Decades of widespread clinical use of the bacterial ribosome A-site targeting aminoglycosides (AGs) enhanced the evolution of resistance to these antibiotics and reduced their clinical efficacy.[1] Three modes of action lead to bacterial resistance to AGs: reduction in the intracellular concentration of the antibiotics by efflux pump proteins or through reduced membrane permeability; structural modifications of the 16S ribosomal RNA leading to reduced target affinity; and deactivation by AG-modifying enzymes (AMEs).[1c, 2] AMEs are divided into three families: AG nucleotidyltransferases (ANTs), AG phosphotransferases (APHs), and AG acetyltransferases (AACs).[1b, 3]
In many cases, AG-resistant bacteria have evolved combinations of resistance mechanisms, a fact that greatly increases the challenge of regaining their clinical efficacy through semi-synthetic modifications. In recent years, several studies demonstrated the potential of exploiting AGs for the development of cationic amphiphilic antimicrobial agents by converting part or all of their pseudo-oligosaccharide alcohols into alkyl or aryl ethers.[4] Some of these amphiphilic analogues demonstrated improved activities against several bacterial strains with resistance to the parent AG antibiotics. In addition to AG-based amphiphiles, several families of cationic amphiphiles including cationic steroids (ceragenins)[5] as well as cationic antimicrobial peptides and peptido-mimetics[6] have been developed and were found to possess potent antimicrobial activity. Unlike most mammalian cell membranes, bacterial membranes are rich in negatively-charged lipids such as cardiolipins and phosphatidylglycerol, which attract cationic amphiphiles through ionic interactions,[5] a fact that may be utilized for selective targeting of bacterial membranes.
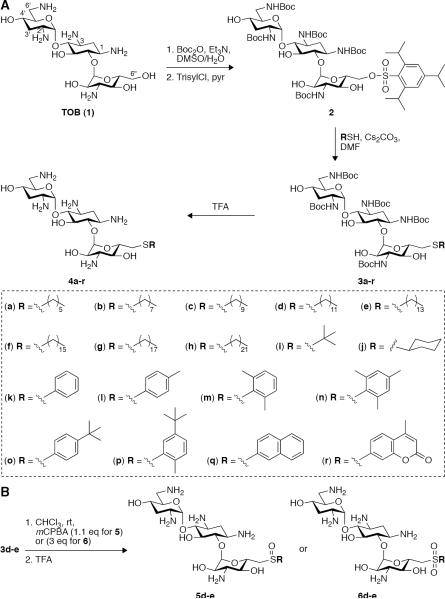
In this manuscript we report the design, synthesis, and antibacterial activity of 18 cationic amphiphiles (4a–r) derived from tobramycin (TOB) (Scheme 1A), a clinically important AG antibiotic that is becoming increasingly compromised by bacterial resistance. We also provide evidence for the mode of action of these derivatives and for the structural requirements for bacterial membranes targeting compared to membranes of red blood cells (RBCs).
Scheme 1.
A. Synthesis of 18 novel 6”-thioether TOB derivatives (4a–r). B. Oxidation of the thioether analogues 3d–e into the corresponding sulfoxides 5d–e and sulfones 6d–e.
We chose to modify the 6”-primary alcohol of TOB and focused on two groups of lypophilic substituents: (i) aliphatic moieties including linear alkyl chains ranging from 6 to 22 carbons in length as well as branched and cyclic alkyls, and (ii) substituted aryl rings. The five amine groups of TOB were protected by Boc groups and the 6”-primary alcohol was selectively converted to the corresponding O-trisyl leaving group to provide compound 2 as previously reported (Scheme 1A).[7] Compound 2 was then reacted with each of the 18 aliphatic and aromatic thiols resulting with the Boc-protected compounds 3a–r in yields ranging from 57 to 94%. Removal of all Boc protecting groups in neat TFA gave the TFA salts of the 6”-thioether TOB derivatives 4a–r with no need for further purification in yields ranging from 74 to 98%.
Compounds 4a–r were screened for their antibacterial activity against 21 Gram-positive and Gram-negative bacterial strains and their minimum inhibitory concentrations (MICs) were determined (Table 1). Amongst the Gram-positive bacteria were pathogenic strains such as methicillin-resistant Staphylococcus aureus (MRSA) (strain C) and vancomycin-resistant Enterococcus (VRE) (strain J) that displayed high levels of resistance to TOB (MIC ≥150 μg/mL). Amongst the Gram-negative were 3 strains of E. coli BL21 (DE3) that we cloned with AMEs: the bi-functional AAC(6')/APH(2”), AAC(3)-IV, and the multi-acetylating Eis, an AAC that confers high levels of resistance to AGs in extensively-drug resistant (XDR) strains of Mycobacterium tuberculosis (Mtb)[8] (strains O, P, and Q, respectively). These three strains had significant to high levels of resistance to TOB (MICs >150, =150, and 18.8 μg/mL, respectively).
Table 1.
Antibacterial activity: MIC values (μg/mL) of the 6”-thioether TOB derivatives 4a–r, 5d–e, and 6d–e compared to the parent drug TOB.
| Gram-positive bacterial strains[a] | Gram-negative bacterial strains[b] | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AG | A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U |
| TOB | 0.3 | 9.4 | >150 | 18.8 | 75 | 1.2 | 9.4 | 18.8 | 2.3 | >150 | 150 | 4.7 | 4.7 | 9.4 | >150 | 150 | 18.8 | 4.7 | 9.4 | 18.8 | 150 |
| 4a | 75 | >150 | >150 | >150 | >150 | 150 | >150 | >150 | 150 | >150 | >150 | 75 | >150 | >150 | >150 | >150 | >150 | 18.8 | 150 | >150 | >150 |
| 4b | 75 | 150 | >150 | 150 | 150 | 37.5 | >150 | 150 | 75 | >150 | >150 | >150 | >150 | >150 | >150 | >150 | >150 | 9.4 | 75 | >150 | >150 |
| 4c | 9.4 | 37.5 | 75 | 150 | 9.4 | 37.5 | >150 | 18.8 | 4.7 | >150 | >150 | 37.5 | 75 | >150 | >150 | >150 | >150 | 2.3 | 9.4 | >150 | >150 |
| 4d | 2.3 | 9.4 | 37.5 | 9.4 | 4.7 | 4.7 | >150 | 4.7 | 9.4 | >150 | 150 | 18.8 | 18.8 | >150 | >150 | 150 | 150 | 4.7 | 18.8 | 37.5 | 150 |
| 4e | 1.2 | 9.4 | 9.4 | 2.3 | 2.3 | 1.2 | 4.7 | 1.2 | 0.3 | 18.8 | 75 | 9.4 | 4.7 | 4.7 | 4.7 | 2.3 | 4.7 | 4.7 | 4.7 | 18.8 | 37.5 |
| 4f | 4.7 | 4.7 | 9.4 | 9.4 | 9.4 | 4.7 | 18.8 | 2.3 | 1.2 | 18.8 | 18.8 | 4.7 | 18.8 | 4.7 | 4.7 | 4.7 | 18.8 | 4.7 | 4.7 | 37.5 | >150 |
| 4g | 9.4 | >150 | 18.8 | >150 | >150 | 75 | >150 | >150 | 9.4 | >150 | 150 | 75 | >150 | >150 | >150 | >150 | >150 | 4.7 | >150 | >150 | >150 |
| 4h | 75 | >150 | >150 | >150 | >150 | 150 | >150 | >150 | 18.8 | >150 | >150 | 18.8 | >150 | >150 | >150 | >150 | >150 | >150 | >150 | >150 | >150 |
| 4i | 1.2 | 150 | >150 | 75 | 75 | 75 | >150 | >150 | 75 | >150 | 150 | 2.3 | 18.8 | >150 | >150 | >150 | 150 | 18.8 | 9.4 | 150 | >150 |
| 4J | 4.7 | 18.8 | >150 | 37.5 | >150 | 9.4 | 150 | 150 | 18.8 | >150 | 150 | 4.7 | 75 | >150 | >150 | >150 | 75 | 9.4 | 18.8 | >150 | >150 |
| 4k | 2.3 | 9.4 | >150 | 18.8 | >150 | 9.4 | 18.8 | >150 | 9.4 | >150 | 75 | 4.7 | 9.4 | 37.5 | >150 | >150 | 75 | 2.3 | 4.7 | 37.5 | >150 |
| 4l | 4.7 | 9.4 | >150 | 18.8 | >150 | 4.7 | 9.4 | 150 | 9.4 | >150 | 75 | 4.7 | 18.8 | 37.5 | 150 | >150 | 37.5 | 1.2 | 2.3 | 37.5 | >150 |
| 4m | 18.8 | 75 | 150 | >150 | 150 | 37.5 | 150 | 37.5 | 37.5 | >150 | 150 | 9.4 | 75 | 150 | >150 | >150 | 150 | 4.7 | 18.8 | >150 | >150 |
| 4n | 75 | 150 | >150 | 150 | 150 | 18.8 | >150 | 75 | 18.8 | >150 | >150 | 37.5 | 150 | >150 | >150 | >150 | >150 | 9.4 | 150 | >150 | >150 |
| 4o | 75 | >150 | >150 | 150 | 150 | >150 | >150 | >150 | 150 | >150 | >150 | >150 | >150 | >150 | >150 | >150 | >150 | 18.8 | 150 | >150 | >150 |
| 4p | 9.4 | >150 | >150 | 150 | 75 | 18.8 | 150 | 75 | 37.5 | >150 | >150 | 37.5 | 150 | >150 | 150 | >150 | >150 | 9.4 | 37.5 | >150 | >150 |
| 4q | 18.8 | 37.5 | >150 | 150 | 150 | 37.5 | 150 | >150 | 37.5 | >150 | >150 | 37.5 | 75 | 150 | >150 | >150 | >150 | 9.4 | 9.4 | 150 | >150 |
| 4r | 37.5 | >150 | >150 | >150 | >150 | >150 | >150 | >150 | >150 | >150 | >150 | 150 | >150 | >150 | >150 | >150 | >150 | 75 | 150 | >150 | >150 |
| 5d | 4.7 | 37.5 | 75 | 18.8 | 9.4 | 4.7 | >150 | 9.4 | 2.3 | 150 | 18.8 | 9.4 | 75 | >150 | >150 | >150 | >150 | 2.3 | 18.8 | 150 | >150 |
| 5e | 2.3 | 18.8 | 18.8 | 4.7 | 2.3 | 1.2 | 75 | 1.2 | 0.3 | 37.5 | 9.4 | 2.3 | 37.5 | 75 | 37.5 | 37.5 | 75 | 4.7 | 9.4 | 37.5 | 75 |
| 6d | 9.4 | 37.5 | 75 | 18.8 | 9.4 | 4.7 | >150 | 9.4 | 2.3 | 75 | 9.4 | 9.4 | 75 | >150 | >150 | >150 | >150 | 2.3 | 18.8 | >150 | >150 |
| 6e | 2.3 | 18.8 | 18.8 | 4.7 | 4.7 | 1.2 | 37.5 | 1.2 | 0.3 | 37.5 | 4.7 | 2.3 | 18.8 | 75 | 37.5 | 75 | 75 | 2.3 | 9.4 | 37.5 | 75 |
S. epidermidis AJCCM228 (A), S. aureus NorA (B), MRSA (C), S. pyogenes serotype M12 (strain MGAS9429) (D), S. mutans UA159 (E), B. subtilis 168 (F), B. subtilis 168 with AAC(6')/APH(2”)-pRB374 (G), B. cereus ATCC11778 (H), B. anthracis 34F2 Sterne strain (I), VRE (J), E. faecalis ATCC29212 (K), and L. monocyfogenes ATCC19115 (L).
E. coli BL21 (DE3) (M), E coli BL21 (DE3) with pET22b (N), E. coli BL21 (DE3) with AAC(6')/APH(2”)-pET22b (O), E. coli BL21 (DE3) with AAC(3)-IV-lnt-pET19b-pps (P), E. coli BL21 (DE3) with Eis (Q), E. coli TolC (R), E. coli MC1061 (S), Shigella clinical isolate 6831 (T), and S. enterica ATCC14028 (U).
The linear aliphatic-chain analogues 4a–h exhibited a parabolic pattern of chain-length dependent antibacterial activity (Figure S45A). Compared to TOB, both the C6- and C8-chain analogues 4a and 4b demonstrated a dramatic loss of antibacterial activity, the C10-chain derivative 4c regained some activity, and the C12-chain analogue 4d demonstrated potent antibacterial activity against several of the strains with resistance to TOB. The greatest improvement in antibacterial activity was observed for the C14- and C16-chain derivatives 4e and 4f, with the most significant effect for 4e that showed marked activity against all of the 21 tested strains. The MIC values of 4e ranged between 0.3 to 18.8 μg/mL against 19 of the 21 tested strains: the exceptions were E. faecalis (K) and S. enterica (U) where only a limited improvement in the antibacterial activity of 4e as compared to TOB was observed (MIC of 4e: 75 and 37.5 μg/mL, respectively, and for TOB: 150 μg/mL). The antibacterial activity dropped again for the C18- and the C22-chain analogues 4g and 4h. With few exceptions, a general drop in the antibacterial activity was observed for the 6”-aromatic-thioethers (4k–r). The more substitution around the aryl ring, the more significant was the loss of antibacterial activity against the tested strains. For example, of the aromatic thioether analogues, 4k with the thiophenyl ring, and 4l with the 4-methyl-thiophenyl ring, demonstrated the best overall antibacterial activity against the tested strains. However, a drop in antibacterial activity was observed for the 2,6-dimethyl-thiophenyl derivative 4m, and a more significant drop was observed for the 2,4,6-trimethyl-thiophenyl analogue 4n. Since thioethers may be susceptible to cellular mediated S-oxidation,[9] we oxidized two of the most potent thioethers (4d–e) to diastereomeric mixtures (~4:1 ratio) of the corresponding sulfoxides (5d–e) and to the sulfones (6d–e) (Scheme 1B). The effect of S-oxidation on the antibacterial activity varied amongst the tested bacterial strains. A reduction in the antibacterial activity of both sulfoxide and sulfone analogues as compared to the parent thioethers was observed for the E. coli BL21 (DE3) strains M–Q and for B. subtilis 168 with AAC(6')/APH(2”)-pRB374 (strain G). In contrast, when tested against E. faecalis (K) and L. monocytogenes (L), all four S-oxidized analogues demonstrated improved antibacterial activities compared to that of the thioethers 4de. For most of the tested strains, S-oxidation did not have a dramatic effect on the antibacterial activity with MIC values identical or one double-dilution higher than those of the thioethers 4d–e. In most of the tested strains, MIC values were identical for the sulfoxides (5d–e) and the corresponding sulfones (6d–e) indicating that the level of S-oxidation has little to no effect on antibacterial activity.
To uncover the reasons for the broad spectrum and improved antimicrobial activity of some of the thioether analogues, we performed several biological tests. The effect of compound 4e on the translation of a luciferase reporter gene was measured in E. coli cell lysates. In lysates of Mycobacterium smegmatis and E. coli, previous studies reported that TOB inhibited luciferase translation at IC50 values of ~20 nM.[10] In our E. coli cell lysate, TOB potently inhibited translation (IC50 = 8.9 ± 1.9 nM), whereas 4e did not reach IC50 value even at 147 nM (measured using the free base forms of TOB and 4e), suggesting that this compound does not target the bacterial ribosome as its major mode of antibacterial activity.
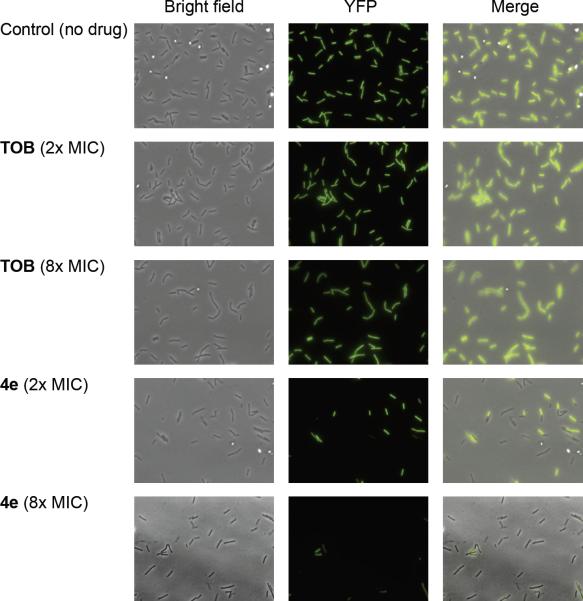
Furthermore, time of kill assays performed on S. mutans UA159 (E) and S. pyogenes (D) revealed that 4e rapidly conferred bacterial cell death as compared to TOB (Figure S45B). At MIC values (2.3 μg/mL for 4e on both strains, and 75 μg/mL (S. mutans) or 18.8 μg/mL (S. pyogenes) for TOB) no viable bacteria remained in the sample treated with 4e after 5 hours of incubation with S. mutans and 3 hours of incubation with S. pyogenes. For both of these strains, almost no reduction in bacterial growth was observed for TOB after the same incubation time. The luciferase translation assay results and the rapid time of kill displayed by 4e suggested that this compound acts by disrupting the bacterial cell membrane. Additional strong evidence for the membrane disruption effect of 4e was obtained by fluorescence microscopy experiments (Figure 1). Fluorescently labeled B. subtilis with constitutive YFP expression (PY79)[11] was incubated for 1 hour with 4e or with TOB at several concentrations. After 1 hour of incubation with TOB at both 2× and 8× the MIC (2.3 and 9.4 μg/mL, respectively), most of the bacterial cells in the sample were viable and maintained good fluorescence. In contrast, a significant drop in fluorescence presumably resulting from the bacterial cell lysis and loss of intracellular content including the YFP was evident after the same incubation time with compound 4e at both 2× and 8× the MIC (4.7 and 18.8 μg/mL, respectively).
Figure 1.
Bright field and epi-fluorescence microscopy. B. subtilis (PY79) cells carrying YFP under an inducible IPTG promoter treated with TOB at 2.3 μg/mL (2× MIC) and 9.4 μg/mL (8× MIC) or with compound 4e at 4.7 μg/mL (2× MIC) and 18.8 μg/mL (8× MIC).
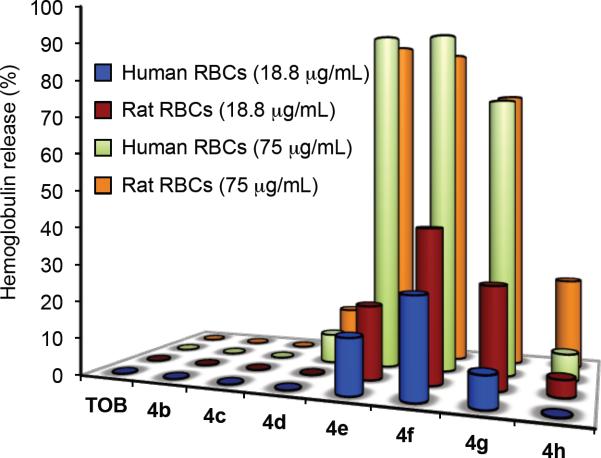
The selectivity of the 6”-thioether derivatives 4b–h towards bacterial membranes was tested using a hemolysis assay on both laboratory rat and human RBCs (Figure 2). The MICs of the most potent thioether analogues ranged between 0.3 to 18.8 μg/mL. Hence, RBCs samples were incubated with analogues 4d–f at a concentration of 75 μg/mL, which is 4–250 times greater than the MIC range and at 18.8 μg/mL, which is 1–60 times the MIC range. At 75 μg/mL, TOB as well as compounds 4b and 4c with the C8- and C10-linear chains caused no measurable hemolysis of rat and human RBCs. Compound 4d with the C12-chain caused 12.6 ± 0.6% hemolysis of rat RBCs and 7.9 ± 1.7% hemolysis of human RBCs. Both compounds 4e (C14-chain) and 4f (C16-chain) caused extensive hemolysis at 75 μg/mL with 93.6 ± 5.5% and 90.2 ± 4.5% of rat RBCs, and 93.7 ± 11.1% and 93.1 ± 5.1% of human RBCs, respectively. Compound 4g (C18-chain) caused 77.2 ± 7.0% hemolysis of rat RBCs and 74.3 ± 9.0% hemolysis of human RBCs. A significant drop in the hemolytic activity was observed for compound 4h (C22-chain), 24.4 ± 5.8% of rat RBCs and 7.1 ± 0.1% of human RBCs.
Figure 2.
Hemolysis tests. Human RBCs (blue “18.8 μg/mL” and green “75 μg/mL”) and rat RBCs (red “18.8 μg/mL” and orange “75 μg/mL”) were incubated with TOB or with analogues 4b–h at concentrations of 18.8 μg/mL or 75 μg/mL for 1 h at 37 °C.
At 18.8 μg/mL, TOB and analogues 4b–d with the C8-, C10-, and C12-linear chains caused no measurable hemolysis of both rat and human RBCs, and compound 4e (C14-chain) caused 19.5 ± 0.3% hemolysis of rat RBCs and 14.3 ± 1.7% hemolysis of human RBCs. Compound 4f (C16-chain) demonstrated the maximal hemolytic effect of 40.4 ± 1.7% (rat RBCs) and 25.4 ± 2.1% (human RBCs), while 4g (C18-chain) caused 26.3 ± 1.9% hemolysis of rat RBCs and 8.0 ± 0.8% hemolysis of human RBCs. At 18.8 μg/mL compound 4h (C22-chain) caused 4.4 ± 0.5% hemolysis of rat RBCs and no measurable hemolysis of human RBCs. Although compound 4f with the C16-chain was one of the most active TOB analogues against the tested bacterial strains, it readily disrupted RBC membranes as well. In contrast, compound 4d (C12-chain) demonstrated potent antimicrobial activity against several of the TOB-resistant bacterial strains and caused little to no measurable hemolysis at the tested concentrations. The hemolysis assay demonstrated that the aliphatic chain length plays a key role in selective targeting of the bacterial membranes versus those of RBCs.
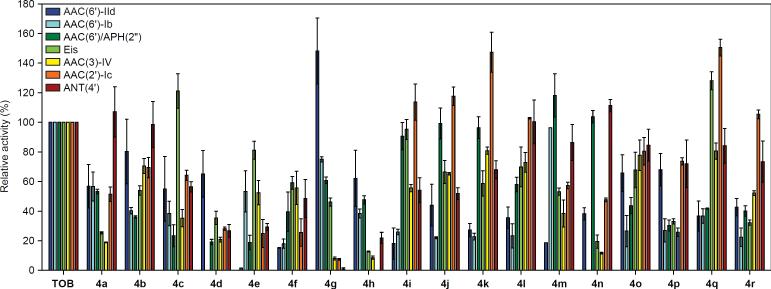
Several amphiphilic AGs based on paromomycin were recently shown to act as inhibitors of the AME APH(3').[12] Moreover, amphiphilic AGs can make their way into the bacterial cell where they are exposed to enzymatic modifications by AMEs. AME modifications (O-phosphorylation, O-adenylation, and N-acetylation), result in a reduction of the overall positive charge of the parent AG. Hence, AME modifications may reduce the affinity of amphiphilic AGs to the negatively-charged bacterial membrane and hamper their antimicrobial activity. Structural information obtained from crystallographic studies of several AMEs indicated that the AG binding pocket is rich in negatively-charged amino acid residues such as glutamic and aspartic acids and that several water molecules are required to stabilize the interactions between AGs and the AMEs' binding pockets.[13] We reasoned that the replacement of the 6”-primary alcohol of TOB with hydrophobic residues would interfere with these hydrophilic-based binding interactions and reduce the ability of the enzymes to modify these molecules. To test this hypothesis, the relative activities of seven different AMEs[14] on compounds 4a–r were compared to that of TOB (Figure 3). While some of the modified compounds served as better substrates for some AMEs, in general a drop in the AME catalytic activity was observed in most of the cases. Analogues 4d–f, which demonstrated the most potent antimicrobial activities, were also the poorest substrates for all of the tested AMEs.
Figure 3.
Bar graph showing the relative initial rates of AME reactions with TOB and the 6”-thioether analogues 4a–r. Rates are normalized to TOB. AMEs tested include AAC(6')-IId (blue), AAC(6')-Ib (cyan), AAC(6')/APH(2”) (dark green), Eis (light green), AAC(3)-IV (yellow), AAC(2')-Ic (orange), and ANT(4') (red).
Hence, bacterial strains containing the tested AMEs will have limited to no ability to inactivate analogues 4d–f through chemical modifications that are catalyzed by these enzymes. This is also true for the S-oxidized compounds 5d–e and 6d–e, which behaved similarly to their non-oxidized counterparts 4d–e (Figure S46).
In conclusion, 18 novel 6”-thioether TOB analogues (4a–r) and 4 S-oxidized compounds (5d–e and 6d–e) have been synthesized and screened for antibacterial activity. The C12-, C14-, and C16-linear chain analogues 4d–f demonstrated potent activity against bacterial strains with high levels of resistance to TOB. We found evidence that the most potent analogue 4e targets bacterial membranes and no longer targets the ribosome as does the parent drug. Hemolysis tests indicated that it is possible to improve the antibacterial activity while reducing the undesired hemolytic effect by altering the length of the aliphatic chain. Finally, thioethers 4d–f served as poor substrates for several AMEs, demonstrating that AMEs have a limited deactivating effect on these AG analogues. The results reported in this manuscript offer guidelines for the design of bacterial membrane targeting amphiphilic AGs.
Supplementary Material
References
- [1].a) Fridman M, Belakhov V, Yaron S, Baasov T. Org. Lett. 2003;5:3575–3578. doi: 10.1021/ol035213i. [DOI] [PubMed] [Google Scholar]; b) Houghton JL, Green KD, Chen W, Garneau-Tsodikova S. Chembiochem. 2010;11:880–902. doi: 10.1002/cbic.200900779. [DOI] [PubMed] [Google Scholar]; c) Wright GD, Berghuis AM, Mobashery S. Adv. Exp. Med. Biol. 1998;456:27–69. [PubMed] [Google Scholar]
- [2].a) Magnet S, Blanchard JS. Chem. Rev. 2005;105:477–498. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]; b) Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. Antimicrob. Agents Chemother. 1999;43:727–737. doi: 10.1128/aac.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kotra LP, Haddad J, Mobashery S. Antimicrob. Agents Chemother. 2000;44:3249–3256. doi: 10.1128/aac.44.12.3249-3256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Ouberai M, El Garch F, Bussiere A, Riou M, Alsteens D, Lins L, Baussanne I, Dufrene YF, Brasseur R, Decout JL, Mingeot-Leclercq MP. Biochim. Biophys. Acta. 2011;1808:1716–1727. doi: 10.1016/j.bbamem.2011.01.014. [DOI] [PubMed] [Google Scholar]; b) Bera S, Zhanel GG, Schweizer F. J. Med. Chem. 2010;53:3626–3631. doi: 10.1021/jm1000437. [DOI] [PubMed] [Google Scholar]; c) Baussanne I, Bussiere A, Halder S, Ganem-Elbaz C, Ouberai M, Riou M, Paris JM, Ennifar E, Mingeot-Leclercq MP, Decout JL. J. Med. Chem. 2010;53:119–127. doi: 10.1021/jm900615h. [DOI] [PubMed] [Google Scholar]; d) Hanessian S, Pachamuthu K, Szychowski J, Giguere A, Swayze EE, Migawa MT, Francois B, Kondo J, Westhof E. Bioorg. Med. Chem. Lett. 2010;20:7097–7101. doi: 10.1016/j.bmcl.2010.09.084. [DOI] [PubMed] [Google Scholar]
- [5].Epand RF, Savage PB, Epand RM. Biochim. Biophys. Acta. 2007;1768:2500–2509. doi: 10.1016/j.bbamem.2007.05.023. [DOI] [PubMed] [Google Scholar]
- [6].a) Findlay B, Zhanel GG, Schweizer F. Antimicrob. Agents Chemother. 2010;54:4049–4058. doi: 10.1128/AAC.00530-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Giuliani A, Rinaldi AC. Cellular and molecular life sciences. 2011;68:2255–2266. doi: 10.1007/s00018-011-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yin LM, Edwards MA, Li J, Yip CM, Deber CM. J. Biol. Chem. 2012;287:7738–7745. doi: 10.1074/jbc.M111.303602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Wang H, Tor Y. Bioorg. Med. Chem. Lett. 1998;8:3665–3670. doi: 10.1016/s0960-894x(98)00663-5. [DOI] [PubMed] [Google Scholar]; b) Wang H, Tor Y. Angew. Chem. Int. Ed. Engl. 1998;37:109–111. [Google Scholar]
- [8].a) Chen W, Biswas T, Porter VR, Tsodikov OV, Garneau-Tsodikova S. Proc. Natl. Acad. Sci. U S A. 2011;108:9804–9808. doi: 10.1073/pnas.1105379108. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Green KD, Chen W, Garneau-Tsodikova S. ChemMedChem. 2012;7:73–77. doi: 10.1002/cmdc.201100332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Wynalda MA, Hutzler JM, Koets MD, Podoll T, Wienkers LC. Drug. Metab. Dispos. 2003;31:878–887. doi: 10.1124/dmd.31.7.878. [DOI] [PubMed] [Google Scholar]; b) Katopodis AG, Smith HA, May SW. J. Am. Chem. Soc. 1988;110:897–899. [Google Scholar]
- [10].a) Hobbie SN, Kalapala SK, Akshay S, Bruell C, Schmidt S, Dabow S, Vasella A, Sander P, Bottger EC. Nucleic Acids Res. 2007;35:6086–6093. doi: 10.1093/nar/gkm658. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sucheck SJ, Wong AL, Koeller KM, Boehr DD, Draker K-A, Sears P, Wright GD, Wong C-H. J. Am. Chem. Soc. 2000;122:5230–5231. [Google Scholar]
- [11].Eldar A, Chary VK, Xenopoulos P, Fontes ME, Loson OC, Dworkin J, Piggot PJ, Elowitz MB. Nature. 2009;460:510–514. doi: 10.1038/nature08150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Szychowski J, Kondo J, Zahr O, Auclair K, Westhof E, Hanessian S, Keillor JW. ChemMedChem. 2011;6:1961–1966. doi: 10.1002/cmdc.201100346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Wolf E, Vassilev A, Makino Y, Sali A, Nakatani Y, Burley SK. Cell. 1998;94:439–449. doi: 10.1016/s0092-8674(00)81585-8. [DOI] [PubMed] [Google Scholar]; b) Wybenga-Groot LE, Draker K, Wright GD, Berghuis AM. Structure. 1999;7:497–507. doi: 10.1016/s0969-2126(99)80066-5. [DOI] [PubMed] [Google Scholar]; c) Sakon J, Liao HH, Kanikula AM, Benning MM, Rayment I, Holden HM. Biochemistry. 1993;32:11977–11984. doi: 10.1021/bi00096a006. [DOI] [PubMed] [Google Scholar]; d) Hon WC, McKay GA, Thompson PR, Sweet RM, Yang DS, Wright GD, Berghuis AM. Cell. 1997;89:887–895. doi: 10.1016/s0092-8674(00)80274-3. [DOI] [PubMed] [Google Scholar]
- [14].a) Green KD, Chen W, Garneau-Tsodikova S. Antimicrob. Agents Chemother. 2011;55:3207–3213. doi: 10.1128/AAC.00312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Porter VR, Green KD, Zolova OE, Houghton JL, Garneau-Tsodikova S. Biochem. Biophys. Re.s Commun. 2010;403:85–90. doi: 10.1016/j.bbrc.2010.10.119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.