Abstract
Rationale
Mitochondria are semi-autonomous cellular organelles with their own genome, which not only supply energy, but also participate in cell death pathways. MicroRNAs (miRNAs) are usually 19–25 nt long, non-coding RNAs, involved in post-transcriptional gene regulation by binding to the 3′-untranslated regions (3′-UTRs) of target mRNA, which impact on diverse cellular processes.
Objective
To determine if nuclear miRNAs translocate into the mitochondria and regulate mitochondrial function with possible pathophysiological implications in cardiac myocytes.
Methods and Results
We find that miR-181c is encoded in the nucleus, assembled in the cytoplasm, and finally translocated into the mitochondria of cardiac myocytes. Immunoprecipitation of Argonaute 2 from the mitochondrial fraction indicates binding of cytochrome c oxidase subunit 1 (mt-COX1) mRNA from the mitochondrial genome with miR-181c. Also, a luciferase reporter construct shows that mi-181c binds to the 3′UTR of mt-COX1. To study whether miR-181c regulates mt-COX1, we overexpressed precursor miR-181c (or a scrambled sequence) in primary cultures of neonatal rat ventricular myocytes (NRVMs). Overexpression of miR-181c did not change mt-COX1 mRNA but significantly decreased mt-COX1 protein, suggesting that miR-181c is primarily a translational regulator of mt-COX1. In addition to altering mt-COX1, over-expression of miR-181c results in increased mt-COX2 mRNA and protein content, with an increase in both mitochondrial respiration and ROS generation in NRVMs. Thus, our data show for the first time that miR-181c can enter and target the mitochondrial genome, ultimately causing electron transport chain complex IV remodeling and mitochondrial dysfunction.
Conclusions
Nuclear miR-181c translocates into the mitochondria and regulates mitochondrial genome expression. This unique observation may open a new dimension to our understanding of mitochondrial dynamics and the role of miRNA in mitochondrial dysfunction.
Keywords: microRNA, mitochondria, cytochrome oxidase, ROS
INTRODUCTION
Mitochondria, in addition to supplying energy, are also involved in many other essential cellular processes, such as signaling, cellular differentiation, growth and cell death 1, 2. Over two thousand proteins are found in the mitochondria but the mitochondrial genome codes for only 13 proteins 3. The majority of mitochondrial proteins are products of nuclear genes and are synthesized in the cytosol, then translocated into the mitochondria 3, 4.
miRNAs are small noncoding RNA molecules that regulate gene expression by inhibiting mRNA translation and/or inducing mRNA degradation 5, 6. Several groups have proposed that miRNAs play critical roles in cardiovascular physiology and disease pathogenesis 7–9. Powerful cardioprotective interventions, such as ischemic preconditioning, also induce changes in miRNAs 10, 11.
The goal of this study was to determine if miRNA, like protein, could translocate into the mitochondria and regulate mitochondrial function with possible pathophysiological implications in cardiac myocytes. Others have found miRNA in the mitochondria of liver cells, HeLa cells, and human myoblasts 12–15, but the significance and functional consequences were not characterized.
MATERIALS AND METHODS
Animals
Sprague-Dawley rats were used in this study. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Johns Hopkins University.
RNA isolation
Hearts from adult male rats were perfused with RNAlater (Qiagen) after washing out the blood 16. Mitochondria were isolated by differential centrifugation as described previously16. RNA was extracted with miRNeasy kit (Qiagen) as per company’s instruction. miRNA enriched RNA were also isolated by Qiagen’s protocol from both heart and the mitochondrial fraction.
Microarray Analysis
Details are provided in the Supplemental Material section.
qRT-PCR
cDNA was made using the miScript Reverse Transcription Kit (Qiagen). PCR was performed using the miScript SYBR green PCR kit (Qiagen) and the iQ5 detector (Bio-Rad). All reactions were performed in triplicate. The data was analyzed following MIQE guidelines.
Cell Culture
NRVMs were isolated from the whole heart of 1- to 2-day-old rats. After isolation, myocytes were transfected using an electroporator (Nucleofector, Amaxa, Gaithersburg, MD) following the protocol for neonatal rat myocytes 17.
Immunoblot Analysis
After blotting gels onto PVDF membranes, membranes were incubated with antibodies that recognize Dicer, Prohibitin, mt-COX-1, Cadherin and alpha-actinin, all from Santa Cruz. The mt-COX2 antibody was from Invitrogen. Immunoreactive proteins were visualized using an enhanced chemiluminescence analysis kit (GE HealthCare).
Mitochondrial Respiration Assay
Mitochondrial respiration was measured polarographically with a Clark-type electrode 16. NRVMs were permeabilized with saponin, inside the sealed chamber and TMPD-Ascorbate was used as a complex IV specific substrate.
ROS Production Assay
H2O2 production from transfected intact NRVMs was monitored fluorimetrically by measurement of oxidation of Amplex red to fluorescent resorufin (Life Technologies, Carlsbad, CA).
RESULTS
Integrity and Purity of Mitochondrial miRNA
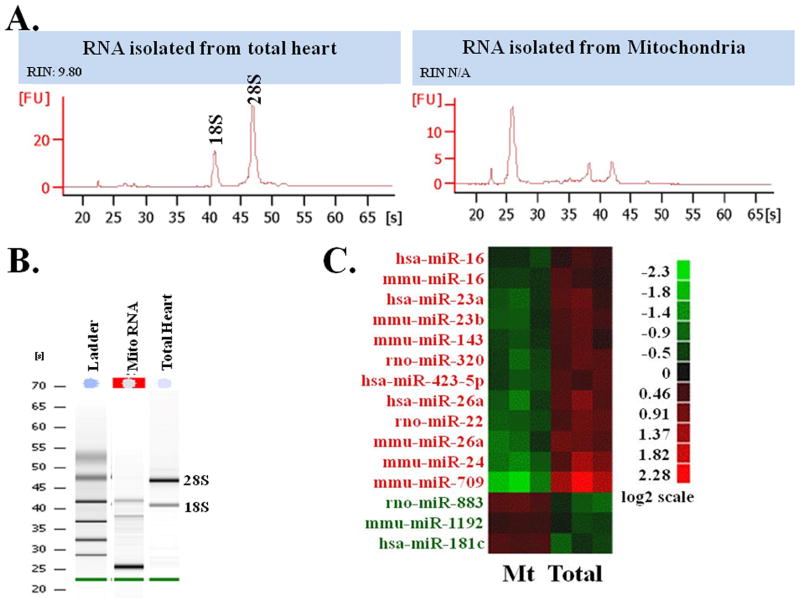
We perfused adult rat hearts with a RNA stabilization reagent, RNAlater, in a Langendorff apparatus 16 and differential centrifugation was carried out to isolate mitochondria 16. We isolated pure and high integrity RNAs from the mitochondria (Fig. 1A & B). Total RNA isolated from heart shows the 18S and 28S rRNA, and the absence of these peaks in the RNA isolated from mitochondria indicates that there is no measurable cytosolic RNA contamination in our mitochondria preparation. Figure 1B also shows that RNA from mitochondria is not degraded. Our RNA isolation procedure delivered a high RNA integrity number (RIN) for the total heart sample. Since the RIN number is calculated from cytosolic rRNA, there is no equivalent measure of mitochondrial RNA integrity, but the peaks in the mitochondrial RNA chromatogram are as sharp as in the total heart sample, suggesting that RNA integrity is similar in all fractions. We used a microarray approach to analyze the miRNA expression profile from total heart and from the mitochondrial fraction (Fig. 1C). Several miRNAs are highly expressed in the mitochondrial fraction compared to whole heart, and in particular, miR-181c was enriched 2-fold in the mitochondrial RNA compared to the total heart RNA (Fig. 1C). Figure 1C includes all miRNAs that are relevant to rat. In some cases the miRNAs were derived from other species, but if the human or mouse miRNA is conserved in the rat, then we included the conserved miRNA data even if it was originally derived from another species.
Figure 1. RNA isolation and identification of miRNA in heart-derived mitochondria.
(A) Capillary electrophoresis of total heart vs. mitochondrial fraction.
(B) Gel electrophoresis of mitochondrial RNA (marked in RED) vs. total heart.
(C) Heat Map representation of the miRNA microarray analysis in Mitochondrial Fraction (Mt) and Total Heart Homogenate (Total), using Affymetrix Chip. The log2 expressions of each gene are mean-centered, based on all the data from that gene.
Mitochondrial localization of miR-181c
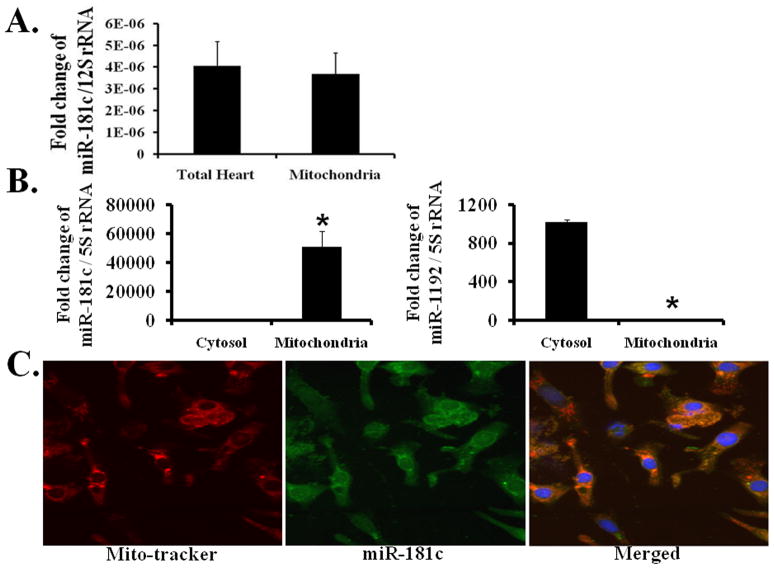
We validated our microarray data using qPCR. 12S rRNA, a mitochondrial gene product 1, served as our internal control. We confirmed that miR-181c was predominantly localized to the mitochondrial fraction; its expression in the mitochondrial fraction was similar to that in the total heart fraction (Fig. 2A). In addition, fluorescence in situ hybridization (FISH) (Fig. 2C) shows co-localization of miR-181c with the mitochondrial marker, mitoTracker red. We also examined miR-181c expression in both cytosolic and mitochondrial fractions (Fig. 2B, left panel), using qPCR and found that miR-181c was enriched primarily in the mitochondrial fraction. We evaluated several other miRNAs as negative controls for mitochondrial localization, and miR-1192 data are included in Fig. 2B (right panel). We used 5S rRNA as an internal control for these sets of qPCR data, as it is known that this gene is present in both the cytosol and mitochondrial fractions 18.
Figure 2. Mitochondrial localization of miR-181c.
qRT-PCR shows that miR-181c is mainly present in the mitochondria.
(A) miR-181c expression is almost the same in total RNA derived from the mitochondrial fraction and the total heart fraction. 12S rRNA is the internal control to normalize these data.
(B) miR-181c expression is mainly detected in the mitochondrial fraction and not in the cytosolic fraction (left panel), whereas miR-1191 is mainly present in the cytosolic fraction (right panel). 5S rRNA is the internal control to normalize these data, as this RNA is present in both cytosol and mitochondrial fractions. *p<0.05 vs. Cytosol.
(C) Fluorescent in situ hybridization demonstrating intramitochondrial localization of miR-181c. Mitochondria are labeled with mito-tracker red (left), miR-181c is green (GFP, middle), and the merged image is shown in the right panel.
Biogenesis of miR-181c
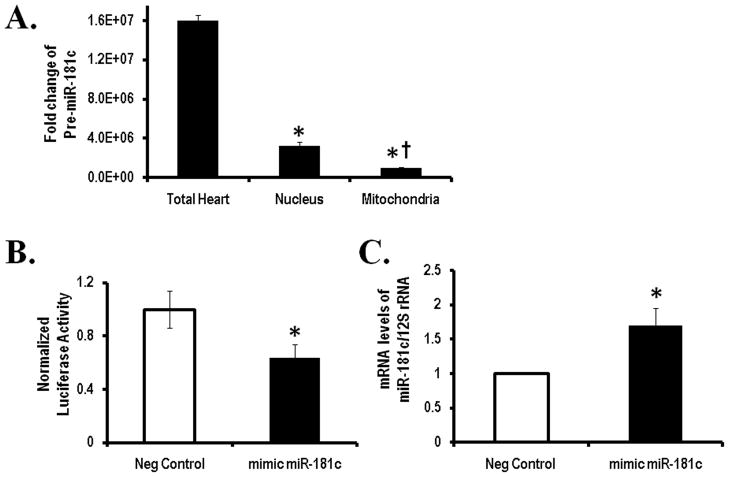
miRNAs derived from nucleus are transcribed as pri-miRNA, cleaved by Drosha to form pre-miRNA and then export from the nucleus to cytoplasm where the complex with Dicer leads to the mature form of miRNAs 5, 6. Since miR-181c is located on chromosome 19, we looked for pre-miR-181c in different cellular compartments to study this processing. We observed that there is little pre-miR-181c in the mitochondria or nuclear fractions compared to the total heart homogenate (Fig. 4A). This suggests that the pre-miR-181c is present predominantly in the cytosolic fraction, and largely absent from the nucleus or mitochondria (Fig. 4A). In addition, Dicer, which generates the mature form of miRNA, is only present in the cytosolic fraction and not in mitochondria (Fig. 3A). These data support the concept that biogenesis of miR-181c is by the conventional pathway using nuclear and cytosolic processing with Drosha and Dicer 5, 6. This is consistent with the hypothesis that after undergoing maturation within the cytosol, miR-181c translocates into the mitochondria.
Figure 4. Biogenesis and target of miR-181c.
(A) qRT-PCR shows the amounts of pre-miRNA-181c in the total heart sample vs. the different fractions. *p<0.05 vs. Total Heart and † p<0.05 vs. Nucleus.
(B) Luciferase activity derived from the COX-1 3′ UTR reporter following transfection into HeLa cells with negative or miR-181c mimic. All values were normalized to Renilla luciferase activity produced from a cotransfected control plasmid. *p<0.05 vs. Neg Control.
(C) Neonatal cardiomyocytes were transfected with precursor miR-181c or a scrambled sequence. qRT-PCR shows increased miR-181c after 48 hours. 12S rRNA served as the internal control to normalize these data. *p<0.05 vs. Neg Control.
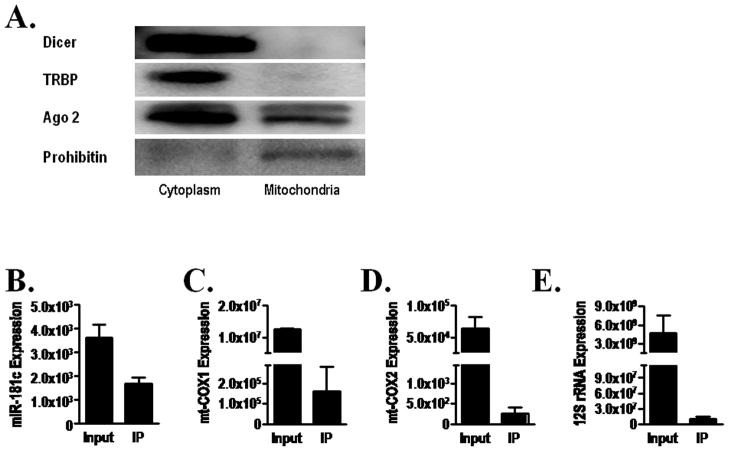
Figure 3. Active involvement of miR-181c in the mitochondrial RISC complex.
(A) Western blot shows that only Ago 2, and not Dicer or TRBP, is present in the mitochondria. Prohibitin is used as a mitochondrial marker.
qRT-PCR shows both (B) miR-181c and (C) mt-COX1 mRNA, and no other mitochondrial gene products, like (D) mt-COX2 mRNA or (E) 12S rRNA; in the Ago 2 immunoprecipitate from the mitochondrial fraction.
Mitochondrial Target of miR-181c
To determine mitochondrial targets of miR-181c, we looked for conventional cytosolic RISC components in the mitochondria and found that only the key component of RISC (RNA-Induced Silencing Complex), Argonaute 2 (Ago2) is present in the mitochondria (Fig. 3A). We then immunoprecipitated Ago2 from the mitochondrial pellet and found that miR-181c was co-immunoprecipitated (Fig. 3B). We also identified mt-COX1 mRNA in the immunoprecipitate (Fig. 3C) but no other mitochondrial RNAs, such as mt-COX2 mRNA or 12S rRNA were found (Fig. 3D and 3E respectively). This suggests that mt-COX1 is a primary target of miR-181c in the mitochondria. To determine the direct effect of miR-181c on the 3′UTR of mt-COX1 mRNA, we cloned the entire 3′UTR of mt-COX1 into the firefly luciferase (f-luc) reporter construct, pG13, and then transfected HeLa cells along with either a scrambled short RNA sequence or miR-181c, and evaluated luciferase activity 20. As shown in Fig. 4B, with the miR-181c mimic group, the luciferase activity significantly decreased, suggesting that the 3′UTR of mt-COX1 mRNA is a binding site of miR-181c.
miR-181c regulation of mt-COX1 translation
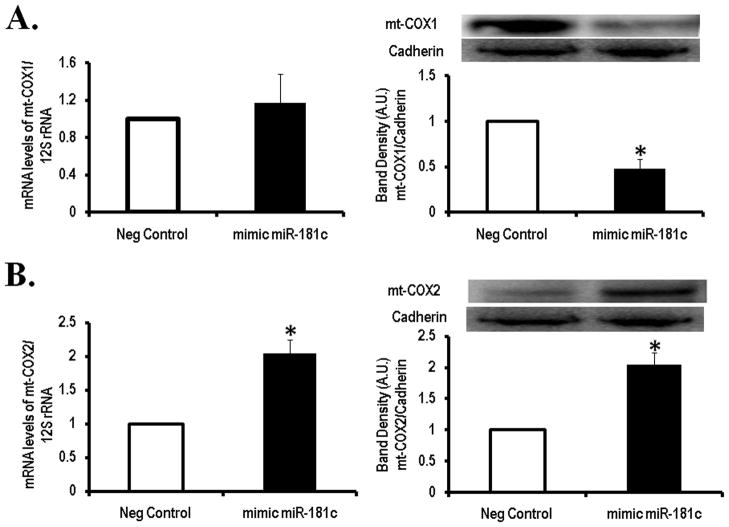
To examine the effects of miR-181c on mt-COX1 we used primary cultures of neonatal rat ventricular myocytes (NRVMs) and overexpressed miR-181c or a scrambled sequence as a control (Fig. 4C). Since it is known that miRNA can both inhibit mRNA translation and/or induce mRNA degradation 5, 6, we looked at both, the mRNA for mt-COX1 and protein expression. No difference was found in mt-COX1 mRNA level between cells overexpressing miR-181c and the control group, using qPCR (Fig. 5A, left panel). Then, we investigated mt-COX1 protein expression and found that overexpression of mir-181c decreased mt-COX1 content compared to control (Fig 5A, right panel). This indicates that miR-181c regulates translation of mt-COX1.
Figure 5. Target of miR-181c in the mitochondrial genome.
(A) Overexpression of miR-181c does not change the steady state level (left panel) of mt-COX1 mRNA but mt-COX1 protein is significantly reduced (right panel). Western blot of mt-COX1 and gel densitometry analysis is shown. Content of mt-COX1 was normalized to Cadherin and 12S rRNA was used as internal control for qRT-PCR. *<p 0.05 vs Neg Control. Mean ± SEM (n=4) are shown.
(B) Overexpression of miR-181c increased mt-COX2 mRNA (left panel) and protein expression (right panel) compared to negative control. Content of mt-COX2 protein was normalized to Cadherin and 12S rRNA was used as internal control for qRT-PCR. *<p 0.05 vs Neg Control. Mean ± SEM (n=4) are shown.
miR-181c effects on mitochondrial transcription
Mitochondrial DNA is a super coiled double-stranded closed circular molecule composed of 37 genes, encoding 2 ribosomal RNAs (12S and 16S rRNAs), 22 transfer RNAs, and 13 mitochondrial proteins 1, all of which are components of the oxidative phosphorylation (OXPHOS) system 1, 18. Mitochondrial RNA synthesis and processing is a highly regulated, coordinated, multi-step process, although not fully characterized 19. RNA synthesis of mitochondrial genes occurs through three transcriptional units, 1 L-strand and 2 H-strands (H1 and H2), cleaved from the D-loop region 18. All 13 OXPHOS mitochondrial genes are sequentially transcribed from the H2-strand 18. This polycistronic primary transcript is then processed by specific endonucleolytic cleavages. Since mt-COX2 and mt-COX3 are directly after mt-COX1 on the polycistronic primary transcript 18, we analyzed their mRNA content to evaluate if mir-181c would affect other OXPHOS components. Indeed, overexpression of miR-181c in NRVMs significantly increased mRNA of mt-COX2 and mt-COX3 (Fig. 5B, left panel and Online Figure VI, respectively). Thus, mir-181c not only regulates mt-COX1 protein translation, but also affects other cytochrome oxidase subunits.
The explanation for an increase in mRNA for mt-COX-2 and mt-COX-3 compared to mt-COX-1 could involve differences in mRNA stability, with an increased rate of degradation of mt-COX1 mRNA relative to mt-COX2 and mt-COX3 mRNA. One mechanism that contributes to mRNA stability is polyadenaylation at the 3′UTR of the cleaved mitochondrial mRNAs 20. Polyadenylation could be inhibited by miR-181c binding to the 3′UTR of COX1 mRNA, leading to accelerated degradation. Since mir-181C does not bind the 3″UTR of mRNA for COX2 and COX3 these two transcripts would not be affected. Recent findings support the role of cleavage and polyadenylation at the 3′UTR on post-transcriptional gene regulation 21, 22. Thus, increased synthesis of COX1 mRNA could be balanced by increased degradation, leading to no change in steady state level of message while increasing mRNA for COX2 and COX3.
We also performed cDNA microarray analysis to assess whether overexpression of miR-181c has other effects on gene expression (Online Figure IV). Although we did not find any significant change in nuclear gene expression, it is possible that there are some changes in the translational process of nuclear genes.
miR-181c causes Complex IV Remodeling
The terminal step in the electron transport chain is the transfer of electrons from cytochrome c, which is located in the intermembrane space, to oxygen to form water, a process that is catalyzed by cytochrome c oxidase (COX), which is complex IV of the electron transport chain. Complex IV is located in the inner mitochondrial membrane 1. mt-COX1, along with mt-COX2 and mt-COX3, are components of complex IV, and are encoded by the mitochondrial genome and synthesized in the mitochondria. Other components of complex IV and cytochrome c are encoded by nuclear genes, synthesized in the cytosol/ER and translocated into the mitochondria. The three mitochondrial DNA-encoded subunits represent the catalytic core of complex IV 23, 24. Since the mRNA for mt-COX2 was increased, we also examined the content of mt-COX2 protein, and as expected, the increase in mRNA was associated with an increase in protein content (Fig 5B). The interplay between mt-COX1, mt-COX2, and mt-COX3 are not yet known, but our data suggest complex IV remodeling. One possible explanation for these findings is that, by inhibiting mt-COX1 translation and reducing mt-COX1 content, miR-181c activates a feed-back loop that increases the transcription of the H2 strand. This increases the synthesis of mRNA for mt-COX1 and mt-COX2, but translational inhibition and enhanced degradation of the mt-COX1 mRNA prevents increased mt-COX1 protein expression whereas mt-COX2 processing is not similarly affected and protein expression increases. A previous study 24 of in-vivo ischemic stress in monkeys also demonstrated a relationship between decreased mt-COX1 and simultaneously increased mt-COX3, which ultimately impaired mitochondrial function. Our results are consistent with the hypothesis that miR-181c is affecting H2-strand transcription and mt-COX1 mRNA stability, as well as inhibiting translation of mt-COX1.
miR-181c regulates Mitochondrial Function
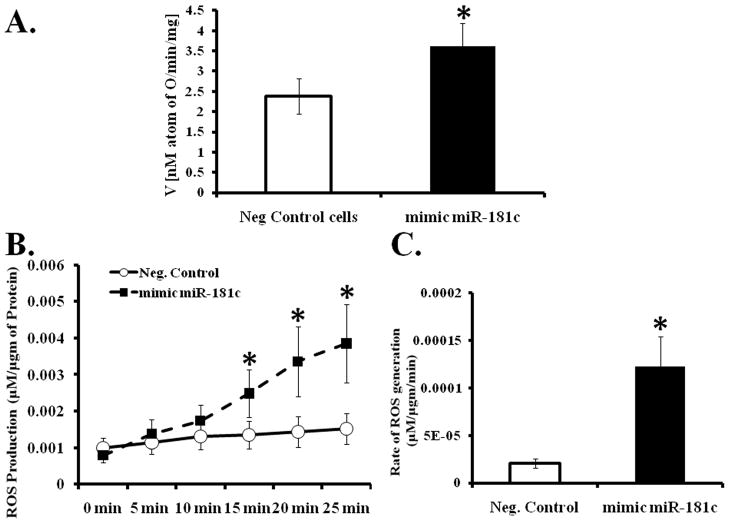
Both mt-COX1 and mt-COX2 are required for mitochondrial respiration 1 and a reduction in cytochrome oxidase activity could affect mitochondrial metabolism, and mitochondrial dysfunction is important in many diseases 18. To determine whether miR-181c regulates mitochondrial energy metabolism, 48 hours after transfection, we selectively permeabilized the plasma membrane of NRVM, and measured O2 consumption using a Clark-type oxygen electrode. To focus specifically on complex IV, we used the complex IV substrate, TMPD/Ascorbate. The rate of O2 consumption is significantly increased in miR-181c transfected NRVMs compared to the negative control group, after adding complex IV substrate (Fig. 6A). To further evaluate how miR-181c alters mitochondrial function, we considered that the increase in oxygen consumption could be related to increased ROS production 2. To test this hypothesis, we measured ROS production in the intact NRVMs using the fluorometric Amplex Red assay. As shown in Figure 6B and 6C, overexpression of miR-181c significantly increased the rate of ROS generation in NRVMs. The data suggest that the increase in oxygen consumption in miR-181c overexpressing NRVMs is due to increased ROS production.
Figure 6. Effect of miR-181c on mitochondrial function.
(A) Myocytes were transfected to overexpress miR-181c, or negative control. Cells were permeabilized and oxygen consumption measured after adding substrate for complex IV, TMPD/Ascorbate. (*p<0.05 vs. Neg Control, n=6)
(B) Time course of ROS generation from myocytes overexpressing miR-181c. (*p<0.05 vs. Neg Control., n=4).
(C) Over-expression of miR-181c significantly increases the rate of ROS production compared to the negative (Neg) control group. *p<0.05 vs. Neg Control n=4.
DISCUSSION
Our study shows for the first time, the existence and functional significance of a specific miRNA in heart-derived mitochondria, miR-181c. miR-181c originates from the nuclear genome, is processed in the cytosol, and translates to the mitochondria. We have also shown that miR-181c regulates mitochondrial energy metabolism by targeting mt-COX1 mRNA, which is a product of the mitochondrial genome. Overexpression of miR-181c results in a loss of mt-COX1 protein and an increase in mt-COX2 and mt-COX3, resulting in complex IV remodeling. The imbalance among the core components of complex IV results in increased ROS production. Thus perturbations induced by miR-181c could have important consequences in myocardial pathophysiology.
miRNAs are small non-coding RNAs, which inhibit the stability and/or translation of a mRNA by binding to the 3′UTRs of target mRNA by forming a multiprotein complex called the RNA-Induced Silencing Complex (RISC) 5, 6. Recently, miRNAs have been found to play a powerful role in various diseases 5, 6,8, 25. In 2005, it was shown that miR-1 plays an important role in the cardiovascular system25, 26. Over the past few years, there have been numerous studies that have pointed to a major role of these small, non-coding single-stranded RNAs in various heart diseases 7–9, 25, 27. Despite an enormous amount data suggesting a pivotal role for miRNA in the regulation of protein synthesis from nuclear genes, little is known about the role of miRNAs in regulating the mitochondrial proteome.
There are five respiratory chain complexes in the inner mitochondrial membrane which generate a proton gradient across the membrane and produce ATP 28. Most of the subunits which form the respiratory chain complexes are encoded by nuclear genes, except for some of the subunits of complex I, III and IV, which are encoded by mitochondrial DNA and synthesized on mitochondrial ribosomes 1, 18, 20. The most critical components of complex IV are the three mitochondrially encoded subunits of cytochrome c oxidase (mt-COX1, mt-COX2, and mt-COX3) and the remaining subunits (IV, Va, Vb, VIa, VIb, VIc, VIIa, VIIb, VIIc and VIII) are encoded by nuclear genes, synthesized in the cytosol, and imported into the mitochondria 1. The mitochondrially encoded subunits, mt-COX1, mt-COX2 and mt-COX3, form the catalytic core of complex IV, and the nuclear encoded subunits have a regulatory role 1. Electron transfer involves cytochrome c donating one electron at a time to CuA, which is bound to mt-COX2. CuA transfers the electron to cytochrome a in mt-COX1, which transfers the electron to heme a3 and CuB, both of which are located in mt-COX1, and then to oxygen to form water 24. As the terminal component of the electron transport chain, complex IV plays a critical role in mitochondrial function 1, 24, 18. The present study has found a very novel regulatory pathway which influences oxygen metabolism via complex IV remodeling, involving miR-181c which is encoded in the nucleus, assembled in the cytoplasm, and finally translocated into the mitochondria. Binding of miR-181c to mt-COX1 mRNA results in an imbalance among the mitochondrially encoded subunits in complex IV, promoting ROS generation.
One of the novel findings from this study is the translocation of miRNA from cytosol into the mitochondria. It has already been shown that RNA produced in the cytoplasm from a nuclear gene can translocate into the mitochondria29. In the same study, the authors concluded that the 5S rRNA resides primarily in the mitochondria once translocated from the cytosol, just like our finding that after maturation in the cytosol, miR-181c translocates into the mitochondria and is retained in that compartment. Our data further suggest that major effect of this translocation is that miR-181c forms a functional RISC complex, is a multi-protein complex that incorporates one strand of a miRNA 5, 6, inside the mitochondria with the key component of RISC, Ago 2, and the mRNA for mt-COX1. A similar compartmentation phenomenon has already been shown for miR-29b30. Because of the affinity of the localization sequence of miR-29b for a nuclear gene, miR-29b translocates into the nucleus from the cytosol and is retained in the nucleus; even though miR-29a and miR-29b are both synthesized in the cytosol 30.
Even though the transcription of mitochondrial DNA has been studied for more than 35 years, several aspects of this process are poorly understood. The process of mitochondrial RNA synthesis and processing is a highly-regulated, multi-step process with significant differences between transcription of nuclear genes and mitochondrial genes 1. The mitochondrial genome consists of a circular DNA molecule and RNA synthesis of mitochondrial genes occurs through three transcription units, 1 L-strand and 2 H-strands (H1 and H2), which are cleaved from the D-loop region 1,20. All the OXPHOS genes are sequentially transcribed from the H2-strand 1,18,20. The polycistronic primary transcript is then processed by specific endonucleolytic cleavage and mitochondrial mRNAs are then polyadenylated by adding ~55 adenylate bases to the 3′ end 31, 32. It has been shown that the polyadenylation process helps to increase mRNA stability 1. The secondary structure of the mitochondrial mRNAs can play an important role in the synthesis of that particular gene. For example, mt-COX3 has a CCA sequence in its 3′ end, and it was reported that the secondary structure controls the synthesis of mature mRNA for mt-COX3 33. Anchoring proteins, such as Pet309, Mss51, Pet 111, etc. then bind to the 5′ end of the mature mRNA and facilitate appropriate placement of the respiratory chain complexes in the inner mitochondrial membrane 18. Because of the polycistronic transcription process in the mitochondria, a microRNA can affect multiple proteins.
In this case, miR-181c in NRVM binds to the 3′UTR of mt-COX1 mRNA, and when miR-181c is overexpressed, mt-COX1 protein expression is reduced, due to either direct steric interference with protein synthesis or the polyadenylation process, which is necessarily to prevent rapid mRNA degradation. As a consequence of the decrease in mt-COX1 protein synthesis, mt-COX1 protein levels fall. This could stimulate a compensatory increase in the synthesis of the H2 strand and mt-COX1, and mt-COX2, and mt-COX3 mRNA synthesis would increase. Without a corresponding increase in the rate of degradation, mt-COX2 and mt-COX3 mRNA and protein would increase, resulting in complex IV remodeling. This would largely be a consequence of the polycistronic transcription process in the mitochondria.
In this case, miR-181c binds to the 3′UTR of mt-COX1 mRNA and inhibits its translation. The polyadenylation of mRNA has been shown to increase mRNA stability. Thus miR-181c could decrease the stability of mt-COX1 mRNA by inhibition of polyadenylation. The decrease in mt-COX1 protein synthesis could stimulate a compensatory increase in the mRNA synthesis of the H2 strand for mt-COX1, mt-COX2, and mt-COX3. While translation of mt-COX1 mRNA would still be inhibited by mir-181c, mt-COX2 and mt-COX3 protein would increase, resulting in complex IV remodeling. This would largely be a consequence of the polycistronic transcription process in the mitochondria.
Mitochondria play an important role in cardiovascular health. Energy production by the mitochondria is critical for cardiac function and mitochondrial ROS production, a byproduct of electron transport, can lead to cellular and mitochondrial dysfunction. Numerous recent studies have shown that increased ROS production can result from damage to electron transport chain components resulting in inefficient electron transfer among the five different complexes, generating superoxide which ultimately leads to mitochondrial dysfunction where mitochondrial oxygen consumption rate is significantly altered 34. It seems that the low levels of ROS generation are important in generating some cardioprotective signaling, whereas, high levels of ROS can lead to oxidative modification of enzymes and proteins and damage to mitochondrial electron transport chain components 34. Being the terminal respiratory complex, complex IV plays an important role in the transfer of electrons from cytochrome c oxidase to its bimetallic catalytic core and then to oxygen 1. This electron transfer reaction helps to generate the mitochondrial membrane potential by pumping protons out of the matrix, and ultimately driving ATP synthesis 1. The mitochondrial genes, mt-COX1, mt-COX2 and mt-COX3, are the components of this catalytic core of complex IV 1. The present study has found a very novel and interesting pathway which influences mitochondrial function by up-regulating ROS production via targeting mt-COX1 and complex IV remodeling. This could have important ramifications for many different cardiovascular disease processes.
SUMMARY AND CONCLUSIONS
Taken together, the results demonstrate that miR-181c has a role in mitochondrial metabolism after translocation by targeting the 3′-UTR region of mt-COX1 mRNA, which ultimately leads to complex IV remodeling. The present study reveals a very novel and interesting aspect of miRNA biology. It has already been shown that miRNA, specifically miR-29b, can translocate into the nucleus from the cytosol and is retained in the nucleus 30. Our study suggests a novel related concept, with translocation of miRNA derived from the nuclear genome into the mitochondria and regulating mitochondrial protein synthesis from mitochondrial genes. Over the past few years, there have been numerous studies that have pointed to the significant role of these small, non-coding single-stranded RNAs in various heart diseases 7–9, 25, 27. This study shows for the first time the existence of a specific miRNA in heart-derived mitochondria, miR-181c, and its effects on mitochondrial function and ROS generation, which could be major factors in various disease processes. Ultimately, better understanding of these processes may lead to the creation of innovative new therapies and diagnostics for heart disease.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
miRNAs have been shown to regulate the expression of ~30% of nuclear genes and to play a significant role in a variety of heart diseases.
Like proteins, RNAs, particularly rRNAs, can also translocate into the mitochondria.
miRNAs have been found in mitochondrial extracts of liver cells, HeLa cells, and human myoblasts.
What New Information Does This Article Contribute?
miRNA derived from the nuclear genome can translocate into mitochondria in the heart.
miRNA can regulate mitochondrial gene expression in cardiac myocytes.
miR-181c, expressed from the nuclear genome, can translocate into mitochondria and affect mitochondrial function and ROS generation by remodeling Complex IV.
Small non-coding RNA (microRNA) can regulate nuclear gene expression and play a major role in many different physiologic and pathologic processes. Although it has been suggested that miRNAs exist in non-heart mitochondria, it is not known whether miRNAs are also present in heart mitochondria, or whether mitochondrial miRNAs exert biological effects. We show here that miRNA derived from the nuclear genome, can translocate into the mitochondria in cardiac myocytes, regulate mitochondrial gene expression and affect mitochondrial function. We found that nuclear miR-181c can translocate into the mitochondria and affect the synthesis of proteins that are encoded by the mitochondrial genome. Increased miR-181c expression results in remodeling of complex IV and increased ROS production. These findings show the potential role of miRs in regulating cardiac function through influencing mitochondrial function.
Acknowledgments
We thank Dr. Peter Munson (Mathematical & Statistical Computing Laboratory, Center for Information Technology, NIH, Bethesda) for his generous advice on the microarray and statistical analysis.
Sources of Funding
This work was supported by a grant from the NIH, HL39752 (CS) and by a grant from the AHA, 0830395N (MF). RW, DL, NR, YY, and EM were supported by the NHLBI Intramural program.
Non-standard Abbreviations and Acronyms
- miRNA
micro RNA
- UTR
UnTranslated Regions
- mt-COX1
cytochrome c oxidase subunit 1
- mt-COX2
cytochrome c oxidase subunit 2
- mt-COX3
cytochrome c oxidase subunit 3
- NRVM
Neonatal Rat Ventricular Myocytes
- ROS
Reactive Oxygen Species
- MIQE
Minimum Information for Publication of Quantitative Real-Time PCR Experiments
- TMPD
N,N,N,N-tetramethyl-p-phenylidene-diamine
- H2O2
Hydrogen peroxide
- RIN
RNA Integrity Number
- qPCR
quantitative real time PCR
- FISH
Fluorescent in-situ hybridization
- RISC
RNA-induced silencing complex
- OXPHOS
oxidative phosphorylation
Footnotes
Disclosures
None
References
- 1.Chen JQ, Cammarata PR, Baines CP, Yager JD. Regulation of mitochondrial respiratory chain biogenesis by estrogens/estrogen receptors and physiological, pathological and pharmacological implications. Biochim Biophys Acta. 2009;1793:1540–1570. doi: 10.1016/j.bbamcr.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy E, Steenbergen C. What makes the mitochondria a killer? Can we condition them to be less destructive? Biochim Biophys Acta. 2011;1813:1302–1308. doi: 10.1016/j.bbamcr.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 4.Soriano ME, Scorrano L. Traveling Bax and forth from mitochondria to control apoptosis. Cell. 2011;145:15–17. doi: 10.1016/j.cell.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Sun H, Wang Y. Restriction of big hearts by a small RNA. Circ Res. 2011;108:274–276. doi: 10.1161/CIRCRESAHA.110.239426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kukreja RC, Yin C, Salloum FN. MicroRNA - New Players in Cardiac Injury and Protection. Mol Pharmacol. 2011;80:558–564. doi: 10.1124/mol.111.073528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin C, Salloum FN, Kukreja RC. A novel role of microRNA in late preconditioning: upregulation of endothelial nitric oxide synthase and heat shock protein 70. Circ Res. 2009;104:572–575. doi: 10.1161/CIRCRESAHA.108.193250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin C, Wang X, Kukreja RC. Endogenous microRNAs induced by heat-shock reduce myocardial infarction following ischemia-reperfusion in mice. FEBS Lett. 2008;582:4137–4142. doi: 10.1016/j.febslet.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bandiera S, Ruberg S, Girard M, Cagnard N, Hanein S, Chretien D, Munnich A, Lyonnet S, Henrion-Caude A. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One. 2011;6:e20746. doi: 10.1371/journal.pone.0020746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. PLoS One. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bian Z, Li LM, Tang R, Hou DX, Chen X, Zhang CY, Zen K. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 2010;20:1076–1078. doi: 10.1038/cr.2010.119. [DOI] [PubMed] [Google Scholar]
- 15.Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das S, Wong R, Rajapakse N, Murphy E, Steenbergen C. Glycogen synthase kinase 3 inhibition slows mitochondrial adenine nucleotide transport and regulates voltage-dependent anion channel phosphorylation. Circ Res. 2008;103:983–991. doi: 10.1161/CIRCRESAHA.108.178970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferlito M, Fulton WB, Zauher MA, Marban E, Steenbergen C, Lowenstein CJ. VAMP-1, VAMP-2, and syntaxin-4 regulate ANP release from cardiac myocytes. J Mol Cell Cardiol. 2010;49:791–800. doi: 10.1016/j.yjmcc.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montoya J, Lopez-Perez MJ, Ruiz-Pesini E. Mitochondrial DNA transcription and diseases: past, present and future. Biochim Biophys Acta. 2006;1757:1179–1189. doi: 10.1016/j.bbabio.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 19.Mick DU, Fox TD, Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat Rev Mol Cell Biol. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 21.Borowski LS, Szczesny RJ, Brzezniak LK, Stepien PP. RNA turnover in human mitochondria: more questions than answers? Biochim Biophys Acta. 2010;1797:1066–1070. doi: 10.1016/j.bbabio.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 22.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capaldi RA, Marusich MF, Taanman JW. Mammalian cytochrome-c oxidase: characterization of enzyme and immunological detection of subunits in tissue extracts and whole cells. Methods Enzymol. 1995;260:117–132. doi: 10.1016/0076-6879(95)60134-1. [DOI] [PubMed] [Google Scholar]
- 24.Wu C, Yan L, Depre C, Dhar SK, Shen YT, Sadoshima J, Vatner SF, Vatner DE. Cytochrome c oxidase III as a mechanism for apoptosis in heart failure following myocardial infarction. Am J Physiol Cell Physiol. 2009;297:C928–934. doi: 10.1152/ajpcell.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res. 2009;104:724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 27.Dorn GW., 2nd MicroRNAs in cardiac disease. Transl Res. 2011;157:226–235. doi: 10.1016/j.trsl.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuznetsov AV, Veksler V, Gellerich FN, Saks V, Margreiter R, Kunz WS. Analysis of mitochondrial function in situ in permeabilized muscle fibers, tissues and cells. Nat Protoc. 2008;3:965–976. doi: 10.1038/nprot.2008.61. [DOI] [PubMed] [Google Scholar]
- 29.Magalhaes PJ, Andreu AL, Schon EA. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol Biol Cell. 1998;9:2375–2382. doi: 10.1091/mbc.9.9.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 31.Bond GL, Prives C, Manley JL. Poly(A) polymerase phosphorylation is dependent on novel interactions with cyclins. Mol Cell Biol. 2000;20:5310–5320. doi: 10.1128/mcb.20.14.5310-5320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirsch M, Penman S. Post-transcriptional addition of polyadenylic acid to mitochondrial RNA by a cordycepin-insensitive process. J Mol Biol. 1974;83:131–142. doi: 10.1016/0022-2836(74)90384-2. [DOI] [PubMed] [Google Scholar]
- 33.Denslow ND, Michaels GS, Montoya J, Attardi G, O’Brien TW. Mechanism of mRNA binding to bovine mitochondrial ribosomes. J Biol Chem. 1989;264:8328–8338. [PubMed] [Google Scholar]
- 34.Murphy E, Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.