Abstract
Microorganisms often form symbiotic relationships with eukaryotes, and the complexity of these relationships can range from those with one single dominant symbiont to associations with hundreds of symbiont species. Microbial symbionts occupying equivalent niches in different eukaryotic hosts may share functional aspects, and convergent genome evolution has been reported for simple symbiont systems in insects. However, for complex symbiont communities, it is largely unknown how prevalent functional equivalence is and whether equivalent functions are conducted by evolutionarily convergent mechanisms. Sponges represent an evolutionarily divergent group of species with common physiological and ecological traits. They also host complex communities of microbial symbionts and thus are the ideal model to test whether functional equivalence and evolutionary convergence exist in complex symbiont communities across phylogenetically divergent hosts. Here we use a sampling design to determine the phylogenetic and functional profiles of microbial communities associated with six sponge species. We identify common functions in the six microbiomes, demonstrating the existence of functional equivalence. These core functions are consistent with our current understanding of the biological and ecological roles of sponge-associated microorganisms and also provide insight into symbiont functions. Importantly, core functions also are provided in each sponge species by analogous enzymes and biosynthetic pathways. Moreover, the abundance of elements involved in horizontal gene transfer suggests their key roles in the genomic evolution of symbionts. Our data thus demonstrate evolutionary convergence in complex symbiont communities and reveal the details and mechanisms that underpin the process.
Keywords: convergent evolution, metagenomics, nitrogen metabolism, eukaryotic-like proteins
Microorganisms form symbiotic relationships with eukaryotes encompassing all evolutionary stages, from simple amoebae to mammals. Symbiotic systems can range in complexity from those with a single dominant microorganism [e.g., Wolbachia in insects (1) or Vibrio in squids (2)] to those with hundreds of obligate or facultative microbial symbionts [e.g., communities in termite hindgut (3) or human colon (4)]. Mechanisms that shape the structure of complex symbiont communities are largely unknown (5); however, recent work on communities of free-living microorganisms indicates that both niche and neutral effects can play a role (6, 7).
Microorganisms can form different kinds of associations with eukaryotic hosts ranging from a facultative epiphytic life on green algae (8) to obligate intracellular symbiosis in insects (9). Symbionts can be acquired horizontally by the host, for example from the seawater for green algae (8) or from food in the human gut (10), and this mode of acquisition will lead to recruitment and selection based on symbiont function. This niche selection can be decoupled from any symbiont taxonomy, thus leading to functionally coherent but phylogenetically divergent communities (11, 12). Symbionts also can be transmitted vertically through reproductive cells and larvae, as has been demonstrated in sponges (13, 14), insects (15), ascidians (16), bivalves (17), and various other animals (18). Vertical transmission generally leads to microbial communities with limited variation in taxonomy and function among host individuals.
Niches with similar selections may exist in phylogenetically divergent hosts that lead comparable lifestyles or have similar physiological properties. Symbionts occupying these equivalent niches therefore might also share functional aspects. This expectation is supported by the recent observation that two phylogenetically distinct bacterial endosymbionts found respectively in sharpshooters or cicadas possess analogous proteins for methionine synthesis (19). In complex symbiont communities, however, it is largely unknown how prevalent functional equivalence is and whether equivalent functions are conducted by evolutionarily convergent mechanisms.
Sponges (phylum Porifera) are basal metazoa that form a major part of the marine benthic fauna across the world’s oceans (20, 21). Despite their evolutionary divergence, sponges have maintained many common physiological characters and ecological roles, including the filter-feeding of planktonic microorganisms and particulate matter (20). Sponges also host complex communities of microbial symbionts, and extensive research over the last decade has documented the phylogenetic diversity and biogeography of sponge-associated microorganisms (22, 23). Symbiont communities in sponges generally are highly specific to host species and are mostly consistent across time and space (22). This stable association can be explained by the vertical transmission of symbionts through larvae (24). However, it has been challenging to identify phylotypes that are common to all sponges and hence represent archetypal symbionts (23). Based on these characteristics, the sponge microbiome represents an ideal model system to test if functional equivalence exists across divergent hosts and to study the mechanisms that shape complex symbiont communities.
Here we use a sampling design to address functional equivalence in complex symbioses by analyzing the phylogenetic and functional composition of symbiont communities in six phylogenetically divergent sponge species. Using a metagenomic approach and comparison with planktonic communities, we identify common functions of sponge symbionts in all six microbiomes. These core functions cover various aspects of metabolism and, importantly, are provided in each sponge species by functionally equivalent symbionts, analogous enzymes, or biosynthetic pathways. Moreover, the abundance of mobile genetic elements suggests that horizontal gene transfer (HGT) has a key role in distributing core functions among symbionts during their coevolutionary association with their host and facilitates functional convergence on the community scale.
Results and Discussion
Overview of Samples and Dataset.
Six sponge species were selected to cover a wide selection of sponge morphologies, incorporate taxonomically diverse species, and encompass a broad geographic range (tropical vs. temperate) (Table 1, Fig. 1, and Materials and Methods). Seawater samples were collected as described by Thomas et al. (26). The microbiomes of 21 samples (comprising six sponges and three water samples, in triplicate) were sequenced via a shotgun strategy, and the resulting reads were assembled, filtered for eukaryotic sequences, and annotated (Materials and Methods and Table S1). We obtained 8,373,475 unique, predicted protein-coding sequences with an average of 398,737 sequences per sample.
Table 1.
Sponge and seawater samples used in the present study
| Sponge/seawater | Cell fraction | Sample ID | Location | Depth (m) | Date |
| Seawater | 0.1–0.8 μm | SW01-A, SW01-B | Bare Island, Sydney | 2 | Oct. 18. 2006 |
| Seawater | 0.8–3 μm | SW08-A | Bare Island, Sydney | 2 | Oct. 18, 2006 |
| Cymbastela coralliophila | <3 μm | Cyr-A, Cyr-B, Cyr-C | Palm Island, Great Barrier Reef | 7–10 | July 29, 2009 |
| Rhopaloeides odorabile | <3 μm | Rho-A, Rho-B, Rho-C | Palm Island, Great Barrier Reef | 7–10 | July 30, 2009 |
| Stylissa sp. 445 | <3 μm | Sty-A, Sty-B, Sty-C | Palm Island, Great Barrier Reef | 7–10 | Aug. 1, 2009 |
| Cymbastela concentrica | <3 μm | Cyn-A, Cyn-B, Cyn-C | Bare Island, Sydney | 7–10 | Sept. 15, 2009 |
| Scopalina sp. | <3 μm | Sco-A, Sco-B, Sco-C | Bare Island, Sydney | 7–10 | Sept. 23, 2009 |
| Tedania anhelans | Total | Ted-A, Ted-B, Ted-C | Bare Island, Sydney | 7–10 | Sept. 17, 2009 |
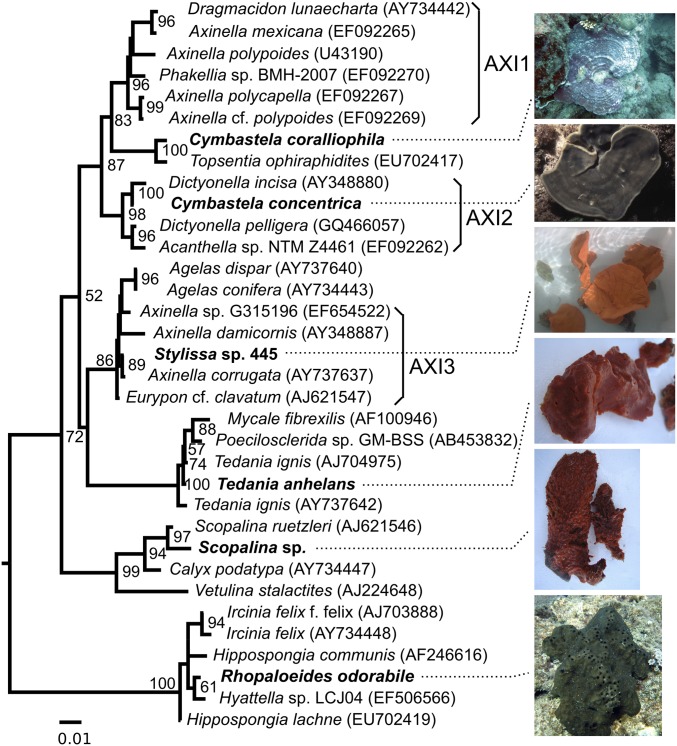
Fig. 1.
Phylogenetic relationship of the sponges used in this study based on 18S rRNA sequences. The maximum-likelihood tree is constructed with a sequence alignment length of 1,694 nt; percentage bootstrapping values (1,000 replications) greater than 50% are shown. The tree is rooted to the coral Acanthogorgia granulata (FJ643593). Sponges from the present study are shown in bold. The Axinella clades are named according to Gazave et al. (25). The photographs of C. coralliophila and R. odorabile were provided by Heidi Luter (Townsville, Australia). The photograph of C. concentrica was provided by Michael Taylor (Auckland, New Zealand).
Sponge Microbial Communities Possess Distinct Phylogenetic and Taxonomic Profiles.
To characterize initially the diversity of the microbial communities associated with the sponge and seawater samples, we reconstructed 16S rRNA gene sequences from the metagenomic datasets (Material and Methods and Table S1). Sixty-two operational taxonomic units (OTUs) at a cutoff distance of 0.03 were generated, and analysis of the 35 most abundant OTUs showed distinct microbial-community profiles among the sponges (Fig. 2). Replicate samples generally were very similar, as is consistent with the previous concept of sponge-specific, stable microbial associations (22). The similarity between replicates also was confirmed by analyzing the community profiles using single-copy genes (SCGs) (SI Results and Discussion and Fig. S1 A–C). The microbial communities of the six sponges represented a wide spectrum of community diversity, evenness, and shared phylotypes (Fig. 2), consistent with other comparative analyses of sponge microbial communities (13, 27). For example, Scopalina sp. and Tedania anhelans were dominated by two very similar phylotypes (less than 0.03 phylogenetic distance) belonging to the Nitrosomonadaceae (Fig. 2 and Fig. S2A), and Rhopaloeides odorabile and Cymbastela coralliophila had more even species distributions with limited overlap (Fig. 2 and Fig. S1 D and E). Both sponge-specific and general marine bacteria and thaumarchaeota were detected in different abundances in these sponges (SI Results and Discussion).
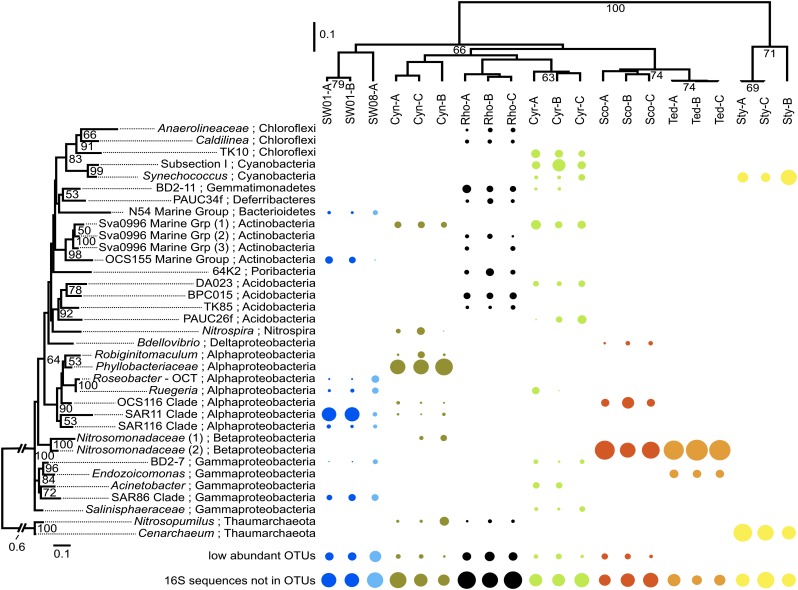
Fig. 2.
Microbial community diversity of sponge and seawater samples. (Right) The relative abundance of the 35 most abundant OTUs (according to the sum of the relative abundance across all samples). Phylogenetic distance cutoff for OTU generation is 0.03. The size of the circle reflects the relative abundance of an OTU in a sample. (Left) Maximum-likelihood tree of the OTUs. Bootstrapping percentages greater than 50% are given (1,000 replications). The tree is rooted with the archaeal clade. Samples are clustered based on the phylogenetic relationships of their OTUs (the top 35 OTUs and the other, low-abundant OTUs) using the weighted Unifrac algorithm with 1,000 rounds of Jackknife values (in percentages) shown in nodes. “16S rRNA sequences not in OTUs” indicates reads that fail to assemble into contigs used for OTU generation.
The different planktonic fractions (i.e., 0.1–0.8 μm, and 0.8–3 μm) showed distinct community profiles compared with the sponge samples and were dominated by organisms from SAR11, SAR86, the Roseobacter clade, Flavobacteriaceae, and the OCS155 Marine Group. These taxa are found frequently in seawater from around the globe (28), as is consistent with the previous notion that sponges contain microbial consortia that are distinct from those of the surrounding seawater (22).
Functional Annotation Reveals Shared Genomic Signatures in Sponge Symbionts.
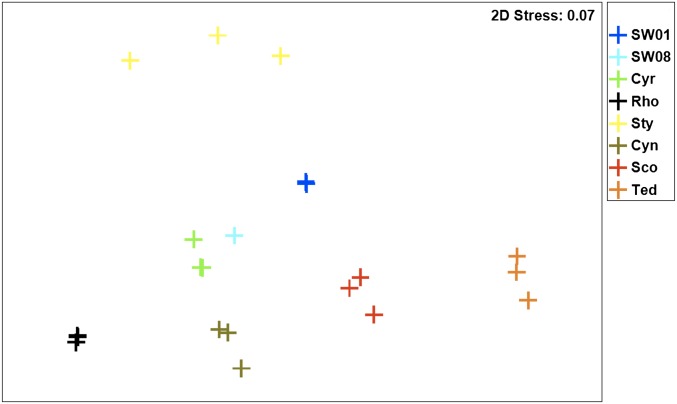
Consistent with the phylogenetic analysis, functional gene annotation based on comparison with the Clusters of Orthologous Group (COG) (29), Protein Family A (Pfam-A) (30), and SEED/Subsystem (31) databases showed that replicate samples were very similar. Microbiomes for each sponge species generally contained distinct gene compositions reflecting specific ecological functions or interactions within their host (Fig. 3 and Fig. S3 A and B). Functional profiles of the community in Stylissa sp. 445 were more distantly related to the other samples because the dominant thaumarchaeal symbionts were poorly represented in the reference databases (Table S1). Interestingly, functional profiles of the communities in Scopalina sp. and T. anhelans were quite distinct, although both were dominated by closely related phylotypes (Fig. 2 and Fig. S2A).
Fig. 3.
Multidimensional scaling (MDS) plots of samples by Bray–Curtis similarity according to COG functional annotation.
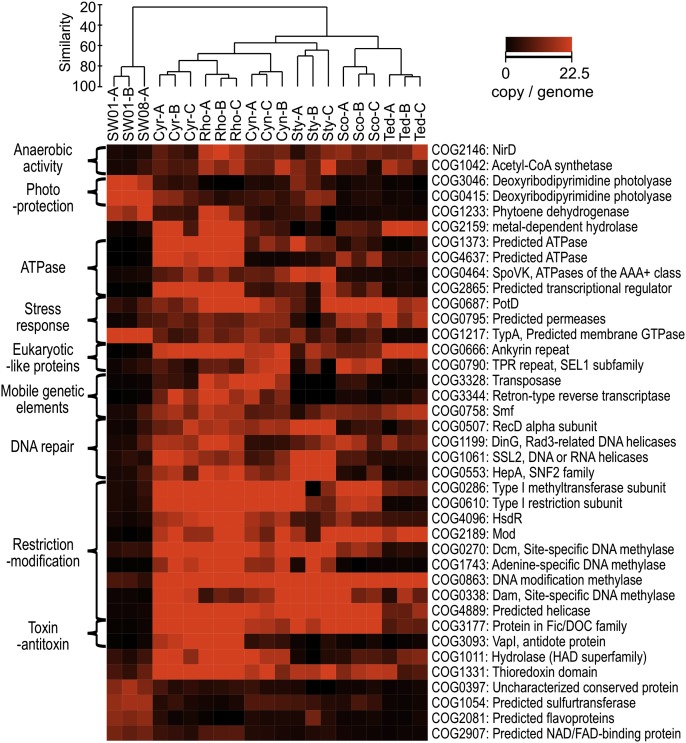
Despite the host-specific functional profiles, statistical analysis (Materials and Methods) identified a range of functional features that distinguish the sponge-associated communities from the microbial communities found in seawater (Fig. 4 and Fig. S3 C and D). None of these differential functions included the 32 protein functions that were found to vary between the planktonic communities of temperate and tropical waters during the large-scale analysis of the Global Ocean Survey (28), supporting the notion that the specificity of sponge symbiotic functions is not an artifact of the specific planktonic references analyzed. The discovery of these common functions in sponge microbiomes indicates similar niches in these divergent hosts. The distinct phylogenetic structure of the microbial communities among these sponges further implies that functionally equivalent symbionts with convergent genomic contents may occupy these niches. These core features are likely to be of general importance for the adaptation of microorganisms to the sponge host environment and revealed some of the basic principles underpinning the symbiotic interactions. These characteristics are discussed in the next sections and in SI Results and Discussion.
Fig. 4.
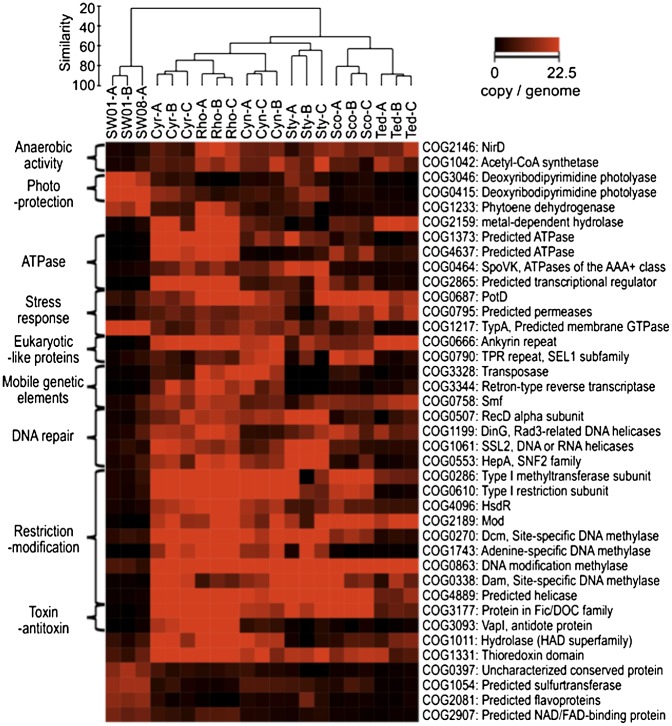
Specific functions abundant in sponge-associated or planktonic microbial communities annotated with COG. The brightness (red) in the heatmap reflects the abundance (copies per genome) of a particular function in a sample. Samples are clustered by Bray–Curtis similarity and average linkage analysis.
Nitrogen Metabolism and Adaptation to Anaerobic Conditions.
The contributions of planktonic and sediment microbial communities to the marine nitrogen cycle is well appreciated (32), but only recently have studies revealed a high rate of nitrogen metabolism in marine microorganisms associated with invertebrate hosts, especially the reef-building corals and sponges (33–35). Nitrogen fixation (36), nitrification (37, 38), anaerobic respiration of ammonium (anammox) (38), and denitrification (38, 39) activities have been analyzed separately in various marine sponges by stable isotope probing or 16S rRNA or functional gene analyses. In the present study, nitrogen metabolism-related functions were enriched significantly in sponge samples (Fig. 4 and Fig. S3 C and D).
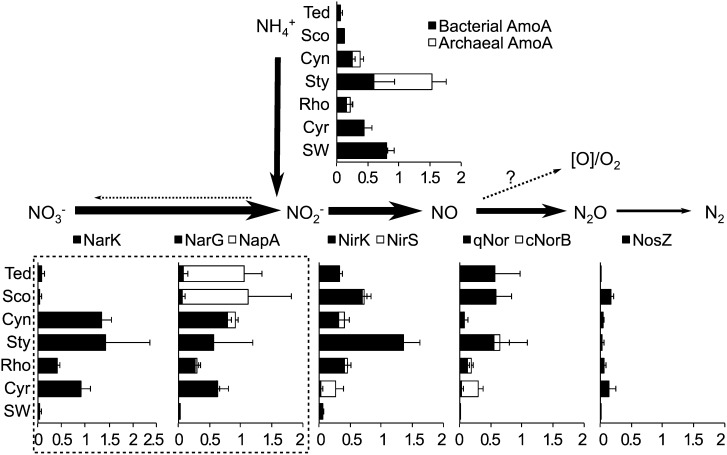
Further detailed annotation provides an opportunity to investigate aspects of the nitrogen cycle in an integrated way across a range of sponge systems. Compared with the planktonic samples, sponge-associated communities generally contained many more genes related to denitrification (Fig. 5). Key enzymes in the first two steps, namely the respiratory nitrate reductase (cytoplasmic NarG or periplasmic NapA) and respiratory nitrite reductase (copper-containing NirS or cytochrome cd1-dependent NirK), were present at 0.3–1.2, and 0.3–1.4 copies per genome, respectively. Interestingly, specific reductase groups were preferentially found in certain sponge symbiont communities. For example, the periplasmic NapA was abundant in the symbiont community of Scopalina sp. and T. anhelans, whereas the cytoplasmic NarG was found more frequently in the other four sponges (Fig. 5). The NarG-rich communities also showed a high number of nitrate/nitrite antiporters (NarK), which import nitrate into the cytoplasm and export nitrite from it (Fig. 5) (40). This result showed that different sponge communities use analogous pathways for denitrification, although it is not clear why these preferences for NarG or NapA exist.
Fig. 5.
Abundance of enzymes in the energy-producing (respiratory) pathways of nitrogen cycling. With the exception of AmoA (Pfam annotation), abundances of enzymes are obtained from SEED/Subsystem annotation (Materials and Methods). Units on the horizontal axis indicate copies per genome. Error bars show SDs.
Further variations in enzyme composition became apparent for the subsequent two steps of denitrification, where generally lower copy numbers of nitric oxide reductase (quinol-dependent qNor or cytochrome c-dependent cNorB) and nitrous oxide reductase (NosZ) were observed, especially in R. odorabile and Cymbastela concentrica. This variation suggests that at least some of the denitrifiers in the sponge microbial communities have incomplete or alternative pathways of nitrate reduction and may accumulate nitric or nitrous oxide (41, 42). Recent studies showed that the methanotrophic bacterium Candidatus Methylomirabilis oxyfera could convert nitric oxide directly to nitrogen and oxygen (43), the latter being used subsequently for methane oxidation. Because oxygen can diffuse only ∼1 mm into sponge tissue, many parts of the sponge become anoxic when pumping stops (44, 45). Dismutation of nitric oxide to produce oxygen might enable the sponge symbionts to maintain aerobic respiration during periods when the host is not actively pumping. However, further analysis would be required to elucidate these theories fully. The organisms putatively involved in denitrification are discussed in SI Results and Discussion.
We also detected genes of both bacterial (PF05145) and archaeal (PF12942) ammonium monooxygenase subunit A (AmoA) (Fig. 5). Abundance varied from very low in Scopalina sp. and T. anhelans to an average of more than one gene copy per genome for Stylissa sp. 445. Although bacterial AmoA appeared to be used exclusively in some sponges, in others (such as Stylissa sp. 445) both bacterial and archaeal ammonium oxidation could take place. In contrast, a recent gene-specific survey of four deep-sea sponges reported that their microbial communities were strongly dominated by the presence of archaeal AmoA rather than the bacterial counterpart (46). Thaumarchaeota phylotypes potentially involved in archaeal ammonia oxidation in our sponges and other aspects of the nitrogen cycle are described in SI Results and Discussion.
Our integrated analysis shows that different sponges host distinct microorganisms that use different enzymes to perform equivalent functions in denitrification and ammonium oxidation. Similar findings regarding the nitrogen cycle also were reported very recently for the microbial communities of three Mediterranean sponges (47). However, not every sponge community in our study encodes for the complete nitrogen cycle. Although respiratory nitrate reduction was ubiquitous, reflecting temporary or permanent anoxic conditions within the interior of sponges (48), many subsequent steps in denitrification reflected incomplete pathways. An alternative to dissimilatory nitrite reduction for supporting anaerobic growth may be acetogenesis. Acetyl-CoA synthetase (ADP forming, COG1042) was abundant in all sponge samples (Fig. 4) and represents the major energy-conserving reaction through substrate level phosphorylation during peptide, pyruvate, and sugar fermentation to acetate (49).
Nutrient Utilization and Nutritional Interactions with the Host.
Nutritional conditions inside a host are notably different from those in the surrounding seawater, as was reflected clearly in the genomic composition of the sponge symbionts. The abundance of creatininase (creatinine amidohydrolase, EC 3.5.2.10, PF02633) and hydantoinases/oxoprolinase (EC 3.5.2.9, PF01968, PF02538, PF05378), which act on the carbon–nitrogen bonds of cyclic amides, demonstrates the ability of sponge symbionts to degrade and use metabolic intermediates such as creatinine, pyrimidines, or 5-oxoproline. Creatine is a nitrogenous organic acid that uses a high-energy phosphate bond to transfer energy between cells of eukaryotic tissue and is considered a metabolically more stable molecule than ATP (50). Most invertebrates, including sponges, synthesize or use creatine (51, 52), which can be converted nonenzymatically and irreversibly to creatinine in vivo (50). This host-originated creatinine is a valuable source of carbon and nitrogen (53), and the two enzymes mentioned above might be crucial for their efficient utilization by sponge symbionts.
Degradation of benzoic compounds by sponge symbionts also was evidenced by the abundance of enzymes from the metal-dependent hydrolase family (COG2159, PF04909), in particular in C. coralliophila, R. odorabile, and T. anhelans (Fig. 4 and Fig. S3C). This family includes 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase (AMCSD), which converts α-amino-β-carboxy-muconate-ɛ-semialdehyde to α-aminomuconate-semialdehyde. AMCSD is involved in the degradation of 2-nitrobenzoic acid, which prokaryotes can use as a sole source of carbon, nitrogen, and energy (54). The proteins belonging to the glyoxalase/bleomycin resistance protein/dioxygenase superfamily (PF00903) found in C. coralliophila, R. odorabile, and C. concentrica (Fig. S3C) were mostly ring-cleavage extradiol dioxygenases and therefore also are likely to be involved in degradation of aromatic compounds (55).
Underpinning those metabolic pathways for potentially host-derived compounds was a generally high abundance of transporters, including the ABC-type transporters with oligomer-binding (TOBE) domains (PF08402), which deliver various substrates (56), and the oligopeptide/dipeptide transporters (PF08352) of the OPN family, which specifically transport oligopeptides, dipeptides, or nickel (Fig. S3C) (57). Interestingly, transport proteins were among the most highly expressed functions in a recent metaproteomic study of the symbionts in C. concentrica (58).
In addition to nutrient acquisition and utilization by the symbiont community, we discovered potential benefits for the host sponge. Sponge symbiont communities were enriched in the function of ThiS (PF02597) and NMT1/THI5-like protein (PF09084) associated with the synthesis of the essential vitamin thiamin pyrophosphate (vitamin B1), which animals must obtain from their diet (Fig. S3C) (59, 60). Genes for thiamine synthesis also were found in the sponge-associated thaumarchaeon Cenarchaeum symbiosum (61), and vitamin B12 synthesis was identified previously as an abundant function in the microbial community of C. concentrica (26) and the genome of Poribacteria sp (42).
In contrast to the planktonic community, sponge symbionts had very few enzymes involved in the breakdown of dimethylsulfoniopropionate (DMSP) (Fig. S3D). DMSP synthesis is estimated to account for ∼1–10% of global marine primary production (62). A large fraction of planktonic bacteria, including members of the Roseobacter and SAR11 clades (Fig. 2), assimilate sulfur from DMSP (62–64). Our metagenomic results suggested that, in contrast to coral-associated bacteria (65), metabolism of DMSP by sponge symbionts is minor, as is consistent with its concentration in marine sponges being generally low and often below the detection limit (66).
This analysis has highlighted some of the common metabolic features of sponge-associated communities and has provided insight and hypotheses on how cometabolism between the sponge host and microorganisms can underpin symbiosis. It also is apparent that sponge symbionts have evolved specific metabolic profiles that are distinct from those of planktonic microorganisms.
Resistance to Environmental and Host-Specific Stress.
The functional comparison also illustrates that sponge symbionts in general have acquired resistance mechanisms that are tailored specifically to the stress experienced within their host environment. Sponge symbionts were enriched in proteins related to stress responses, such as the universal stress protein (USP) (PF00582) and PotD (COG0687) (Fig. 4 and Fig. S3C). In Escherichia coli, UspA is expressed in response to a wide variety of stressors, including nutrient starvation and exposure to heat, acid, heavy metals, oxidative agents, osmotic stress, antibiotics, and uncouplers of oxidative phosphorylation (67). PotD is a periplasmic protein involved in the uptake of polyamines, such as putrescine, spermidine, and cadaverine (68). Intracellular polyamines are linked to the fitness, survival, and pathogenesis of many bacteria living in host environments (69). Further signal-transduction proteins and regulators with a potential role in stress response are discussed in the SI Results and Discussion.
Sponges or their symbionts are well known for their production of antimicrobial compounds that potentially protect against fouling, predation, and competition (70). Permanent symbionts need to defend themselves against this chemical stress, and the abundance of predicted permeases YjgP/YjgQ (COG0795) likely contributes to this protection (Fig. 4). This family contains LptF and LptG, which transport lipopolysaccharides (LPS) from the inner membrane to the cell surface in Gram-negative bacteria (71). LPS locates in the outer membrane and can serve as a selective permeability barrier against many toxic chemicals, such as detergents and antibiotics (72).
Filter-feeding sponges also can accumulate a high concentration of heavy metals, including mercury (73). As a defense against this toxin, sponge symbionts showed a high abundance of mercuric reductase proteins (Subsystem: Mercuric Reductase), which catalyze the reduction of Hg(II) to elemental mercury Hg(0) and act in the detoxification of the immediate environment (74). Supporting this genomic prediction is the observation that all microbial isolates from an Indian Ocean sponge possessed high levels of resistance against mercury (75).
Eukaryotic-Like Proteins and Their Potential Interaction with the Host.
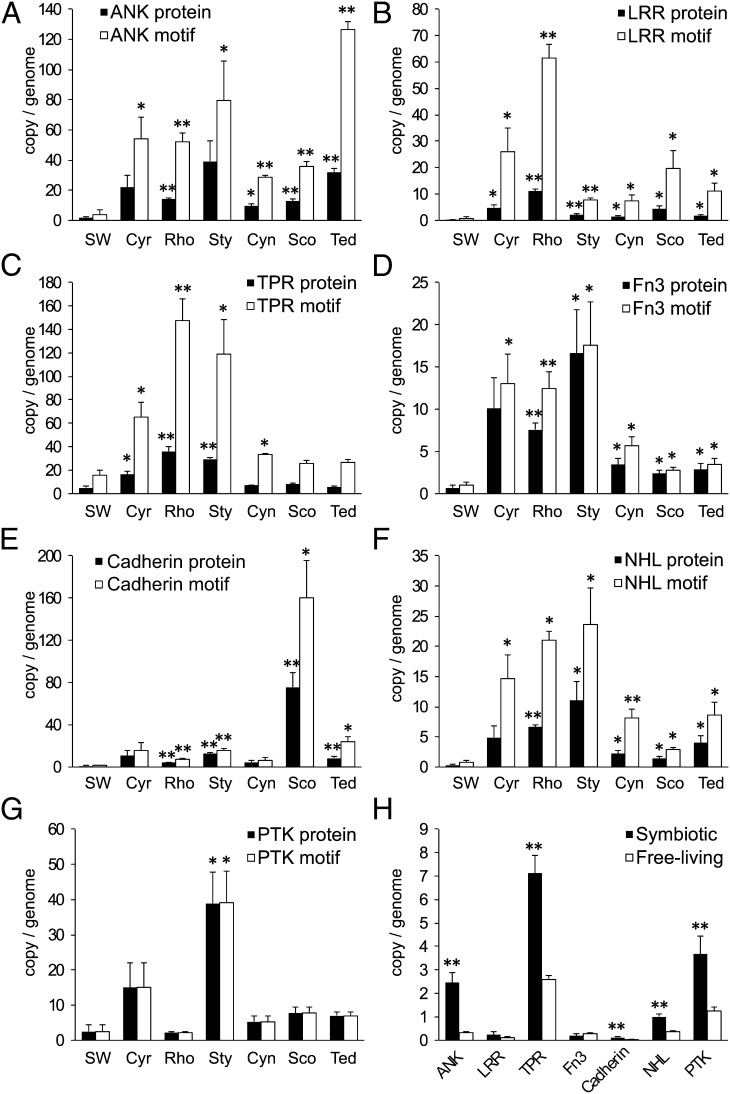
A recent metagenomic analysis of the microbial community of C. concentrica found that sponge symbionts were enriched in eukaryotic-like proteins (ELPs) and, in particular, in proteins containing ankyrin repeats (ANKs) and tetratricopeptide repeats (TPRs) (26). A similar observation was reported subsequently in the sponge-associated Poribacteria genome (42). These classes of proteins often are found in facultative or obligate symbionts and are postulated to modulate host behavior by interfering with eukaryotic protein–protein interactions that are mediated by these repeat domains. For example, recent work provided evidence that Legionella pneumophila used ANK-containing proteins (ARPs) to interfere with cytoskeletal processes of their amoebal host (76). Such molecular interference may be critical for sponge symbionts that need to escape phagocytosis by their host (26). The likely importance of these proteins was highlighted further by the detection of expressed bacterial ARPs in the sponge C. concentrica (58).
All six sponge metagenomes were rich not only in proteins with ANKs (COG0666, PF00023) and TPRs (COG0790, PF00515, PF07719) but also in proteins with leucine-rich repeats (LRR) (PF00560) and NHL repeats (PF01436) (Figs. 4 and 6 and Fig. S3C). LRR proteins are essential for virulence in the pathogen Yersinia pestis (77) and can activate host-cell invasion by the pathogen Listeria monocytogenes (78). NHL domains occur in a variety of proteins and potentially function in protein–protein interactions (79). Fibronectin domain III proteins (PF00041) and cadherins (PF00028), which likely are involved in adhesion to the host cells, also were abundant (Fig. 6 D and E). Fibronectin proteins can bind to integrins to mediate cell–cell contact and possible colonization (80), whereas cadherins can play a role in cellular uptake of bacteria into eukaryotic host cells (81). Many of the ELPs detected in the dataset also have predicted signal peptides and thus may function extracellularly in sponges (Fig. S4). The abundant type IV secretion systems (Fig. S4) in sponge symbionts also might play a role in delivering those ELPs into the host cells, as has been shown for L. pneumophila (76).
Fig. 6.
Abundance of ELPs in seawater versus sponge samples (A–G) and in free-living versus symbiotic species in the Integrated Microbial Genomes database (accessed November 25, 2011) (H). Abundance is normalized by copies per genome of each sample. “Abundance of motifs” refers to the number of repeats; “abundance of proteins” refers to the number of proteins with the repeat; the ratio of abundance of motifs to abundance of repeats gives an estimate of the average number of motifs per protein. Error bars show SDs. Tests between each sponge and the seawater group were performed at 95% confidence interval, and significant differences are marked. *P ≤ 0.05 but > 0.01; **P ≤ 0.01.
Although ELPs as a class generally are more abundant in sponge microorganisms than in the surrounding seawater, unweighted Unifrac clustering based on the pair-wise alignment showed very little sequence similarity between the ELPs of different symbiont communities (Fig. S5). In addition, the ELPs have very limited sequence similarity to proteins from eukaryotes (including the sponge Amphimedon queenslandica), further indicating that sponge symbionts have divergent and highly specific sets of ELPs.
Together, these data show that sponge symbionts generally contain a large abundance and variety of ELPs that likely mediate interactions with their hosts, implying that each symbiont community may undertake very specific ELP-mediated interactions and communications with their host.
Genomic Evolution Through Horizontal Gene Transfer.
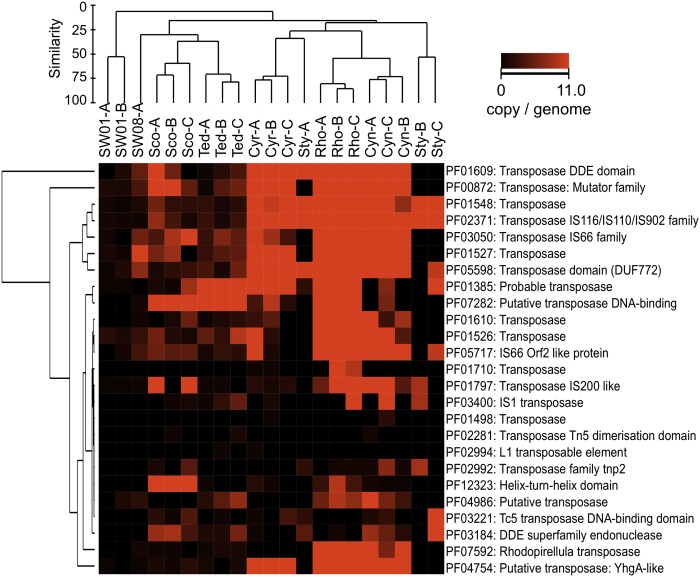
Mobile genetic elements (MGEs), such as transposons, plasmids, and prophages, mediate HGT, which facilitates the evolutionary adaptation of microbial populations to specific niches (82) and acts as a key driver in the evolution of the human gut microbiome (83). Compared with the planktonic bacterial community, the six sponge communities investigated here had a high abundance of systems involved in HGT, including transposase (COG3328, PF02371, PF01609, PF00872, PF05598), conjunctive transfer systems (COG3451, Subsystem: Type 4 secretion and conjugative transfer, Subsystem: Conjugative transfer related cluster), and retroid elements containing reverse transcriptase (COG3344, PF00078) and integrase (PF00665) (Fig. 4 and Fig. S3 C and D). Genetic systems for transformation (COG0758) also were abundant in some species. A high likelihood of genetic exchange and rearrangement in the microbial communities of sponges was supported further by an overrepresentation of DNA recombination and repair enzymes (SI Results and Discussion). Besides this general tendency, each species has a specific set of HGT systems. For example, R. odorabile was especially rich in transposases, conjugative elements, retroid elements, and a subpopulation of phages, whereas Stylissa sp. 445 had a lower number of transposases but a remarkable abundance of T4-like phages (Fig. 4, Fig. S3 C and D, and SI Results and Discussion).
To explore further the hypothesis that different communities have different set of MGEs, the samples were clustered according to the abundance of 25 detected transposases or transposon-related proteins/domains as annotated by the Pfam database (Fig. 7). Although the clustering revealed very similar transposase profiles among sponge replicates, each sponge community had its own distinct set of transposase systems. There was no evidence for biogeography (i.e., tropical communities were not more similar to each other than to the temperate-water sponges), a finding that could indicate a limited intercommunity (i.e., between different sponge species) dispersal of these MGEs within a region. We therefore propose that transposons might play a very specific role in exchanging genetic material within communities. These mobile elements potentially can allow all community members to share traits that are specific for the adaptation to their common host (e.g., via conjugation or transformation) and consequently facilitate the evolution of functional convergence inside a symbiont community. Alternatively, transposons could be involved in disrupting nonessential genes that no longer are required for a bacterial/archaeal symbiont as it evolves into a stable association with the host (Conclusion).
Fig. 7.
Abundance and diversity of transposases. The brightness (red) in the heatmap reflects the abundance (copies per genome) of a particular transposase in a sample. Samples are clustered by Bray–Curtis similarity and average general algorithm. Transposase entries are clustered with Euclidian distance and complete linkage.
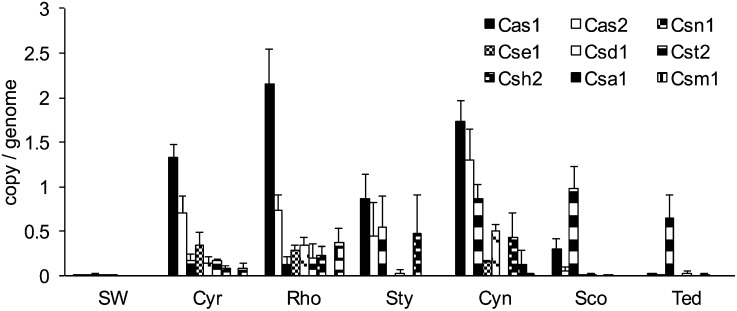
High rates of HGT can be detrimental to the cell (84), because HGT erodes genomic integrity (85). Moreover, the high filter-feeding rates of sponges make them particularly vulnerable to phage attack from the plankton (26). Phage-mediated transduction can lead to lysis and death of the bacterial cell (86). Therefore, one would expect that effective mechanisms would be required to control excessive genetic exchange and to minimize the introduction of foreign DNA in sponge microbial communities. Indeed, restriction-modification (R-M) systems, clustered, regularly interspaced, short, palindromic repeats (CRISPRs), and CRISPR-associated (CAS) proteins were abundant and diverse across all sponge datasets (Figs. 4 and 8 and Figs. S3 C and D and S6A).
Fig. 8.
Abundances of CAS proteins in subfamilies.
R-M and toxin–antitoxin (T–A) systems also are often considered selfish elements that are involved in MGE competition and an “arms-race” between the chromosomes and the MGEs (87, 88). The evolutionary accumulation of these systems in sponge symbionts further supports the postulated high rate of HGT inside symbiont communities of sponges (see above and SI Results and Discussion).
CRISPRs are recently discovered inheritable and adaptive immune systems that provide resistance against the integration of extrachromosomal DNA. Aided by CAS proteins, CRISPRs can acquire short DNA sequences from the invading phage or plasmid and incorporate that sequence into an array of spacer sequences (89). These spacers can be transcribed and hybridized to invading DNA, which then is degraded by CAS proteins with nuclease function (89). To understand better the diversity of CRISPRs in sponge-associated communities, the potential CRISPR arrays from the metagenomic datasets were extracted (SI Materials and Methods). In total, 203 CRISPR arrays were detected under stringent filtering criteria from five sponge metagenomes with an average of 0.28–0.74 CRISPR copies per genome (Fig. S6A). No CRISPR was detected in the samples of Stylissa sp 445; this result likely is related to the presence of abundant cyanophages in this species (SI Results and Discussion and Fig. S7). CRISPRs also were virtually absent in seawater samples.
Clustering of repeats and spacers at a cutoff of 50% similarity showed almost no overlap between the different sponge species (Fig. S6 B and C). Because spacer regions are “historical” records of current and past phage infection (90), the lack of overlap indicates that microbial communities from the same geographic location, such as the Great Barrier Reef, have experienced attacks by distinct viral populations. This finding is consistent with the distinct bacterial and archaeal communities within each sponge species (Fig. 2), because many phages have a high degree of host specificity (91). However, different replicates with very similar species composition contained very few common spacers, reflecting the dynamic nature of phage infection and suggesting that phage defense might involve small-scale temporal or spatial variation (92). A search of potential targets for spacers gave further evidence for the local adaptation of CRISPRs (Fig. S6D and SI Results and Discussion).
The diverse nature of CRISPRs also was underpinned by a variety of CAS proteins (93, 94). CAS proteins have been classified, based on the sequenced microbial genome, into a core set, eight subtype-specific groups, and a receptor activity-modifying (RAMP) module-related group (94). Proteins from the core set and all subtypes were detected in the sponge microbiomes and occurred in different abundances for each sponge species (Fig. 8). Surprisingly, a low abundance (or absence) of Cas1 and Cas2 proteins was observed in the microbial communities of Scopalina sp. and T. anhelans. Cas1 and Cas2 are considered hallmarks of a functional CRISPR array and were proposed to have an essential function during the spacer-acquisition step (94). Interestingly, the lack of Cas1 and Cas2 coincided with a high abundance of Csn1 (Fig. 8). Csn1 is a multidomain protein thought to possess multiple functions that otherwise are performed by individual proteins of other subtypes. In all known genomic arrangements, the gene for Csn1 is located exclusively upstream of the genes for Cas1 and Cas2, which themselves are always upstream of and directly adjacent to the CRISPR array (94, 95). In the T. anhelans dataset, however, we found one contig where the Csn1 gene is directly upstream of the CRISPR array (Fig. S8B). This deviation from the canonical Cas1/Cas2-based CRISPR arrangement highlights potential variation in CRISPR structure in sponge-associated microorganisms. It also is worthwhile noting that almost all current Csn1/Nmeni-containing CRISPRs are found in genomes of vertebrate pathogens and commensals (94), although their functional potential is not clear.12
Conclusion
Our analysis shows that, despite large phylogenetic differences, recognizable core functions exist in symbiont communities from phylogenetically divergent but functionally related sponges. Thus, symbiont communities in divergent hosts can possess a degree of functional equivalence. The specific symbiotic functions identified here are consistent with the current understanding of the biological and ecological roles of sponge-associated microorganisms and have provided insight into symbiont functions (e.g., creatinine metabolism). Detailed metagenomic analysis of the present study thus has advanced the understanding of the interactions between complex symbiont communities and their eukaryotic hosts, thereby contributing to an enhanced appreciation of the sponge holobionts (96).
Both niche selection and neutral hypothesis have been used to model community structures of free-living and host-associated microorganisms (5, 6, 11). The initial symbiont acquisition, the potential for vertical symbiont transmission, and the evolution of obligate relationships between sponges and microorganisms (22) most likely have had a major influence on these two types of processes for community assembly. Initially, different types of free-living microorganisms likely entered into associations with sponge hosts. These early associations may have been less selective and more random (i.e., neutral), and hence different sponge species would have acquired different phylogenetic clades of microorganisms. This scenario is consistent with the distinct taxonomic profiles in different sponge species observed here and in other studies (22). As the symbiotic relationship evolves, and vertical transmission occurs (13, 24), symbionts maintain or acquire functions that stabilize their interaction with their host. Thus, for different host species with similar functional niches, symbionts will converge functionally. Our detailed analysis of six microbiomes indeed revealed that many of these sponge-specific functions are fulfilled by phylogenetically distinct symbionts as well as by analogous enzymes and biosynthetic pathways (e.g., the CAS proteins or the types of nitrate reductase proteins involved in denitrification). Therefore symbiont communities in divergent hosts have evolved different genomic solutions to perform the same function or to occupy the same niche. However, this evolutionary adaptation to the sponge environment does not necessarily restrict any particular symbiont from interacting with other sponge species. In fact, the acquisition of general features for a sponge-associated lifestyle (such as CRISPRs or ELPs) could allow certain symbionts to be transmitted horizontally to multiple sponge species. Such a scenario is consistent with the observation that some symbiont taxa are found in multiple sponges (22).
The highly abundant and diverse MGEs detected in sponge symbionts may play key roles in these evolutionary processes in three ways. First, MGEs can mediate HGT and distribute essential core functions, such as stress resistance, ELPs, and phage defense, among community members. This process would facilitate the evolutionary adaptation of specific symbionts to the host environment (97). As a consequence, individual genomes from different phylogenetic lineages should become more similar to each other, as is consistent with recent genomic observations in mammalian gut bacteria (98). Second, adaptation to a host environment no longer may require all functions of free-living bacteria, and removal of nonessential genes can be mediated by a dramatic increase in transposon density (99). Examples of this process might include the loss of photolyase genes in sponge symbionts (Fig. S9B and SI Results and Discussion). Such a process would be the same as the reduction in genomic functionality observed in facultative and obligate symbiosis of simple systems (100). Third, individual genomes could use MGEs to eliminate functions that already are provided by other genomes and whose benefit might be shared within the community. This process would result in niche specialization of individual phylotypes, for example, through nutritional interdependence (101). As a consequence of this specialization, different communities would evolve or use different members (and hence different genes) for specific tasks, as is consistent with the functional equivalence concept introduced above. At the genomic level of a population, this process also would result in a high level of heterogeneity, which already has been noted during the assembly of two sponge symbiont genomes (42, 61). This genomic evolution of sponge symbionts also appears to be an ongoing process, because transposable elements are actively expressed in contemporary microbial communities of C. concentrica (58).
Future studies involving the reconstruction of genomes from metagenomic data and single-cell sequencing techniques (42) would offer additional evolutionary insights into sponge symbiosis and might reveal further principles of functional equivalence, evolutionary convergence, and specialization during the complex coevolution with an ancient animal host.
Materials and Methods
Details of the materials and methods used in this study are available in SI Materials and Methods. Briefly, sponge and water samples were collected in triplicate on the sites shown in Table 1. Metagenomic DNA was extracted from size-fractionated microbial samples and was shotgun sequenced with the Roche 454 Titanium pyrosequencing platform. Phylogenetic profiles were generated based on reconstructed 16S rRNA genes and SCGs. Functional profiles based on the comparison against COG (29), Pfam-A (v24.0) (30), and SEED/Subsystem (31) databases were normalized to the average genome copy. Such metagenomic gene profiles are suitable for describing the ecological and evolutionary adaptation of entire microbial communities (102). Functions that were enriched in the sponge group, as compared with the planktonic samples, were determined using MetaStats (103), with modifications.
Supplementary Material
Acknowledgments
We thank Kerensa McElroy, Martin Thompson, Shaun Nielsen, and Dr. Pui Yi Yung (University of New South Wales) and Dr. Dirk Erpenbeck (Ludwig Maximilians University Munich) for technical support and Patricia Sutcliffe and Dr. Merrick Ekins (Queensland Museum) for sponge identification. We also thank Kirsty Collard (University of New South Wles), Rose Cobb, Rochelle Soo, and Dr. Chris Battershill (Australian Institute of Marine Science) for assistance in sample collection. Sequencing data were produced by The J. Craig Venter Institute’s Joint Technology Center under the leadership of Yu-Hui Rogers and assistance from Matt Lewis. This work was supported by the Australian Research Council, the Gordon and Betty Moore Foundation, the Centre for Marine Bio-Innovation, and the J. Craig Venter Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this article are available through the Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis Web site, http://camera.calit2.net (project ID CAM_PROJ_BotanyBay).
This article is a PNAS Direct Submission.
See Author Summary on page 10765 (volume 109, number 27).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203287109/-/DCSupplemental.
References
- 1.Serbus LR, Casper-Lindley C, Landmann F, Sullivan W. The genetics and cell biology of Wolbachia-host interactions. Annu Rev Genet. 2008;42:683–707. doi: 10.1146/annurev.genet.41.110306.130354. [DOI] [PubMed] [Google Scholar]
- 2.McFall-Ngai M. Host-microbe symbiosis: The squid-Vibrio association—a naturally occurring, experimental model of animal/bacterial partnerships. Adv Exp Med Biol. 2008;635:102–112. doi: 10.1007/978-0-387-09550-9_9. [DOI] [PubMed] [Google Scholar]
- 3.Hongoh Y. Toward the functional analysis of uncultivable, symbiotic microorganisms in the termite gut. Cell Mol Life Sci. 2011;68:1311–1325. doi: 10.1007/s00018-011-0648-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchesi JR. Prokaryotic and eukaryotic diversity of the human gut. Adv Appl Microbiol. 2010;72:43–62. doi: 10.1016/S0065-2164(10)72002-5. [DOI] [PubMed] [Google Scholar]
- 5.Robinson CJ, Bohannan BJ, Young VB. From structure to function: The ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ofiteru ID, et al. Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA. 2010;107:15345–15350. doi: 10.1073/pnas.1000604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chisholm RA, Pacala SW. Niche and neutral models predict asymptotically equivalent species abundance distributions in high-diversity ecological communities. Proc Natl Acad Sci USA. 2010;107:15821–15825. doi: 10.1073/pnas.1009387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke C, Thomas T, Lewis M, Steinberg P, Kjelleberg S. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 2011;5:590–600. doi: 10.1038/ismej.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López-Sánchez MJ, et al. Evolutionary convergence and nitrogen metabolism in Blattabacterium strain Bge, primary endosymbiont of the cockroach Blattella germanica. PLoS Genet. 2009;5:e1000721. doi: 10.1371/journal.pgen.1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA. 2011;108:14288–14293. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webster NS, et al. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol. 2010;12:2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp KH, Eam B, Faulkner DJ, Haygood MG. Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl Environ Microbiol. 2007;73:622–629. doi: 10.1128/AEM.01493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran NA, Baumann P. Bacterial endosymbionts in animals. Curr Opin Microbiol. 2000;3:270–275. doi: 10.1016/s1369-5274(00)00088-6. [DOI] [PubMed] [Google Scholar]
- 16.Kojima A, Hirose E. Transmission of cyanobacterial symbionts during embryogenesis in the coral reef ascidians Trididemnum nubilum and T. clinides (Didemnidae, Ascidiacea, Chordata) Biol Bull. 2012;222:63–73. doi: 10.1086/BBLv222n1p63. [DOI] [PubMed] [Google Scholar]
- 17.Brissac T, Gros O, Merçot H. Lack of endosymbiont release by two Lucinidae (Bivalvia) of the genus Codakia: Consequences for symbiotic relationships. FEMS Microbiol Ecol. 2009;67:261–267. doi: 10.1111/j.1574-6941.2008.00626.x. [DOI] [PubMed] [Google Scholar]
- 18.McFall-Ngai MJ. Unseen forces: The influence of bacteria on animal development. Dev Biol. 2002;242:1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- 19.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor MW, Thacker RW, Hentschel U. Genetics. Evolutionary insights from sponges. Science. 2007;316:1854–1855. doi: 10.1126/science.1144387. [DOI] [PubMed] [Google Scholar]
- 21.Bell JJ. The functional roles of marine sponges. Estuar Coast Shelf Sci. 2008;79:341–353. [Google Scholar]
- 22.Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt S, et al. Assessing the complex sponge microbiota: Core, variable and species-specific bacterial communities in marine sponges. ISME J. 2012;6:564–576. doi: 10.1038/ismej.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U. Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microbiol. 2008;74:7694–7708. doi: 10.1128/AEM.00878-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gazave E, et al. Polyphyly of the genus Axinella and of the family Axinellidae (Porifera: Demospongiaep) Mol Phylogenet Evol. 2010;57:35–47. doi: 10.1016/j.ympev.2010.05.028. [DOI] [PubMed] [Google Scholar]
- 26.Thomas T, et al. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 2010;4:1557–1567. doi: 10.1038/ismej.2010.74. [DOI] [PubMed] [Google Scholar]
- 27.Taylor MW, Schupp PJ, Dahllöf I, Kjelleberg S, Steinberg PD. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ Microbiol. 2004;6:121–130. doi: 10.1046/j.1462-2920.2003.00545.x. [DOI] [PubMed] [Google Scholar]
- 28.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatusov RL, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falkowski PG. Evolution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the ocean. Nature. 1997;387:272–275. [Google Scholar]
- 33.Beman JM, Roberts KJ, Wegley L, Rohwer F, Francis CA. Distribution and diversity of archaeal ammonia monooxygenase genes associated with corals. Appl Environ Microbiol. 2007;73:5642–5647. doi: 10.1128/AEM.00461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kneip C, Lockhart P, Voss C, Maier UG. Nitrogen fixation in eukaryotes—new models for symbiosis. BMC Evol Biol. 2007;7:55. doi: 10.1186/1471-2148-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiore CL, Jarett JK, Olson ND, Lesser MP. Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 2010;18:455–463. doi: 10.1016/j.tim.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson CR, Fay P. Nitrogen fixation in coral reef sponges with symbiotic cyanobacteria. Nature. 1979;279:527–529. [Google Scholar]
- 37.Bayer K, Schmitt S, Hentschel U. Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ Microbiol. 2008;10:2942–2955. doi: 10.1111/j.1462-2920.2008.01582.x. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann F, et al. Complex nitrogen cycling in the sponge Geodia barretti. Environ Microbiol. 2009;11:2228–2243. doi: 10.1111/j.1462-2920.2009.01944.x. [DOI] [PubMed] [Google Scholar]
- 39.Schläppy ML, et al. Evidence of nitrification and denitrification in high and low microbial abundance sponges. Mar Biol. 2010;157:593–602. doi: 10.1007/s00227-009-1344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moir JWB, Wood NJ. Nitrate and nitrite transport in bacteria. Cell Mol Life Sci. 2001;58:215–224. doi: 10.1007/PL00000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philippot L. Denitrifying genes in bacterial and Archaeal genomes. Biochim Biophys Acta. 2002;1577:355–376. doi: 10.1016/s0167-4781(02)00420-7. [DOI] [PubMed] [Google Scholar]
- 42.Siegl A, et al. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 2011;5:61–70. doi: 10.1038/ismej.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ettwig KF, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 44.Hoffmann F, Rapp HT, Zöller T, Reitner J. Growth and regeneration in cultivated fragments of the boreal deep water sponge Geodia barretti Bowerbank, 1858 (Geodiidae, Tetractinellida, Demospongiae) J Biotechnol. 2003;100:109–118. doi: 10.1016/s0168-1656(02)00258-4. [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann F, Larsen O, Rapp HT, Osinga R. Oxygen dynamics in choanosomal sponge explants. Mar Biol Res. 2005;1(2):160–163. [Google Scholar]
- 46.Radax R, Hoffmann F, Rapp HT, Leininger S, Schleper C. Ammonia-oxidizing archaea as main drivers of nitrification in cold-water sponges. Environ Microbiol. 2012;14:909–923. doi: 10.1111/j.1462-2920.2011.02661.x. [DOI] [PubMed] [Google Scholar]
- 47.Ribes M, et al. Functional convergence of microbes associated with temperate marine sponges. Environ Microbiol. 2012;14:1224–1239. doi: 10.1111/j.1462-2920.2012.02701.x. [DOI] [PubMed] [Google Scholar]
- 48.Hoffmann F, et al. Oxygen dynamics and transport in the Mediterranean sponge Aplysina aerophoba. Mar Biol. 2008;153:1257–1264. doi: 10.1007/s00227-008-0905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siebers B, Schönheit P. Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr Opin Microbiol. 2005;8:695–705. doi: 10.1016/j.mib.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 50.Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol Rev. 2000;80:1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- 51.Ellington WR, Suzuki T. Early evolution of the creatine kinase gene family and the capacity for creatine biosynthesis and membrane transport. Subcell Biochem. 2007;46:17–26. doi: 10.1007/978-1-4020-6486-9_2. [DOI] [PubMed] [Google Scholar]
- 52.Van Pilsum JF, Stephens GC, Taylor D. Distribution of creatine, guanidinoacetate and the enzymes for their biosynthesis in the animal kingdom. Implications for phylogeny. Biochem J. 1972;126(2):325–345. [PubMed] [Google Scholar]
- 53.Yoshimoto T, et al. Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism. J Mol Biol. 2004;337:399–416. doi: 10.1016/j.jmb.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 54.Hasegawa Y, et al. A novel degradative pathway of 2-nitrobenzoate via 3-hydroxyanthranilate in Pseudomonas fluorescens strain KU-7. FEMS Microbiol Lett. 2000;190:185–190. doi: 10.1111/j.1574-6968.2000.tb09284.x. [DOI] [PubMed] [Google Scholar]
- 55.Moran GR. 4-Hydroxyphenylpyruvate dioxygenase. Arch Biochem Biophys. 2005;433:117–128. doi: 10.1016/j.abb.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 56.Koonin EV, Wolf YI, Aravind L. Protein fold recognition using sequence profiles and its application in structural genomics. Adv Protein Chem. 2000;54:245–275. doi: 10.1016/s0065-3233(00)54008-x. [DOI] [PubMed] [Google Scholar]
- 57.Dassa E, Bouige P. The ABC of ABCS: A phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol. 2001;152:211–229. doi: 10.1016/s0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 58.Liu M, Fan L, Zhong L, Kjelleberg S, Thomas T. Metaproteogenomic analysis of a community of sponge symbionts. ISME J. 2012 doi: 10.1038/ismej.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Webb ME, Marquet A, Mendel RR, Rébeillé F, Smith AG. Elucidating biosynthetic pathways for vitamins and cofactors. Nat Prod Rep. 2007;24:988–1008. doi: 10.1039/b703105j. [DOI] [PubMed] [Google Scholar]
- 60.Begley TP, Chatterjee A, Hanes JW, Hazra A, Ealick SE. Cofactor biosynthesis—still yielding fascinating new biological chemistry. Curr Opin Chem Biol. 2008;12:118–125. doi: 10.1016/j.cbpa.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hallam SJ, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howard EC, et al. Bacterial taxa that limit sulfur flux from the ocean. Science. 2006;314:649–652. doi: 10.1126/science.1130657. [DOI] [PubMed] [Google Scholar]
- 63.Vila M, et al. Use of microautoradiography combined with fluorescence in situ hybridization to determine dimethylsulfoniopropionate incorporation by marine bacterioplankton taxa. Appl Environ Microbiol. 2004;70:4648–4657. doi: 10.1128/AEM.70.8.4648-4657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malmstrom RR, Kiene RP, Cottrell MT, Kirchman DL. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl Environ Microbiol. 2004;70:4129–4135. doi: 10.1128/AEM.70.7.4129-4135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raina JB, Dinsdale EA, Willis BL, Bourne DG. Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol. 2010;18:101–108. doi: 10.1016/j.tim.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 66.Van Alstyne K, Schupp P, Slattery M. The distribution of dimethylsulfoniopropionate in tropical Pacific coral reef invertebrates. Coral Reefs. 2006;25(3):321–327. [Google Scholar]
- 67.Nyström T, Neidhardt FC. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol Microbiol. 1994;11:537–544. doi: 10.1111/j.1365-2958.1994.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 68.Igarashi K, Kashiwagi K. Polyamine transport in bacteria and yeast. Biochem J. 1999;344:633–642. [PMC free article] [PubMed] [Google Scholar]
- 69.Shah P, Swiatlo E. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol. 2008;68:4–16. doi: 10.1111/j.1365-2958.2008.06126.x. [DOI] [PubMed] [Google Scholar]
- 70.Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 71.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hansen IV, Weeks JM, Depledge MH. Accumulation of copper, zinc, cadmium and chromium by the marine sponge Halichondria panicea Pallas and the implications for biomonitoring. Mar Pollut Bull. 1995;31(1–3):133–138. [Google Scholar]
- 74.Barkay T, Wagner-Döbler I. Microbial transformations of mercury: Potentials, challenges, and achievements in controlling mercury toxicity in the environment. Adv Appl Microbiol. 2005;57:1–52. doi: 10.1016/S0065-2164(05)57001-1. [DOI] [PubMed] [Google Scholar]
- 75.Selvin J, Shanmugha Priya S, Seghal Kiran G, Thangavelu T, Sapna Bai N. Sponge-associated marine bacteria as indicators of heavy metal pollution. Microbiol Res. 2009;164:352–363. doi: 10.1016/j.micres.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 76.Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Evdokimov AG, Anderson DE, Routzahn KM, Waugh DS. Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: A leucine-rich repeat protein with the shortest repeating unit. J Mol Biol. 2001;312:807–821. doi: 10.1006/jmbi.2001.4973. [DOI] [PubMed] [Google Scholar]
- 78.Marino M, Braun L, Cossart P, Ghosh P. Structure of the lnlB leucine-rich repeats, a domain that triggers host cell invasion by the bacterial pathogen L. monocytogenes. Mol Cell. 1999;4:1063–1072. doi: 10.1016/s1097-2765(00)80234-8. [DOI] [PubMed] [Google Scholar]
- 79.Slack FJ, Ruvkun G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem Sci. 1998;23:474–475. doi: 10.1016/s0968-0004(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 80.Hoffmann C, Ohlsen K, Hauck CR. Integrin-mediated uptake of fibronectin-binding bacteria. Eur J Cell Biol. 2011;90:891–896. doi: 10.1016/j.ejcb.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 81.Lecuit M, et al. A role for alpha-and beta-catenins in bacterial uptake. Proc Natl Acad Sci USA. 2000;97:10008–10013. doi: 10.1073/pnas.97.18.10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moliner C, Fournier PE, Raoult D. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol Rev. 2010;34:281–294. doi: 10.1111/j.1574-6976.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- 83.Smillie CS, et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 84.Doolittle WF, Kirkwood TB, Dempster MA. Selfish DNAs with self-restraint. Nature. 1984;307:501–502. doi: 10.1038/307501b0. [DOI] [PubMed] [Google Scholar]
- 85.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 87.Yarmolinsky MB. Programmed cell death in bacterial populations. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]
- 88.Garcia-Pino A, et al. Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation. J Biol Chem. 2008;283:30821–30827. doi: 10.1074/jbc.M805654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 90.Vale PF, Little TJ. CRISPR-mediated phage resistance and the ghost of coevolution past. Proc Biol Sci. 2010;277:2097–2103. doi: 10.1098/rspb.2010.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu J, Chen F, Hodson RE. Distribution, isolation, host specificity, and diversity of cyanophages infecting marine Synechococcus spp. in river estuaries. Appl Environ Microbiol. 2001;67:3285–3290. doi: 10.1128/AEM.67.7.3285-3290.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11:457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- 93.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLOS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 96.Webster NS, Taylor MW. Marine sponges and their microbial symbionts: Love and other relationships. Environ Microbiol. 2012;14:335–346. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- 97.Chia N, Goldenfeld N. Dynamics of gene duplication and transposons in microbial genomes following a sudden environmental change. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;83:021906. doi: 10.1103/PhysRevE.83.021906. [DOI] [PubMed] [Google Scholar]
- 98.Zaneveld JR, Lozupone C, Gordon JI, Knight R. Ribosomal RNA diversity predicts genome diversity in gut bacteria and their relatives. Nucleic Acids Res. 2010;38:3869–3879. doi: 10.1093/nar/gkq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 100.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 101.Pfeiffer T, Bonhoeffer S. Evolution of cross-feeding in microbial populations. Am Nat. 2004;163:E126–E135. doi: 10.1086/383593. [DOI] [PubMed] [Google Scholar]
- 102.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 103.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLOS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]