Abstract
In the course of apoptosis, activated caspases cleave ∼500 to ∼1,000 different proteins in a mammalian cell. The dynamics of apoptosis involve a number of previously identified, caspase-generated proapoptotic protein fragments, defined as those that increase the probability of apoptosis. In contrast to activated caspases, which can be counteracted by inhibitor of apoptosis proteins, there is little understanding of antiapoptotic responses to proapoptotic protein fragments. One possibility is the regulation of proapoptotic fragments through their selective degradation. The previously identified proapoptotic fragments Cys-RIPK1, Cys-TRAF1, Asp-BRCA1, Leu-LIMK1, Tyr-NEDD9, Arg-BID, Asp-BCLXL, Arg-BIMEL, Asp-EPHA4, and Tyr-MET bear destabilizing N-terminal residues. Tellingly, the destabilizing nature (but not necessarily the actual identity) of N-terminal residues of proapoptotic fragments was invariably conserved in evolution. Here, we show that these proapoptotic fragments are short-lived substrates of the Arg/N-end rule pathway. Metabolic stabilization of at least one such fragment, Cys-RIPK1, greatly augmented the activation of the apoptosis-inducing effector caspase-3. In agreement with this understanding, even a partial ablation of the Arg/N-end rule pathway in two specific N-end rule mutants is shown to sensitize cells to apoptosis. We also found that caspases can inactivate components of the Arg/N-end rule pathway, suggesting a mutual suppression between this pathway and proapoptotic signaling. Together, these results identify a mechanistically specific and functionally broad antiapoptotic role of the Arg/N-end rule pathway. In conjunction with other apoptosis-suppressing circuits, the Arg/N-end rule pathway contributes to thresholds that prevent a transient or otherwise weak proapoptotic signal from reaching the point of commitment to apoptosis.
Keywords: arginylation, ATE1, proteolysis, UBR1
Perturbations of apoptosis, a specific kind of programmed cell death, play major roles in human diseases, including cancer, disorders of immunity, and neurodegenerative syndromes. Apoptosis is mediated by proteases called caspases, which are activated in response to extracellular signals or upon intracellular stresses (reviewed in refs. 1–12). Apoptosis involves the activation of initiator caspases (e.g., caspase-8). This and other initiator caspases cleave and activate effector caspases, including caspase-3. By making sequence-specific cuts in many cellular proteins, caspases either abolish or alter the functions of these proteins (13, 14). Activation of caspases can result in death of a cell, cell differentiation, or other effects, depending on physiological context (2, 3, 5).
After induction of apoptosis in a mammalian cell, ∼500 to ∼1,000 different proteins are cleaved by caspases (4, 5, 8). A small but biologically significant subset of the resulting fragments comprises polypeptides with proapoptotic activity. Such fragments are defined, operationally, as those that can be shown to increase the probability of apoptosis compared with full-length precursors of these fragments. Previous studies identified about 20 proapoptotic fragments of mammalian proteins (references in SI Results and SI Materials and Methods). In contrast to activated caspases, which can be counteracted by inhibitor of apoptosis (IAP) proteins (15, 16), there is little understanding of antiapoptotic responses to caspase-generated proapoptotic fragments. We report here that the Arg/N-end rule pathway down-regulates a number of proapoptotic fragments through their selective degradation.
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue. Regulated degradation of specific proteins by the N-end rule pathway mediates a number of functions, including the sensing of heme, nitric oxide, and oxygen; elimination of misfolded proteins; signaling by G proteins; and regulation of peptide import, DNA repair, chromosome cohesion/segregation, fat metabolism, spermatogenesis, neurogenesis, cardiovascular development, and many processes in plants (reviewed in refs. 17–22). The N-end rule pathway polyubiquitylates proteins that contain specific degradation signals (degrons), thereby targeting these proteins for degradation by the 26S proteasome. Degrons recognized by the N-end rule pathway include a set called N-degrons. The main determinant of an N-degron is a destabilizing N-terminal residue of a substrate protein. Recognition components of the N-end rule pathway, called N-recognins, are specific E3 Ub ligases that can target N-degrons (17, 23). The N-end rule pathway consists of two branches, the Ac/N-end rule and the Arg/N-end rule pathways. The Ac/N-end rule pathway recognizes proteins with N-terminally acetylated residues (24). The Arg/N-end rule pathway targets specific unacetylated N-terminal residues (Fig. 1A) (17, 25). The primary destabilizing N-terminal residues Arg, Lys, His, Leu, Phe, Tyr, Trp, and Ile are directly recognized by E3 N-recognins, whereas N-terminal Asp, Glu, Asn, Gln, and Cys function as destabilizing residues through their preliminary modifications. These modifications include N-terminal arginylation (Nt-arginylation) by the ATE1 arginyl-transferase (R-transferase) (Fig. 1A) (17, 26–30).
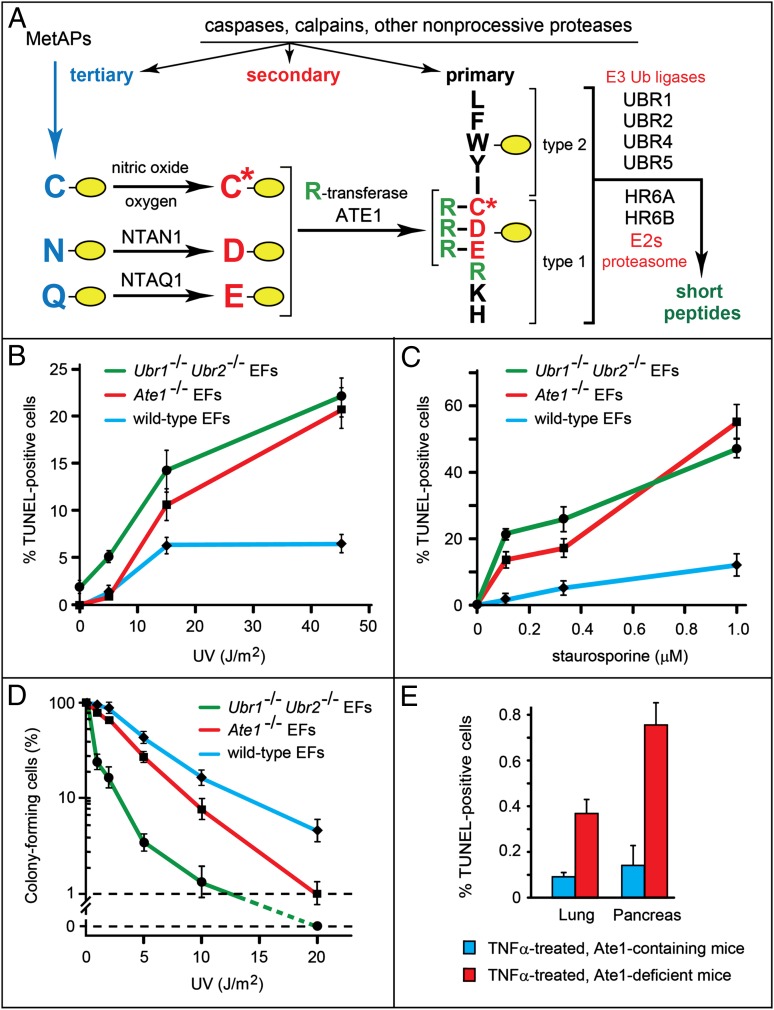
Fig. 1.
Partial ablations of the Arg/N-end rule pathway sensitize cells to apoptosis. (A) The mammalian Arg/N-end rule pathway (17). N-terminal residues are indicated by single-letter abbreviations for amino acids. Yellow ovals denote the rest of a protein substrate. Primary, secondary, and tertiary denote mechanistically distinct subsets of destabilizing N-terminal residues. C* denotes oxidized N-terminal Cys (either Cys-sulfinate or Cys-sulfonate). (B) Percentage of apoptotic (TUNEL-positive) cells produced by UV irradiation in WT, Ate1−/−, and Ubr1−/− Ubr2−/− mouse EF cell lines (◆, ■, and ●, respectively). (C) Same as B but for induction of apoptosis by staurosporine. (D) Same as B but for the colony-forming ability of treated cells. (E) Percentage of TUNEL-positive cells in the lungs and pancreases of ATE1-containing and -deficient mice that had been treated with TNFα (Fig. S4); ∼30,000 cells in lung and ∼15,000 cells in pancreas were scored for TUNEL-positive cells vs. total (DAPI-stained) cells. SEMs are indicated. Statistical analyses were performed using the t test (P < 0.0001 for lung, P < 0.00004 for pancreas).
Because cleavage sites in proteins that give rise to proapoptotic fragments coevolved with other elements of proapoptotic and antiapoptotic circuits, one possibility is that proapoptotic protein fragments are regulated through their selective degradation. Indeed, as shown below, at least 10 of the previously identified proapoptotic fragments are short-lived substrates of the Arg/N-end rule pathway. Metabolic stabilization of at least one such fragment was found to greatly augment the activation of the apoptosis-inducing effector caspase-3. In agreement with this understanding, even a partial ablation of the Arg/N-end rule pathway is shown to make cells hypersensitive to apoptosis. We also found that caspases can cleave and inactivate components of the Arg/N-end rule pathway, suggesting a mutual suppression between the Arg/N-end rule and proapoptotic signaling. Together, these results identified a mechanistically specific and functionally broad antiapoptotic role of the Arg/N-end rule pathway.
Results
Conservation of Destabilizing N-Terminal Residues in Proapoptotic Fragments.
By surveying the literature for the previously identified proapoptotic mammalian protein fragments, we found that, of 20 such fragments, 16 of them bear N-terminal residues that are destabilizing in the Arg/N-end rule pathway (Fig. 1A and Fig. S1). These N-terminal residues have been unambiguously mapped in studies cited in SI Results. None of these studies considered a possible regulation of proapoptotic fragments by the Arg/N-end rule pathway.
We also found that the N-terminal residues of these proapoptotic fragments (they are P1′ residues in the cleavage sites of the full-length precursors) were conserved among vertebrates (Figs. S1 and S2), with one seeming exception: the P1′ residue of the caspase cleavage site in the tumor suppressor BRCA1 is Asp in humans and mice but Ile in water mice, Glu in rabbits, and Asn in bats (Fig. S2C). Tellingly, however, all of these P1′ residues of BRCA1 in different mammals are destabilizing in the Arg/N-end rule (Fig. 1A and Fig. S2C). A constraint of this kind would be expected if a short in vivo half-life of the proapoptotic fragment (rather than the exact identity of its N-terminal residue) was a fitness-increasing property of this fragment maintained by selection during evolution. The retention of destabilizing P1′ residues in the precursors of proapoptotic fragments (Figs. S1 and S2) is noteworthy for another reason as well. More than 90% of the mapped caspase cleavage sites in cellular proteins contain, at their P1′ positions, small residues such as Gly, Ser, Thr, and Ala (4, 8). These residues are not recognized by the Arg/N-end rule pathway (Fig. 1A). Thus, remarkably, the destabilizing P1′ residues in the precursors of proapoptotic fragments (let alone the evolutionary conservation of these residues) (Figs. S1 and S2) would not be expected on a priori grounds given the preponderance of smaller P1′ residues in caspase substrates at large (4, 8). Together, these findings (Figs. S1 and S2) strongly suggested adaptive (fitness-increasing) origins of destabilizing P1′ residues in the precursors of proapoptotic fragments.
Proapoptotic Fragments as Short-Lived Arg/N-End Rule Substrates.
The initial uncertainty about whether a protein with a destabilizing N-terminal residue is an efficacious substrate of the Arg/N-end rule pathway stems from the multideterminant organization of an N-degron. This degradation signal must contain not only a sterically accessible destabilizing N-terminal residue but also a suitably located disordered segment of a target protein and a ubiquitylatable internal Lys residue (17).
To determine whether the proapoptotic fragments (Fig. S1) were, in fact, degraded by the Arg/N-end rule pathway, we used the extensively validated Ub reference technique (URT) derived from the Ub fusion technique (17, 31, 32). Cotranslational cleavage of a URT-based Ub fusion by deubiquitylases produces, at the initial equimolar ratio, both a test protein with a desired N-terminal residue and a reference fDHFR-UbR48, a flag-tagged derivative of the mouse dihydrofolate reductase (Fig. 2A). In URT-based pulse-chase assays, the labeled test protein is quantified by measuring its levels relative to the levels of a stable reference at the same time point during a chase (Fig. 2A). In addition to being more accurate than pulse-chases without a built-in stable reference, URT makes it possible to measure the degradation of the test protein before the chase (i.e., during the pulse) (24, 31, 32).
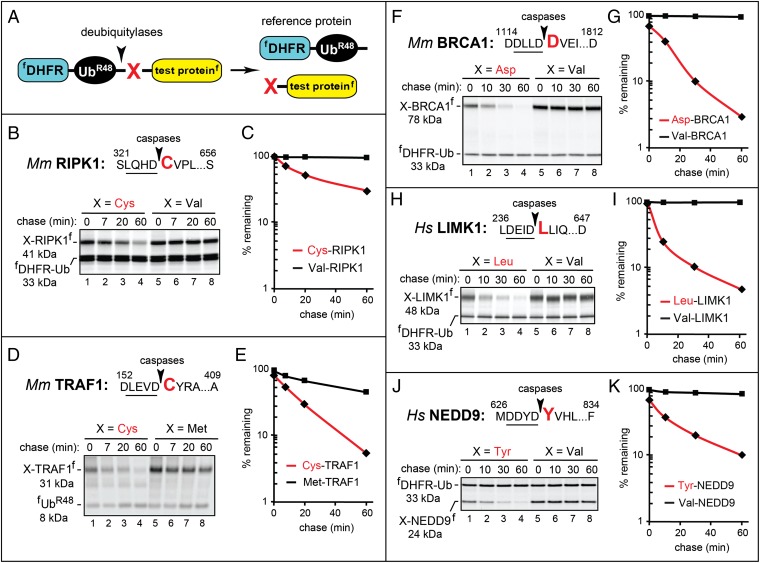
Fig. 2.
Cys-RIPK1, Cys-TRAF1, Asp-BRCA1, Leu-LIMK1, and Tyr-NEDD9 as short-lived N-end rule substrates. Molecular masses of the test proteins were ∼1 kDa larger than their indicated (natural) sizes, because these fragments contained the ∼1 kDa flag epitope. (A) The URT assay (31, 32). (B) The caspase cleavage site in mouse RIPK1 is indicated by an arrowhead, with the P1′ residue shown in red. Residue numbers are the numbers of the first shown residue and the last residue of full-length RIPK1. Cys326-RIPK1f (produced from fDHFR-UbR48-Cys326-RIPK1f) and Val326-RIPK1f were expressed in reticulocyte extract and labeled with 35S-Met/Cys for 10 min at 30 °C, followed by a chase, immunoprecipitation with anti-flag antibody, SDS/PAGE, and autoradiography. (C) Quantification of B using the reference protein fDHFR-UbR48. (D) Same as B but with mouse X157-TRAF1f (X = Cys, Met). In these pulse chases, the reference was flag-tagged fUbR48 instead of fDHFR-UbR48. (E) Quantification of D. (F) Same as in B but with mouse X1119-BRCA1f (X = Asp, Val). (G) Quantification of F. (H) Same as in B but with human X241-LIMK1f (X = Leu, Val). (I) Quantification of H. (J) Same as in B but with human X631-NEDD9f (X = Tyr, Val). (K) Quantification of J [◆, a natural proapoptotic fragment; ■, an otherwise identical fragment with N-terminal Val (or Met in D and E)].
URT-based 35S-pulse chases with 10 proapoptotic fragments were performed in a transcription-enabled rabbit reticulocyte extract, which contains the Arg/N-end rule pathway and has been extensively used to analyze this pathway (Figs. 1A, 2, and 3 and Fig. S1) (17). All of the examined proapoptotic fragments have been shown to be generated in vivo by activated caspases except for Arg71-BID and Asp61-BCLXL, which are produced by activated calpains during cell death mediated by an increase in Ca2+ (Fig. S1). Specific URT fusions (Fig. 2A) C-terminally tagged with the flag epitope were labeled with 35S-Met/Cys in reticulocyte extract for 10 min at 30 °C, followed by a chase, immunoprecipitation with a monoclonal anti-flag antibody, SDS/PAGE, autoradiography, and quantification (Figs. 2 and 3). The logic of these assays (17) involves a comparison between the degradation rates of a protein bearing a destabilizing N-terminal residue and an otherwise identical protein with an N-terminal residue such as Val, which is not recognized by the Arg/N-end rule pathway (Fig. 1A).
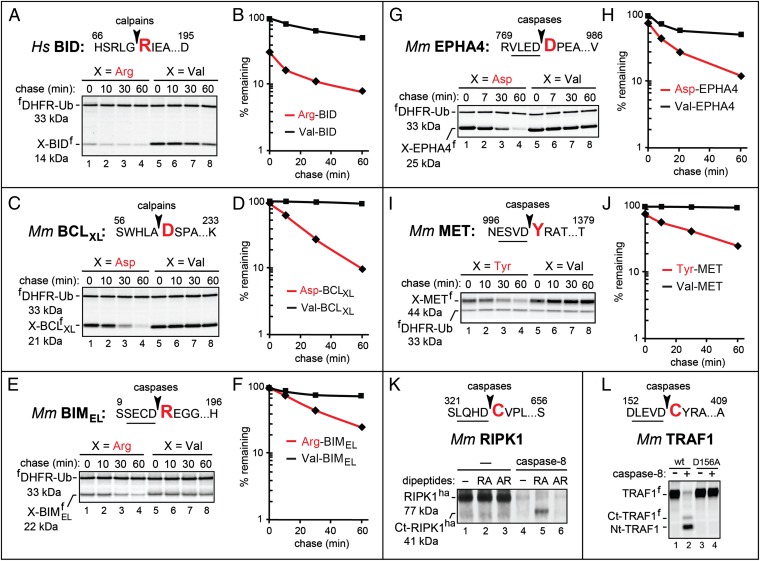
Fig. 3.
Arg-BID, Asp-BCLXL, Arg-BIMEL, Asp-EPHA4, and Tyr-MET as short-lived N-end rule substrates. (A) Human Arg71-BIDf (produced from fDHFR-UbR48-Arg71-BIDf), the otherwise identical Val71-BIDf, and the calpain cleavage site in BID. Fig. 2 shows notations and pulse-chase procedures. (B) Quantification of A. (C) Same as in A but with mouse X61-BCLXLf (X = Asp, Val). (D) Quantification of C. (E) Same as in A but with mouse X14-BIMELf (X = Arg, Val). (F) Quantification of E. (G) Same as in A but with mouse X774-EPHA4f (X = Asp, Val). (H) Quantification of G. (I) Same as in A but with mouse X1001-METf (X = Tyr, Val). (J) Quantification of I. (K) Cleavage of RIPK1 by caspase-8. Mouse RIPK1ha was expressed in reticulocyte extract for 45 min at 30 °C, followed by the addition of cycloheximide and either recombinant human caspase-8 or buffer alone. The samples were incubated for 30 min followed by SDS/PAGE and immunoblotting with anti-ha antibody. Lane 1, RIPK1ha; lanes 2 and 3, same as lane 1 but with the Arg-Ala and Ala-Arg, respectively, at 5 mM added the start of assays; lanes 4–6, same as lanes 1–3, respectively, but with caspase-8 as well. Note the caspase-generated C-terminal RIPK1 fragment migrating at the Mr of the 41-kDa Cys326-RIPK1ha in lane 5 (with 5 mM Arg-Ala) but not in lane 4 (no added dipeptide) or lane 6 (with 5 mM Ala-Arg). (L) Cleavage of TRAF1 by caspase-8. Mouse TRAF1f and its caspase-resistant mutant TRAF1D156A(f) were expressed in reticulocyte extract for 1 h at 30 °C in the presence of [35S]methionine, followed by the addition of cycloheximide, unlabeled methionine and cysteine, and either caspase-8 or buffer alone. Samples were incubated for another 30 min, followed by SDS/PAGE and autoradiography. Lanes 1 and 3, TRAF1f and TRAF1D156A(f), respectively; lanes 2 and 4, same as lanes 1 and 3, respectively, but with caspase-8. Note the much lower (degradation-mediated) level of the 35S-labeled, caspase-generated C-terminal TRAF1 fragment denoted as Ct-TRAF1f (it migrates at the expected Mr of the Cys157-TRAF1 fragment) compared with the N-terminal fragment (denoted as Nt-TRAF1f), despite the higher content of 35S-Met in Ct-TRAF1f.
Brief descriptions of the 10 proapoptotic fragments below (Figs. 2 and 3) are complemented by the detailed references and descriptions in SI Results. The proapoptotic fragments Cys326-RIPK1 (its precursor is a protein kinase and regulator of apoptosis), Asp1119-BRCA1 (its precursor is an E3 Ub ligase and a tumor suppressor), Leu241-LIMK1 (its precursor is a protein kinase), Tyr631-NEDD9 (its precursor is a scaffolding protein), Asp61-BCLXL (its precursor is an antiapoptotic protein), Arg14-BIMEL (its precursor is a regulator of apoptosis), and Tyr1001-MET (its precursor is a dependence receptor) were short-lived in reticulocyte extract, whereas the otherwise identical fragments bearing the N-terminal Val residue (which is not recognized by the Arg/N-end rule pathway) were either stable or nearly stable (Figs. 2 and 3). These results indicated that the seven cited proapoptotic fragments contained Arg/N-degrons and furthermore, that their Arg/N-degrons were either the sole or major degradation signals in these proteins. Similarly, the proapoptotic fragments Cys157-TRAF1 (its precursor is a regulator of apoptosis), Arg71-BID (its precursor is proapoptotic as well but less than Arg71-BID), and Asp774-EPHA4 (its precursor is a dependence receptor) were also short-lived in reticulocyte extract. Their counterparts bearing N-terminal Val (or Met in the case of X157-TRAF1) were considerably longer lived but still unstable, indicating the presence of both Arg/N-degrons and internal degrons in these three proapoptotic fragments (Figs. 2 and 3).
In sum, we examined 10 of 16 proapoptotic fragments as possible Arg/N-end rule substrates (Fig. S1) and found that all 10 of them were, in fact, short-lived substrates of the Arg/N-end rule pathway (Figs. 2 and 3). In a majority of these fragments, their Arg/N-degrons were their sole degrons at least in reticulocyte extract.
Stabilization of the Cys-RIPK1 Proapoptotic Fragment Greatly Augments the Activation of Apoptosis-Inducing Caspase-3.
The human Cys325-RIPK1 fragment was a short-lived Arg/N-end rule substrate both in reticulocyte extract (Fig. 2 B and C) (half-life of ∼20 min) and T-Rex-293 human cells (Fig. S3 A and B) (initial half-life of ∼15 min). These cell lines were designed to stably express either Cys325-RIPK1 or Val325-RIPK1 using a doxycycline (Dox)-inducible promoter and site-specific integration of transgenes. In contrast to the short-lived mouse or human Cys-RIPK1 fragment, the otherwise identical Val-RIPK1 was either stable (in reticulocyte extract) (Fig. 2 B and C) or degraded at a much lower rate (in T-Rex-293 cells) (Fig. S3 A and B).
The proapoptotic activity of the Cys325-RIPK1 fragment involves the promotion of activation of the initiator caspase-8 (33). One role of caspase-8 is the activation of the effector caspase-3, a major step to apoptosis (5). Although both the short-lived Cys325-RIPK1f fragment and the otherwise identical long-lived Val325-RIPK1f could be observed to activate caspase-3 in T-Rex-293 cells, the levels of caspase-3 activation were strikingly higher in cells that expressed the long-lived Val325-RIPK1 (Fig. 4A). Assays identical to the ones in Fig. 4A but with measurements of overtly apoptotic cells (instead of activated caspase-3) indicated a greatly increased proapoptotic activity of the stabilized Val325-RIPK1f fragment compared with its short-lived Cys325-RIPK1f counterpart.
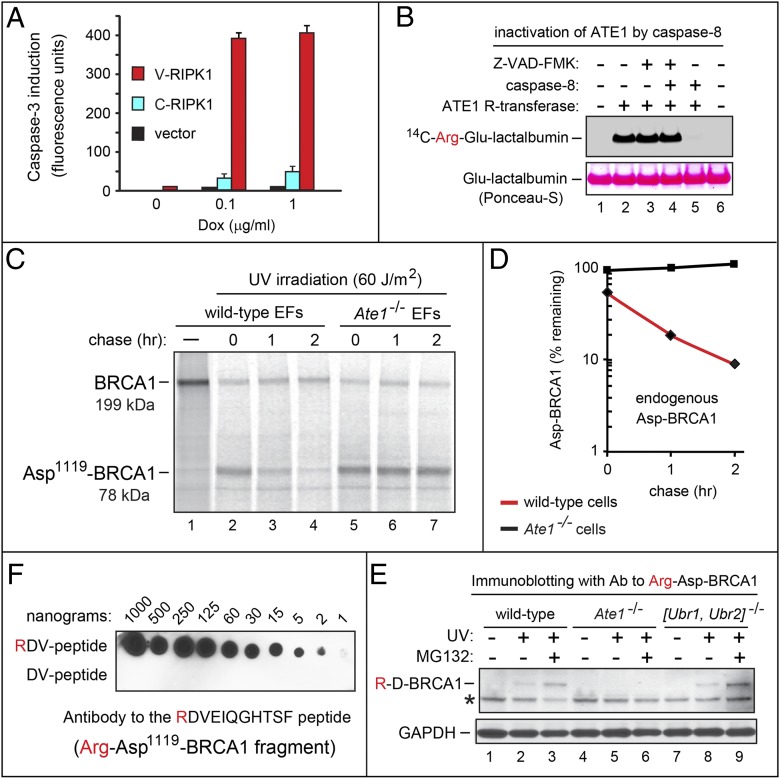
Fig. 4.
Cys-RIPK1, Cys-BRCA1, ATE1, and the Arg/N-end rule pathway. (A) Activation of caspase-3 in human T-Rex-293–based cell lines stably expressing human X325-RIPK1f fragments (X = Cys, Val) from a Dox-inducible promoter. Expression of X325-RIPK1f was induced with Dox over 24 h followed by the addition of cycloheximide to a subtoxic (2.5 μg/mL) concentration, 12 h of additional incubation, and measurements, in triplicate, of caspase-3 activity in cell extracts. SDs are indicated. Black bars, an integrated vector alone; blue bars, Cys325-RIPK1f; red bars, Val325-RIPK1f. (B) Functional inactivation of the ATE1 R-transferase by caspase-8. Purified mouse ATE1 (12.5 μg/mL) was treated with recombinant caspase-8 (0.1 μg/mL) or buffer alone for 30 min at 25 °C in the presence or absence of the pan-caspase inhibitor Z-VAD-FMK. Reactions were stopped, followed by the addition of Glu-lactalbumin (an Nt-arginylation reporter), 14C-l-arginine, and other components of ATE1 arginylation assay, incubation for 30 min at 30 °C, SDS/PAGE, and autoradiography (Upper). (Lower) Bands of total lactalbumin stained with Ponceau-S. (C) WT and Ate1−/− mouse EF cell lines were UV-irradiated (60 J/m2), followed by incubation for 4 h and the labeling of cells with 35S-Met/Cys for 60 min. Caspase inhibitors Z-DEVD-FMK and Z-VAD-FMK, as well as unlabeled Met and Cys were then added, followed by a chase, immunoprecipitation of cell extracts with antibody to BRCA1, SDS/PAGE, and autoradiography. (D) Quantification of data in C. ◆, Asp-BRCA1 in UV-irradiated WT cells; ■, Asp-BRCA1 in UV-irradiated Ate1−/− cells. (E) Immunoblotting of mouse cell extracts with antibody to Arg-Asp1119-BRCA1 fragment. Lanes 1–3, WT EF cells untreated, UV-irradiated, or UV-irradiated and treated with MG132; lanes 4–6, same as lanes 1–3 but with Ate1−/− EF cells; lanes 7–9, same as lanes 1–3 but with Ubr1−/− Ubr2−/− EF cells. The band of Arg-Asp1119-BRCA1 is indicated. *A cross-reacting protein. (F) A dot blot assay for antibody to the Arg-Asp1119-BRCA1 fragment. Indicated amounts of the RDVEIQGHTSFC or the DVEIQGHTSFC peptide were probed with the affinity-purified antibody to RDVEIQGHTSFC (in the text).
To assess the previously shown production of the proapoptotic Cys-RIPK1 fragment by caspase-8 (33–35), the full-length, C-terminally ha-tagged mouse RIPK1ha was expressed in reticulocyte extract in the presence or absence of active caspase-8. Degradation of Arg/N-end rule substrates can be selectively inhibited by dipeptides such as Arg-Ala and Leu-Ala, which bear type 1 (basic) or type 2 (bulky hydrophobic) destabilizing N-terminal residues, respectively, and interact with type 1/2 substrate-binding sites of N-recognins (Fig. 1A) (17). In the absence of caspase-8, RIPK1ha remained intact (Fig. 3K, lanes 1–3). In contrast, virtually no RIPK1ha was retained in the presence of caspase-8 (Fig. 3K, lane 4 compared with lanes 1–3) because of the formation of the Cys326-RIPK1ha fragment (33–35) and its degradation by the Arg/N-end rule pathway (Fig. 2 B and C). As would be expected, the addition of the Arg-Ala (but not the Ala-Arg) dipeptide to reticulocyte extract partially stabilized a C-terminal fragment of RIPK1 that migrated at the Mr of Cys326-RIPK1ha (Fig. 3K, lane 5 compared with lanes 4 and 6).
In Vivo Stabilization of the Asp-BRCA1 Fragment in the Absence of Arginylation.
Mouse BRCA1, an E3 Ub ligase and a tumor suppressor, is cleaved by caspases during apoptosis, resulting in the proapoptotic 78-kDa Asp1119-BRCA1 fragment (SI Results and Figs. S1 and S2C). Similarly to several other proapoptotic Arg/N-end rule substrates (Fig. S1), the degradation of Asp1119-BRCA1 requires its preliminary Nt-arginylation by the ATE1 R-transferase (Fig. 1A). The half-life of Asp1119-BRCA1 in reticulocyte extract was ∼6 min, whereas Val1119-BRCA1 was stable (Fig. 2 F and G).
In contrast to functionally overlapping E3 N-recognins of the mammalian Arg/N-end rule pathway (Fig. 1A), the upstream R-transferase is encoded by a single Ate1 gene. Taking advantage of this fact, we asked whether caspase-mediated production of Asp1119-BRCA1 from endogenous BRCA1 in living cells is accompanied by degradation of this fragment by the Arg/N-end rule pathway. A mouse embryonic fibroblast (EF) cell line and its Ate1−/− counterpart that lacked Nt-arginylation (Fig. 1A) (17, 26) were treated with apoptosis-inducing UV irradiation, followed by incubation for 4 h and labeling with [35S]methionine/cysteine for 60 min. Caspase inhibitors Z-DEVD-FMK and Z-VAD-FMK (the latter is a pan-caspase inhibitor) were then added, followed by a chase, immunoprecipitation of cell extracts with antibody to BRCA1, SDS/PAGE, and autoradiography (Fig. 4 C and D). Caspase inhibitors were added to halt the production of Asp1119-BRCA1 from BRCA1, so that the 35S-labeled Asp1119-BRCA1 fragment could be followed during chase.
The bulk of the 199-kDa BRCA1 was cleaved or otherwise degraded after UV irradiation (Fig. 4C, lane 1 compared with lanes 2–7). The previously identified endogenous Asp1119-BRCA1 fragment (SI Results) migrated at the expected Mr and was present at the beginning of chase in both Ate1−/− and WT EF cells (Fig. 4C). However, the prechase levels of Asp1119-BRCA1 were reproducibly approximately twofold higher in Ate1−/− cells, suggesting a significant degradation of Asp1119-BRCA1 during 35S-labeling in WT cells (Fig. 4 C and D). Moreover, whereas 35S-labeled Asp1119-BRCA1 was short-lived in WT cells during the chase (half-life of ∼25 min), it was stable in Ate1−/− cells, which lacked Nt-arginylation (Fig. 4 C and D).
In another approach, we prepared a rabbit antibody to RDVEIQGHTSFC, the Nt-arginylated N-terminal sequence of the Asp1119-BRCA1 fragment (except for C-terminal Cys, which was used to conjugate peptide to a carrier protein). This antibody was affinity-purified both positively (against RDVEIQGHTSFC) and negatively (against DVEIQGHTSFC). The resulting antibody exhibited a striking specificity for the Nt-arginylated RDVEIQGHTSFC peptide (Fig. 4F). Immunoblotting extracts of WT, Ate1−/−, and double-mutant Ubr1−/− Ubr2−/− EF cells with this antibody produced results in agreement with pulse-chase findings (Fig. 4 C and D). Specifically, whereas virtually no Arg-Asp1119-BRCA1 fragment was detectable in untreated WT EF cells, apoptosis-inducing UV irradiation of these cells resulted in a detectable level of the (short-lived) Arg-Asp1119-BRCA1 fragment, with an additional (expected) increase of this fragment in the presence of MG132, a proteasome inhibitor (Fig. 4E, lanes 1–3). In contrast and in agreement with specificity of this antibody, no Arg-Asp1119-BRCA1 could be detected in extracts of Ate1−/− EF cells (which lacked Nt-arginylation) under any conditions (Fig. 4E, lanes 4–6), although the Asp1119-BRCA1 fragment was present in these cells after UV irradiation, which could be seen using a pan-BRCA1 antibody (Fig. 4C). The results with Ubr1−/− Ubr2−/− EF cells, which lacked two of four E3 Ub ligases of the Arg/N-end rule pathway and retained Nt-arginylation (Fig. 1A), were qualitatively similar to the results with WT EF cells (Fig. 4E, lanes 7–9).
We conclude that, despite significant differences in the molecular environment (Asp1119-BRCA1 in reticulocyte extract vs. endogenous Asp1119-BRCA1 in apoptotic mouse cells), the kinetics of cleavage that generate Asp1119-BRCA1 (cotranslational vs. posttranslational), and the nature of proteases involved (deubiquitylases vs. caspases), the Asp1119-BRCA1 fragment was a short-lived Arg/N-end rule substrate in either the heterologous or natural setting (Figs. 2 F and G and 4 C and D).
Partial Ablations of the Arg/N-End Rule Pathway Make Cells Hypersensitive to Apoptosis.
If the degradation of proapoptotic protein fragments counteracts cell death, even a partial stabilization of these fragments would be expected to increase the sensitivity of cells to apoptosis. One line of evidence of this kind, described above, was a greatly increased proapoptotic activity of a metabolically stabilized derivative of the proapoptotic Cys325-RIPK1 fragment (Fig. 4A).
Of the 10 proapoptotic fragments identified here as Arg/N-end rule substrates, 5 fragments (Asp-BRCA1, Cys-RIPK1, Cys-TRAF1, Asp-BCLXL, and Asp-EPHA4) are targeted for degradation through their Nt-arginylation (Figs. 1A, 2, 3, and 4 and Figs. S1, S2, and S3). An Ate1−/− EF cell line, which lacked Nt-arginylation (26), was indeed hypersensitive to apoptosis induced by either UV irradiation or staurosporine, a kinase inhibitor known to induce apoptosis (Fig. 1 B–D). The hypersensitivity of Ate1−/− cells to apoptosis was detected using either the TUNEL assay for fragmented DNA (Fig. 1 B and C) or a growth assay for the ability of treated cells to form colonies (Fig. 1D). To preclude the possibility that other, non-Ate1 mutations in these Ate1−/− cells might underlie the observed phenotype, we repeated the above experiments (Fig. 1 B–D) with a second Ate1−/− EF cell line produced from independently derived Ate1−/− embryos and obtained the same results. In addition and also importantly, primary early-passage (nontransformed) Ate1−/− EF cells were similar to the postcrisis Ate1−/− cell lines in their hypersensitivity to apoptosis (Fig. 1 B–D compared with Fig. S3C). We also examined a Ubr1−/− Ubr2−/− EF cell line, which retained Nt-arginylation but had a weakened Arg/N-end rule pathway because of the ablation of two (of at least four) N-recognins (Fig. 1A) (17, 22). Similarly to Ate1−/− EFs (which lacked Nt-arginylation but retained N-recognins), Ubr1−/− Ubr2−/− EFs were hypersensitive to apoptosis induced by either UV or staurosporine (Fig. 1 B–D).
Together with the Cys/Val-RIPK1 metabolic stabilization results (Fig. 4A), the findings with specific Arg/N-end rule mutants (Fig. 1 B–D) indicate that the Arg/N-end rule pathway is a repressor of apoptosis, at least in part through its ability to destroy proapoptotic fragments (Figs. 2 and 3). Importantly, the apoptosis hypersensitivity of two distinct Arg/N-end rule mutants was not accompanied by any increase of cell death (relative to WT controls) in the absence of proapoptotic treatments.
Our previous work has produced Ate1flox/- mouse strains in which one copy of Ate1 was inactive, whereas the other (floxed) copy of Ate1 could be inactivated in adult mice through a transient activation of Cre recombinase (30). The Cys-RIPK1 fragment promotes apoptosis through a positive feedback that facilitates the activation of caspase-8, which in turn, cleaves RIPK1 to generate more Cys-RIPK1 (33–35). As shown here, the Cys-RIPK1 fragment is a short-lived Arg/N-end rule substrate that is targeted for degradation through Nt-arginylation (Figs. 1A and 2 B and C and Figs. S1 and S2A). We compared the levels of endogenous RIPK1 in ATE1-containing and ATE1-deficient mice that had been treated for 12 h with TNFα. A remarkable all-or-none pattern was discovered in TNFα-responsive tissues: whereas the levels of RIPK1 remained unchanged in the lungs and pancreases of TNFα-treated ATE1-containing mice, full-length RIPK1 virtually disappeared from identically treated ATE1-deficient mice (Fig. S4A, lanes 1–6 and 13–18). Similar but not as striking patterns were observed with spleens (Fig. S4A, lanes 7–12). No changes in RIPK1 were observed in the brains of TNFα-treated mice of either genotype, presumably because of the relative lack of TNFα access to the brain.
Although a RIPK1 fragment that migrated at the Mr of the 41-kDa Cys326-RIPK1 (Fig. 2 B and C and Fig. S2A) could be detected in pancreases of TNFα-treated ATE1-deficient mice, the low level of this fragment suggested instability of Cys326-RIPK1, even in ATE1-lacking cells (Fig. S4A, lanes 19 and 20 compared with lanes 17 and 18, which are the lower-exposure counterparts of lanes 19 and 20). The presence of an internal degron in Cys-RIPK1, in addition to its N-degron, was consistent with residual instability of the human Val-RIPK1 fragment (compared with Cys-RIPK1) in T-Rex-293 cells (Fig. S3B), whereas only the Arg/N-degron of Cys-RIPK1 was active in reticulocyte extract (Fig. 2 B and C). A mutually nonexclusive possibility is that the absence of Nt-arginylation in ATE1-deficient mice not only augments the RIPK1/caspase-8 positive feedback but also increases, through a mechanism to be understood, the TNFα-induced processive degradation of full-length RIPK1, a conditionally short-lived protein (6, 33, 36). In contrast to results with ATE1-deficient mouse tissues (Fig. S4A), a RIPK1 fragment that migrated at the Mr of the 41-kDa Cys326-RIPK1 was readily detectable in Ate1−/− mouse EF cells that had been treated with both TNFα and a subtoxic level of cycloheximide, a regimen that induces apoptosis in EF cells (33) (Figs. S3 D and E and S5B). In addition, full-length RIPK1 was decreased but retained in both Ate1−/− and WT EF cells treated with TNFα-cycloheximide (Figs. S3 D and E and S5B).
The TNFα-induced loss of RIPK1 from the lungs and pancreases of ATE1-deficient mice (Fig. S4A) was accompanied by an approximately fourfold increase of apoptotic (TUNEL-positive) cells in these tissues compared with ATE1-containing mice (Fig. 1E and Fig. S4B). The complete or nearly complete disappearance of RIPK1 from the pancreases, lungs, and spleens of TNFα-treated ATE1-deficient (but not ATE1-containing) mice (Fig. S4A) suggested that most ATE1-deficient cells in thus affected tissues may become TUNEL-positive if examined significantly later than 12 h after TNFα treatment. Such a test would be a technically difficult proposition, given the increased toxicity of TNFα to ATE1-deficient mice. Even by 12 h after TNFα treatment, the majority of acinar cells in the pancreases of ATE1-deficient (but not ATE1-containing) mice exhibited signs of damage, an alteration that may be causally linked to the also observed decrease of tubulin in the pancreases of TNFα-treated Ate1-deficient mice (Fig. S4A, lanes 16 and 18 compared with lane 13; each lane corresponds to a sample from an individual mouse). The positive feedback that involves RIPK1, the Cys-RIPK1 fragment, and caspase-8 (33–35) is likely to play a role in the striking effect of TNFα on the levels of RIPK1 in the absence of Nt-arginylation (Fig. S4A). A detailed understanding of this robustly reproducible all-or-none effect, which is likely to involve more than one RIPK1 circuit, is the subject of future studies.
Caspases Can Down-Regulate the Arg/N-End Rule Pathway.
Given the observed antiapoptotic activity of the Arg/N-end rule pathway, might there be a mutual suppression between this pathway and proapoptotic effectors such as, for example, activated caspases? To address this possibility, we measured the Nt-arginylation of added Glu-lactalbumin (an Nt-arginylation reporter) in extracts from mouse NIH 3T3 cells that had been treated either with TNFα alone (50 ng/mL), a subtoxic level of cycloheximide (10 μg/mL), or both, the latter treatment being an apoptosis-inducing one (33). Treatments of cells with cycloheximide alone reproducibly increased Nt-arginylation approximately twofold after 24 h (Fig. S5A). This effect may be caused by a down-regulation, through a cycloheximide-mediated decrease in translation, of a short-lived repressor of the ATE1 R-transferase. In contrast, the treatment with cycloheximide plus TNFα decreased Nt-arginylation by ∼2.3-fold after 24 h, which is in agreement with the results of ATE1-specific immunoblotting of cell extracts that indicated the in vivo degradation of ATE1 during apoptosis (Fig. S5 A and B). ATE1 and E3 N-recognins, such as UBR1 (Fig. 1A), contain caspase cleavage sites (Fig. S5C). Indeed, purified mouse ATE1 was found to be both cleaved and functionally inactivated by added caspase-8 (Fig. 4B and Fig. S5D). The UBR1 E3 N-recognin in an extract from mouse 3T3 cells was also cleaved by caspase-8 and/or other activated caspases in the extract. These cleavages, which remain to be examined for their effects on UBR1 activity, could be inhibited by a pan-caspase inhibitor (Fig. S5E). In sum, there is a mutual suppression between the Arg/N-end rule pathway and proapoptotic signaling.
Discussion
The previously identified proapoptotic protein fragments Cys-RIPK1, Cys-TRAF1, Asp-BRCA1, Leu-LIMK1, Tyr-NEDD9, Arg-BID, Asp-BCLXL, Arg-BIMEL, Asp-EPHA4, and Tyr-MET (Figs. S1 and S2) are produced in cells by activated caspases (eight fragments) or activated calpains (two fragments). They were shown here to be short-lived substrates of the Arg/N-end rule pathway (Figs. 2, 3, and 4 and Figs. S1 and S2). The complete set of proapoptotic fragments that are also short-lived Arg/N-end rule substrates is almost certainly larger than the 10 substrates identified in the present study, because several other previously described proapoptotic fragments remain to be examined for their degradation by the Arg/N-end rule pathway (Fig. S1). In addition, more of such fragments are likely to be identified in the future. By destroying these fragments, the Arg/N-end rule pathway down-regulates their proapoptotic activities and thereby inhibits reactions that lead to cell death.
We also found that the metabolic stabilization of a specific proapoptotic fragment such as Cys-RIPK1 (through its conversion to the otherwise identical Val-RIPK1 fragment) greatly augments the activation of the apoptosis-inducing effector caspase-3 in human cells as well as their eventual apoptosis (Fig. 4 A and B), indicating that the antiapoptotic activity of the Arg/N-end rule pathway is likely to be a significant contributor to the physiological control of apoptosis. This conclusion is also supported by conceptually independent evidence provided by Arg/N-end rule mutants. Mouse cells in which the Arg/N-end rule pathway was weakened in two entirely different ways (through the ablation of either Nt-arginylation or two of four N-recognins) were hypersensitive to apoptosis induced by TNFα, staurosporine, or UV irradiation (Fig. 1 B–E and Figs. S3C and S4B). This finding would be expected if the Arg/N-end rule pathway is a repressor of apoptosis at least in part through its ability to destroy proapoptotic fragments (Figs. 2 and 3). It remains to be determined whether the antiapoptotic function of the Arg/N-end rule pathway is confined to this mechanism or involves additional processes as well.
As described in Results, the destabilizing nature of N-terminal residues of the proapoptotic fragments (these residues are at P1′ positions in the cleavage sites of the corresponding precursor proteins) is fully conserved among vertebrates, either through direct conservation of P1′ identities (Figs. S1 and S2) or, in the case of BRCA1, despite a drift of the exact identity of the P1′ residue. In the latter case, the identity of the P1′ residue changed during the evolution of mammals but remained destabilizing in the Arg/N-end rule pathway (Fig. S2C). A constraint of this kind would be expected if a short in vivo half-life of a proapoptotic fragment (rather than the exact identity of a P1′ residue in a protein precursor) was a fitness-increasing property of this fragment maintained by selection during evolution. More than 90% of the mapped caspase cleavage sites in cellular proteins contain, at their P1′ positions, small residues such as Gly, Ser, Thr, and Ala (4, 8), which are not recognized by the Arg/N-end rule pathway (Fig. 1A). Therefore, destabilizing P1′ residues in the precursors of proapoptotic fragments (let alone the evolutionary conservation of these residues) (Figs. S1 and S2) would not be expected on a priori grounds, given the preponderance of smaller P1′ residues in caspase substrates at large (4, 8). In sum, the evolutionary patterns of destabilizing P1′ residues (future N-terminal residues) in the precursors of proapoptotic fragments, including the retention of their destabilizing activity despite a drift of their identity (Fig. S2C), strongly suggest that the metabolic instability of proapoptotic fragments is their functionally important attribute.
Protease-generated proapoptotic fragments likely act as components of positive feedbacks that amplify specific aspects of apoptosis, including caspase activation. For most proapoptotic fragments (Fig. S1), the mechanisms of their previously shown proapoptotic activity are either unknown or surmised, at best, in general terms. Cys-RIPK1 (Figs. 2 B and C and 3K and Figs. S1, S2A, S3 A, B, D, and E, and S4A) is a partial exception. This fragment can be produced from RIPK1 by caspase-8 and other caspases (33–36). Full-length RIPK1 is a conditionally short-lived kinase and a multifunctional regulator of not only apoptosis but also programmed necrosis (necroptosis) and antiviral responses that do not involve cell death (6, 33). The caspase-generated Cys-RIPK1 fragment (Fig. S2A) promotes apoptosis, at least in part, by accelerating the activation of caspase-8, which cleaves RIPK1 to generate more Cys-RIPK1. The result is a proapoptotic positive feedback (33–35).
Previous work has shown that a Drosophila IAP protein (DIAP1) is cleaved by the DRICE caspase, resulting in the Asn21-DIAP1 fragment that is degraded by the Arg/N-end rule pathway (37). However, in contrast to proapoptotic fragments (Fig. S1), both full-length DIAP1 and the Asn21-DIAP1 fragment are antiapoptotic proteins. The biological role of the degradation of Asn21-DIAP1 by the Arg/N-end rule pathway remains to be understood.
We found that the ATE1 R-transferase is degraded in apoptotic cells, that caspases can cleave both ATE1 and the UBR1 N-recognin, and that these cleavages can functionally inactivate at least R-transferase, suggesting a dynamic of mutual suppression between the Arg/N-end rule pathway and proapoptotic signaling (Figs. 1A and 4B and Fig. S5). Biological ramifications of this antagonism are the subject of future studies. A parsimonious interpretation is that caspase-mediated inhibition of the Arg/N-end rule pathway would increase late in apoptosis, when a robust proapoptotic signaling begins to succeed in outplaying the antiapoptotic activity of the Arg/N-end rule pathway and other antiapoptotic circuits strongly enough for proapoptotic processes to reach the point of no return.
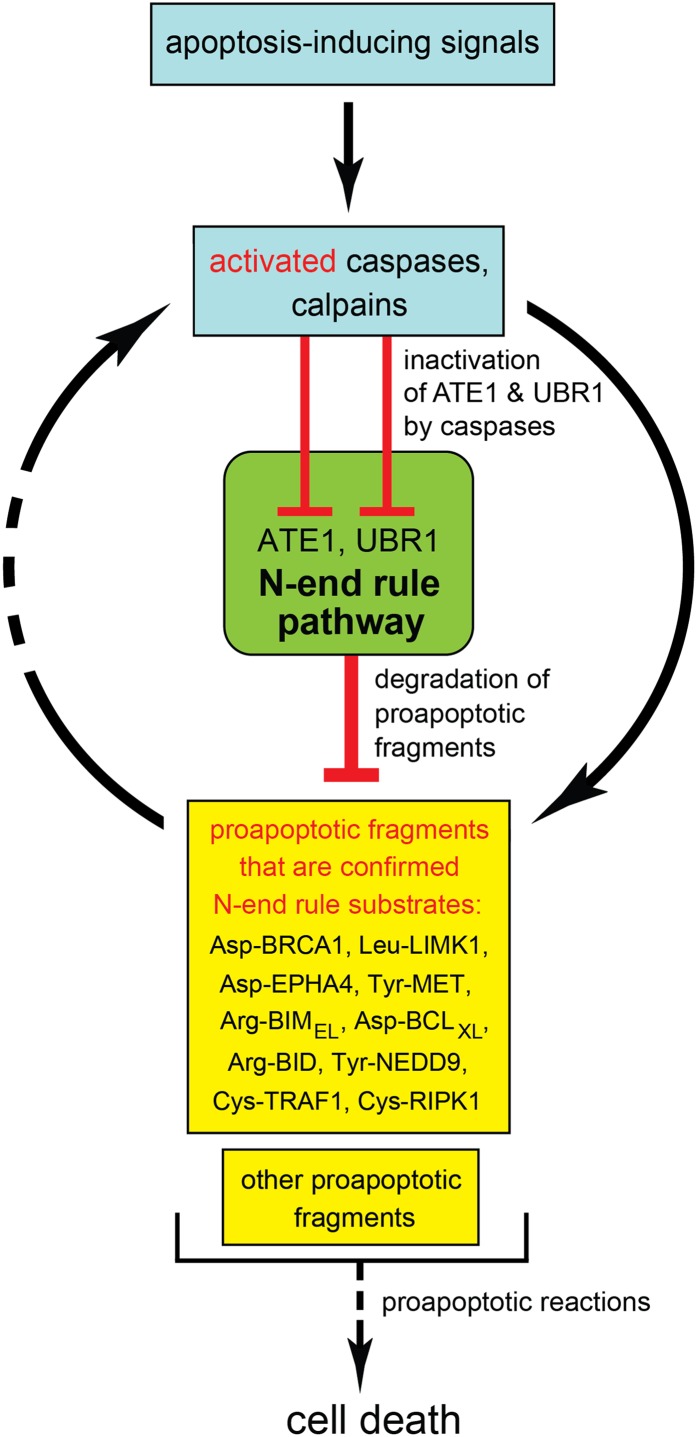
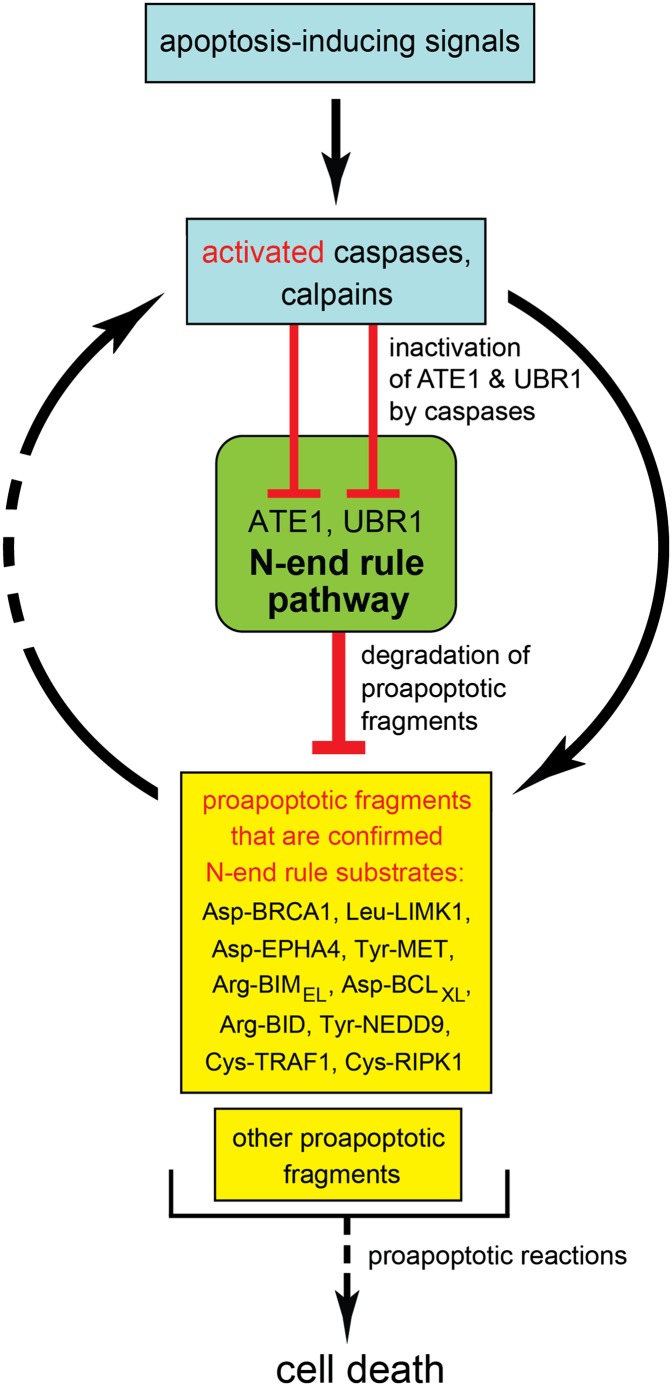
For clarity, the working model of the layer of antiapoptotic control described in the present work focuses solely on apoptosis vis á vis the Arg/N-end rule pathway (Fig. 5). Our results indicate that the Arg/N-end rule pathway inhibits apoptosis at least in part through selective degradation of proapoptotic fragments. In conjunction with other apoptosis-suppressing circuits, the Arg/N-end rule pathway contributes to thresholds that prevent a transient or otherwise weak proapoptotic signal from reaching the point of commitment to apoptosis. Other antiapoptotic effectors include IAP proteins and the NF-κB transcriptional regulon (5, 15, 16). IAPs inhibit and/or degrade activated caspases, whereas the Arg/N-end rule pathway destroys specific proapoptotic fragments other than caspases (Fig. 5 and Fig. S1).
Fig. 5.
The Arg/N-end rule pathway as a regulator of apoptosis. Key aspects of this working model are the demonstrated degradation of proapoptotic Arg/N-end rule substrates (Asp-BRCA1, Leu-LIMK1, Tyr-NEDD9, Arg-BID, Asp-BCLXL, Arg-BIMEL, Asp-EPHA4, Tyr-MET, Cys-TRAF1, and Cys-RIPK1) by the Arg/N-end rule pathway and the counteracting inhibition of this pathway by activated caspases. In conjunction with other antiapoptotic circuits, the selective degradation of proapoptotic protein fragments by the Arg/N-end rule pathway acts to generate thresholds that prevent a transient or otherwise weak proapoptotic signal from reaching the point of commitment to apoptosis.
This understanding (Fig. 5) both accounts for and is in agreement with previous findings. Specifically, spermatocytes of mice that lack either the UBR2 N-recognin or the ATE1 R-transferase undergo apoptosis during meiosis, resulting in male infertility (30, 38). Johanson–Blizzard syndrome (JBS) is caused by inactivating mutations in both copies of UBR1, which encodes an N-recognin (Fig. 1A) (23, 39). Clinical features of JBS are consistent with our finding that even a partial ablation of the Arg/N-end rule pathway sensitizes cells to apoptosis (Fig. 1 B–E and Fig. S3C). For example, nasal wing aplasia (near-absence of nostrils) in JBS patients is likely to result from apoptosis of cells that normally form nasal wings. Insufficiency and inflammation of exocrine pancreas in JBS are likely to be caused by apoptosis of acinar cells (17, 23). The antiapoptotic activity of the Arg/N-end rule pathway suggests that its up-regulation may facilitate malignant phenotypes in mammalian cell lineages. Indeed, the gene for UBR5 (EDD), one of N-recognins (Fig. 1A), is often amplified in ovarian carcinoma, and it is an adverse prognostic factor in this disease (40).
Regulation of apoptosis and its derangements in cancer, neurodegenerative syndromes, metabolic disorders, and malfunctions of immunity are among the hallmarks of these diseases. Our results have shown that the Arg/N-end rule pathway is a mechanistically specific repressor of programmed cell death (Fig. 5). This discovery may allow a modulation of apoptosis in clinically relevant settings by altering the activity of the Arg/N-end rule pathway. The targeting of some N-degrons in proapoptotic protein fragments involves not only specific Ub ligases (N-recognins) but also preceding steps, including the effects of nitric oxide and oxygen, the activity of the NTAN1 and NTAQ1 Nt-amidases, and the activity of the ATE1 R-transferase (Fig. 1A and Fig. S1) (17). The resulting possibility of augmenting or inhibiting specific branches of the Arg/N-end rule pathway opens up pharmaceutical opportunities to alter the properties of apoptotic circuits.
Materials and Methods
Mutants in the Arg/N-End Rule Pathway.
Mouse strains included conditionally ATE1-deficient Ate1flox/-, CaggCreER mice, in which the remaining copy of Ate1 could be inactivated through a transient activation of Cre recombinase (30). Untransformed cells as well as cell lines included T-Rex-293 human cell lines stably expressing test proteins; primary (precrisis) mouse Ate1−/− EF cells and established cell lines; and a Ubr1−/− Ubr2−/− EF cell line. They are described in SI Materials and Methods.
Pulse-Chase, Caspase, Apoptosis, Arginylation, and Immunoblotting Assays.
Assays with the transcription/translation-enabled reticulocyte extract, in vivo pulse-chase assays, and other technical details of this study are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank B. Wadas for helpful comments on the manuscript, N. Malkova for advice and assistance with mouse experiments, J. Li for his contribution to producing anti-Ate1 antibody, and E. Udartseva for excellent technical assistance. We also thank the present and former members of the Varshavsky laboratory for their assistance and advice. This work was supported by National Institutes of Health Grants DK039520, GM031530, and GM085371 (to A.V.) and the March of Dimes Foundation.
Footnotes
The authors declare no conflict of interest.
See Author Summary on page 10757 (volume 109, number 27).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207786109/-/DCSupplemental.
References
- 1.Yuan J, Horvitz HR. A first insight into the molecular mechanisms of apoptosis. Cell. 2004;116(2 Suppl):S53–S56. doi: 10.1016/s0092-8674(04)00028-5. [DOI] [PubMed] [Google Scholar]
- 2.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strasser A, Cory S, Adams JM. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 2011;30:3667–3683. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 5.Green DR. Means to an End: Apoptosis and Other Cell Death Mechanisms. Plainview, NY: Cold Spring Harbor Laboratory Press; 2011. [Google Scholar]
- 6.Green DR, Oberst A, Dillon CP, Weinlich R, Salvesen GS. RIPK-dependent necrosis and its regulation by caspases: A mystery in five acts. Mol Cell. 2011;44:9–16. doi: 10.1016/j.molcel.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsson M, Zhivotovsky B. Caspases and cancer. Cell Death Differ. 2011;18:1441–1449. doi: 10.1038/cdd.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pop C, Salvesen GS. Human caspases: Activation, specificity, and regulation. J Biol Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138:838–854. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor RC, Cullen SP, Martin SJ. Apoptosis: Controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 11.Salvesen GS, Riedl SJ. Caspase mechanisms. Adv Exp Med Biol. 2008;615:13–23. doi: 10.1007/978-1-4020-6554-5_2. [DOI] [PubMed] [Google Scholar]
- 12.Hara MR, Snyder SH. Cell signaling and neuronal death. Annu Rev Pharmacol Toxicol. 2007;47:117–141. doi: 10.1146/annurev.pharmtox.47.120505.105311. [DOI] [PubMed] [Google Scholar]
- 13.Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agard NJ, et al. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Natl Acad Sci USA. 2012;109:1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gyrd-Hansen M, Meier P. IAPs: From caspase inhibitors to modulators of NF-κB, inflammation and cancer. Nat Rev Genet. 2010;10:561–574. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- 16.Srinivasula SM, Ashwell JD. IAPs: What’s in a name? Mol Cell. 2008;30:123–135. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dougan DA, Micevski D, Truscott KN. The N-end rule pathway: From recognition by N-recognins, to destruction by AAA+ proteases. Biochim Biophys Acta. 2012;1823:83–91. doi: 10.1016/j.bbamcr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Graciet E, Wellmer F. The plant N-end rule pathway: Structure and functions. Trends Plant Sci. 2010;15:447–453. doi: 10.1016/j.tplants.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Varshavsky A. Discovery of cellular regulation by protein degradation. J Biol Chem. 2008;283:34469–34489. doi: 10.1074/jbc.X800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: Prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Tasaki T, Kwon YT. The mammalian N-end rule pathway: New insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Hwang C-S, et al. Ubiquitin ligases of the N-end rule pathway: Assessment of mutations in UBR1 that cause the Johanson–Blizzard syndrome. PLoS One. 2011;6:e24925. doi: 10.1371/journal.pone.0024925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang C-S, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang C-S, Shemorry A, Auerbach D, Varshavsky A. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat Cell Biol. 2010;12:1177–1185. doi: 10.1038/ncb2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon YT, et al. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 27.Hu R-G, et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 28.Hu R-G, Wang H, Xia Z, Varshavsky A. The N-end rule pathway is a sensor of heme. Proc Natl Acad Sci USA. 2008;105:76–81. doi: 10.1073/pnas.0710568105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Piatkov KI, Brower CS, Varshavsky A. Glutamine-specific N-terminal amidase, a component of the N-end rule pathway. Mol Cell. 2009;34:686–695. doi: 10.1016/j.molcel.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brower CS, Varshavsky A. Ablation of arginylation in the mouse N-end rule pathway: Loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS One. 2009;4:e7757. doi: 10.1371/journal.pone.0007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varshavsky A. Ubiquitin fusion technique and related methods. Methods Enzymol. 2005;399:777–799. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T, Varshavsky A. Degradation signals in the lysine-asparagine sequence space. EMBO J. 1999;18:6017–6026. doi: 10.1093/emboj/18.21.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y, Devin A, Rodriguez Y, Liu Z-G. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13:2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JW, Choi E-J, Joe CO. Activation of death-inducing signaling complex (DISC) by pro-apoptotic C-terminal fragment of RIP. Oncogene. 2000;19:4491–4499. doi: 10.1038/sj.onc.1203796. [DOI] [PubMed] [Google Scholar]
- 35.Martinon F, Holler N, Richard C, Tschopp J. Activation of a pro-apoptotic amplification loop through inhibition of NF-kappaB-dependent survival signals by caspase-mediated inactivation of RIP. FEBS Lett. 2000;468:134–136. doi: 10.1016/s0014-5793(00)01212-6. [DOI] [PubMed] [Google Scholar]
- 36.Rajput A, et al. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by caspase-8-mediated cleavage of the RIP1 protein. Immunity. 2011;34:340–351. doi: 10.1016/j.immuni.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Ditzel M, et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 38.Kwon YT, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zenker M, et al. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson–Blizzard syndrome) Nat Genet. 2005;37:1345–1350. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 40.O’Brien PM, et al. The E3 ubiquitin ligase EDD is an adverse prognostic factor for serous epithelial ovarian cancer and modulates cisplatin resistance in vitro. Br J Cancer. 2008;98:1085–1093. doi: 10.1038/sj.bjc.6604281. [DOI] [PMC free article] [PubMed] [Google Scholar]