Abstract
Crystal structures of human epidermal growth factor receptor (EGFR) with bound ligand revealed symmetric, doubly ligated receptor dimers thought to represent physiologically active states. Such complexes fail to rationalize negative cooperativity of epidermal growth factor (EGF) binding to EGFR and the behavior of the ligandless EGFR homolog ErbB2/HER2, however. We report cell-based assays that provide evidence for active, singly ligated dimers of human EGFR and its homolog, ErbB4/HER4. We also report crystal structures of the ErbB4/HER4 extracellular region complexed with its ligand Neuregulin-1β that resolve two types of ErbB dimer when compared to EGFR:Ligand complexes. One type resembles the recently reported asymmetric dimer of Drosophila EGFR with a single high-affinity ligand bound and provides a model for singly ligated human ErbB dimers. These results unify models of vertebrate and invertebrate EGFR/ErbB signaling, imply that the tethered conformation of unliganded ErbBs evolved to prevent crosstalk among ErbBs, and establish a molecular basis for both negative cooperativity of ligand binding to vertebrate ErbBs and the absence of active ErbB2/HER2 homodimers in normal conditions.
Human epidermal growth factor receptor (EGFR) and its homologs, known as ErbBs or HERs, are essential receptor tyrosine kinases that mediate cell proliferation and differentiation during animal development and are the targets of multiple cancer therapies (1). EGFR is the archetype of single-pass membrane-spanning receptors thought to transmit signals by ligand-induced dimerization (2, 3), and structural studies show that ligand binding to human EGFR promotes rearrangement of its four extracellular domains from a tethered to an extended conformation in which a loop, termed the dimerization arm, becomes exposed and mediates formation of symmetric receptor dimers (4) (Fig. 1A). At odds with a ligand-induced dimerization model of EGFR signaling, however, are recent studies showing dimers of human EGFR in the absence of ligand (5–8) as well as negative cooperativity when epidermal growth factor (EGF) binds to EGFR (9). Curiously, the single Drosophila EGFR homolog adopts an extended conformation in the absence of ligand and forms asymmetric receptor dimers with a single high-affinity ligand bound (10, 11), suggesting different mechanisms may regulate EGFR activation in Drosophila and humans.
Fig. 1.
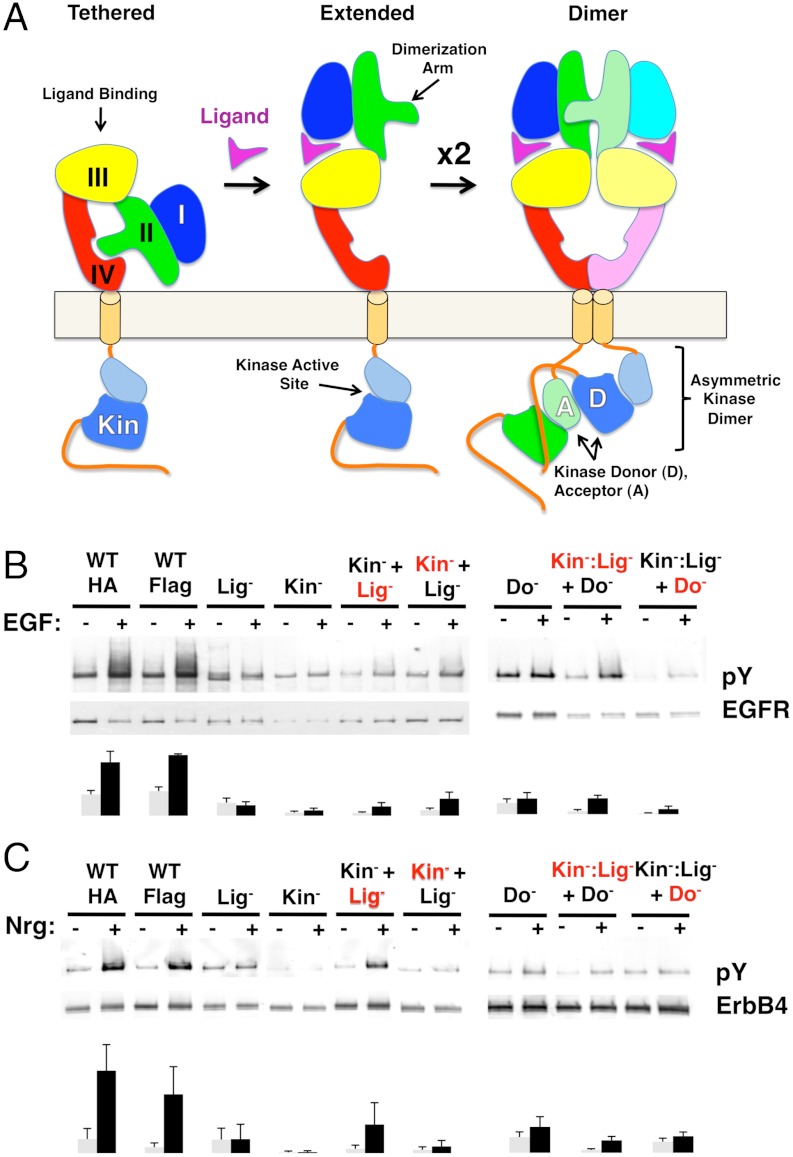
Evidence for singly ligated ErbB signaling dimers. (A) Schematic diagram showing tethered, extended, and dimeric conformations of EGFR with sites of function-targeting mutations indicated. (B) Antiphosphotyrosine and anti-EGFR Western blots of tagged full-length EGFR immunoprecipitated from stably transfected CHO cells. Wild-type (WT) EGFR was tagged with either hemagglutinin (HA) or Flag peptides, EGFR bearing an inactivating mutation in its kinase active site (Kin-) was tagged with HA, and EGFR bearing a mutation in its ligand-binding site (Lig-) was tagged with Flag. Mutation in the Kinase donor site (Do-) and combination of the Kinase- and Ligand-targeting mutations on a single EGFR (Kin-: Lig-) were also tested. Serum-starved cells were either untreated (−) or treated (+) with EGF for 5 minutes. Each WT or mutant EGFR was transfected singly; the Kin- and Lig- variant EGFRs were cotransfected as were the (Kin-: Lig-) and (Do-) variants. When cotransfected, the tag used for immunoprecipitation prior to Western blotting is indicated in red. (C) Similar experiments using ErbB4 and its ligand Neuregulin 1β (Nrg) are shown. The lanes shown were run on the same gel, but rearranged electronically to match the order of experiments in (B). Bar graphs represent quantitation of bands from at least 3 independent experiments.
We report here evidence for active, singly ligated homodimers of human EGFR and its homolog, ErbB4. We also report the crystal structure of the ErbB4 extracellular region bound to its ligand Neuregulin-1β, which allows resolution of two types of human EGFR/ErbB dimers, one of which resembles the asymmetric Drosophila EGFR dimer and appears to reflect a singly ligated ErbB dimer state. These results compel reappraisal of canonical views of ligand-induced dimerization and show that several previously anomalous properties of human EGFR and its homologs represent vertebrate innovations on a core signaling mechanism present in invertebrates.
Results and Discussion
We reasoned that if singly ligated dimers of human EGFR exist as implied by negative cooperativity (9), an EGFR variant incapable of binding ligand may remain able to participate in signaling dimers. To test this idea, we introduced debilitating amino-acid substitutions into the ligand-binding site of one EGFR variant and the kinase active site of another. These variants show negligible ligand-dependent phosphorylation when expressed individually in CHO cells, but coexpression restores phosphorylation in response to ligand as judged by both general and specfic antiphosphotyrosine antibodies (Fig. 1B and SI Appendix, Fig. S1). The simplest explanation for this observation is that ligand-binding deficient receptors are able to pair with kinase-deficient receptors to form active, singly ligated EGFR dimers. Similar results were obtained for ErbB4/HER4 (Fig. 1C). Amino-acid substitutions in the ErbB4 dimerization arm in the context of either ligand-binding or kinase-activity deficient ErbB4 variants eliminates responsiveness when cotransfected (SI Appendix, Fig. S2), implicating dimerization arms from both partners in formation of singly ligated ErbB4 dimers. Participation of unliganded ErbBs in a signaling dimer despite burial of the dimerization arm in the tethered conformation likely reflects favorable energetics of the interreceptor dimer interface relative to the tethered state within a preformed dimer.
An essential feature of EGFR activation is an asymmetric dimer of EGFR kinase domains in which the C-terminal region of a “donor” kinase contacts the N-terminal region of an “acceptor” kinase and stimulates it (12) (Fig. 1A), and the question arises whether the extracellular asymmetry of singly ligated EGFR dimers is coupled to this intracellular asymmetry. The ability of ligand-binding deficient EGFR to be activated by kinase-dead EGFR demonstrates that unliganded EGFRs can function as the acceptor kinase (Fig. 1B). To determine if ligand-binding deficient EGFR can function as a donor kinase, debilitating amino-acid substitutions were simultaneously introduced into the ligand-binding and kinase active sites of one EGFR and into the kinase donor site of another EGFR. Neither variant showed ligand-dependent phosphorylation when expressed on its own, but weak, ligand-dependent phosphorylation was observed when coexpressed (Fig. 1B). Similar results were obtained for ErbB4 (Fig. 1C). This observation suggests that unliganded EGFRs can serve as both a donor and an acceptor kinase and that extracellular asymmetry is not absolutely coupled to intracellular asymmetry, consistent with studies suggesting a loose linkage between ligand binding and kinase activation (13). A recent report using a luciferase fragment complementation assay showed that normal activation of EGFR/ErbB2 heterodimers required the EGFR kinase to be active, suggesting that the liganded partner (EGFR) could initially only function as an acceptor kinase and that extra- and intracellular asymmetry are coupled (14). In this case the intracellular kinases differ (vs. EGFR or ErbB4 homodimers), which may contribute to additional stabilization of the EGFR kinase in the acceptor role in the absence of phosphorylation. It will be interesting to determine if this is indeed the case or whether other factors underlie this apparent difference. The sites of all tested amino-acid substitutions are listed in SI Appendix, Table S1. None of these sites impaired cell surface expression as judged by cell-surface biotinylation, and EGFR expression levels were estimated by Western blot (SI Appendix, Fig. S3 and Table S2). Curiously, an original ligand-binding mutation introduced in EGFR, D355R, failed to express on the cell surface unless cotransfected with kinase-deficient EGFR. This observation suggests that EGFR molecules interact early in biogenesis and that this interaction can rescue otherwise nonviable forms of EGFR.
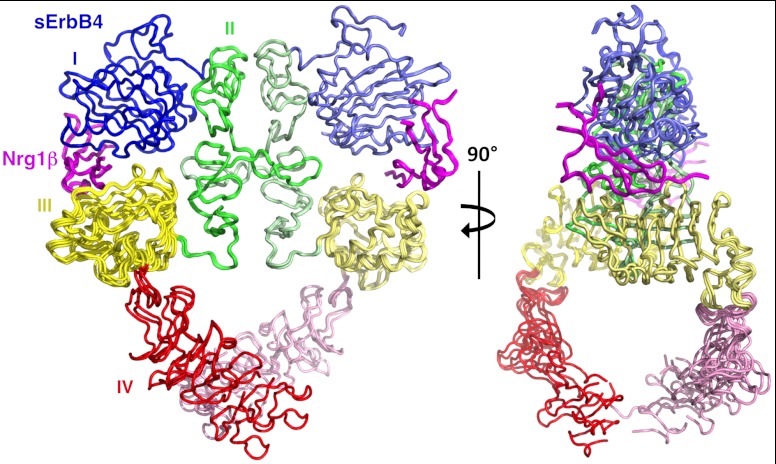
The presence of active, singly ligated EGFR dimers on the cell surface raises the question of why crystal structures of the human EGFR extracellular region with ligand bound reveal symmetric, doubly ligated EGFR dimers (15, 16). To address this issue, we sought additional crystal structures of liganded ErbB extracellular regions and report here the 3.0 Å crystal structure of the human ErbB4 extracellular region (sErbB4) complexed with Neuregulin-1β (Nrg) Table 1. Three independent 2∶2 sErbB4:Nrg dimers are present in the crystallographic asymmetric unit (Fig. 2). Domains I-III of the six sErbB4 subunits superimpose well among themselves (pairwise rmsds of 0.2–0.5 Å for Cαs) and with homologous regions of high-affinity ligand complexes of both human and Drosophila EGFR extracellular regions (sEGFR) (pairwise rmsds of 1.1–1.9 Å for Cαs) (Fig. 2 and SI Appendix, Fig. S4, and Table S3) (11, 13, 15, 16). The C-terminal juxtamembrane regions of domain IV are closely apposed when well ordered, suggesting interactions between transmembrane regions in active receptor dimers, but regions of domain IV homologous to those involved in direct dimer contacts in EGFR:EGF complexes (13) are mostly disordered and few specific intersubunit domain IV contacts are resolvable.
Table 1.
X-ray data collection and refinement statistics
| ErbB4-Nrg1β | |
| Data collection | |
| Wavelength | 0.98 |
| Space group | P21 |
| Unit cell dimensions | |
| a (Å) | 85.7 |
| b (Å) | 223.5 |
| c (Å) | 146.9 |
| β (°) | 99.7 |
| No. of unique reflections | 103,807 |
| Completeness (%) | 98.9 (87.0) |
| < l/σ > | 8.6 (1.3) |
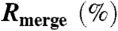 |
11.6 (68.5) |
| Redundancy | 3.7 (2.7) |
| Refinement | |
| Resolution (Å) | 50–3.0 |
 |
19.0 |
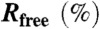 |
22.7 |
| rmsd | |
| Bond angle (Å) | 0.01 |
| Bond angle (°) | 1.26 |
Values in parentheses are for the highest-resolution shell.
Fig. 2.
The sErbB4:Nrg1β structure. Orthogonal views of a worm diagram of the three independent sErbB4:Nrg1β dimers in the crystallographic asymmetric unit following superposition of domains I, II, and III of sErbB4. Domains I, II, III, and IV are colored blue, green, yellow, and red, respectively, and Nrg1β is colored magenta. Lighter hues are used for the rightmost sErbB4 subunit.
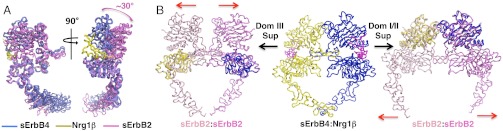
Although consisting of nearly symmetric 2∶2 ligand:receptor complexes in each case, comparison of sErbB4:Nrg1β and human sEGFR:ligand dimers reveals two distinct dimer interfaces (Fig. 3). Dimers of sErbB4:Nrg1β and sEGFR:EGF (13, 16) are similar to one another but differ from TGFα−bound dimers of a truncated form of EGFR (tEGFR) comprising the N-terminal 3 extracellular domains (15) (Fig. 3). Superposition of a single receptor subunit of the tEGFR:TGFα dimer with a single subunit of either the sErbB4:Nrg1β or sEGFR:EGF dimers reveals the opposite ErbB subunits to differ by a 29° scissor-like rotation about the dimerization arms. This rotation disrupts dimer contacts made by the N-terminal regions of domain II in the tEGFR:TGFα complex, notably those mediated by two contiguous loops formed by residues 190–208 (187–205 in ErbB4). These loops are flush across the tEGFR:TGFα dimer interface but staggered in the sErbB4:Nrg1β and sEGFR:EGF dimers (Figs. 3 and 4).
Fig. 3.
Two types of vertebrate ErbB dimer interaction. (Top) Orthogonal views of worm diagrams of sErbB4:Nrg1β and sEGFR:EGF (13) dimers following superposition of domains I, II, and III (see Fig. 1A for domain nomenclature). One receptor subunit is colored yellow, the other blue; Nrg1β is colored magenta. (Bottom) Orthogonal views of worm diagrams of the tEGFR:TGFα complex (15) colored as in the top panel. The position of domain IV has been modeled on each subunit based on the domain III/IV relationship in the sEGFR:EGF complex and apo-sEGFR structures (SI Appendix, Fig. S5). The modeled domain IV of one subunit is colored black and the other gray. The modeled regions are enclosed in a dashed red box with clashing regions indicated. Red asterisks mark the dimer interaction site mediated by the N-terminal regions of domain II, which differs in the two dimer types. Yellow and blue lines approximate the long axes of the receptor subunits to illustrate the relative scissoring of the subunits in the two dimer types.
Fig. 4.
The tEGFR:TGFα domain II dimer interface is similar to the interface in Drosophila sEGFR:Spitz complexes and modeled ErbB2-containing heterodimers. (Top row) In the leftmost panel, a “side” view of an sErbB4:Nrg1β dimer (equivalent to the Top Right image of Fig. 2) with one sErbB4 subunit colored blue, another yellow, and Nrg1β magenta is shown. Moving rightwards, one subunit of the EGFR:EGF (13), tEGFR:TGF〈 (15), or Drosophila EGFR:Spitz (11) complexes has been superposed on domains I, II, and III of the yellow sErbB4 subunit. In the far right panel, domain III of sErbB2 (22) has been superimposed on domain III of the blue sErbB4 subunit. Domains I and II of the non-ErbB4 receptors are colored red, and domains III and IV are colored light green. Colored arrows indicate shifts in unsuperposed subunit domains relative to sErbB4 subunit domains, which in the case of tEGFR, Drosophila EGFR, and ErbB2 align the domain I/II interface regions directly opposite the corresponding regions of the opposite receptor subunit. (Bottom row) “Top” views of the superpositions shown in the Top row following a 90° rotation about a horizontal axis in the plane of the page. The superposed receptor subunits are colored yellow, the unsuperposed sErbB4 subunit is colored blue, and domains I and II of the unsuperposed EGFR or ErbB2 subunits are colored red. Red and green asterisks mark the two loops encompassed by ErbB4 residues 187–205 that are staggered in ErbB4 dimers but directly opposed in tEGFR, Drosophila EGFR, and modeled ErbB2-containing dimers.
The different ErbB dimers can be explained by truncation of domain IV in the tEGFR:TGFα complex (15). The orientation of domain IV relative to domain III is conserved in EGFR structures whether liganded, unliganded, Drosophila, or human (SI Appendix, Fig. S5), and modeling domain IV onto each subunit of the tEGFR:TGFα dimer shows that its presence would result in severe intersubunit clashes (Fig. 3). Accommodating domain IV in sErbB4:Nrg1β and sEGFR:EGF complexes while maintaining interreceptor dimerization arm contacts necessitates the scissor-like rotation of receptor subunits relative to their orientation in the tEGFR:TGFα dimer. A flush, tEGFR:TGFα-like arrangement of domain II loops is also observed in the asymmetric dimer of Drosophila sEGFR, in which only one receptor subunit has high affinity ligand bound (Fig. 4) (11). In this case, the absence of a high-affinity ligand in one subunit effectively uncouples the relative orientation of the domain I/II and III/IV pairs and allows domain I and the N-terminal region of domain II of this subunit to shift and form the flush domain II interface without requiring domain IV clashes (Fig. 4). These results highlight the importance of the relative position of the distinct dimer contact regions in domains II and IV in forming optimal ErbB dimers (Fig. 1A, Fig. 2) as their relative position, and thus the nature of possible dimer contacts, changes when high-affinity ligand is bound.
Spontaneous formation of the flush dimer contact in human tEGFR dimers indicates that it is almost certainly more stable than the staggered contact observed in dimers of sErbB4 and sEGFR. The flush dimer would thus preferentially form in singly ligated dimers of EGFR in which the relative positions of domains II and IV are uncoupled in the unliganded subunit. This observation, coupled with the results of our cell-based assays, strongly implies that asymmetric Drosophila sEGFR-like dimers are conserved in human ErbBs (SI Appendix, Fig. S6). This conservation is satisfying from an evolutionary perspective and provides a structural rationale for negative cooperativity of ligand binding to EGFR (9). Binding of ligand to the unliganded subunit of singly ligated ErbB dimers requires conversion from a flush interface to the less stable staggered dimer interface, which necessarily reduces the apparent affinity of the second receptor for ligand relative to the first and results in negative cooperativity and a weaker receptor dimer (9). The fact that intracellular regions are required for negative cooperativity likely reflects the importance of these regions for stabilizing receptor dimers in the absence of ligand (17). A theoretical study recently suggested interactions with the cell membrane may also induce a Drosophila EGFR-like dimer in human EGFR (18).
Our cell-based assays imply that singly ligated EGFR dimers are signaling competent (Fig. 1). The fact that the Q194A mutation, which preferentially targets dimer contacts in the tEGFR:TGFα complex vs. the 2∶2 sEGFR:EGF complex (SI Appendix, Fig. S7), does not impair EGFR signaling (19) suggests that doubly ligated EGFR dimers are also signaling competent. Additionally, the actual and modeled positions of domain IV are similar in 2∶2 ErbB:ligand complexes and the asymmetric Drosophila sEGFR dimer, which we take to be an approximate model of singly ligated human EGFR dimers (11, 13) (SI Appendix, Fig. S8), suggesting equivalent arrangements of transmembrane and intracellular regions in both vertebrate dimer types.
Why, then, are asymmetric Drosophila sEGFR-like dimers not observed in crystals of human sEGFR or sErbB4 complexed with ligand? The presence of asymmetric dimers in crystals of Drosophila sEGFR indicates that the extra stability of the asymmetric dimer interface more than compensates for the extra stability available from converting the low-affinity ligand interaction to a high-affinity interaction. That vertebrate ErbBs proceed to a weaker, symmetric dimer interface with two high-affinity ligand-receptor interactions implies that the energetic balance between high- and low-affinity dimer or ligand interactions has shifted during evolution, at least for the extracellular regions. One source for this shift is apparent. The tethered conformation of vertebrate ErbB extracellular regions, which buries the dimerization arm, necessarily reduces the ability of soluble, unliganded ErbB extracellular regions to dimerize with liganded partners. The presence of asymmetric, singly ligated ErbB dimers in cells but not in crystals of isolated ErbB extracellular regions thus underscores the importance of human EGFR intracellular region-mediated preformed dimers, which are likely needed to stabilize asymmetric, singly ligated dimers (17).
ErbB2/HER2 is an atypical ErbB that appears specialized to participate in asymmetric dimers. ErbB2/HER2 is the only EGFR homolog without known canonical ligands and serves as a universal ErbB heterodimerization partner (20, 21). Unlike all other unliganded human ErbBs, the ErbB2/HER2 extracellular region adopts a constitutively extended conformation in which its dimerization arm is exposed (Fig. 5). This conformation rationalized ErbB2’s role as a promiscuous heterodimerization partner but failed to explain the absence of ErbB2 homodimers in normal conditions (22, 23). As noted by earlier authors (19), ligand binding to human ErbBs not only promotes conversion from the tethered to the extended conformation but also a change in conformation of domain II, which bridges ligand-binding domains I and III and bends to allow optimal contacts with ligand. By stabilizing formation of the asymmetric dimer, the change from a straight to a bent conformation in domain II in one dimer subunit appears to be the ligand-dependent on-off switch for Drosophila EGFR (11), which lacks the tethered conformation (10).
Fig. 5.
The fixed extended conformation of ErbB2 precludes formation of canonical homodimers. (A) Orthogonal views of worm diagrams of the six sErbB4:Nrg1β subunits (sErbB4 is colored blue and Nrg1β yellow) following superposition of domain III of sErbB4 with domain III of three independent sErbB2 structures (pink) (22, 23). (B) Worm diagrams of a superposition of sErbB2 (light and dark pink) on domain III of an sErbB4:Nrg1β dimer (one sErbB4 subunit colored yellow and the other blue) (Left ); only domain III of the sErbB4:Nrg1β complex is shown for clarity, and red arrows indicate the shift in position of domains I and II of the ErbB2 subunits relative to their position in the sErbB4:Nrg1β complex. The sErbB4:Nrg1β complex is shown in the middle panel. In the Right most panel, domain I and the N-terminal part of domain II of ErbB2 is superposed on the corresponding regions of each subunit of the sErbB4:Nrg1β complex; only the superposed regions of the sErbB4:Nrg1β complex are shown for clarity. Red arrows indicate the shift in position of domains III and IV of the ErbB2 subunits relative to the position of the corresponding domains in the sErbB4:Nrg1β complex. The human sErbB2 structure was used for the superposition (22).
A direct contact between ErbB2 domains I and III that occludes canonical ligand binding surfaces and fixes the extended conformation also fixes a “straight” conformation of ErbB2 domain II. In this conformation, the relative orientation of ErbB2 domain II and IV dimer contact regions is optimally aligned to pair with a liganded partner and serve as the unliganded subunit in asymmetric, singly ligated ErbB heterodimers but precludes formation favorable ErbB2 homodimers (Figs. 4 and 5). Superposing ErbB2 extracellular regions on either domain II or domain IV contact regions of EGFR/ErbB dimers demonstrates that ErbB2 is not capable of simultaneously forming favorable domain II dimer contacts and bringing the juxtamembrane regions of domain IV into close proximity (Fig. 5), which appears to be a feature of ErbB signaling (4, 13, 24). Unlike Drosophila EGFR, ligand binding is not required for ErbB2 to participate in active signaling complexes owing to the presence of ligand-binding homologs, which allowed ErbB2 to evolve a fixed straight domain II conformation and specialize as a heterodimerization partner. ErbB2 activation in cases of pathological overexpression thus seems likely to be mediated by intracellular regions (25, 26). Curiously, a weak dimer of unliganded dmEGFR that looks similar to the leftmost dimer in Fig. 5B was observed in crystals and may serve as a model for weak, unliganded, and presumably inactive ErbB extracellular region dimers (10).
Participation of unliganded ErbBs in active signaling complexes prompts reassessment of the role of the tethered conformation, which was first interpreted as keeping ErbBs “off” in the absence of ligand (4, 27, 28). It is now apparent that the straight conformation of domain II is sufficient for this purpose, as evidenced by the absence of a tethered conformation in Drosophila EGFR and the failure of tether mutations in human EGFR to result in receptor activation (10, 29, 30). In an organism with multiple EGFR homologs, however, the ability of an unliganded ErbB to participate in a signaling complex means that ligand binding to any ErbB could activate all coexpressed ErbBs. Such promiscuous activation is observed for ErbB2/HER2, for example, which is the only vertebrate ErbB not to adopt a tethered conformation. An additional inhibitory mechanism was thus needed to prevent indiscriminate ErbB responses to individual ligands in species with multiple ErbB homologs. By precluding unliganded EGFR, ErbB3, or ErbB4 from pairing with liganded forms of other ErbBs, the tethered conformation fulfills this role and likely facilitated diversification of ErbB function. The tethered conformation of human EGFR (28), ErbB3 (27), and ErbB4 (31) and the fixed, ligandless conformation of ErbB2 (22, 23) thus appear to have arisen following the appearance of multiple ErbB homologs as elaborations on the core signaling mechanism present in Drosophila EGFR. As tethered ErbBs appear able to convert to a signaling competent extended-straight conformation when dimerized with a liganded partner, which ErbB dimers form in the absence of ligand will govern the nature of ErbB responses and is an important avenue for future investigation. The stability of doubly-ligated ErbBs may have arisen to allow heterodimerization of ErbBs when ligands for both are present.
ErbBs have evolved many mechanisms to safeguard and modulate their potent activity. The presence of inactive, singly ligated, and doubly ligated human ErbB dimers confers several advantages over a ligand-induced dimerization activation mechanism. Inactive dimers present a barrier to activation through random dimerization, and the presence of singly and doubly ligated dimers furnishes a mechanism to tune responses to different concentrations or affinities of ligands (11). The results presented here show how specific intra- and intermolecular conformations combine to govern ErbB activity and lead to a unifying model of ErbB activation that rationalizes previously puzzling properties of EGFR and its homologs.
Materials and Methods
Generation of ErbB-expressing Cell Lines.
Genes encoding ErbB mutants were generated by QuikChange mutagenesis (Stratagene) using PfuTurbo AD (Agilent) and sequenced. Variant ErbB genes were then subcloned with their native signal sequences into pSSX, a version of pSGHV0 modified to eliminate the growth hormone tag and add C-terminal Flag or HA tags (32). CHO-S cells (Invitrogen) were maintained in adherent culture in DMEM:F12 supplemented with 5% FBS. Stably transfected cell lines were created using FuGENE (Roche) according to manufacturer’s instructions. Cells were cotransfected with a total of 1 μg DNA per ml culture and 0.1 μg per ml of pCDNA 3.1 (Invitrogen), which contains the neomycin resistance gene. After 24 hours, fresh medium containing 1 mg/ml G418 was added, and the cells fed every three days until colonies appeared. Colonies were picked, expanded, and screened for ErbB expression by Western using the appropriate tag antibody. Antibodies used for immunoprecipitation or Western detection were Flag-M2 (Sigma), 3F10 anti-HA (Roche), anti-EGFR (Santa Cruz sc-71033), anti-EGFR pY1068 (Abcam EP774Y), anti-ErbB4 (Santa Cruz sc-283), and 4G10 antiphosphotyrosine (Millipore). The number of receptors per cell was estimated by comparison of anti-EGFR band intensities of cell lysates compared to intensities of known amounts of purified tEGFR (33); cell lines with approximately equal expression of the ErbB variants were chosen for stimulation assays.
ErbB Activity Assays.
ErbB-expressing cell lines were plated in 2 wells of a six-well plate at 0.2 × 106 cells per well and grown 24 hr. On the day of the assay, cells were washed three times with 2 ml Ham’s F12 supplemented with 1 mg/ml BSA, then serum-starved for at least 3 hours at 37 °C in the same medium. EGF or Nrg1β was added to one of the two wells at a final concentration of 100 ng ml-1, and the plates incubated at 37 °C for 5 minutes. The wells were then washed once with ice-cold phosphate-buffered saline, and 250 μl of RIPA buffer (50 mM Tris pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with 1 mM activated Na3VO4, 1 mM PMSF, and Benzonase nuclease (Sigma) was added directly to the wells. The cells were allowed to lyse for 30 minutes with gentle rocking, and the appropriate antibody was added to the lysate (using 0.5 μg/ml for anti-Flag and 0.1 μg/ml for anti-HA). Lysates were next added directly to 20 μl Protein G Sepharose 4 Fast Flow (GE), and allowed to bind overnight at 4 °C. Finally, beads were washed 3 times with 1 ml of RIPA buffer supplemented with 1 mM activated Na3VO4 and eluted by adding 20 μl of 5× SDS-PAGE loading buffer containing 10% fresh β-mercaptoethanol and incubating for 30 minutes at room temperature. Eluted proteins were separated on 4% to 20% Tris-Glycine SDS-PAGE gels (Invitrogen), transferred to PVDF membranes, and probed with the antiphosphotyrosine antibody 4G10. A portion of crude lysate was reserved, run separately, and probed with antibodies against HA, Flag, EGFR, or ErbB4 to assess total receptor expression.
Expression of sErbB4 and Nrg-1β.
A cDNA encoding residues 1–615 (numbering from the mature N-terminus) of the JM-a isoform of human ErbB4 (sErbB4) was subcloned into the pSGHV0 expression vector (32), transfected into Lec1 Chinese hamster ovary cells (34), and a cell line expressing approximately 1 mg/liter of sErbB4 selected. pSGHV0 directs expression of target proteins as fusion proteins with human growth hormone at the N-terminus followed by an octahistidine tag and a tobacco etch virus (TEV) protease recognition site. Following concentration and dialysis of sErbB4-conditioned medium, the sErbB4 fusion protein was purified using Ni-NTA chromatography, cleaved by TEV protease, deglycosylated using endoglycosidases H and F, and further purified using anion-exchange and size-exclusion chromatographies. Approximately 0.7–0.8 mg of purified sErbB4 was obtained per liter of starting medium. A gene encoding the 55 amino-acid EGF repeat from human Neuregulin-1β (GTSHLVKCAE KEKTFCVNGG ECFMVKDLSN PSRYLCKCPN EFTGDRCQNY VMASF) was synthesized, subcloned into the pET32 expression vector, and expressed as a fusion protein with Thioredoxin, a histidine tag, and a TEV protease recognition site at its N-terminus in Origami cells (Novagen). The fusion protein was purified from lysates using Ni-NTA chromatography, cleaved using TEV protease, and further purified using cation exchange and size-exclusion chromatographies. A complex of sErbB4 and Nrg-1β was prepared by mixing a molar excess of Nrg-1β with sErbB4 and purifying the complex using size-exclusion chromatography.
Crystallization, X-ray Data Collection, and Structure Determination.
The purified sErbB4:Nrg-1β complex was dialyzed into 2.5 mM Tris pH 8.0, 25 mM NaCl and concentrated to 4 mg/ml. Crystals were grown at 20 °C by the hanging drop vapor diffusion method. 2 μl protein solution was mixed with 1 μl of a solution of 8% PEG 6000, 0.1 M Mg(OAc)2, and 0.1 M MES pH 6.0, and 0.5 μl Hampton Silver Bullet Bio condition F1 (0.25% w/v β-nicotinamide adenine dinucleotide phosphate tetrasodium salt, 0.25% w/v adenosine 5′-triphosphate disodium salt hydrate, 0.25% w/v N-acetyl-D-galactosamine, 0.25% w/v gentamicin sulfate salt hydrate, 0.02 M HEPES sodium pH 6.8) and 0.5 μl Hampton Silver Bullet Bio condition F7 (0.20% w/v thymine, 0.20% w/v sodium pyrophosphate tetrabasic decahydrate, 0.20% w/v D-glyceric acid calcium salt dihydrate, 0.20% w/v β-cyclodextrin, 0.20% w/v myo-inositol, 0.02 M HEPES sodium pH 6.8). Crystals grew as needles or blocks with a maximum size of 50 × 50 × 70 μm3. Crystals were soaked in 20% (v/v) PEG 200 prior to flash freezing in liquid nitrogen. X-ray diffraction data were collected from a single crystal at 23-ID-D-GM/CA of the advanced photon source at Argonne National Laboratory.
X-ray diffraction data were integrated, scaled, and merged using HKL2000 (35), and the structure determined by molecular replacement using the program MOLREP (36) with a single receptor:ligand subunit of the EGFR:TGFα complex used as a search model (15), which easily identified all six receptor subunits in the crystallographic asymmetric unit. Individual domains of the unliganded sErbB4 structure (31) were then superposed on the EGFR domains and fragments of domain IV placed in electron density as they became apparent during refinement. Refinement was carried out using the programs REFMAC (37), PHENIX (38), and autoBUSTER (39) alternated with model building using COOT (40). Final X-ray data collection and refinement statistics are presented in SI Appendix, Table S3.
Supplementary Material
ACKNOWLEDGMENTS.
We thank S. Gabelli and M. Bianchet for assistance with crystallography, T. Gilbert for assistance with cell-based assays, W. Yang and P. Cole for comments on the MS, and M. Becker and N. Venugopalan of GM/CA CAT at the Advanced Photon Source (APS) for beamline assistance. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the APS was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. This work supported by R01GM099321 from the National Institutes of Health (D.J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3U7U).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201114109/-/DCSupplemental.
References
- 1.Burgess AW. EGFR family: Structure physiology signalling and therapeutic targets. Growth Factors. 2008;26:263–274. doi: 10.1080/08977190802312844. [DOI] [PubMed] [Google Scholar]
- 2.Heldin CH. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y, Schlessinger J. Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry. 1987;26:1443–1451. doi: 10.1021/bi00379a035. [DOI] [PubMed] [Google Scholar]
- 4.Burgess AW, et al. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- 5.Chung I, et al. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464(7289):783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 6.Gadella TW, Jr, Jovin TM. Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J Cell Biol. 1995;129:1543–1558. doi: 10.1083/jcb.129.6.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moriki T, Maruyama H, Maruyama IN. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J Mol Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- 8.Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 9.Macdonald JL, Pike LJ. Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proc Natl Acad Sci USA. 2008;105:112–117. doi: 10.1073/pnas.0707080105. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarado D, Klein DE, Lemmon MA. ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor. Nature. 2009;461(7261):287–291. doi: 10.1038/nature08297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarado D, Klein DE, Lemmon MA. Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell. 2010;142:568–579. doi: 10.1016/j.cell.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Lu C, et al. Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol Cell Biol. 2010;30:5432–5443. doi: 10.1128/MCB.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macdonald-Obermann JL, Piwnica-Worms D, Pike LJ. Mechanics of EGF receptor/ErbB2 kinase activation revealed by luciferase fragment complementation imaging. Proc Natl Acad Sci USA. 2012;109:137–142. doi: 10.1073/pnas.1111316109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrett TP, et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 16.Ogiso H, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 17.Macdonald-Obermann JL, Pike LJ. The intracellular juxtamembrane domain of the epidermal growth factor (EGF) receptor is responsible for the allosteric regulation of EGF binding. J Biol Chem. 2009;284:13570–13576. doi: 10.1074/jbc.M109.001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tynan CJ, et al. Human epidermal growth factor receptor (EGFR) aligned on the plasma membrane adopts key features of Drosophila EGFR asymmetry. Mol Cell Biol. 2011;31:2241–2252. doi: 10.1128/MCB.01431-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson JP, et al. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell Biol. 2005;25:7734–7742. doi: 10.1128/MCB.25.17.7734-7742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Citri A, Skaria KB, Yarden Y. The deaf and the dumb: The biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284:54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 21.Holbro T, Hynes NE. ErbB receptors: Directing key signaling networks throughout life. Annu Rev Pharmacol Toxicol. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- 22.Cho HS, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 23.Garrett TP, et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell. 2003;11:495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 24.Burke CL, Lemmon MA, Coren BA, Engelman DM, Stern DF. Dimerization of the p185neu transmembrane domain is necessary but not sufficient for transformation. Oncogene. 1997;14:687–696. doi: 10.1038/sj.onc.1200873. [DOI] [PubMed] [Google Scholar]
- 25.Aertgeerts K, et al. Structural analysis of the mechanism of inhibition and allosteric activation of the kinase domain of HER2 protein. J Biol Chem. 2011;286:18756–18765. doi: 10.1074/jbc.M110.206193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slamon DJ, et al. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 27.Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- 28.Ferguson KM, et al. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 29.Mattoon D, Klein P, Lemmon MA, Lax I, Schlessinger J. The tethered configuration of the EGF receptor extracellular domain exerts only a limited control of receptor function. Proc Natl Acad Sci USA. 2004;101:923–928. doi: 10.1073/pnas.0307286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker F, et al. CR1/CR2 interactions modulate the functions of the cell surface epidermal growth factor receptor. J Biol Chem. 2004;279:22387–22398. doi: 10.1074/jbc.M401244200. [DOI] [PubMed] [Google Scholar]
- 31.Bouyain S, Longo PA, Li S, Ferguson KM, Leahy DJ. The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proc Natl Acad Sci USA. 2005;102:15024–15029. doi: 10.1073/pnas.0507591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leahy DJ, Dann CE, Longo P, Perman B, Ramyar KX. A mammalian expression vector for expression and purification of secreted proteins for structural studies. Protein Expr Purif. 2000;20:500–506. doi: 10.1006/prep.2000.1331. [DOI] [PubMed] [Google Scholar]
- 33.Qiu C, et al. In vitro enzymatic characterization of near full length EGFR in activated and inhibited states. Biochemistry. 2009;48:6624–6632. doi: 10.1021/bi900755n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanley P. Chinese hamster ovary cell mutants with multiple glycosylation defects for production of glycoproteins with minimal carbohydrate heterogeneity. Mol Cell Biol. 1989;9:377–383. doi: 10.1128/mcb.9.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 37.Vagin AA, et al. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.