Abstract
The rapid emergence of bacterial strains resistant to multiple antibiotics is posing a growing public health risk. The mechanisms underlying the rapid evolution of drug resistance are, however, poorly understood. The heterogeneity of the environments in which bacteria encounter antibiotic drugs could play an important role. E.g., in the highly compartmentalized human body, drug levels can vary substantially between different organs and tissues. It has been proposed that this could facilitate the selection of resistant mutants, and recent experiments support this. To study the role of spatial heterogeneity in the evolution of drug resistance, we present a quantitative model describing an environment subdivided into relatively isolated compartments with various antibiotic concentrations, in which bacteria evolve under the stochastic processes of proliferation, migration, mutation and death. Analytical and numerical results demonstrate that concentration gradients can foster a mode of adaptation that is impossible in uniform environments. It allows resistant mutants to evade competition and circumvent the slow process of fixation by invading compartments with higher drug concentrations, where less resistant strains cannot subsist. The speed of this process increases sharply with the sensitivity of the growth rate to the antibiotic concentration, which we argue to be generic. Comparable adaptation rates in uniform environments would require a high selection coefficient (s > 0.1) for each forward mutation. Similar processes can occur if the heterogeneity is more complex than just a linear gradient. The model may also be applicable to other adaptive processes involving environmental heterogeneity and range expansion.
Keywords: first passage processes, stochastic modeling, evolutionary ecology
Worldwide, bacteria exhibiting resistance to multiple antibiotics have become a pressing public health problem. Resistant strains have consistently emerged a few years after the introduction of new antibiotics, and an increasing number of strains can evade multiple classes of antimicrobial drugs. Even though antibiotic resistance evolves right under our eyes and is well documented, the principles underlying its rapid evolution are still poorly understood (1, 2).
Many factors are likely to contribute to the rapid evolution of antibiotic resistance. One is the mere size of bacterial populations: a tuberculosis cavity, for instance, can contain 107–109 bacilli (3). This situation is exacerbated by mutator strains, which have over-all increased mutation rates (4), and stress-induced elevation of mutation rates (5). The selection of rare resistant mutants is thought to be facilitated by low drug concentrations, which may occur after a treatment or when a treatment regimen is not strictly adhered to (6). Once enzymes providing some degree of resistance have emerged, they can be efficiently transferred to other bacteria by mobile elements such as plasmids, transposons, and integrons (1).
Here, we explore whether spatial heterogeneity could facilitate the evolution of antibiotic resistance. In people and livestock treated with antibiotics, pharmaco-kinetic parameters vary between different organs and tissues (7). As a result, antibiotic concentrations are not spatially homogeneous (8). In addition, bacteria migrate between both treated and untreated patients, who have a spectrum of immune responses. Antibiotic resistance therefore naturally evolves in heterogeneous environments.
In itself, the idea that environmental heterogeneity could promote the evolution of drug resistance is not new. Over a decade ago, it was proposed that heterogeneity could assist the evolution of drug resistance of HIV (9). Models suggested that, in homogeneous environments, the drug concentration has to be in a narrow range near the minimal inhibitory concentration (MIC) of the virus (called the selective window) for an effective selection of resistance: if the concentration is too high, both the wild type and feasible mutants are inhibited, whereas if it is too low, the wild type may out-compete the mutant (9). However, if the environment consists of two compartments, in one of which the drug does not penetrate well (a sanctuary or reservoir), the selective window is greatly enlarged. Samples from postmortem tissues of AIDS patients indeed suggest that compartmentalization in the central nervous system plays a role in the evolution of drug-resistant HIV strains (10).
A similar effect could favor the evolution of antibiotic resistance in bacteria (8, 11, 12). Often, several mutations are required for a bacterium to obtain a medically relevant resistance level (13). In a homogeneous drug concentration, a single bacterium has to rapidly acquire these mutations to survive the treatment. If more than 2 specific mutations are required, this is unlikely (see SI Text). Heterogeneous environments, however, could provide sanctuaries, allowing these mutations to be selected one by one. Such ideas have led to the concept of “resistance-selective environments” as environments that favor the evolution of antibiotic resistance (11, 12).
Similar ideas have emerged independently in a different context. In ecology, a habitat is called a sink if mortality exceeds reproduction, so that a population is maintained only owing to constant immigration from a habitat where reproduction exceeds mortality, called a source (14–16). The genotypes of immigrants into the sink are sampled from the standing genetic variation of the source, and are poorly adapted to the sink. Such dynamics could foster adaptation to the sink conditions (14–16). Indeed, the idea of a sanctuary introduced above largely coincides with the notion of a source. Not surprisingly, source–sink dynamics have recently been associated with the evolution of virulence (17–19), insecticide resistance (20), and antibiotic resistance (21). We recently developed a stochastic model of adaptation in source–sink ecologies and derived mathematically how the rate of adaptation to the sink conditions depends on the population size as well as the rates of migration, mutation, reproduction, and death (22).
Recent experiments vividly illustrated the potential role of environmental heterogeneity in promoting antibiotic resistance (23). In these experiments, a microfluidic device was used to set up a smoothly varying concentration profile of Ciproflaxacin in a two-dimensional landscape of connected hexagonal wells. Surprisingly, after bacteria were inoculated in the center of the device, resistance to Ciproflaxacin evolved in just 10 h, due to four single-nucleotide substitutions in three genes. Even though the mechanism that allowed the rapid accumulation of these mutations is unknown, the concentration gradient seems essential, as resistance did not evolve when the concentration was uniform.
Even though the importance of spatial heterogeneity for the evolution of antibiotic resistance has been suggested in the literature, to our knowledge no attempts have been made to quantitatively study possible mechanisms in mathematical models. Here we formulate such a model, called the staircase model, for the first time. In its simplest form, this model describes adaptation in an antibiotic gradient, but many of its qualitative features also apply to more general kinds of spatial heterogeneity. To correctly capture the importance of rare stochastic events, the model is fully probabilistic. Solution of the model demonstrates that spatial heterogeneity provides a mode of adaptation that is impossible in uniform environments and could strongly facilitate the evolution of antibiotic resistance. Analytical calculations and numerical simulations were used to characterize this mode of evolution. We present explicit mathematical results for the adaptation rate as a function of parameters. These results show that the speed of this mode of evolution increases sharply if the bacterial growth rate depends abruptly on the drug concentration, e.g., for a mesa-shaped fitness landscape. To place these results in perspective, we compare them to a generic, spatially uniform model of evolution on a “Mt. Fuji”-type fitness landscape. This comparison shows that similar adaptation rates can be obtained in homogeneous environments, but only if each mutation required for resistance carries a large fitness advantage.
Model
Fig. 1 illustrates the staircase model. We consider an environment that is divided into a series of relatively isolated compartments, inhabited by bacteria. In the figure, these compartments are plotted horizontally. Each compartment is associated with a fixed antibiotic drug concentration, which increases from left to right. The genotype of each bacterium is characterized by a positive integer g, plotted vertically. Importantly, bacteria with a larger g are more resistant (i.e., have a higher MIC), and hence can proliferate in compartments with higher drug concentrations: a bacterium with genotype g = 3 can proliferate in compartments 1 to 3, but not in compartments 4 and up. This defines a diagonal “staircase” separating the area of the diagram in which bacteria can proliferate (above the staircase) from the region where they cannot (below it).
Fig. 1.
The staircase model. We consider an environment divided into several compartments (plotted horizontally) in which bacteria proliferate (grow), migrate, mutate, and die. The bacterial genotypes are characterized by an integer g (plotted vertically) reflecting their of antibiotic resistance level. Given its genotype and location, each bacterium inhabits a unique “tile” on the diagram. Mutations move bacteria one tile up or down, and migration moves them left or right. Importantly, the concentration of an antibiotic drug increases from left to right. As a result, bacteria can grow in compartment i only if they have a genotype g≥i, i.e., if they live above the diagonal “staircase”.
Each bacterium can proliferate, migrate, die (possibly due to clearance by the immune system), and mutate, all in a stochastic manner. Migration occurs between neighboring compartments, at a rate ν per bacterium. Bacteria die at a rate δ. Forward mutations, at a rate μf, increase g by one, corresponding to an increased resistance. Backward mutations do the opposite, at a rate μb. Bacterial proliferation (or “growth”) takes on the logistic form; the density-dependent proliferation rate of a bacterium of genotype g in compartment i is:
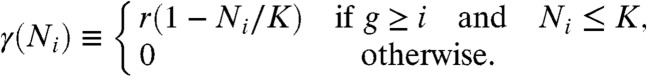 |
[1] |
Here, Ni is the population size in compartment i (including all genotypes), K is the carrying capacity of the compartments, and r is referred to as the “growth rate”. (Note that γ(Ni) ≈ r at population sizes well below K). For simplicity, we have assumed that r is the same for all genotypes and compartments above the staircase, and 0 below it. This all-or-none behavior will be relaxed below, where we will show that it does not affect the qualitative results as long as r is well above the death rate δ. The logistic growth introduces competition between all bacteria living in the same compartment, irrespective of their genotype, but not between those in different compartments. It also ensures that the population in each compartment remains finite: Growth, death and migration balance when the population reaches a size N ≈ [1 - δ′/r]K, where δ′ ≡ δ + 2ν is the total rate at which bacteria disappear from a compartment, by death or migration.
Results
How would an initially non-resistant population (g = 1) evolve under the aforementioned processes? To answer this question for a wide range of parameter values, we have used both computer simulations (kinetic Monte Carlo) and analytical theory.
Climbing the Staircase.
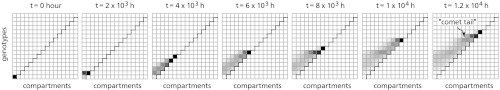
Fig. 2 shows snapshots of a typical simulation; see also Movie S1 for the entire dynamical process. The darkness of each “tile” indicates the number of bacteria of the corresponding genotype and compartment; also see Fig. S1. Initially, only compartment 1 is inhabited, and all bacteria have type 1. However, in the course of the simulation, the population gradually spreads up- and rightward. Mutants emerge with increasing resistance, allowing the population to invade more and more compartments. A moving front forms that steadily climbs the staircase.
Fig. 2.
Snapshots of the population evolving antibiotic resistance. Shades of gray indicate the population density at each tile. The population, initially non-resistant and hence confined to compartment 1, adapts and expands simultaneously, in a probabilistic, stepwise fashion. While the population front “climbs the staircase”, a “comet tail” trails behind. Parameters: K = 105, r = 1/h, μf = 10-7/h, μb = 10-4/h, δ = 10-1/h, ν = 10-3/h.
Importantly, the evolution process exhibited in Fig. 2 is fundamentally different from the notion of a population climbing a fitness landscape. In a typical cartoon representation of a fitness landscape (Fig. S2 A and B), the vertical axis represents fitness (often identified with the growth rate) and the horizontal axis a genotype space; populations climb “fitness slopes” because individuals higher up produce more offspring on average. In contrast, neither of the axes in Fig. 2 represents fitness. Because the growth rate is fixed above the staircase and 0 below it (see Eq. 1), the space-dependent fitness landscape is actually a mesa-shaped plateau (Fig. S2C). The adapting population spreads along the edge of this plateau, despite the absence of a fitness slope.
If there is no fitness slope, why does the population adapt? As will be shown quantitatively below, a mutant with an increased resistance can, by chance, end up in a previously unpopulated compartment and found an entirely new colony. Each adaptation therefore allows the population to expand to an additional compartment, and conversely, the adaptation is successful only because it allows the population to expand its range. As a result, the population grows as it adapts.
As Movie S1 shows, the population front advances in a stepwise fashion, where a step is the colonization of a new compartment. Each step is triggered by the arrival of a new, more resistant mutant in the next uncolonized compartment. After this “founder” arrives, a typical infection ensues: unless, by chance, the founder’s lineage quickly goes extinct, an exponentially growing population is established that rapidly fills up the compartment’s carrying capacity. Another step is taken only when the next, rare mutant invades the next compartment.
As the front ascends the staircase, a “comet tail” trails behind. This arises because resistant strains tend to wander backward (leftward), where they compete with less resistant strains. Even though the more resistant strains have no growth advantage in these compartments, they eventually drive their competitors to extinction owing to their continued immigration. Indeed, in time very few bacteria with g < 3 remain, even in compartment 1 (see Fig. S1).
The Adaptation Steps: Source–sink Dynamics at the Front.
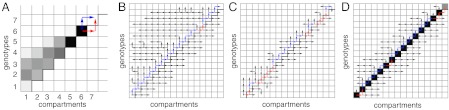
Fig. 3A is an enlargement of a snapshots from Fig. 2. The population has adapted to the drug concentration in compartment 6, but cannot yet invade compartment 7. Bacteria constantly migrate from compartment 6, where reproduction exceeds mortality, to compartment 7, where mortality exceeds reproduction. This means that compartments 6 and 7 display source–sink dynamics (introduced above). In fact, at any time, the population front displays source–sink dynamics. As a result, the adaptation processes in the staircase model resemble those in source–sink systems (22).
Fig. 3.
Dynamics of the model. (A) Snapshot of a simulation. At this time, the population has adapted to the drug concentrations in compartments ≤ 6. The next adaptation step most likely unfolds in one of two ways, indicated with blue and red arrows. The adaptation rate critically depends on the rates of these two “paths”. (B) The spread of the bacteria for the simulation in Fig. 1. For each tile, an arrow indicates the origin of its first inhabitant. Colors indicate for each adaptation step whether the red or the blue path was taken. The blue path is dominant. (C) The red path becomes more likely if the mutation rate and the carrying capacity are high. Here, μfK = 102; now the red and blue paths occur with equal probability. (D) If drug resistance comes at a large fitness cost s, the blue path is repressed. Here, s = 0.2; both the population density at the end of the simulation and the spread of the population are shown. No comet tail is found. (Parameters: as in Fig. 1).
An adaptation step will occur when a founder, with genotype 7, finds compartment 7. This can come about in many ways, but few are feasible. Red and blue arrows indicate the two most likely “paths”. These each involve only one mutation and one migration event. Other paths require more events: e.g., a bacterium of type 5 in compartment 5 could acquire two mutations and migrate two steps to the right. But assuming mutation and migration rates are low, this is much less likely. The blue path starts with a forward mutation in compartment 6. The resulting mutant has no advantage there; yet, it or one of its descendants has a chance to move to compartment 7. Such a scenario, in which an initially neutral mutation becomes beneficial by a fortunate change of environment, is known as the Dykhuizen–Hartl effect (24, 25). In the red path, the order is reversed: a bacterium first migrates to compartment 7; there it cannot grow, but may obtain a mutation before it is cleared. Both paths can produce a founder, but the one that tends to do it faster is traveled more often.
When properly modified, the theory of source–sink systems can predict the rates of both paths and their relative likelihoods (22). (Analytical results and derivations are provided in the SI Text). The theory indicates that two situations have to be distinguished. If forward mutations are rare (that is, μfN ≪ δ′), the blue path is faster than the red one, and adaptation typically follows the blue path. We call this as the mutation-limited regime. If instead forward mutations occur frequently (μfN≫δ '), the blue and red paths become equally fast and equally likely (the migration-limited regime). The red path is never faster than the blue path.
Fig. 3B displays, for the simulation shown in Fig. 2, a “historical record” of the spread of the population. For each tile, an arrow indicates how its first occupant arrived: by immigration (horizontal arrows) or by mutation (vertical ones). The arrows along the staircase are colored to visualize for each adaptation step whether the blue or the red path was taken. The parameters are in the mutation-limited regime, where the theory predicts the blue path to be faster. Indeed, the red path is taken only once. Quantitatively, the theory predicts the blue path to be 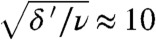 times faster; we repeated the simulation 50 times and confirmed that the red path was taken in 11 ± 1% of the steps. Fig. 3C shows similar data for a simulation with parameters in the migration-limited regime, where both paths should be equally fast; as expected, the red path was taken in 47 ± 3% of the steps.
times faster; we repeated the simulation 50 times and confirmed that the red path was taken in 11 ± 1% of the steps. Fig. 3C shows similar data for a simulation with parameters in the migration-limited regime, where both paths should be equally fast; as expected, the red path was taken in 47 ± 3% of the steps.
The Adaptation Rate.
After a few steps, the rate with which the front advances becomes constant (see Fig. S3). This steady-state rate v is a natural measure of the adaptation rate. How fast can this mode of adaptation be, and how does this depend on the parameters?
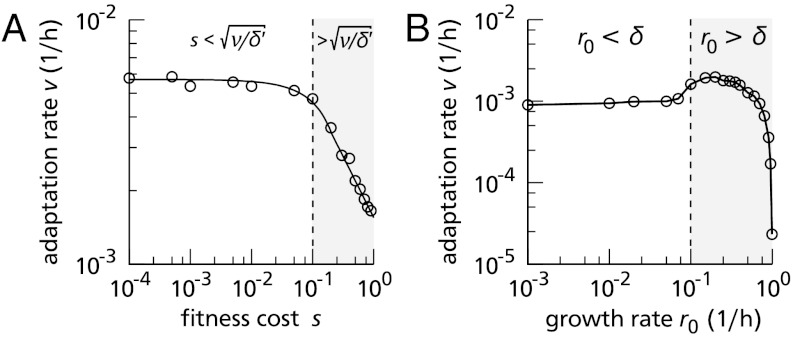
Fig. 4 shows the adaptation rate, obtained by simulation, for a large range of parameters. Because analytical calculations (discussed below) indicate that the adaptation rate depends on μf and K only through their product μfK, it is plotted as a function of μfK, for several values of the migration rate (Fig. 4A) and the death rate (Fig. 4B). Importantly, the results establish that adaptation in the staircase model can be very fast. Assuming reasonable colony sizes, of the order K = 108 (3), forward mutation rates as low as μf = 10-8/h (estimates range from 10-6 to 10-9 per cell division (3, 5, 26)), and a low migration rate ν ≈ 10-3/h, advantageous mutations can accumulate at a rate of 1 in 40 h. The waiting time can be reduced further, to mere hours, if the mutation rate is elevated by stress-induced mutagenesis (5), hypermutators (27), and/or if the migration rate is larger.
Fig. 4.
The adaptation rate. Symbols represent averages over 50 simulations, 15 steps each; the errors are smaller than the symbols. Lines represent theoretical predictions. (A) The adaptation rate increases with μf, K, and ν, and can become remarkably high. Two scaling regimes are visible. (B) The adaptation rate decreases with the death rate provided μfK is small. Unless specified, parameters are as in Fig. 1.
To further characterize the rate of adaptation, we use a model of evolution in a homogeneous environment guided by a “Mt. Fuji”-type fitness landscape as a benchmark. We imagine a well-mixed population of bacteria behaving exactly as in the staircase model: they grow logistically, mutate forward and backward, and die, all stochastically at the specified rates. (Migration has no effect in this model and is neglected). We assume that each forward mutation carries a selection coefficient s, leading to the fitness landscape shown in Fig. S4A. In this model, the population again obtains a constant adaptation rate (see Fig. S4B). In Fig. S4C the adaptation rates for a broad range of parameters, obtained by simulation, are plotted against s (black symbols). Also shown are the adaptation rates of the staircase model, for the same parameters (horizontal lines and red symbols). This comparison allows us to characterize the adaption rate in the staircase model by calculating the selection coefficient required to obtain the same rate in the homogeneous model. The adaption rate of the staircase model is seen to correspond to a high selection coefficient of s > 0.1 for the wide range of parameters tested. This result is based on a low migration rate ν ≈ 10-3/h (as used above); increasing this rate only increases the corresponding selection coefficient. This demonstrates that adaptation in a homogeneous environment can also be effective, but requires large selective advantages for each mutation to achieve the same adaptation rate. Ultimately, this is because a spatial structure strongly reduces the competition experienced by beneficial mutants. In uniform Mt. Fuji models, large time scales are associated with the fixation of mutants. In the staircase model, this process is circumvented, because a founder can rapidly establish a new population in a new compartment without having to out-compete less resistant strains.
Fig. 4 also shows predictions from analytical calculations (curves). To calculate the rate of adaptation, we exploited an important observation. We mentioned that each adaptation step requires the arrival of a new founder in the next not-yet-colonized compartment, which then establishes an exponentially growing colony. The observation is that the adaptation rate is usually not limited by this fast exponential growth phase, but by the waiting time before the founder arrives (exceptions are discussed below). The adaptation rate thus follows from the average waiting time before the founder’s arrival, which can be calculated analytically using the theory of first passage processes (28) (see SI Text).
In both Fig. 4
A and B, two regimes can be distinguished: at low values of μfK (the mutation-limited regime discussed above) the adaptation rate is proportional with μfK, while at high values (the migration-limited regime) it scales as 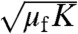 . Indeed, the analytical theory confirms that
. Indeed, the analytical theory confirms that
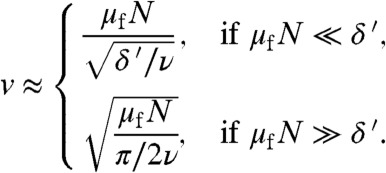 |
[2] |
(Remember that N is proportional to K). These equations also show that the adaptation rate is proportional to 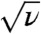 (unless ν ≈ δ′), and that in the migration-limited regime it depends only weakly on the death rate, whereas in the mutation-limited regime it scales approximately as
(unless ν ≈ δ′), and that in the migration-limited regime it depends only weakly on the death rate, whereas in the mutation-limited regime it scales approximately as 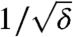 . As evident from Fig. 4, the theory and the simulation results are in excellent agreement; only when the adaptation is very fast (v ≈ 10-1/h) serious deviations occur. This is no surprise: in this very fast regime, the exponential growth phase becomes a substantial part of the adaptation process and can no longer be neglected.
. As evident from Fig. 4, the theory and the simulation results are in excellent agreement; only when the adaptation is very fast (v ≈ 10-1/h) serious deviations occur. This is no surprise: in this very fast regime, the exponential growth phase becomes a substantial part of the adaptation process and can no longer be neglected.
There is one important caveat, however. If the growth rate r of the founder is close to its death rate, the exponential growth phase becomes slow, because growth and death of the founder’s population almost cancel; indeed, the time scale of this phase is Te ≈ ln N/(r - δ′). In this regime, the theory above breaks down. More importantly: when the growth of the founder’s colony becomes slow, the adaptation rate slows down too. The adaptation rates presented above therefore require that r is substantially greater than δ′, so that Te is small (see Discussion).
Including a Cost of Resistance.
Drug resistance often requires adaptations that carry a fitness cost in the absence of the drug (13, 29). To examine the effect of such a cost, we slightly modify the model. So far, in compartment i, all bacteria with genotype g≥i had the same growth rate r. Instead, we now assume that bacteria with g > i (more resistant than necessary) grow a factor 1 - s more slowly than those with g = i. The cost s can be interpreted as a local selection coefficient.
We included the fitness cost in our theory (see SI Text and Movie S2) (22). A fitness cost only alters the blue path. The result shows, however, that the effect is small provided 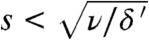 . That is, unless the migration rate is tiny compared to the death rate, a large fitness cost is required to significantly hamper adaptation. We have tested this prediction with simulations at various values of s, presented in Fig. 5A. The results confirm that the adaptation rate is only mildly affected unless
. That is, unless the migration rate is tiny compared to the death rate, a large fitness cost is required to significantly hamper adaptation. We have tested this prediction with simulations at various values of s, presented in Fig. 5A. The results confirm that the adaptation rate is only mildly affected unless 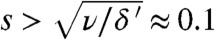 , which in population genetics is considered a very large fitness cost, in particular at large population sizes (here, K = 105) (30).
, which in population genetics is considered a very large fitness cost, in particular at large population sizes (here, K = 105) (30).
Fig. 5.
Consequences of a resistance cost or a finite growth rate below the staircase. Circles present simulation results; lines are smooth interpolations. (A) The adaptation rate is insensitive to the cost s unless 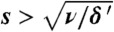 , where δ ' ≡ δ + 2ν. (B) It is also insensitive to the growth rate of the bacteria below the staircase, r0. The regime r0 > δ is outside the scope of this study, because there adaptation is not required to colonize all compartments. Nevertheless, we see that the adaptation rate first increases, but then rapidly decreases as r0 approaches r; in this limit a founder, experiences strong competition from less-resistant immigrants, which grow at a very similar rate r0 ≈ r. (Parameters are as in Fig. 1 except that μf = 10-6/h in Fig. 4A).
, where δ ' ≡ δ + 2ν. (B) It is also insensitive to the growth rate of the bacteria below the staircase, r0. The regime r0 > δ is outside the scope of this study, because there adaptation is not required to colonize all compartments. Nevertheless, we see that the adaptation rate first increases, but then rapidly decreases as r0 approaches r; in this limit a founder, experiences strong competition from less-resistant immigrants, which grow at a very similar rate r0 ≈ r. (Parameters are as in Fig. 1 except that μf = 10-6/h in Fig. 4A).
To examine the maximal effect of a resistance cost, we consider the case where genotypes g > i do not grow at all. Dynamically, the blue and red paths then become very similar, except that the roles of mutation and migration are reversed. Indeed, they are now equally fast (see SI Text). It can then be shown that the adaptation is slowed down by at most a factor 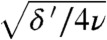 . For the parameters in Fig. 5A this amounts to a factor of 5. The effect of a fitness cost is therefore generally mild, unless s is very large and the migration rate is very low compared to the death rate.
. For the parameters in Fig. 5A this amounts to a factor of 5. The effect of a fitness cost is therefore generally mild, unless s is very large and the migration rate is very low compared to the death rate.
The cost of resistance does influence the comet tail. A comet tail arises when strains migrating leftwards out-compete strains with a lower resistance. In the presence of a high resistance cost, however, overly resistant strains have a reduced growth rate and will no longer be able to replace their less resistant cousins. This happens when the rate of immigration, νN, is more than compensated by the loss in total growth rate suffered due to the fitness cost, which is approximately rsN(1 - N/K) when the immigrants reach fixation. Thus we directly obtain that the comet tail is suppressed if s > ν/δ′ (a precise derivation is given in the SI Text). Then, the population is restricted to a narrow band above the staircase, and every compartment becomes populated with specialists adapted precisely to the local antibiotic concentration, as in Fig. 3D.
A Finite Growth Rate Below the Staircase.
Thus far, we have assumed that the genotypes below the staircase cannot grow at all, which is probably not realistic. We therefore study how the dynamics change if bacteria below the staircase do grow, albeit at a reduced rate r0. If r0 > δ, the bacteria do not need to adapt to start a growing population in all compartment, because their growth rate exceeds their death rate from the start. Since this is not the scenario we set out to study, we are interested in the regime r0 ≤ δ.
Fig. 5B shows the adaptation rate as a function of r0, obtained by simulations. Surprisingly, the adaptation rate is hardly affected as long as r0 is smaller than the death rate. In this case an immigrant is indeed likely to die before it can grow. Only when r0 approaches δ does the adaptation speed up; but even at r0 = δ (the dashed line) the effect is less than a factor of two. This suggests that the growth rate below the staircase is of minor importance. This turns out to be true quite generally, as we derive in the SI Text. Essentially, even though increasing r0 speeds up the red path, given the constraint r0 < δ it will never become faster than the blue path (in the absence of a resistance cost). At best, both paths can become equally fast, in which case the total rate of adaptation will be doubled.
An Increased Death Rate Below the Staircase.
We have so far assumed that the antibiotic interfered with the growth of the bacteria. Some antibiotics, however, affect a bacterium’s death rate instead. How does an increased death rate for bacteria below the staircase affect adaptation?
Increasing the death rate below the staircase, now called δ0, will inhibit the red path: if δ0 is high, immigrants tend to die before they have a chance to grow or mutate. Therefore, when δ0 is increased, the red path is gradually eliminated, until only the blue path remains. At the same time we have seen that, in the absence of a resistance cost, the red path is typically slower than the blue one to begin with, so that the rate of adaptation is dominated by the rate of the blue path. At best, the red path is initially as fast as the blue one, in which case eliminating it leads to a two-fold slowdown. The adaptation rate must therefore be insensitive to changes in δ0.
Fig. S5 shows results for the special case in which the growth rate below the staircase is not affected at all by the drug (r0 = r), but the death rate is. For otherwise fixed parameters, we vary δ0. As predicted, the adaptation rate is very insensitive to δ0. The theoretical rate of the red path is also plotted, and decreases rapidly with δ0. Only if δ0 is minimal (δ0 ≈ r0) does the red path contribute somewhat to the adaptation rate, resulting in just a slight increase.
All-to-all Migration.
In all the above cases, migration was possible between neighboring compartments only. This is reasonable if the compartments are lined up in a quasi-one-dimensional concentration gradient, as realized in the experiments of (23). This is not necessarily always the case, however. E.g., transmission of bacteria from one organ to another or from one patient to another need not follow the order of drug concentrations. To demonstrate that a strict ordering of the compartments is not required for the qualitative results described above, we briefly consider a variation of the model in which migration is possible between any two compartments.
It is beyond the scope of this article to fully characterize this model, but simulation results can provide a proof of principle; see Supporting Fig. S6 and Movie S3. As before, the population quickly adapts and expands, following the familiar stepwise pattern. Such simulations establish that no linear ordering of the compartments is required for fast adaptation and range expansion to occur.
However, because migration is now possible between any two compartments, the dynamics become different at the quantitative level. Besides the blue and red paths—still the shortest paths to adaptation—the all-to-all migration introduces many alternatives. The most resistant strain at a given time will spread rapidly to all other compartments. The next mutation can then occur in any of those, from where the mutant has a chance to migrate to the uncolonized compartment with the lowest drug concentration and establish a colony. Even though each path of this type requires at least three events (e.g., migration, mutation, and again migration), many such paths are possible, and together they cannot be ignored. An interesting consequence is that each subsequent adaptation step is slightly different, because the number of already colonized compartments increases; as a result, the adaptation rate increases in time, as shown in Fig. S3.
Discussion
We presented a stochastic model to study the evolution of antibiotic resistance in the presence of a gradient of antibiotic. The model combines ideas from several fields, including the concepts of sanctuaries, the selective window, the notion of resistance-selective environments, and the theories of source–sink dynamics and range expansion. The model was purposefully kept simple: thus the roles of growth, death, migration, mutation, population size, fitness cost of resistance, and spatial geometry could be characterized quantitatively. It serves as the starting point for more realistic and complex models in future studies.
Our results show that gradients of antibiotics can lead to a mode of adaptation that is qualitatively different from evolution in uniform environments. A key point is that, in heterogeneous environments, adaptation and range expansion are likely coupled. This fundamentally distinguishes the staircase model from other models with spatially structured populations (31–36), or models of evolution at the front of growing colonies (37). This has important consequences. First, it allows more resistant mutants, with an expanded range, to evade competition by invading previously inaccessible habitats. Second, compartments with a low antibiotic concentration act as sanctuaries, allowing mutations to be selected one by one, whereas in homogeneous environments all mutations are required at once to survive a drug treatment. In this sense, heterogeneity truly provides a staircase to resistance. Third, because the population expands until the drug concentration at the border of its range is just above the current MIC, this concentration is always conveniently in the selective window of the next mutation. Nevertheless, similar adaptation rates can also be obtained in uniform environments, provided each mutation required for resistance is under strong selection (s > 0.1).
However, an important caveat was found: to obtain a fast adaptation rate, the growth rate of the founder needs to be substantially higher than its death rate. In the staircase model, the growth rate abruptly drops from its maximum value r at i ≤ g to its minimal value r0 at i > g (the plateau-like fitness landscape of Fig. S2C). As a result, the founder can have a growth rate r substantially higher than δ′ in compartment i and a growth rate r0 smaller than δ′ in compartment i + 1, as illustrated by the red lines in Fig. S7A. If the growth rate is a smoother function (black lines), the founder’s growth rate is expected to be closer to the death rate (compare the sizes of the two double-headed arrows), which could considerably slow down adaptation. To test how the growth of real bacteria depends on the antibiotic concentration, we have measured the growth rate of an Escherichia coli strain subjected to various concentrations of the antibiotic chloramphenicol (Cm). This strain (EQ92, see SI Text) harbored weak constitutive expression of Cm Acetyltransferase (CAT), providing partial resistance to Cm. The results, plotted in Fig. S7B, indicate that growth is halted abruptly when [Cm] > 0.5 mM. Such an abrupt dependence of bacterial growth on drug concentration may arise generically due to a global, growth-rate dependent positive feedback loop (38) caused by a generic negative effect of antibiotics on gene expression (39) (including the expression of drug resistance such as CAT or other endogenous mechanisms such as multidrug efflux pumps), and the negative effect of this expression on the efficacy of antibiotics (manuscript in preparation). Given the caveat discussed above, this positive feedback loop may contribute to a fast adaptation. Yet, decisive tests will have to come from evolution experiments; setups using microfluidic devices seem ideally suited to provide a proof of concept in controlled laboratory conditions (23).
Lastly, we note that although the staircase model was conceived to study the evolution of antibiotic resistance, its formulation is quite general and may be applicable to other evolutionary processes that involve changes in a species’ range. In the course of evolution, species have adapted to a huge variety of conditions and now populate virtually every corner of the earth. To understand the evolutionary processes that produced this diversity, a thorough understanding of the role of adaptive range expansion seems crucial. We hope that the staircase model will contribute to this understanding as well.
Materials and Methods
All simulations were performed using the standard kinetic Monte Carlo algorithm. For the effect of antibiotic on the growth of partially resistant bacteria, we constructed an E. coli K-12 strain constitutively expressing CAT from the chromosome. This strain was grown in minimal medium containing various sublethal amounts of Cm. (See SI Text for details.)
Supplementary Material
ACKNOWLEDGMENTS.
We thank Herbert Levine, Shumo Liu, David Kessler, Sidney Redner, Weiqun Peng, and Matthew Scott for insightful discussions, and Zhongge Zhang for contributing strain EQ92. This work was supported by the Center for Theoretical Biological Physics (NSF PHY-0822283), by the NIH (RO1GM-095903), and via a subcontract by the NCI Physical Science-Oncology program (1 U54 CA143803). JBD also acknowledges the support of a NSF Graduate Research Fellowship.
Note.
While this work was under review, a related manuscript was brought to our attention: Greulich P, Waclaw B, Allen RJ, “Interplay between mutational pathway and drug gradient controls time to evolution of drug resistance,” arXiv:1202.5431v3.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117716109/-/DCSupplemental.
References
- 1.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 2.MacLean RC, Hall AR, Perron GG, Buckling A. The population genetics of antibiotic resistance: Integrating molecular mechanisms and treatment contexts. Nat Rev Genet. 2010;11:405–414. doi: 10.1038/nrg2778. [DOI] [PubMed] [Google Scholar]
- 3.Sharma SK, Mohan A. Multidrug-resistant tuberculosis—A menace that threatens to destabilize tuberculosis control. Chest. 2006;130:261–272. doi: 10.1378/chest.130.1.261. [DOI] [PubMed] [Google Scholar]
- 4.LeClerc JE, Li BG, Payne WL, Cebula TA. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 5.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olofsson SK, Geli P, Andersson DI, Cars O. Pharmacodynamic model to describe the concentration-dependent selection of cefotaxime-resistant Escherichia coli. Antimicrob Agents Chemother. 2005;49:5081–5091. doi: 10.1128/AAC.49.12.5081-5091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott AM, Berning SE, Iseman MD, Peloquin CA. Failure of drug penetration and acquisition of drug-resistance in chronic tuberculous empyema. Tuber Lung Dis. 1995;76:463–467. doi: 10.1016/0962-8479(95)90016-0. [DOI] [PubMed] [Google Scholar]
- 8.Baquero F, Negri MC. Selective compartments for resistant microorganisms in antibiotic gradients. Bioessays. 1997;19:731–736. doi: 10.1002/bies.950190814. [DOI] [PubMed] [Google Scholar]
- 9.Kepler TB, Perelson AS. Drug concentration heterogeneity facilitates the evolution of drug resistance. Proc Natl Acad Sci USA. 1998;95:11514–11519. doi: 10.1073/pnas.95.20.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkala EJ, He J, West JT, Wood C, Petito CK. Compartmentalization of HIV-1 in the central nervous system: Role of the choroid plexus. AIDS. 2005;19:675–684. doi: 10.1097/01.aids.0000166090.31693.aa. [DOI] [PubMed] [Google Scholar]
- 11.Baquero F, Negri MC, Morosini MI, Blazquez J. Antibiotic-selective environments. Clin Infect Dis. 1998;27:S5–S11. doi: 10.1086/514916. [DOI] [PubMed] [Google Scholar]
- 12.Baquero F, Canton R. In: Antimicrobial Drug Resistance, Infectious Disease. Mayers DL, editor. Vol 1. Totowa, NJ: Humana Press; 2009. pp. 9–32. [Google Scholar]
- 13.Lipsitch M. The rise and fall of antimicrobial resistance. Trends Microbiol. 2001;9:438–444. doi: 10.1016/s0966-842x(01)02130-8. [DOI] [PubMed] [Google Scholar]
- 14.Holt RD, Gomulkiewicz R. The evolution of species’ Niches: A population dynamic perspective. In: Othmer HG, Adler FR, Dallon JC, editors. Case Studies in Mathematical Modeling—Ecology, Physiology, and Cell Biology. Englewood Cliffs, NJ: Prentice Hall; 1997. pp. 25–50. [Google Scholar]
- 15.Ronce O, Kirkpatrick M. When sources become sinks: Migrational meltdown in heterogeneous habitats. Evolution. 2001;55:1520–1531. doi: 10.1111/j.0014-3820.2001.tb00672.x. [DOI] [PubMed] [Google Scholar]
- 16.Holt RD, Gomulkiewicz R, Barfield M. The phenomology of niche evolution via quantitive traits in a ‘black-hole’ sink. Proc Biol Sci. 2003;270:215–224. doi: 10.1098/rspb.2002.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chattopadhyay S, et al. Haplotype diversity in “source-sink” dynamics of Escherichia coli urovirulence. J Mol Evol. 2007;64:204–214. doi: 10.1007/s00239-006-0063-5. [DOI] [PubMed] [Google Scholar]
- 18.Sokurenko EV, Gomulkiewicz R, Dykhuizen DE. Opinion—Source-sink dynamics of virulence evolution. Nat Rev Microbiol. 2006;4:548–555. doi: 10.1038/nrmicro1446. [DOI] [PubMed] [Google Scholar]
- 19.Dennehy JJ, Friedenberg NA, McBride RC, Holt RD, Turner PE. Experimental evidence that source genetic variation drives pathogen emergence. Proc Biol Sci. 2010;277:3113–3121. doi: 10.1098/rspb.2010.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caprio MA. Source-sink dynamics between transgenic and non-transgenic habitats and their role in the evolution of resistance. J Econ Entomol. 2001;94:698–705. doi: 10.1603/0022-0493-94.3.698. [DOI] [PubMed] [Google Scholar]
- 21.Perron GG, Gonzalez A, Buckling A. Source-sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proc Biol Sci. 2007;274:2351–2356. doi: 10.1098/rspb.2007.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermsen R, Hwa T. Sources and sinks: a stochastic model of evolution in heterogeneous environments. Phys Rev Lett. 2010;105:28410(4). doi: 10.1103/PhysRevLett.105.248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang QC, et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science. 2011;333:1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 24.Dykhuizen D, Hartl DL. Selective neutrality of 6PGD allozymes in E. coli and the effects of genetic background. Genetics. 1980;96:801–817. doi: 10.1093/genetics/96.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge University Press; 1983. [Google Scholar]
- 26.Kohler T, MicheaHamzehpour M, Plesiat P, Kahr AL, Pechere JC. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2540–2543. doi: 10.1128/aac.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babic M, Bonomo RA. In: Antimicrobial Drug Resistance, Infectious Disease. Mayers DL, editor. Vol 1. Humana Press; 2009. pp. 65–74. [Google Scholar]
- 28.Redner S. A Guide to First-Passage Processes. Cambridge, UK: Cambridge University Press; 2001. [Google Scholar]
- 29.Andersson DI. The biological cost of mutational antibiotic resistance: Any practical conclusions? Curr Opin in Microbiol. 2006;9:461–465. doi: 10.1016/j.mib.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Hartl DL. A Primer of Population Genetics. 3rd Ed. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- 31.Fisher RA. The wave of advance of advantageous genes. Ann Eugen. 1937;7:355–369. [Google Scholar]
- 32.Caprio MA, Tabashnik BE. Gene flow accelerates local adaptation among finite populations—Simulating the evolution of insecticide resistance. J Econ Entomol. 1992;85:611–620. [Google Scholar]
- 33.Lipsitch M, Levin BR. Population dynamics of tuberculosis treatment: Mathematical models of the roles of non-compliance and bacterial heterogeneity in the evolution of drug resistance. Int J Tuberc Lung Dis. 1998;2:187–199. [PubMed] [Google Scholar]
- 34.Smith DL, Dushoff J, Perencevich EN, Harris AD, Levin SA. Persistent colonization and the spread of antibiotic resistance in nosocomial pathogens: Resistance is a regional problem. Proc Natl Acad Sci USA. 2004;101:3709–3714. doi: 10.1073/pnas.0400456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leimar O, Doebeli M, Dieckmann U. Evolution of phenotypic clusters through competition and local adaptation along an environmental gradient. Evolution. 2008;62:807–822. doi: 10.1111/j.1558-5646.2008.00334.x. [DOI] [PubMed] [Google Scholar]
- 36.Debarre F, Lenormand T, Gandon S. Evolutionary epidemiology of drug-resistance in space. Plos Comput Biol. 2009;5:e1000337. doi: 10.1371/journal.pcbi.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hallatschek O, Hersen P, Ramanathan S, Nelson DR. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci USA. 2007;104:19926–19930. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: Origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.