129SvEv background with MHC-Gαq40 transgenic mice in an FVB/N background. It has been shown that MHC-Gαq40 transgenic mice (see “Experimental Procedures”) develop cardiac hypertrophy in adulthood associated with induction of fetal gene expression and reduced cardiac contractility (29). Immunoblotting analysis showed a ∼5-fold overexpression of Gαq in the hearts of ATE1−/−;MHC-Gαq40 embryos at E15.5 compared with littermate controls (Fig. 5A), whereas no differences were observed for the liver, lung, and brain (Fig. 5B).
FIGURE 5.
Generation of ATE1−/−;MHC-Gαq40 embryos overexpressing Gαq in hearts. A, immunoblotting of Gαq in hearts of various mutant embryos at E13.5 whose genotypes are indicated. B, same as in A except that livers, lungs, and brains were used. C, gross morphology of embryos at E14.5 and E15.5 from ATE1+/− and MHC-Gαq40 parents. Vascular defects of ATE1−/− embryos in this study were indistinguishable from those observed in the previous study (7) in which PECAM-1 staining revealed poorly developed blood vessels. Note that cardiac overexpression of Gαq does not rescue ATE1−/− embryos from circulation defects. Arrowheads indicate hemorrhages. Scale bars, 3 mm. D, close-up views of yolk sacs of embryos shown in C. Arrow, the main artery. Scale bar, 1 μm.
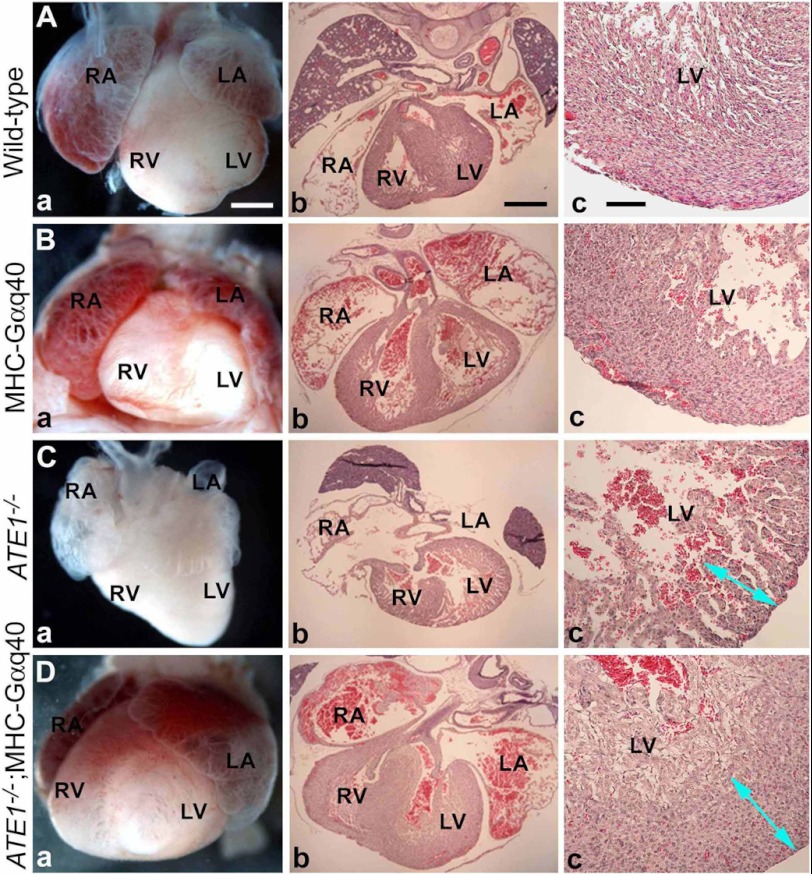
Cardiac overexpression of Gαq did not cause a significant difference in the gross morphology of ATE1−/− and ATE1−/−;MHC-Gαq40 embryos when observed at E14.6 through E16.5 (Fig. 5, C and D). In both genotypes, local hemorrhages indicative of circulation defects were obvious at E14.5 and became severe at E15.5 through E16.5. Overall, the morphological phenotypes (Fig. 5, C and D) observed in ATE1−/− and ATE1−/−;MHC-Gαq40 embryos in the C57/129;FVB background were indistinguishable from those of embryos in the C57/129 background that have been characterized in the previous (7) and current (supplemental Figs. 2 and 3) studies. Importantly, when embryonic hearts were harvested and morphologically examined, cardiac overexpression of Gαq did rescue significantly ATE1−/− hearts from cardiac defects. For instance, in sharp contrast to E16.5 ATE1−/− hearts (n = 5) morphologically arrested at ∼E14.5 (Fig. 6, C versus A), ATE1−/−;MHC-Gαq40 hearts (n = 8) at the same stage showed virtually normal morphology relative to wild-type and MHC-Gαq40 embryos (Fig. 6, D versus A and B). In addition, cross-sections of E16.5 ATE1−/−;MHC-Gαq40 hearts showed significant rescue effects for thin myocardium, VSD, trabeculation, and atrial septal defect (Fig. 6D and data not shown) relative to control ATE1−/− hearts. These results suggest that overexpression of Gαq in the heart significantly rescues ATE1−/− embryos from cardiac defects.
FIGURE 6.
Overexpression of Gαq subunit in heart improves cardiac development in ATE1−/− embryos. Shown are gross morphology (panels a), cross-sections (panels b), and close-up views of left ventricles (panels c) of embryonic hearts. Genotypes of these hearts are shown to the left: wild-type (A), MHC-Gαq40 (B), ATE1−/− (C), and ATE1−/−;MHC-Gαq40 (D) embryos. Scale bars, 300 (panels a and b) and 100 μm (panels c). RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle. Arrowheads in panel c indicate the thickness of left ventricular walls. Note that the ventricular wall of the ATE1−/−;MHC-Gαq40 heart (D) is comparable with that in wild-type heart (A).
Vascular Defects in ATE1-deficient Mouse Embryos Are Independent from Cardiac Defects
Despite cardiac rescue by Gαq overexpression, ATE1−/−;MHC-Gαq40 embryos still died around E15.5 and E16.5 with no obvious difference in timing and morphology compared with control ATE1−/− embryos, suggesting that cardiac defects are not the primary cause of death in ATE1-deficient embryos. To determine whether vascular defects observed in ATE1−/− embryos are independent from cardiac defects, we examined the gross morphology of embryos at E14.5 through E16.5. ATE1−/−;MHC-Gαq40 embryos (n = 16) invariably developed morphological defects indistinguishable from vascular defects in the ATE1−/− yolk sac and embryos proper observed in a previous (7) and this (n = 15) study (Fig. 5, C and D). Although we do not exclude the possibility of subtle changes in vascular integrity, these results suggest that vascular defects in ATE1−/− embryos may be the primary cause of death and independent from cardiac defects.
Degradation of RGS4 Spatiotemporally Correlates to ATE1 Distribution in Mouse Embryos and Is Sensitive to Oxygen Availability
The substrates of arginylation that may underlie cardiovascular defects in ATE1−/− embryos include a set of structurally related RGS proteins (RGS4, RGS5, and RGS16) that can act as GTPase-activating proteins for Gαq (11, 12, 22). RGS4 and RGS5 have been characterized as regulators of G-protein signaling in the heart and blood vessels, respectively (35–38). In cultured cells, the degradation of RGS4 can be mediated by arginylation-dependent N-end rule ubiquitylation (11, 22) or an internal degron (non-N-degron)-based ubiquitylation by an unknown E3 ligase (data not shown) depending on cell types and states.
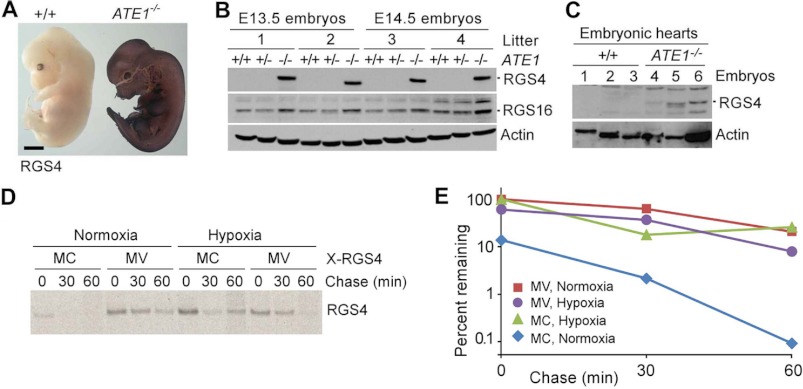
To determine the spatiotemporal relationship between RGS4 and ATE1, we performed whole-mount immunostaining of RGS4 in +/+ and ATE1−/− embryos at E12.5 and E13.5. RGS4 was barely detectable in +/+ embryos but was drastically accumulated in ATE1−/− embryos (Fig. 7A and data not shown), indicating strong arginylation-dependent degradation of endogenous RGS4 in growing embryos. Immunoblotting analysis of whole embryos and embryonic hearts showed an accumulation of RGS4 in the absence of ATE1 (Fig. 7, B and C) without a significant change in transcription (supplemental Fig. 8A). Immunostaining on cross-sections revealed a strong correlation between RGS4 and ATE1 in all cell types examined that express RGS4 (data not shown), including hearts (supplemental Fig. 8B). RGS4-positive cells were relatively enriched along the migratory pathway of neural crest cells, including dorsal root ganglia, sympathetic ganglia, muscle lineage, and developing alveolus (supplemental Fig. 9). In these cells, the expression of ATE1 was also prominent (Ref. 7 and data not shown). Consistently, previous studies suggested that both RGS5 and ATE1 are prominently expressed in arteries (7, 38, 39). These results indicate that ATE1 plays a role in homeostasis of G-protein signaling in hearts and other tissues through regulated proteolysis of multiple RGS proteins. The rapid degradation of RGS4 also explains why its expression at the protein level currently remains elusive in the hearts even though an abundant mRNA expression correlates to cardiac proliferation and hypertrophy (40, 41).
FIGURE 7.
Characterization of RGS4 turnover in +/+ and ATE1−/− embryos. A, whole-mount immunohistochemistry of RGS4 in +/+ and ATE1−/− embryos at E12.5. Scale bar, 1 mm. B, immunoblotting of RGS4 and RGS16 in extracts of +/+ and ATE1−/− embryos. C, immunoblotting of RGS4 in extracts of +/+ and ATE1−/− embryonic hearts at E13.5. D, pulse-chase analysis of RGS4 (MC) and C2V-RGS4 (MV) in normoxia and hypoxia (0.1% O2). The transfected cells were labeled for 12 min with [35S]Met/Cys followed by anti-RGS4 immunoprecipitation, SDS-PAGE analysis, and autoradiography. E, quantitation of data in D using a PhosphorImager.
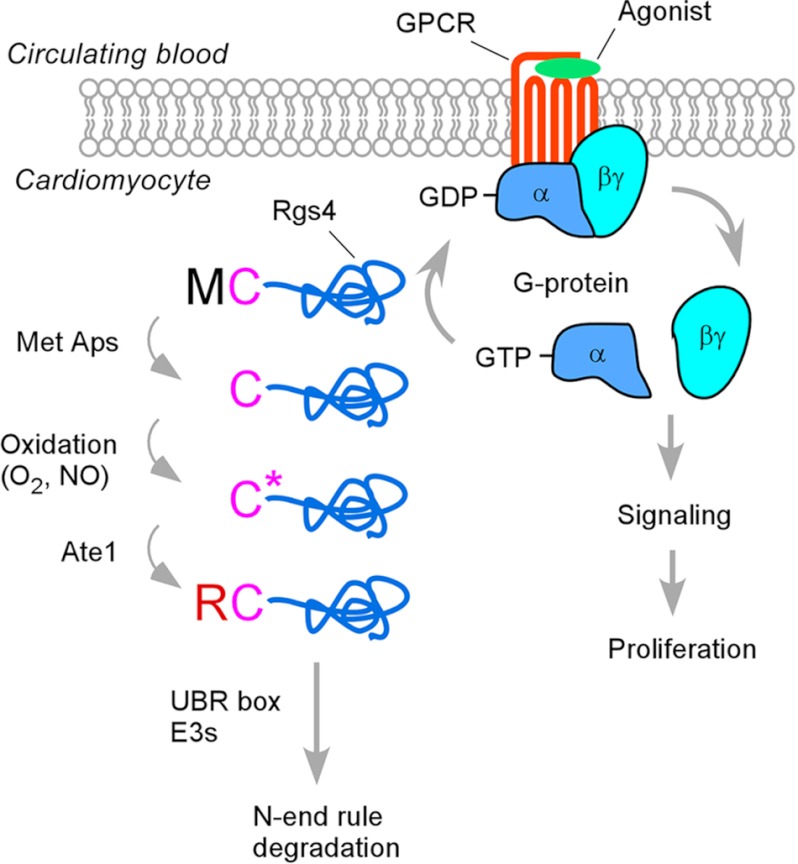
RGS4 has the N-terminal Met-Cys sequence in which the Cys-2 residue is a degradation signal (degron) for the N-end rule pathway (11). We have observed previously that the Cys-2 residue of RGS4 is conjugated with 48 Da following the removal of the initiator Met residue. Given that a mass of 48 daltons corresponds to three oxygen atoms, the degradation of RGS4 may be sensitive to oxygen availability in circulating blood. To determine the effect of oxygen availability on in vivo degradation of RGS4, we used mouse embryonic fibroblasts because cardiomyocytes are resistant to transient transfection. Mouse embryonic fibroblasts were transiently transfected with a plasmid expressing RGS4 and cultured in normoxia or hypoxia (0.1% O2). We labeled newly synthesized RGS4 proteins with [35S]Met/Cys for 12 min, inhibited protein synthesis with cycloheximide, and chased the decay of 35S-labeled RGS4 molecules using immunoprecipitation of RGS4. Consistent with the previous observation (11), in normoxia, RGS4 was rapidly degraded with a drastically reduced level (∼14%) of the zero time point compared with C2V-RGS4 in which a mutation of the pro-N-degron Cys-2 to Val inhibits arginylation (Fig. 7, D and E). Notably, in hypoxia, normally short lived RGS4 was significantly stabilized as compared with the half-lives of C2V-RGS4 in normoxia and hypoxia (Fig. 7, D and E). A previous study reported the hypoxia-sensitive degradation for a set of short lived proteins carrying the Met-Cys sequence (for a review, see Ref. 3), including mammalian RGS5 (11) and the ethylene response factor group VII transcription factors (e.g. hypoxia response elements 1 and 2 and ras-related protein 2.12 (RAP2.12)) of the plant Arabidopsis (42, 43). These results suggest that oxidation of the pro-N-degron Cys of RGS4 may contribute to sensing and reacting to oxygen availability in circulating blood by altering G-protein signaling (Fig. 8).
FIGURE 8.
Model in which ATE1 R-transferases regulate homeostasis of G-protein signaling in cardiomyocytes through arginylation of RGS4 (and RGS5 and RGS16 as well). In this model, N-terminal Met is cotranslationally cleaved off by Met aminopeptidases (Met Aps), exposing the pro-N-degron Cys-2 at the N terminus. In normally growing embryos, the Cys-2 residue is oxidized into CysO2(H) or CysO3(H) and subsequently arginylated by ATE1 R-transferases, producing the N-degron Arg, which is recognized by N-recognins containing the UBR box. The degradation of RGS4 through the N-end rule pathway leads to the activation of G-protein signaling and thus cell proliferation. However, if the Cys-2 residue is not readily oxidized, for example in ischemia, RGS4 is accumulated and turns down G-protein signaling. This mechanism may represent a sensor of oxygen or its derivative in the cardiovascular system.
The Arabidopsis and human genomes encode at least 206 and 502 proteins with the Met-Cys motif (9, 42, 43). Thus, ATE1-dependent arginylation may be relevant to the majority of these Met-Cys proteins, representing a unique proteome whose functions include sensing oxygen and other cellular stresses through oxidation and arginylation of the pro-N-degron Cys.
DISCUSSION
In the current study, we studied the physiological function of the arginylation branch of the N-end rule pathway in embryonic hearts and cardiomyocytes. We report that thin myocardium and VSD of ATE1−/− embryonic hearts first observed at E12.5 is primarily caused by a specific impairment of proliferation in cardiomyocytes but not cardiac fibroblasts, consistent with the prominent expression of ATE1 in cardiomyocytes relative to cardiac fibroblasts. Our results suggest that ATE1-deficient hearts and cardiomyocytes therein, but not cardiac fibroblasts, are impaired in the PLC/PKC-MEK1-ERK axis of Gαq-activated protein signaling. By overexpressing Gαq in ATE1−/− hearts using the MHC promoter, we were able to rescue significantly ATE1−/− embryos from thin myocardium and VSD. Of note, cardiac overexpression of Gαq did not noticeably affect vascular defects, demonstrating that cardiac and vascular defects of ATE1−/− embryos are largely independent from each other and cell-autonomous and that vascular defects may be the primary cause of death in ATE1−/− embryos. The impaired G-protein signaling is attributed in part to failure to mediate arginylation-dependent degradation of RGS4 (and RGS5 and RGS16 as well) known to function as a GTPase-activating protein for GPCR-activated Gαq during cardiac G-protein signaling. Given the biochemical property and physiological function of RGS4 as a negative regulator of Gαq in the heart models (35, 36), it is reasonable to speculate that abnormal accumulation of RGS4 (and RGS5 and RGS16 as well) impairs G-protein signaling in cardiomyocytes, contributing to the growth arrest of myocardium during embryogenesis. However, it should be noted that ATE1 has been implicated in a variety of physiological processes, including arginylation of many cellular proteins (see the Introduction). Therefore, there are likely to be additional molecular mechanisms that contribute to cardiovascular defects in ATE1−/−embryos.
The mammalian heart consumes 3–20 times more O2 than the brain (44) and thus requires a constant supply of O2 for its function. For example, coronary artery disease with consequent myocardial ischemia and necrosis is a leading cause of heart failure worldwide. Although O2 is a major determinant of cardiac gene expression and numerous cellular processes, little is known about its role in cardiovascular signaling and the mechanism by which the heart senses its concentration to modulate intracellular processes. Our results indicate hypoxia-sensitive, arginylation-dependent degradation of RGS4 (this study) and RGS5 (11), consistent with the finding that ATE1-marked β-galactosidase and RGS5 mRNAs are prominently expressed in artery relative to veins (7, 37–39, 45). In mouse embryonic fibroblasts, the degradation of RGS4 involves the cleavage of N-terminal Met, which exposes the pro-N-degron Cys at the N terminus (11) (Fig. 8). The exposed Cys-2 is conjugated with a mass of 48 Da that is thought to represent oxidation to CysO2(H) and subsequent conversion to CysO3(H), whose structure is similar to the arginylation-permissive pro-N-degron Asp (7, 11, 12) (supplemental Fig. 1). Therefore, it is likely that the reduced availability of O2 inhibits the Cys-2 oxidation prior to arginylation by ATE1. These observations suggest that in hearts and blood vessels under normal physiological conditions in which cells are exposed to sufficient O2 and NO RGS4 with the pro-N-degron Cys-2 is constitutively degraded to maintain G-protein signaling, allowing cells to sense extracellular ligands to a maximal level (Fig. 8). However, when O2 (or other molecules that induce Cys-2 oxidation) in circulating blood is not sufficient, for example in ischemia caused by cardiac arrest or other cellular stresses, these substrates are rapidly accumulated in a real time basis to turn down GPCR signaling, decoupling cells from extracellular proliferation signals (Fig. 8). Thus, arginylation-induced proteolysis may function as a cellular stress response to maintain homeostasis in GPCR signaling in the heart (via RGS4) and blood vessels (via RGS5).
Acknowledgments
We are grateful to Dong Hoon Han for administrative support, Dong Oh Moon for critical discussions, and the staff of the animal facility at the University of Pittsburg for the care and maintenance of mice.
This work was supported, in whole or in part, by National Institutes of Health Grants HL083365 (to Y. T. K.) and HL067050 (to Y. S. L.). This work was also supported by World Class University Grant R31-2008-000-10103-0 (to Y. T. K.), the World Class Institute (to B. Y. K.), Ministry of Education, Science, and Technology Grants 2011-0007990 and 2011-0030938 (to M. J. L.), and Ministry of Health and Welfare Grant A111227 (to M. J. L.).

This article contains supplemental Figs. 1–9.
- ATE1 or R-transferase
- arginyl-tRNA-protein transferase
- E
- embryonic day
- PLC
- phospholipase C
- CysO2(H)
- Cys sulfinic acid
- CysO3(H)
- Cys sulfonic acid
- VSD
- ventricular septal defect
- PTA
- persistent truncus arteriosus
- MHC
- α-myosin heavy chain
- GPCR
- G-protein-coupled receptor
- RGS
- regulator of G-protein signaling
- UBR
- ubiquitin protein ligase E3 component N-recognin.
REFERENCES
- 1. Sriram S. M., Kim B. Y., Kwon Y. T. (2011) The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 12, 735–747 [DOI] [PubMed] [Google Scholar]
- 2. Sriram S. M., Kwon Y. T. (2010) The molecular principles of N-end rule recognition. Nat. Struct. Mol. Biol. 17, 1164–1165 [DOI] [PubMed] [Google Scholar]
- 3. Tasaki T., Sriram S. M., Park K. S., Kwon Y. T. (2012) The N-end rule pathway. Annu. Rev. Biochem., DOI: 10.1146/annurev-biochem-051710-093308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bachmair A., Finley D., Varshavsky A. (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 [DOI] [PubMed] [Google Scholar]
- 5. Kaji H., Novelli G. D., Kaji A. (1963) A soluble amino acid-incorporating system from rat liver. Biochim. Biophys. Acta 76, 474–477 [PubMed] [Google Scholar]
- 6. Kwon Y. T., Kashina A. S., Varshavsky A. (1999) Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol. Cell. Biol. 19, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kwon Y. T., Kashina A. S., Davydov I. V., Hu R. G., An J. Y., Seo J. W., Du F., Varshavsky A. (2002) An essential role of N-terminal arginylation in cardiovascular development. Science 297, 96–99 [DOI] [PubMed] [Google Scholar]
- 8. Balzi E., Choder M., Chen W. N., Varshavsky A., Goffeau A. (1990) Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J. Biol. Chem. 265, 7464–7471 [PubMed] [Google Scholar]
- 9. Tasaki T., Kwon Y. T. (2007) The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem. Sci. 32, 520–528 [DOI] [PubMed] [Google Scholar]
- 10. Gonda D. K., Bachmair A., Wünning I., Tobias J. W., Lane W. S., Varshavsky A. (1989) Universality and structure of the N-end rule. J. Biol. Chem. 264, 16700–16712 [PubMed] [Google Scholar]
- 11. Lee M. J., Tasaki T., Moroi K., An J. Y., Kimura S., Davydov I. V., Kwon Y. T. (2005) RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 15030–15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu R. G., Sheng J., Qi X., Xu Z., Takahashi T. T., Varshavsky A. (2005) The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 [DOI] [PubMed] [Google Scholar]
- 13. Kwon Y. T., Reiss Y., Fried V. A., Hershko A., Yoon J. K., Gonda D. K., Sangan P., Copeland N. G., Jenkins N. A., Varshavsky A. (1998) The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 7898–7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tasaki T., Mulder L. C., Iwamatsu A., Lee M. J., Davydov I. V., Varshavsky A., Muesing M., Kwon Y. T. (2005) A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 25, 7120–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tasaki T., Zakrzewska A., Dudgeon D. D., Jiang Y., Lazo J. S., Kwon Y. T. (2009) The substrate recognition domains of the N-end rule pathway. J. Biol. Chem. 284, 1884–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brower C. S., Varshavsky A. (2009) Ablation of arginylation in the mouse N-end rule pathway: loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS One 4, e7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leu N. A., Kurosaka S., Kashina A. (2009) Conditional Tek promoter-driven deletion of arginyltransferase in the germ line causes defects in gametogenesis and early embryonic lethality in mice. PLoS One 4, e7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurosaka S., Leu N. A., Zhang F., Bunte R., Saha S., Wang J., Guo C., He W., Kashina A. (2010) Arginylation-dependent neural crest cell migration is essential for mouse development. PLoS Genet. 6, e1000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yoshida S., Ito M., Callis J., Nishida I., Watanabe A. (2002) A delayed leaf senescence mutant is defective in arginyl-tRNA:protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 32, 129–137 [DOI] [PubMed] [Google Scholar]
- 20. Graciet E., Walter F., Maoiléidigh D. O., Pollmann S., Meyerowitz E. M., Varshavsky A., Wellmer F. (2009) The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc. Natl. Acad. Sci. U.S.A. 106, 13618–13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spradling A. C., Stern D., Beaton A., Rhem E. J., Laverty T., Mozden N., Misra S., Rubin G. M. (1999) The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153, 135–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davydov I. V., Varshavsky A. (2000) RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J. Biol. Chem. 275, 22931–22941 [DOI] [PubMed] [Google Scholar]
- 23. Ditzel M., Wilson R., Tenev T., Zachariou A., Paul A., Deas E., Meier P. (2003) Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell Biol. 5, 467–473 [DOI] [PubMed] [Google Scholar]
- 24. Decca M. B., Bosc C., Luche S., Brugière S., Job D., Rabilloud T., Garin J., Hallak M. E. (2006) Protein arginylation in rat brain cytosol: a proteomic analysis. Neurochem. Res. 31, 401–409 [DOI] [PubMed] [Google Scholar]
- 25. Corbett E. F., Michalak M. (2000) Calcium, a signaling molecule in the endoplasmic reticulum? Trends Biochem. Sci. 25, 307–311 [DOI] [PubMed] [Google Scholar]
- 26. Karakozova M., Kozak M., Wong C. C., Bailey A. O., Yates J. R., 3rd, Mogilner A., Zebroski H., Kashina A. (2006) Arginylation of β-actin regulates actin cytoskeleton and cell motility. Science 313, 192–196 [DOI] [PubMed] [Google Scholar]
- 27. Hu R. G., Brower C. S., Wang H., Davydov I. V., Sheng J., Zhou J., Kwon Y. T., Varshavsky A. (2006) Arginyltransferase, its specificity, putative substrates, bidirectional promoter, and splicing-derived isoforms. J. Biol. Chem. 281, 32559–32573 [DOI] [PubMed] [Google Scholar]
- 28. Wong C. C., Xu T., Rai R., Bailey A. O., Yates J. R., 3rd, Wolf Y. I., Zebroski H., Kashina A. (2007) Global analysis of posttranslational protein arginylation. PLoS Biol. 5, e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. D'Angelo D. D., Sakata Y., Lorenz J. N., Boivin G. P., Walsh R. A., Liggett S. B., Dorn G. W., 2nd (1997) Transgenic Gαq overexpression induces cardiac contractile failure in mice. Proc. Natl. Acad. Sci. U.S.A. 94, 8121–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee M. J., Pal K., Tasaki T., Roy S., Jiang Y., An J. Y., Banerjee R., Kwon Y. T. (2008) Synthetic heterovalent inhibitors targeting recognition E3 components of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 100–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kwon Y. T., Xia Z., An J. Y., Tasaki T., Davydov I. V., Seo J. W., Sheng J., Xie Y., Varshavsky A. (2003) Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol. Cell. Biol. 23, 8255–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kwon Y. T., Xia Z., Davydov I. V., Lecker S. H., Varshavsky A. (2001) Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3α) of the N-end rule pathway. Mol. Cell. Biol. 21, 8007–8021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Srivastava D., Olson E. N. (2000) A genetic blueprint for cardiac development. Nature 407, 221–226 [DOI] [PubMed] [Google Scholar]
- 34. Akhter S. A., Luttrell L. M., Rockman H. A., Iaccarino G., Lefkowitz R. J., Koch W. J. (1998) Targeting the receptor-Gq interface to inhibit in vivo pressure overload myocardial hypertrophy. Science 280, 574–577 [DOI] [PubMed] [Google Scholar]
- 35. Owen V. J., Burton P. B., Mullen A. J., Birks E. J., Barton P., Yacoub M. H. (2001) Expression of RGS3, RGS4 and Giα2 in acutely failing donor hearts and end-stage heart failure. Eur. Heart J. 22, 1015–1020 [DOI] [PubMed] [Google Scholar]
- 36. Rogers J. H., Tsirka A., Kovacs A., Blumer K. J., Dorn G. W., 2nd, Muslin A. J. (2001) RGS4 reduces contractile dysfunction and hypertrophic gene induction in Gαq overexpressing mice. J. Mol. Cell. Cardiol. 33, 209–218 [DOI] [PubMed] [Google Scholar]
- 37. Hamzah J., Jugold M., Kiessling F., Rigby P., Manzur M., Marti H. H., Rabie T., Kaden S., Gröne H. J., Hämmerling G. J., Arnold B., Ganss R. (2008) Vascular normalization in Rgs5-deficient tumours promotes immune destruction. Nature 453, 410–414 [DOI] [PubMed] [Google Scholar]
- 38. Bondjers C., Kalén M., Hellström M., Scheidl S. J., Abramsson A., Renner O., Lindahl P., Cho H., Kehrl J., Betsholtz C. (2003) Transcription profiling of platelet-derived growth factor-B-deficient mouse embryos identifies RGS5 as a novel marker for pericytes and vascular smooth muscle cells. Am. J. Pathol. 162, 721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cho H., Kozasa T., Bondjers C., Betsholtz C., Kehrl J. H. (2003) Pericyte-specific expression of Rgs5: implications for PDGF and EDG receptor signaling during vascular maturation. FASEB J. 17, 440–442 [DOI] [PubMed] [Google Scholar]
- 40. Adams L. D., Geary R. L., McManus B., Schwartz S. M. (2000) A comparison of aorta and vena cava medial message expression by cDNA array analysis identifies a set of 68 consistently differentially expressed genes, all in aortic media. Circ. Res. 87, 623–631 [DOI] [PubMed] [Google Scholar]
- 41. Patten M., Stübe S., Thoma B., Wieland T. (2003) Interleukin-1β mediates endotoxin- and tumor necrosis factor α-induced RGS16 protein expression in cultured cardiac myocytes. Naunyn Schmiedebergs Arch. Pharmacol. 368, 360–365 [DOI] [PubMed] [Google Scholar]
- 42. Gibbs D. J., Lee S. C., Isa N. M., Gramuglia S., Fukao T., Bassel G. W., Correia C. S., Corbineau F., Theodoulou F. L., Bailey-Serres J., Holdsworth M. J. (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Licausi F., Kosmacz M., Weits D. A., Giuntoli B., Giorgi F. M., Voesenek L. A., Perata P., van Dongen J. T. (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422 [DOI] [PubMed] [Google Scholar]
- 44. Giordano F. J. (2005) Oxygen, oxidative stress, hypoxia, and heart failure. J. Clin. Investig. 115, 500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wieland T., Mittmann C. (2003) Regulators of G-protein signalling: multifunctional proteins with impact on signalling in the cardiovascular system. Pharmacol. Ther. 97, 95–115 [DOI] [PubMed] [Google Scholar]