Background: The mechanism by which mycobacteria maintain ribosome stability during latent infection is unknown.
Results: Mycobacterium smegmatis uses a DosR-regulated S30AE domain protein, RafH, to maintain the associated (stable) ribosome during the hypoxia model of persistence.
Conclusion: Ribosomal stability, mediated by RafH, is essential for survival and emergence from hypoxia.
Significance: Learning how mycobacteria stabilize ribosomes to survive hypoxia is crucial for understanding latent tuberculosis.
Keywords: Bacteria, Bacterial Pathogenesis, Bacterial Signal Transduction, Infectious Diseases, Tuberculosis, Latent Tuberculosis, Non-replicating Persistence, Latency, Hypoxia, Stationary Phase
Abstract
It is thought that during latent infection, Mycobacterium tuberculosis bacilli are retained within granulomas in a low-oxygen environment. The dormancy survival (Dos) regulon, regulated by the response regulator DosR, appears to be essential for hypoxic survival in M. tuberculosis, but it is not known how the regulon promotes survival. Here we report that mycobacteria, in contrast to enteric bacteria, do not form higher-order structures (e.g. ribosomal dimers) upon entry into stasis. Instead, ribosomes are stabilized in the associated form (70S). Using a strategy incorporating microfluidic, proteomic, and ribosomal profiling techniques to elucidate the fate of mycobacterial ribosomes during hypoxic stasis, we show that the dormancy regulator DosR is required for optimal ribosome stabilization. We present evidence that the majority of this effect is mediated by the DosR-regulated protein MSMEG_3935 (a S30AE domain protein), which is associated with the ribosome under hypoxic conditions. A Δ3935 mutant phenocopies the ΔdosR mutant during hypoxia, and complementation of ΔdosR with the MSMEG_3935 gene leads to complete recovery of dosR mutant phenotypes during hypoxia. We suggest that this protein is named ribosome-associated factor under hypoxia (RafH) and that it is the major factor responsible for DosR-mediated hypoxic survival in mycobacteria.
Introduction
The vast majority of individuals infected with Mycobacterium tuberculosis are asymptomatic and harbor bacilli in a latent infection that can reactivate to cause acute tuberculosis, often decades after initial infection (1).
During latent infection, M. tuberculosis bacilli are retained within granulomas (2), where they can survive for several decades (3). However, the physiological state in which they exist is poorly defined. There is evidence that the M. tuberculosis granuloma may be a low-oxygen environment (4), and so a significant body of research has been concerned with understanding the hypoxic adaptations of M. tuberculosis (5–7).
The dormancy survival regulon, regulated by the response regulator DosR, comprises 48 genes and appears to be essential for hypoxic survival in many mycobacterial species, including M. tuberculosis (8–11), Mycobacterium bovis BCG (12), and Mycobacterium smegmatis (13). DosR is controlled by two heme-containing sensor histidine kinases, DosS and DosT, that are responsive to redox potential, oxygen tension, nitric oxide, and carbon monoxide (14–20). The requirement for DosR activity in in vitro persistence models is well established (11–13, 21) but remains controversial in vivo (22, 23). Recent data indicate a role of the DosR regulon in adaptation to low-energy conditions, possibly via an effect on metabolism or at the level of biosynthesis through maintenance of ATP homeostasis and redox balance during hypoxia (11, 24, 25). However, the reason why loss of DosR leads to loss of viability during hypoxia in mycobacteria is not known.
To be able to re-emerge from the non-replicating persistent state, mycobacteria must maintain a level of macromolecular integrity compatible with sufficient functionality to resume growth, and it seems likely that stabilization of key cellular components, rather than de novo synthesis, will be an important factor in this. The bacterial ribosome is a 2.5-MDa complex comprising three RNA molecules and over 50 proteins made at great energetic expense to the cell. Given that in mycobacteria ribosomal genes are down-regulated concomitantly with the cessation of growth (26) and that there is limited chemical energy available to the non-growing cell (11, 24), it is probable that mycobacteria stabilize their ribosomes during prolonged stasis. The role of the stringent response in mycobacterial adaptation to nutrient stress survival, including hypoxia, supports this hypothesis (24, 27). RelA, in concert with CarD, mediates the down-regulation of ribosomal RNA synthesis and ribosomal protein genes during starvation (28). Consequently, ΔrelA M. tuberculosis has five times more ribosomes than wild-type bacteria in carbon-limited conditions, outlining the importance of this adaptation for the maintenance of sustainable energy levels (27).
Studies of ribosomal stability during the transition from active growth to stasis have been carried out primarily in Escherichia coli. When E. coli cells stop growing, part of the ribosomal population is degraded, whereas a proportion undergoes dimerization, leading to the formation of 100S ribosomal dimers that are not translationally active and are considered to be in a hibernation state (29–32). As such, ribosome stabilization could be important in mycobacterial persistence and, consequently, latent infection. We hypothesize that during non-replicating persistence and, consequently, during latent infection, mycobacterial ribosomes are stabilized to be activated and re-engage in translation upon resuscitation and exit from the persistent state. Therefore, we set out to investigate the stability of mycobacterial ribosomes, focusing on the hypoxic persistence model (33).
We demonstrate that, unlike enteric bacteria, mycobacterial ribosomes do not dimerize upon cessation of growth and that the dormancy regulator DosR controls ribosome stability during the hypoxic stationary phase. We present evidence for a dissociation-dependent process of ribosomal degradation and evidence to support a role for DosR control of ribosome stability in hypoxic mycobacteria through its control of a hypoxia-regulated, ribosome-associated protein.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Media
The strains used during this study are listed in supplemental Table S2. M. smegmatis was routinely cultured at 37 °C in LB medium (0.5% (w/v) NaCl, 0.5% (w/v) yeast extract, 1.0% (w/v) tryptone) supplemented with 0.05% (w/v) Tween 80. For carbon starvation and hypoxic growth experiments, the cells were grown in Hartmans-de Bont medium as described previously (13, 34, 35). Exponential (log) phase cells were harvested at A600 of 0.5. Stationary phase cells and cells grown under hypoxic or carbon-limiting conditions were harvested 48 h post-inoculation unless stated otherwise. For the heat shock experiments, cultures were grown into hypoxic stationary phase, and the cultures were transferred to a 55 °C incubator at 48 h post-inoculation. For culture on solid medium we added 2% (w/v) agar to LB medium prior to autoclaving. Bacterial viability was determined by plating 20-μl serial dilutions onto solid media, and growth was enumerated after 3 days.
RNA Isolation, Biosynthesis, and rRNA Profiling
Cells from 50 ml of bacterial culture were harvested by centrifugation (6000 × g, 10 min at 4 °C), and RNA was isolated using TRIzol as described previously (36). RNA concentration was determined spectrophotometrically. RNA synthesis was assessed by adding L-[5,6-3H]uracil (PerkinElmer Life Sciences) to a final concentration of 0.5 μCi/ml to cultures. The RNA was extracted as described above and quantified spectrophotometrically prior to adding 3 ml of UltimaGold scintillation liquid (PerkinElmer Life Sciences). Radioactivity was measured using a Wallac scintillation counter (PerkinElmer Life Sciences). RNA integrity was assayed using the Agilent RNA 6000 Nano Chip kit (Agilent) and analyzed by a 2100 Bioanalyzer instrument (Agilent). RNA samples were prepared for analysis according to the instructions of the manufacturer.
Crude Lysate Preparation and Ribosomal Profiling
Cultures to be grown into normoxic stationary phase were grown in 2.5-liter glass conical flasks (Pyrex) containing either 1 liter of Hartmans-de Bont or LB medium, hypoxic cultures were grown in flasks containing 1.7 liters of LB medium sealed with a tight-fitting rubber bung that has been perforated and modified to fit a 33-mm SubaSeal air-tight rubber septum (Sigma). Each bung was further sealed with silicone grease and wrapped in parafilm to minimize the diffusion of oxygen across the seal. At least 6 liters of culture were harvested by centrifugation (10,000 × g, 15 min at 4 °C), and the pellet was resuspended in ribosomal buffer (10 mm Tris-HCl (pH 7.4), 70 mm KCl, 10 mm MgCl2) at the concentration of 1.0 g wet weight/ml and lysed by a single passage through a French press (10,000 psi). After cell breakage, the cellular debris and unbroken cells were removed by centrifugation (30,000 × g, 30 min at 4 °C). Ribosomal profiling was performed as described previously (37).
FPLC Purification of Ribosomes
Ribosomes were purified as described previously (37) using an AKTA-FPLC liquid chromatography system (GE Healthcare) using a 0.6-ml mixer unit and either a 100-μl or 5-ml sample loop. Two quaternary amine convective interaction media (QA) monolithic discs (BIA separations) encased within a polyoxymethylene casing (BIA separations) were used for the separation.
iTRAQ4 Labeling of Ribosomal Peptides
The protein concentration in ribosomal samples was determined using a Coomassie Plus protein assay (Thermo). 100 μg of protein was acetone-precipitated and resuspended in 20 μl of dissolution buffer provided in the iTRAQ labeling kit (Applied Biosystems). The samples were labeled with the isobaric 8plex iTRAQ reagent (Applied Biosystems) according to the protocol of the manufacturer. Two FPLC-purified samples were analyzed for each of the following conditions during the compositional analysis: wild-type M. smegmatis log phase, wild-type carbon starvation, wild-type hypoxia, and ΔdosR hypoxia. Peptides were then purified using an iCAT cation exchange cartridge (Applied Biosystems) to remove chemicals that may interfere with mass spectrometry. The eluent was vacuum-dried in a SpeedVac, resuspended in 100 μl of distilled water, and used for mass spectrometric analysis.
Mass Spectrometric Analysis of iTRAQ-labeled Ribosomal Peptides
The samples were analyzed on an Applied Biosystems QTrap MS coupled to an Agilent 1100 LC stack, and the system was controlled using Analyst software (Version 1.5.1 AB SCIEX). The Agilent stack consisted of a binary pump, capillary pump, well plate autosampler, and a column oven with an integrated six-port valve. The system was configured to load samples onto a trap column (Agilent Zorbax SB, 5 μm × 0.3 mm × 35 mm) using the binary pump. The trap column was washed and then switched into the capillary flow. Peptides were separated on a capillary column (Agilent SB, 5 μm 0.3 mm × 150 mm column). The LC was interfaced to the MS with a turbo ion spray source. The loading/washing solvent was H2O containing 3% CH3CN and 0.3% COOH at a flow rate of 50 μl/min, and the resolving solvent was a gradient system of 0% B to 30% B over 90 min at a flow rate of 5 μl/min (A: 94.9% H2O, 5% CH3CN, 0.1% COOH; B: 94.9% CH3CN, 5% H2O, 0.1% COOH). The column oven was heated to 40 °C, and the valve was switched to direct the flow from the trap on to the resolving column after a 60-min wash. Typically, the MS parameters were set as follows: Curtain gas, 10 psi; GS1, 20 psi; GS2, 20 psi; interface heater on; TEM, 150 °C; and DP, 65.
Data were collected with an IDA method consisting of a survey scan (350 m/z to 1200 m/z), an enhanced resolution scan, and four enhanced product ion scans (100 m/z to 1500 m/z). Dynamic background subtraction was used prior to ion selection. The four most abundant doubly or triply charged ions were selected for the product ion scans. To increase the depth of sample coverage, multiple runs of the same sample in which the mass range of the initial survey scan was divided into distinct mass windows were performed. The mass ranges used were 325–400, 400–500, 500–550, 550–600, 600–650, 650–700, 700–800, 800–900, and 900–1200 m/z. The resulting spectra were analyzed and quantified by ProteinPilot software (version 2.0.1, revision 67476, Applied Biosystems/MDS SCIEX). Specifically, spectra were assigned with the Paragon algorithm (default parameters) using a custom FASTA protein database containing all predicted proteins encoded by the M. smegmatis genome and a collection of common contaminants (database released on 11/07/2007 by Applied Biosystems). Proteins were identified and quantified using the ProGroup algorithm (Unused ProtScore of less than 1.3–95% confidence). The quoted significance values were obtained with the inbuilt statistical analysis tool. ProteinPilot-calculated ratios of label abundance for the measured peptides were used to plot the distribution of peptides from the 30S and the 50S ribosomal subunits. The significance of the difference of the distributions was calculated using a two-tailed, unpaired Student's t test.
MSMEG_3935 and MSMEG_1878 detection Using Multiple Reaction Monitoring MS
Using the configuration described above but operating the QTrap in “Triple Quadrupole” mode, an multiple reaction monitoring acquisition method on the basis of the detection of specific mother ion → daughter ion transitions (see supplemental Table S4) was developed. Although maintaining the same solvents, the resolving gradient was modified to reduce the run time (0–40% B over 40 min, wash time reduced to 5 min). We tested six peptides identified in silico using the MIDAS Workflow Designer (AB SCIEX, see AB SCIEX Tech Note, 2007, Publication 114TN38-01) as possible candidates for multiple reaction monitoring analysis. Three unique peptides with good signal intensity and well defined chromatography were used for the analysis of each protein (shown in bold in supplemental Table S4). Only ions with ion transitions corresponding to these peptides were recorded. The data were analyzed with the Analyst software.
Construction of the MSMEG_3935 Mutant and Complementation
Mutant M. smegmatis strains were obtained following the protocol established as in Ref. 38. Briefly, a linear construct was generated so that it contained a hygromycin cassette flanked by ∼500 bp homologous to the sequence found up- and downstream of MSMEG_3935 in M. smegmatis. The construct was amplified by PCR and electroporated into M. smegmatis mc2155 cells harboring the pJV53 plasmid (see supplemental Table S2) and, following recovery, plated onto 7H10 medium containing hygromycin (20 μg/ml). Genomic DNA was purified from potential mutants (39) and assessed for the presence of the hygromycin cassette as well as the insertion of HygR into the correct genetic locus by PCR. The MSMEG_3935 mutant was complemented with a single copy of MSMEG_3935 under the control of a tetracycline-regulatable promoter. The regulatable vector pTETR2 was created on the basis of the system developed by Ehrt et al. (40, 41). The pMC1s and pME1sL1 vectors provided by Sabine Ehrt were adapted to include the tetracycline repressor and the gene of interest on a single integrating plasmid, and the tetracycline repressor was replaced with a codon-optimized version with improved gene silencing (40).
Complementation of ΔdosR in M. smegmatis
An M. smegmatis ΔdosR mutant (13) was complemented with the M. tuberculosis dosR under the control of its own promoter in the integrating pMV306 vector. The M. tuberculosis dosR gene was amplified from genomic DNA using the primers dosRF TCTAGAATAAGGACTAACGGCCCTCA and dosRR AAGCTTTCATGGTCCATCACCGGG, incorporating the restriction sites XbaI and HindIII, respectively, which were used in the subsequent cloning into pMV306 (42).
RESULTS
Survival and Biosynthesis during Long-term Hypoxia in M. smegmatis
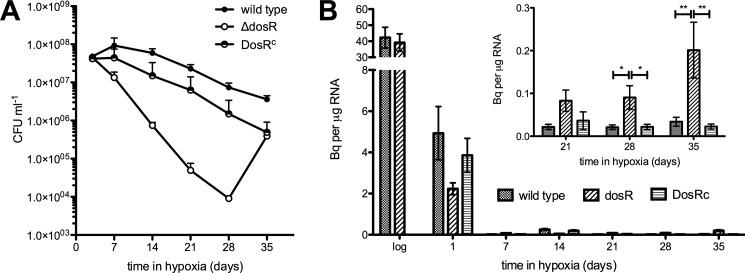
We determined the viability of the wild type, ΔdosR, and ΔdosR complemented with the dosR gene from M. tuberculosis (DosRc) strains of M. smegmatis during prolonged hypoxia and found that our results were consistent with those in the literature (13). ΔdosR had a severe survival defect in hypoxic conditions, and we were able to complement this phenotype with the dosR gene from M. tuberculosis (Fig. 1A), supporting the functional overlap between DosR from M. smegmatis and M. tuberculosis (8, 9, 13). The regrowth of the ΔdosR mutant after 5 weeks in hypoxic stasis is not well understood and seems to be specific to M. smegmatis, but it is not due to the acquisition of a suppressor mutation (13). Indeed, data suggest that ΔdosR mutant cells may spontaneously reactivate from a “non-culturable” dormant state (43). We estimated the biosynthetic activity of these strains by measuring [3H]uracil incorporation into RNA and found a decrease of over 90% early after the onset of hypoxic stasis (Fig. 1B). Given the long labeling period required for these experiments, these data are likely to reflect turnover as well as de novo synthesis The levels of incorporation thereafter were several orders of magnitude smaller than during active growth suggesting that metabolic activity of the culture as a whole is very low. Interestingly levels of incorporation were significantly higher in the ΔdosR strain compared with the wild type, in particular after 21 days of hypoxia. This increase was reversed in the complemented strain. This effect was even more pronounced when the average incorporation per colony forming unit was calculated (see supplemental Fig. S1).
FIGURE 1.
The effect of DosR on survival and RNA synthesis of M. smegmatis during hypoxic stationary phase. A, cultures of M. smegmatis mc2155 wild-type, ΔdosR, and ΔdosR/DosRc were grown into hypoxic stationary phase, and their viability was determined by plating at the times indicated. CFU, culture-forming units. B, RNA biosynthesis was measured by determining [3H]uracil incorporation for 24 h. Bq = becquerels. Each data point is the average of measurements from three independent cultures. Error bars are mean ± S.D. *, p < 0.05, Bonferroni post-test following two-way analysis of variance. **, p < 0.001, Bonferroni post-test following two-way analysis of variance.
DosR Is Needed for Ribosomal Stability during Hypoxia in M. smegmatis
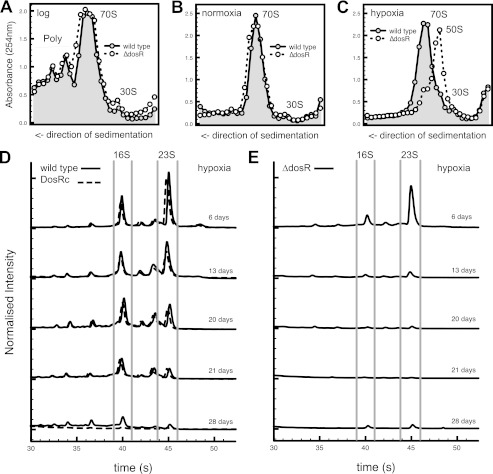
We carried out ribosomal profiling of the wild type and ΔdosR M. smegmatis during exponential growth and normoxic and hypoxic stationary phase in LB medium (Fig. 2). The transition from growth to normoxic stasis was characterized by the disappearance of polysomes and the apparent stabilization of ribosomes in the associated form. No formation of 100S ribosome dimers was apparent. The profiles of the ΔdosR strain were comparable with the wild type and the DosRC strain during exponential growth and in normoxic stationary phase. In contrast, upon entry into hypoxic stationary phase, there were clear differences between the wild type and ΔdosR mutant strain (Fig. 2C). 70S ribosomes were much less abundant in the ΔdosR mutant strain, with the 50S subunit being the dominant species. Levels of the 30S subunit were very low in comparison with the 50S subunit. These data suggest a preferential loss of the 30S ribosomal subunit in hypoxic stationary phase. No evidence of higher-order ribosomal structures reminiscent of the 100S dimer of enteric bacteria was observed during hypoxia (30). We next analyzed the stability of rRNA during prolonged hypoxia in the wild type, ΔdosR, and DosRc strains using microfluidics-based separation. After 6 days in hypoxic stationary phase, both 16S and 23S rRNA are present and resolved (Fig. 2, D and E). After this time, we observed a progressive degradation of the rRNA pool in the wild type and DosRc. We were able to detect the presence of multiple intermediate RNA species (not observed during active growth) that could be indicative of discrete rRNA cleavage events. Specific stepwise breakdown of rRNA is documented in enteric bacteria (44). The degradation of rRNA in the ΔdosR strain occurred more rapidly, with the majority of the culture devoid of rRNA within the first 2 weeks of hypoxia. RNA fragmentation and degradation was specific to hypoxia. Prolonged stasis under carbon starvation conditions did not lead to significant loss of rRNA integrity in neither the wild type nor ΔdosR strains (supplemental Fig. S2). It is important to note that rRNA levels closely paralleled the changes seen in ribosome profiling (supplemental Fig. S3).
FIGURE 2.
Loss of DosR has a profound effect on ribosome and ribosomal RNA stability. Cultures of M. smegmatis mc2155 wild-type and the ΔdosR mutant were grown at 37 °C to mid-exponential phase (A), normoxic stationary phase (B), or hypoxic stationary phase (C). Cells were harvested, lysed, and ribosomes were isolated by ultracentrifugation and fractionated through a linear sucrose gradient (15–40%). Then the A254 and refraction were determined per fraction and plotted. D and E, for ribosomal RNA stability experiments, cultures were grown to hypoxic stationary phase at 37 °C in LB. RNA was purified using the TRIzol reagent and analyzed with RNA Nano 6000 chips on an Agilent 2100 bioanalyzer. Traces shown are the average of three independent experiments and were normalized to account for chip variability.
Ribosomal Subunit Ratio Is Significantly Altered by Stasis in M. smegmatis
In the absence of changes to the higher-order organization of ribosomes, we turned to protein compositional analysis to better understand the role that DosR might play in ribosome stability. Using quantitative proteomics (iTRAQ), we compared the composition of ribosomes obtained from actively growing, hypoxic (wild type and ΔdosR) and carbon-starved cultures of M. smegmatis. We were able to detect and quantify all but two ribosomal proteins: L31 and L34. Importantly, all ribosomal proteins were detected in all samples, indicating that complete loss of ribosomal proteins during stasis was unlikely.
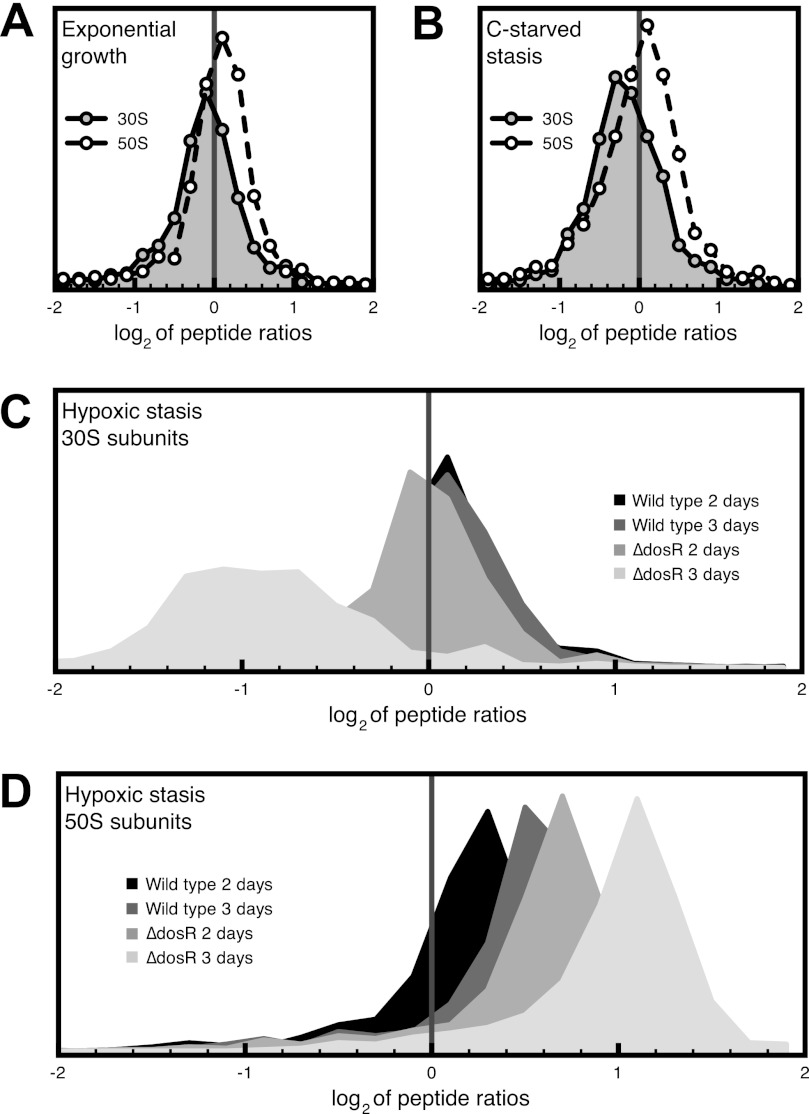
There was an increase in the relative abundance of large ribosome subunit (50S) proteins during hypoxic stasis when compared with active growth. Similarly, the relative levels of small subunit (30S) proteins were found to be lower during carbon starvation (supplemental Table S1). To assess the significance of this finding, we compared the ratios of all ribosomal peptides from stationary phase samples (stationary phase/exponential phase) to those of the control set (exponential phase/exponential phase). We only analyzed peptides that were identified with at least 95% confidence and were found to come from either 50S or 30S ribosomal subunits. By doing so, we were able to determine that overall, 50S subunit peptides were significantly more abundant in all hypoxic samples (Student's t test, p < 0.01) (Fig. 3). In the case of ribosomes from ΔdosR cultures harvested 3 days after the onset of hypoxic stasis, we were able to detect a significant decrease (2-fold) of 30S ribosomal peptides (Student's t test, p < 0.01) (Fig. 3C). Taken together, these observations pointed to a stasis-induced bias toward 50S subunits, which occurs more slowly in wild-type cultures than in the ΔdosR mutant. To illustrate this effect, we plotted the distribution of the logarithm of peptide ratios (treatment/exponential phase (Fig. 3)). We expected the logarithm of the ratios to be normally distributed and centered around the mean (μ) of 0 in the case where there is no difference between the two samples. An observed mean significantly different from 0 would be indicative of a difference in the levels of that subunit for those two samples. Of particular interest is the time-dependent progressive increase in the ratio of 50S peptides, which orders hypoxic samples as follows: μ = 0 < wild type (2 days of hypoxia) < wild type (3 days of hypoxia) < ΔdosR (2 days of hypoxia) < ΔdosR (3 days of hypoxia) < μ = 1.1.
FIGURE 3.
Hypoxic stasis results in the progressive degradation of the ribosome that is manifested by an imbalance in the 30S/50S ratio. The abundance of ribosomal peptides (confidence score of at least 95% in ProteinPilot) from each biological sample was compared with the corresponding abundance in control samples (actively growing M. smegmatis). The logarithms of the ratios of peptides corresponding to either 30S or 50S ribosomal subunits were pooled, and their distribution was plotted (data were split into 25 bins). A, comparison of ribosomal peptides from actively growing cells. B, carbon-starved ribosomes versus exponential phase ribosomes. C, hypoxic stasis of the indicated strain and time versus exponential phase for 30S ribosome peptides. D, hypoxic stasis of the indicated strain and time versus exponential phase for 50S ribosomal peptides.
The only exception to the trend of increasing relative abundance of large subunit proteins was protein L9, which was significantly underrepresented in all samples from hypoxic stasis (p < 0.01). Overall, iTRAQ analysis of ribosomes obtained from hypoxic stationary phase cells confirmed the 50S bias observed during ribosomal profiling of hypoxic cultures (Fig. 2).
We were able also to analyze the levels of known ribosomally associated proteins: elongation factor Tu and trigger factor. Both were significantly (p < 0.05) less abundant in ΔdosR hypoxic samples (2.40-fold and 3.91-fold, respectively) when compared with log phase cultures (supplemental Table S1). The same was true for wild-type hypoxic samples where EF-Tu and trigger factor were significantly (p < 0.05) less abundant (4.95-fold and 2.87-fold, respectively) when compared with active growth (supplemental Table S1). In addition to known ribosomally associated proteins, we detected two hypothetical proteins, MSMEG_1878 and MSMEG_3935, that associated exclusively with stationary phase ribosomes in wild-type bacteria. In silico sequence analysis indicates the presence of an S30AE (ribosomal protein S30AE/Sigma 54 modulation protein) domain in MSMEG_1878 and MSMEG_3935 (supplemental Fig. S4). The domain was predicted by Pfam (63) to be present between residues 32–131 (E value, 1.3 × 10−22) in MSMEG_1878 and between residues 12–99 (E value, 6.2 × 10−6) in MSMEG_3935. These are the only two S30AE proteins encoded by the M. smegmatis genome. Homologues of MSMEG_1878 are present in all mycobacterial genomes sequenced to date, including Mycobacterium leprae, where it is the sole S30AE protein. There is at least one MSMEG_3935 homologue in most sequenced mycobacterial genomes, with the exception of Mycobacterium abscessus, Mycobacterium avium, and M. leprae (45). Phylogenetic analysis of mycobacterial S30AE proteins points to the presence of two clusters (see supplemental Fig. S4). Group I contains MSMEG_1878 and the M. tuberculosis homologue Rv3241c and its members are closely related to S30AE domain proteins from other bacteria, except that the mycobacterial proteins have C-terminal extensions of >100 amino acids. Group II contains MSMEG_3935, and the M. tuberculosis homologue Rv0079 and the sequences of its members are less conserved outside of mycobacteria, apart from some firmicutes.
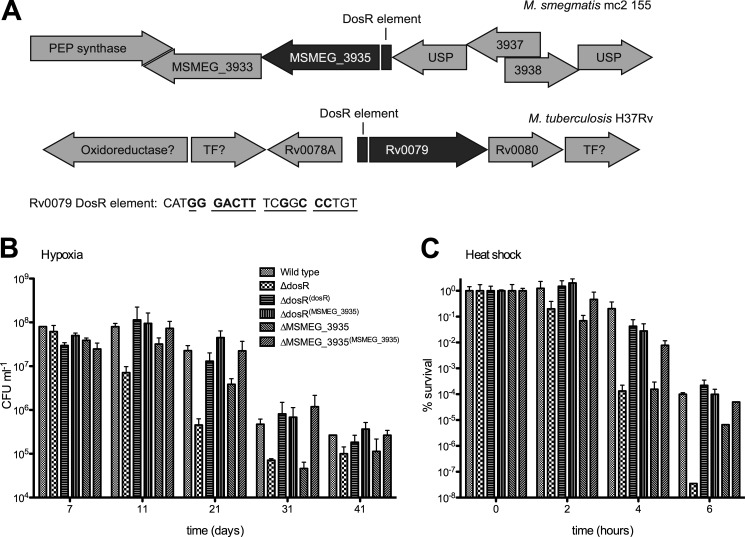
The low abundance of peptides derived from MSMEG_1878 and MSMEG_3935 in stationary phase samples, and their absence in exponentially growing cells precluded a quantitative and statistically significant analysis. Nonetheless, qualitative analysis of the apparent differences in their levels in samples obtained from wild-type and ΔdosR hypoxic cultures may be biologically very relevant. The M. tuberculosis homologue of MSMEG_3935, Rv0079, is a member of the DosR regulon. Importantly, MSMEG_3935 appears to have a putative DosR binding site 82 bp upstream of the start codon (Fig. 4) that is very similar to that of Rv0079 found 87 bp upstream of the start codon (9). This finding complements the observations presented in Fig. 2 and leads us to hypothesize that DosR plays a specific role in ribosomal stability via the action of MSMEG_3935.
FIGURE 4.
Loss of MSMEG_3935 is the major factor leading to the ΔdosR mutant phenotype. A, genetic context of MSMEG_3935 and Rv0079. PEP, synthase-phosphoenolpyruvate synthase; TF, transcription factor; USP, universal stress protein. The numbers indicate the standard annotation of genes whose function is unknown. The figure is not drawn to scale. MSMEG_3935 has a putative DosR-responsive element upstream of its start codon. The DosR response of element of MSMEG_3935 has a high degree of sequence identity of that of Rv0079 (underlined). Letters in bold are in agreement with the consensus determined by Park et al. (9). B, viability of the mc2155 wild type, ΔdosR, and ΔMSMEG_3935 mutants and the complemented strains ΔdosR(dosR), ΔdosR(MSMEG_3935), andΔMSMEG_3935(MSMEG_3935) during hypoxic stationary phase was determined. Triplicate cultures were grown in LB media + 0.05% Tween 80, and viability was determined by plating at the times indicated. Error bars represent mean ± S.D. CFU, colony-forming units. C, viability of the mc2155 wild type, ΔdosR, and ΔMSMEG_3935 mutants and the complemented strains ΔdosR/DosRc, ΔdosR/MSMEG_3935c, andΔMSMEG_3935/MSMEG_3935c during 55 °C heat shock was determined. Triplicate cultures were grown at 37 °C in LB media + 0.05% Tween 80 into hypoxic stationary phase. After 1 day in stationary phase, cultures were transferred to 55 °C, and viability was determined by plating at the times indicated. Error bars represent mean ± S.D.
S30AE Domain Proteins MSMEG_3935 and MSMEG_1878 Are Associated with the Mycobacterial Ribosome Exclusively during Stasis
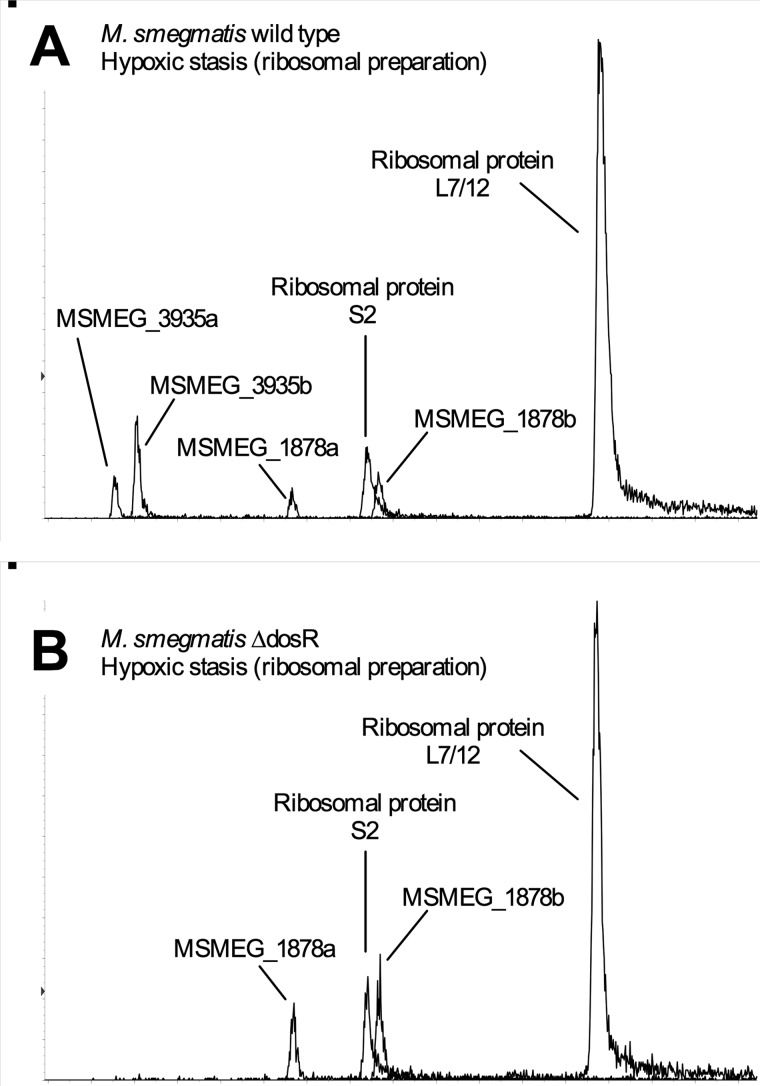
Because of the low signal obtained from MSMEG_1878 and MSMEG_3935 peptides during our iTRAQ analysis, we validated our results by using the more sensitive complementary approach of multiple reaction monitoring mass spectrometry. (46) Using this technique, which targets unique tryptic peptides from proteins, MSMEG_3935 and MSMEG_1878 were found not to associate with ribosomes from actively growing cells. The interaction of MSMEG_1878 was observed during both hypoxic and carbon-starved stationary phase, whereas MSMEG_3935 was detected to associate only with ribosomes from hypoxic cultures, and, crucially, this association was DosR-dependent, as no MSMEG_3935 could be detected in ribosomes isolated from ΔdosR during hypoxia. (Fig. 5) These data indicate that MSMEG_3935 is DosR-regulated. We considered the interesting possibility that the lack of induction of MSMEG_3935 in the ΔdosR mutant might explain the loss of viability of mycobacteria during hypoxic stationary phase.
FIGURE 5.
MS analysis using multiple reaction monitoring indicates that MSMEG_3935 is absent from hypoxic M. smegmatis ΔdosR cultures. MSMEG_3935 is absent from M. smegmatis ΔdosR ribosomes during hypoxia. Ribosomes from hypoxic stationary phase cultures of M. smegmatis wild type (A) and M. smegmatis ΔdosR (B) were isolated and analyzed by tandem mass spectrometry using the multiple reaction monitoring method. The transitions used to detect peptides from MSMEG_3935, MSMEG_1878, and ribosomal proteins L7/12 and S2 are listed in supplemental Table S4.
Loss of MSMEG_3935 is the major factor leading to the dosR mutant phenotype
We generated a ΔMSMEG_3935 strain and assayed the stability of its rRNA during prolonged hypoxia (see Fig. S5). The mutant exhibited decreased levels of 16S and 23S rRNA when compared with wild type. We wanted to establish whether MSMEG_3935 plays a role hypoxic survival of M. smegmatis and if so to determine its contribution to the hypoxia phenotype of ΔdosR. The ΔMSMEG_3935 mutant was complemented in trans by MSMEG_3935 expressed from a Tet regulatable promoter. We introduced the same construct into the ΔdosR mutant to determine the contribution of MSMEG_3935 to the ΔdosR mutant phenotypes (9–13). ΔMSMEG_3935 phenocopies the ΔdosR mutant during hypoxic stasis (Fig. 4), suggesting that loss of MSMEG_3935 is the major contributing factor to the hypoxic survival defect of ΔdosR mutant strains. Crucially, expressing MSMEG_3935 in trans in the ΔdosR mutant led to complete sparing of its hypoxic survival defect, as the survival of the ΔdosR/3935 strain was not significantly different from DosRc or the wild-type M. smegmatis.
Although not tested in other mycobacteria, in M. smegmatis, loss of dosR leads to increased sensitivity to heat shock at 55 °C (13). When tested, ΔMSMEG_3935 proved to be equally sensitive to heat shock as ΔdosR. Consistently with the rescue of hypoxic survival, expressing MSMEG_3935 in trans can complement the heat shock phenotype of ΔdosR, suggesting that MSMEG_3935 mediates the known in vitro phenotypes of the ΔdosR mutant (Fig. 4C).
DISCUSSION
The marked decrease in RNA biosynthesis and turnover in hypoxic cultures (Fig. 1B, inset) indicated that in the absence of biosynthesis the mycobacterial cell would need to rely on stabilization to maintain its complement of RNA molecules. In fact, stabilization of mycobacterial mRNA has been reported during stasis (34). The ΔdosR cells show a higher level of [3H]uracil incorporation than the wild type or DosRc during hypoxia, suggesting higher RNA turnover. The physiological implications include an increased level of energy consumption as well as a possible decrease in the stability of RNA. Both could explain the ΔdosR hypoxic phenotype. The importance of nucleic acid turnover to maintain hypoxic survival is supported by the finding that a purF mutant, defective in de novo purine synthesis, has a hypoxic survival phenotype that is remarkably similar to a dosR mutant (13, 35).
In enteric bacteria and Staphylococcus aureus ribosome stabilization occurs through the formation of 100S ribosomal dimers (30, 47), but such higher-order ribosome structures, including ribosome dimers, do not form in mycobacteria (Fig. 2). We identified the associated ribosome as the predominant species during normoxic, hypoxic, and carbon-starved stasis, suggesting that stabilization occurs at the level of 70S monomers in mycobacteria. There is a time-dependent depletion of the M. smegmatis ribosomes during hypoxic stasis. It seems that the degradation occurs first by the dissociation of 70S ribosomes, with a coinciding accumulation of the 50S but not the 30S subunit, suggesting that the small subunit is more susceptible to degradation. This process continues until no 70S ribosomes can be detected and only a small proportion of 50S remains. This course of events may reflect a more general mechanism, as the same sequence of changes has been observed during many stress conditions, leading to ribosomal degradation in E. coli (44, 48–50). In all these studies, 16S rRNA appeared to be less stable than 23S rRNA, leading to a disparity in the level of subunits that resulted from the dissociation of 70S ribosome. The “50S subunit bias” was particularly evident from our quantitative proteomics analysis. There was a clear correlation between the relative increase in the levels of the 50S subunit and the time cells were in hypoxic stasis (Fig. 3D). This effect was particularly pronounced in the ΔdosR mutant, which was found to undergo faster ribosome destabilization during hypoxia using ribosomal profiling, rRNA profiling, and compositional analysis. These observations provide significant evidence in support of ribosomal association as the major mechanism of ribosomal stability in stationary phase M. smegmatis.
Furthermore, we observed that hypoxic ribosomes contained significantly less ribosomal protein L9 than ribosomes from actively growing cells. L9 protrudes from the ribosome structure (51) and interacts with mRNA, thus preventing mistranslation because of mRNA slippage (52). The levels of the two ribosomally associated factors EF-Tu (needed to shuttle aminoacylated-tRNA (53)) and Trigger factor (a chaperone for nascent proteins (54)) were also found to be lower during hypoxia than during active growth. We do not know at present whether the absence of L9 from hypoxic cultures is a result of lowered expression or its reduced incorporation into or its selective removal from the ribosome. Nevertheless, the absence of L9 would lead to a severe decrease in the translational fidelity of the ribosome, whereas the reduction of associated EF-Tu and Trigger factor indicate reduced translational activity. An additional possibility is that thus modified ribosomes enhance the translation of mRNAs required for survival during prolonged hypoxia (22). An analogous mechanism to the stress-induced cleavage of 16S rRNA by MazF leads to an increased affinity of modified ribosomes for leaderless mRNAs, thus presenting a new paradigm of posttranscriptional regulation of gene expression (55). Although further work is required to validate the selective translation hypothesis, it does seem possible that in non-growing mycobacteria, associated 70S ribosomes have a decreased level of translational activity. A model for ribosome stabilization in M. smegmatis on the basis of the data presented here is shown in Fig. 6. Overall, unlike E. coli or the Gram-positive bacterium S. aureus, M. smegmatis does not resort to ribosomal dimerization for stability but, rather, stabilizes associated 70S ribosomes.
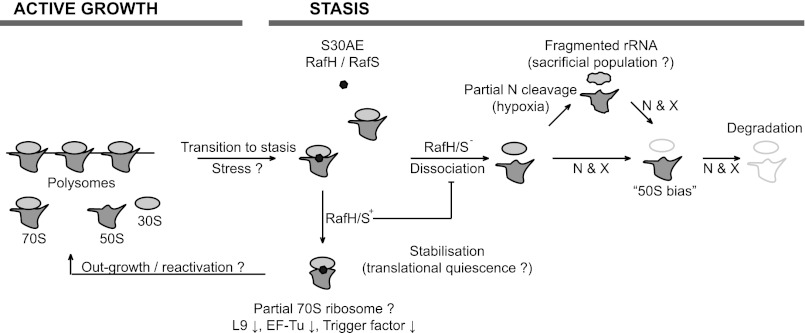
FIGURE 6.
Model of ribosomal stability during stasis. Active growth is characterized by the presence of actively translating ribosomes present either as 70S monomers or polysomes or free 50S and 30S subunits as they progress through the ribosomal cycle. Upon the cessation of growth (or exposure of stress), the S30AE domain proteins (RafH and RafS) become ribosome-attached and promote the associated form of the ribosome (70S), therefore leading to its stabilization and perhaps translational quiescence. We suggest that the S30AE-bound subpopulation of ribosomes is protected to allow the outgrowth of bacteria following stasis. Ribosomes that are not bound by S30AE proteins are more likely to expose the interface of the subunits, which serves as the substrate for endo- and exoribonucleases. Cleavage is likely to occur in a specific manner, leading to the fragmentation of rRNA (see Fig. 2). Although ribosomes may continue to exist with fragmented rRNA, perhaps as a “sacrificial population” to carry out the translation necessary to survive prolonged stasis, they eventually begin to disappear, leading to the complete degradation of the ribosome, breaking down first the 30S subunit leading to a 50S bias followed, finally, by the loss of the 50S subunit. S30AE, denotes RafH (MSMEG_3935) or RafS (MSMEG_1878); N, specific endonucleases; X, specific exonucleases; L9, large ribosomal subunit protein L9; EF-Tu, elongation factor Tu.
Loss of DosR has a destabilizing effect on the ribosome and rRNA during hypoxia (Figs. 2 and 3). The stability of rRNA is closely related to viability. Davis et al. (48) stipulated that ribosomal degradation during starvation is not the result of cell death but rather its cause. Our data show that rRNA stability and viability are interdependent and that the loss of one leads to the loss of the other. In the case of hypoxia, viability falls simultaneously with the loss of rRNA stability. However, the exact causality is not clear. Changes in the composition of ribosomes during hypoxia provide important clues to a direct role for DosR in ribosomal stability. ΔdosR ribosomes contain the entire complement of integral ribosomal proteins. Therefore, the apparent loss of stability is unlikely to be intrinsic to the ribosomal particle. This is an important conclusion, as impaired ribosomal subunit assembly is known to lead to a decrease in the stability of the entire ribosome (56). Importantly, the “50S bias” and rRNA fragmentation both occur in ΔdosR during hypoxia just as in the wild-type strain. However, they do so more rapidly and to a greater extent. During hypoxia, ΔdosR ribosomes are therefore subject to the same processes as wild-type ribosomes. However, they seem to be more susceptible to these destabilizing factors than their wild-type counterparts. As association is pivotal for the maintenance of ribosomal stability, the loss of an association-promoting interaction could lead to loss of stability. We identified MSMEG_3935 as a possible mediator of ribosomal association. Genetic deletion of this member of the DosR regulon led to reduced viability during prolonged hypoxia as well as an apparent decrease in rRNA stability. It is important to note that not all single deletions of DosR regulon genes lead to the same phenotype. Individual universal stress protein deletions were shown to have no effect on hypoxic survival in M. tuberculosis (57). Studies of the DosR regulon have to date provided little or no clear mechanistic insight into the roles of DosR regulon proteins. The reason why loss of DosR leads to loss of hypoxic viability in mycobacteria is not known. However, the remarkable phenotype of the ΔMSMEG_3935 mutant provides an explanation. Both with respect to hypoxic survival and heat shock, the ΔMSMEG_3935 mutant phenocopies the ΔdosR mutant. Furthermore, providing MSMEG_3935 in trans fully restores the wild-type phenotypes to ΔdosR. These data indicate convincingly that loss of MSMEG_3935 is responsible for the phenotypes of ΔdosR in M. smegmatis and, we argue, also in M. bovis BCG and M. tuberculosis.
S30AE domain proteins have been best studied in E. coli where they are involved in ribosomal function and stability. The two S30AE domain proteins of E. coli are cold shock protein Y, whose induction interferes with the ribosome cycle and stabilizes the 70S ribosome (58), and hibernation-promoting factor, which is essential for the maturation of 90S ribosomal particles to 100S dimers during stasis (31). The action of both E. coli S30AE proteins is to promote the inherently more stable associated form of the ribosome (44), hence they can be thought of as ribosome-stabilizing factors.
Hibernation-promoting factor and protein Y in E. coli and light-repressed transcript A (LrtA) in the cyanobacterium Synechococcus PCC 7002 (another S30AE domain protein) are de facto inhibitors of translation (31, 59, 60). Their expression and ribosomal association is induced during periods that are not favorable to bacterial growth. Their activity would allow the cell to control translational activity without having to alter the ribosomal pool at great biosynthetic and energetic cost. Do MSMEG_3935 and MSMEG_1878 share this function? Their expression pattern during stasis would support this hypothesis. MSMEG_3935 and its homologue in M. tuberculosis, Rv0079, are induced during hypoxia (9, 61), and MSMEG_1878 is expressed more highly during carbon-limited conditions in M. smegmatis (61), as is its homologue in M. tuberculosis, Rv3241c (62). We have established during this study that they do associate to the ribosome exclusively during stasis: hypoxic stasis in the case of MSMEG_3935 and both hypoxic and carbon-starved stasis in the case of MSMEG_1878. Furthermore, loss of MSMEG_3935 has a detrimental effect on survival during hypoxia. It is worthwhile noting that the detected levels of MSMEG_3935 and MSMEG_1878 do not match those of the ribosomal proteins to which they associate. Therefore, MSMEG_3935/MSMEG_1878-associated ribosomes account for only a proportion of the total ribosomal population. The direct action of either of these proteins on ribosomal activity remains to be explored further. The role of MSMEG_1878 remains to be determined, but its function in M. smegmatis is probably distinct, as MSMEG_1878, although associated with the ribosome during hypoxia, does not compensate for the absence of MSMEG_3935. For this reason, we suggest to rename MSMEG_3935 to RafH (ribosome-associated factor during hypoxia) and MSMEG_1878 to RafS (ribosome-associated factor during stasis).
This work was supported by the Wellcome Trust.

This article contains supplemental Figs. S1–S5 and Tables S1–S4.
- iTRAQ
- Isobaric tag for relative and absolute quantitation
- iCAT
- isotope coded affinity tag
- MIDAS
- multiple reaction monitoring initiated detection and sequencing
- IDA
- information dependent acquisition.
REFERENCES
- 1. Vynnycky E., Fine P. E. (1997) The annual risk of infection with Mycobacterium tuberculosis in England and Wales since 1901. Int. J. Tuberc. Lung Dis. 1, 389–396 [PubMed] [Google Scholar]
- 2. Russell D. G. (2007) Who puts the tubercle in tuberculosis? Nat. Rev. Microbiol. 5, 39–47 [DOI] [PubMed] [Google Scholar]
- 3. Lillebaek T., Dirksen A., Vynnycky E., Baess I., Thomsen V. Ø., Andersen A. B. (2003) Stability of DNA patterns and evidence of Mycobacterium tuberculosis reactivation occurring decades after the initial infection. J. Infect. Dis. 188, 1032–1039 [DOI] [PubMed] [Google Scholar]
- 4. Via L. E., Lin P. L., Ray S. M., Carrillo J., Allen S. S., Eum S. Y., Taylor K., Klein E., Manjunatha U., Gonzales J., Lee E. G., Park S. K., Raleigh J. A., Cho S. N., McMurray D. N., Flynn J. L., Barry C. E., 3rd. (2008) Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76, 2333–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barry C. E., 3rd, Boshoff H. I., Dartois V., Dick T., Ehrt S., Flynn J., Schnappinger D., Wilkinson R. J., Young D. (2009) The spectrum of latent tuberculosis. Rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wayne L. G., Sohaskey C. D. (2001) Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55, 139–163 [DOI] [PubMed] [Google Scholar]
- 7. Timm J., Post F. A., Bekker L. G., Walther G. B., Wainwright H. C., Manganelli R., Chan W. T., Tsenova L., Gold B., Smith I., Kaplan G., McKinney J. D. (2003) Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. U.S.A. 100, 14321–14326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sherman D. R., Voskuil M., Schnappinger D., Liao R., Harrell M. I., Schoolnik G. K. (2001) Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. U.S.A. 98, 7534–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park H. D., Guinn K. M., Harrell M. I., Liao R., Voskuil M. I., Tompa M., Schoolnik G. K., Sherman D. R. (2003) Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Voskuil M. I., Visconti K. C., Schoolnik G. K. (2004) Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis 84, 218–227 [DOI] [PubMed] [Google Scholar]
- 11. Leistikow R. L., Morton R. A., Bartek I. L., Frimpong I., Wagner K., Voskuil M. I. (2010) The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J. Bacteriol. 192, 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boon C., Dick T. (2002) Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184, 6760–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Toole R., Smeulders M. J., Blokpoel M. C., Kay E. J., Lougheed K., Williams H. D. (2003) A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J. Bacteriol. 185, 1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dasgupta N., Kapur V., Singh K. K., Das T. K., Sachdeva S., Jyothisri K., Tyagi J. S. (2000) Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 80, 141–159 [DOI] [PubMed] [Google Scholar]
- 15. Roberts D. M., Liao R. P., Wisedchaisri G., Hol W. G., Sherman D. R. (2004) Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279, 23082–23087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Honaker R. W., Leistikow R. L., Bartek I. L., Voskuil M. I. (2009) Unique roles of DosT and DosS in DosR regulon induction and Mycobacterium tuberculosis dormancy. Infect. Immun. 77, 3258–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ioanoviciu A., Yukl E. T., Moënne-Loccoz P., de Montellano P. R. (2007) DevS, a heme-containing two-component oxygen sensor of Mycobacterium tuberculosis. Biochemistry 46, 4250–4260 [DOI] [PubMed] [Google Scholar]
- 18. Kumar A., Toledo J. C., Patel R. P., Lancaster J. R., Jr., Steyn A. J. (2007) Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U.S.A. 104, 11568–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sousa E. H., Tuckerman J. R., Gonzalez G., Gilles-Gonzalez M. A. (2007) DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 16, 1708–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yukl E. T., Ioanoviciu A., de Montellano P. R., Moënne-Loccoz P. (2007) Interdomain interactions within the two-component heme-based sensor DevS from Mycobacterium tuberculosis. Biochemistry 46, 9728–9736 [DOI] [PubMed] [Google Scholar]
- 21. Voskuil M. I., Schnappinger D., Visconti K. C., Harrell M. I., Dolganov G. M., Sherman D. R., Schoolnik G. K. (2003) Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rustad T. R., Harrell M. I., Liao R., Sherman D. R. (2008) The enduring hypoxic response of Mycobacterium tuberculosis. PLoS ONE 3, e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rustad T. R., Sherrid A. M., Minch K. J., Sherman D. R. (2009) Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol. 11, 1151–1159 [DOI] [PubMed] [Google Scholar]
- 24. Gengenbacher M., Rao S. P. S., Pethe K., Dick T. (2010) Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology 156, 81–87 [DOI] [PubMed] [Google Scholar]
- 25. Rao S. P., Alonso S., Rand L., Dick T., Pethe K. (2008) The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 105, 11945–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reddy T. B., Riley R., Wymore F., Montgomery P., DeCaprio D., Engels R., Gellesch M., Hubble J., Jen D., Jin H., Koehrsen M., Larson L., Mao M., Nitzberg M., Sisk P., Stolte C., Weiner B., White J., Zachariah Z. K., Sherlock G., Galagan J. E., Ball C. A., Schoolnik G. K. (2009) TB database. An integrated platform for tuberculosis research. Nucleic Acids Res. 37, D499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Primm T. P., Andersen S. J., Mizrahi V., Avarbock D., Rubin H., Barry C. E., 3rd (2000) The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182, 4889–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stallings C. L., Stephanou N. C., Chu L., Hochschild A., Nickels B. E., Glickman M. S. (2009) CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138, 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilson D. N., Nierhaus K. H. (2004) The how and Y of cold shock. Nat. Struct. Mol. Biol. 11, 1026–1028 [DOI] [PubMed] [Google Scholar]
- 30. Wada A., Igarashi K., Yoshimura S., Aimoto S., Ishihama A. (1995) Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem. Biophys. Res. Commun. 214, 410–417 [DOI] [PubMed] [Google Scholar]
- 31. Ueta M., Ohniwa R. L., Yoshida H., Maki Y., Wada C., Wada A. (2008) Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J. Biochem. 143, 425–433 [DOI] [PubMed] [Google Scholar]
- 32. Yoshida H., Maki Y., Kato H., Fujisawa H., Izutsu K., Wada C., Wada A. (2002) The ribosome modulation factor (RMF) binding site on the 100S ribosome of Escherichia coli. J. Biochem. 132, 983–989 [DOI] [PubMed] [Google Scholar]
- 33. Wayne L. G., Hayes L. G. (1996) An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64, 2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smeulders M. J., Keer J., Speight R. A., Williams H. D. (1999) Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181, 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keer J., Smeulders M. J., Williams H. D. (2001) A purF mutant of Mycobacterium smegmatis has impaired survival during oxygen-starved stationary phase. Microbiology 147, 473–481 [DOI] [PubMed] [Google Scholar]
- 36. Rustad T. R., Roberts D. M., Reiling L. P., Sherman D. R. (2009) in Mycobacteria Protocols (Parish T. B., Brown A. C., eds.) pp. 13–23, Humana Press [Google Scholar]
- 37. Trauner A., Bennett M. H., Williams H. D. (2011) Isolation of bacterial ribosomes with monolith chromatography. PLoS ONE 6, e16273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Kessel J. C., Hatfull G. F. (2007) Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 [DOI] [PubMed] [Google Scholar]
- 39. Belisle J. T., Sonnenberg M. G. (1998) Isolation of genomic DNA from mycobacteria. Methods Mol. Biol. 101, 31–44 [DOI] [PubMed] [Google Scholar]
- 40. Klotzsche M., Ehrt S., Schnappinger D. (2009) Improved tetracycline repressors for gene silencing in mycobacteria. Nucleic Acids Res. 37, 1778–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ehrt S., Guo X. V., Hickey C. M., Ryou M., Monteleone M., Riley L. W., Schnappinger D. (2005) Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stover C. K., de la Cruz V. F., Fuerst T. R., Burlein J. E., Benson L. A., Bennett L. T., Bansal G. P., Young J. F., Lee M. H., Hatfull G. F. (1991) New use of BCG for recombinant vaccines. Nature 351, 456–460 [DOI] [PubMed] [Google Scholar]
- 43. Shleeva M., Mukamolova G. V., Young M., Williams H. D., Kaprelyants A. S. (2004) Formation of “non-culturable” cells of Mycobacterium smegmatis in stationary phase in response to growth under suboptimal conditions and their Rpf-mediated resuscitation. Microbiology 150, 1687–1697 [DOI] [PubMed] [Google Scholar]
- 44. Zundel M. A., Basturea G. N., Deutscher M. P. (2009) Initiation of ribosome degradation during starvation in Escherichia coli. RNA 15, 977–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K. F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. (2006) From genomics to chemical genomics. New developments in KEGG. Nucleic Acids Res. 34, D354–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Unwin R. D., Griffiths J. R., Whetton A. D. (2009) A sensitive mass spectrometric method for hypothesis-driven detection of peptide post-translational modifications. Multiple reaction monitoring-initiated detection and sequencing (MIDAS). Nat. Protoc. 4, 870–877 [DOI] [PubMed] [Google Scholar]
- 47. Ueta M., Wada C., Wada A. (2010) Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells 15, 43–58 [DOI] [PubMed] [Google Scholar]
- 48. Davis B. D., Luger S. M., Tai P. C. (1986) Role of ribosome degradation in the death of starved Escherichia coli cells. J. Bacteriol. 166, 439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. El-Sharoud W. M., Niven G. W. (2007) The influence of ribosome modulation factor on the survival of stationary-phase Escherichia coli during acid stress. Microbiology 153, 247–253 [DOI] [PubMed] [Google Scholar]
- 50. Niven G. W. (2004) Ribosome modulation factor protects Escherichia coli during heat stress, but this may not be dependent on ribosome dimerisation. Arch. Microbiol. 182, 60–66 [DOI] [PubMed] [Google Scholar]
- 51. Selmer M., Dunham C. M., Murphy F. V., 4th, Weixlbaumer A., Petry S., Kelley A. C., Weir J. R., Ramakrishnan V. (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 [DOI] [PubMed] [Google Scholar]
- 52. Atkins J. F., Björk G. R. (2009) A gripping tale of ribosomal frameshifting. Extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol. Biol. Rev. 73, 178–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krab I. M., Parmeggiani A. (1998) EF-Tu. A GTPase odyssey. Biochim. Biophys. Acta 1443, 1–22 [DOI] [PubMed] [Google Scholar]
- 54. Deuerling E., Schulze-Specking A., Tomoyasu T., Mogk A., Bukau B. (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400, 693–696 [DOI] [PubMed] [Google Scholar]
- 55. Vesper O., Amitai S., Belitsky M., Byrgazov K., Kaberdina A. C., Engelberg-Kulka H., Moll I. (2011) Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Uicker W. C., Schaefer L., Koenigsknecht M., Britton R. A. (2007) The essential GTPase YqeH is required for proper ribosome assembly in Bacillus subtilis. J. Bacteriol. 189, 2926–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hingley-Wilson S. M., Lougheed K. E., Ferguson K., Leiva S., Williams H. D. (2010) Individual Mycobacterium tuberculosis universal stress protein homologues are dispensable in vitro. Tuberculosis 90, 236–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vila-Sanjurjo A., Schuwirth B. S., Hau C. W., Cate J. H. (2004) Structural basis for the control of translation initiation during stress. Nat. Struct. Mol. Biol. 11, 1054–1059 [DOI] [PubMed] [Google Scholar]
- 59. Agafonov D. E., Kolb V. A., Spirin A. S. (2001) Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2, 399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tan X., Varughese M., Widger W. R. (1994) A light-repressed transcript found in Synechococcus PCC 7002 is similar to a chloroplast-specific small subunit ribosomal protein and to a transcription modulator protein associated with sigma 54. J. Biol. Chem. 269, 20905–20912 [PubMed] [Google Scholar]
- 61. Berney M., Cook G. M. (2010) Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS ONE 5, e8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Betts J. C., Lukey P. T., Robb L. C., McAdam R. A., Duncan K. (2002) Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731 [DOI] [PubMed] [Google Scholar]
- 63. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]