Abstract
Circadian clocks govern a wide range of cellular and physiological functions in various organisms. Recent evidence suggests distinct functions of local clocks in peripheral mammalian tissues such as immune responses and cell cycle control. However, studying circadian action in peripheral tissues has been limited so far to mouse models, leaving the implication for human systems widely elusive. In particular, circadian rhythms in human skin, which is naturally exposed to strong daytime-dependent changes in the environment, have not been investigated to date on a molecular level. Here, we present a comprehensive analysis of circadian gene expression in human epidermis. Whole-genome microarray analysis of suction-blister epidermis obtained throughout the day revealed a functional circadian clock in epidermal keratinocytes with hundreds of transcripts regulated in a daytime-dependent manner. Among those, we identified a circadian transcription factor, Krüppel-like factor 9 (Klf9), that is substantially up-regulated in a cortisol and differentiation-state-dependent manner. Gain- and loss-of-function experiments showed strong antiproliferative effects of Klf9. Putative Klf9 target genes include proliferation/differentiation markers that also show circadian expression in vivo, suggesting that Klf9 affects keratinocyte proliferation/differentiation by controlling the expression of target genes in a daytime-dependent manner.
Keywords: glucocorticoids, skin cancer
Biological rhythms regulate cellular and physiological processes ranging from milliseconds to years. A well-studied timing system is the circadian (∼24-h) clockwork that allows organisms to anticipate diurnal variations in environmental conditions such as light, food availability, oxidative stress, pathogen exposure, or temperature. In mammals, the central circadian pacemaker resides in the suprachiasmatic nucleus (SCN), located in the anterior hypothalamus. Oscillations of SCN neurons are cell-autonomous, self-sustained, and synchronized to external time cues (Zeitgebers) such as light (1). The SCN, in turn, synchronizes peripheral clocks by systemic time cues such as neuronal input, hormonal signaling (e.g., cortisol), body temperature, and possibly many others (2). Interestingly, most cells in peripheral tissues also possess cell-autonomous clockworks with a similar molecular makeup to SCN neurons. These peripheral clocks are thought to generate or amplify daytime-dependent physiological and metabolic functions in a tissue-specific manner by circadian regulation of clock-controlled genes (3, 4).
On a molecular level, circadian oscillation is generated by interlocked transcriptional–translational feedback loops. The transcription factor dimer CLOCK/BMAL1 drives expression of target genes such as Periods (Per1-3) and Cryptochromes (Cry1-2) by binding to E-box elements in their promoters. The negative feedback is formed by PER/CRY protein complexes that shuttle back into the nucleus, where they block CLOCK/BMAL1-mediated transactivation, thereby inhibiting their own transcription (5). In a second feedback loop, the orphan nuclear receptor REV-ERBα rhythmically represses Bmal1 transcription, presumably adding to the robustness of the circuitry. The molecular clock, but also rhythms of systemic cues such as hormones or temperature, can mediate circadian expression of tissue-specific output genes either directly or by controlling transcription factors that in turn drive rhythmic expression of target genes. Transcriptional analysis in mice revealed that large subsets of genes (2–10%) in a given tissue are rhythmically expressed (6). This allows tissue-specific circadian control of various physiological functions including cell growth, metabolic processes or immune function (7–9). Consequently, disruption of the circadian system can lead to severe pathological conditions such as metabolic syndrome (10), diabetes (11), and cancer (12). Although mouse models have greatly advanced our understanding of the connections between circadian rhythms, physiology and disease studies of the molecular mechanisms of circadian action in human tissues are still in its infancy.
In particular, circadian rhythms of human skin have received only little attention. Because the epidermis forms the outermost barrier between body and environment, it is naturally exposed to diurnal environmental variations such as UV radiation, temperature, or pathogen exposure. Accordingly, some skin functions such as barrier recovery and sebum secretion show circadian variations (13). However, on a molecular level, it is still unclear whether a functional circadian clock is operative in human epidermis and how this clock might contribute to rhythmic skin function.
Here, we report genome-wide identification of rhythmic gene expression in human epidermis using a three-point sampling strategy. We find significant daytime-dependent gene expression in hundreds of epidermal transcripts. In vitro studies further revealed that many of these genes including canonical clock genes are driven by a keratinocyte clock. We identified Krüppel-like factor 9 (Klf9) as a circadian transcription factor that regulates the rhythmic expression of several output genes. We found that Klf9 is expressed in a cortisol dependent and differentiation-specific manner. Gain- and loss-of-function experiments revealed a strong antiproliferative effect of Klf9 in keratinocytes in vitro. Thus, our results demonstrate a cell autonomous circadian clock in human epidermis and point to a role for the circadian transcription factor Klf9 for rhythmic modulation of target genes with implications for dermatological homeostasis and disease.
Results
Functional Circadian Clock in Human Epidermis.
Circadian rhythms in peripheral tissues are largely dependent on local molecular clocks driving rhythmic gene expression that result in timed cellular and physiological functions. To test whether such a clock is operative in human epidermis in vivo, we obtained epidermal biopsies from 20 healthy human subjects (see Fig. S1 A and B for control parameters) throughout the activity phase (0930 hours to 1930 hours) using the suction-blister method (see Material and Methods).
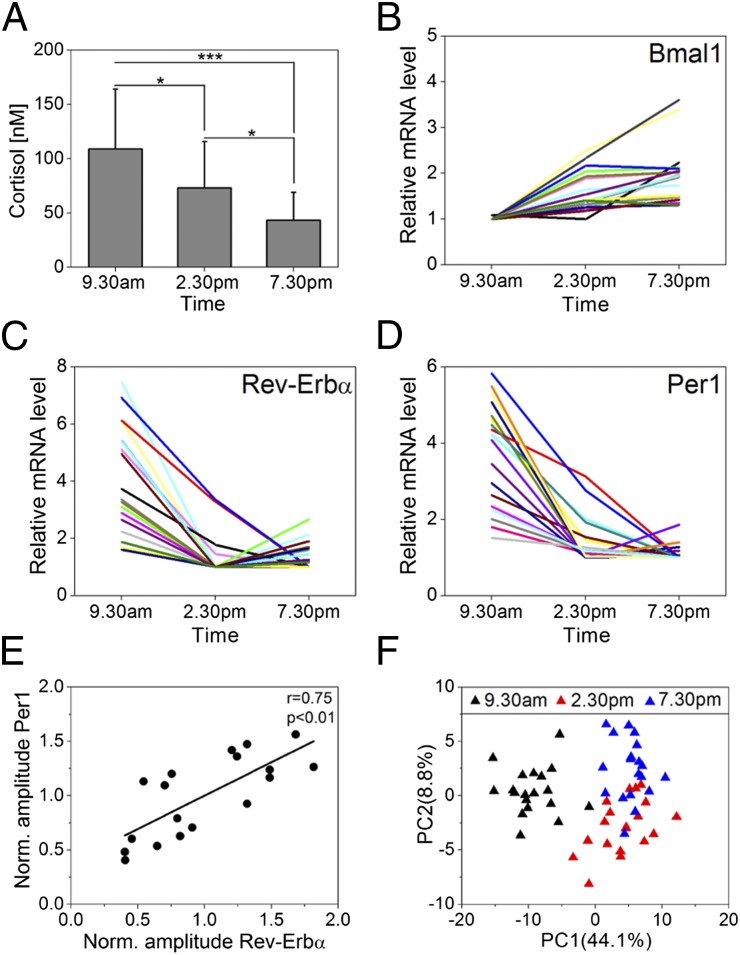
For each subject, we determined cortisol levels in saliva samples and found, as expected, significant diurnal variations (Fig. S1C). Cortisol is known to contribute to the synchronization process of peripheral oscillations (14, 15). To test whether cortisol may exert a similar function in the epidermis, we analyzed cortisol levels in epidermal suction-blister fluids (SBF) collected at 0930 hours, 1430 hours, and 1930 hours from 20 subjects. Again, we detected a strong decrease in suction-blister cortisol and cortisone levels throughout the day (Fig. 1A and Fig. S1 D, H, and I). To assess potential rhythmic gene expression in the epidermis, we performed whole genome microarray analysis of suction-blister epidermis samples from 19 subjects. In total 18,224 annotated genes were found to be expressed with highly significant diurnal regulation of Bmal1, Per1, and Rev-Erbα: essential genes of the core clock circuitry (Fig. 1 B–D). Per1 and Rev-Erbα displayed high expression levels in the morning compared with midday and evening in all subjects, whereas Bmal1 expression increases during the day. This phase relation was as expected from the molecular makeup of the circadian clockwork. In general, substantial interindividual variances were detected in “amplitudes” (defined as maximum minus minimum expression in the analyzed time frame) of daytime-dependent genes similar to other studies using human blood mononuclear cells or oral mucosa as human tissue source (16–18). This is likely primarily attributable to interindividual differences in the epidermal oscillator, because we find a strong correlation of Per1 and Rev-Erbα amplitudes between individual subjects (Fig. 1E and Fig. S1 F and G).
Fig. 1.
Circadian gene expression in human epidermis. (A) Cortisol levels in suction-blister liquid obtained at indicated times were determined in 20 individuals. Mean values ± SD are given. *P < 0.05; ***P < 0.001 (Mann–Whitney U test). (B–D) Genome-wide microarray analysis was performed using suction-blister epidermis of 19 subjects at indicated times. Bmal1 (B), Rev-Erbα (C), and Per1 (D) expression of each subject are shown relative to minimum. (E) Normalized (relative to mean of all subjects) amplitudes (maximum minus minimum expression) of Per1 and Rev-Erbα were correlated. The Pearson’s correlation coefficient (r = 0.75) and the corresponding P value (P < 0.01) indicate a strong linear correlation. (F) Principal component analysis (PCA) of microarrays was performed for the 250 top-ranking genes according to q value. The relative variance in gene expression (relative to total variance) comprised in PC1 and PC2 is indicated.
To identify putative clock-controlled genes, we performed statistical analysis of microarray data from 19 subjects revealing 294 annotated genes that showed significantly (q < 0.05) different expression levels throughout the day (Fig. S2 and Table S1B). Principal component analysis between microarrays led to a clear clustering according to daytime (Fig. 1F). Most regulated genes showed peak expression in the morning (Fig. S1E), similar to Per1 and Rev-Erbα (E-box phase), suggesting similar regulatory mechanisms. These data (together with theoretical considerations; see Fig. S1J and Table S2) clearly demonstrate that our three-time-point experimental setup allows identifying daytime-dependent variations in gene expression in human epidermis. Among the genes ranking highest in the statistical analysis, we found several core clock genes such as Bmal1 and Npas2, as well as E-box driven genes Per1-3, Cry2, Rev-Erbα, and Rev-Erbβ [Fig. S3A; for quantitative (q)PCR validation, see Fig. S3 D and E].
In addition, a set of transcription factors, rate limiting enzymes and cell cycle regulators was identified within the top ranking genes (Fig. S3 B and C). Kyoto Encyclopedia of Genes and Genomes pathway analysis identified highly significant daytime-dependent cellular processes, including “circadian rhythms,” “DNA replication,” “metabolic processes,” and “cell cycle” (Table S1A), indicating that a substantial part of epidermal physiology is likely under circadian control.
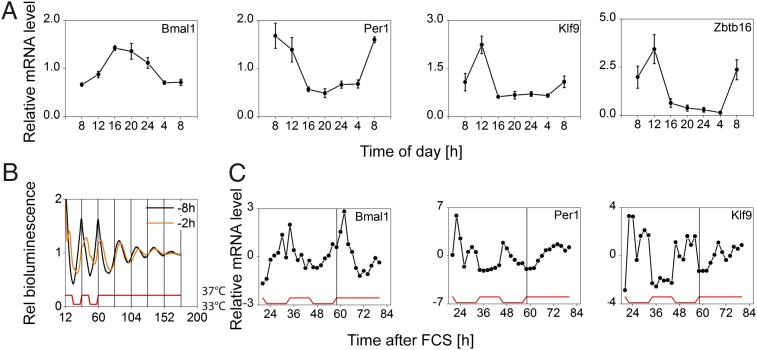
To further validate our three-time-point sampling strategy, we performed two additional studies. First, suction blisters from six subjects were obtained in 12-h intervals over 2 consecutive days (0930 hours and 2130 hours each day). In this setup, we obtained similar results for the expression dynamics of clock and clock-controlled genes (including the transcription factors Klf9 and Zbtb16) (Fig. S4A). Moreover, we obtained punch biopsies from six subjects every 4 h for a full 24-h cycle. Again, we found highly rhythmic transcript levels for clock and clock-controlled genes with a similar phase compared with the three-point sampling strategy (Fig. 2A and Fig. S4 B and C).
Fig. 2.
Epidermis clock in vivo and in vitro. (A) Gene expression in punch-biopsy epidermis obtained at indicated times is shown relative to 24-h mean expression (mean of six subjects ± SEM; except Zbtb16: n = 5). Bmal1, Per1, Zbtb16: P < 0.001; Klf9: P = 0.001. (B) Circadian rhythms of Bmal1 reporter activity in primary keratinocytes. Note that instant entrainment is achieved if FCS is administered 8 h before the temperature cycles (black curve). This synchronization paradigm was used for subsequent experiments. (C) Cells were harvested in regular 2-h intervals starting 24 h after FCS for 60 h and mRNA levels of depicted genes were determined. Expression levels are given relative to mean expression. Bmal1, Per1: P < 0.001; Klf9: P = 0.001. (For P values during entrainment and free run, see Table S1C). Statistical analysis for circadian gene expression was performed using CircWave software (43).
Together, these data indicate a functional circadian clock in human epidermis that likely controls various output genes with implications for cellular functions. Moreover, circadian gene expression in human epidermis can be accurately determined using a simple three-point sampling strategy.
Cell-Autonomous Circadian Rhythms in Cultured Primary Keratinocytes.
In mouse liver, a small subset of rhythmic genes is not (or not only) driven by the local liver clock but by systemic factors, such as temperature or cortisol (19). To discriminate these possibilities for some of our identified rhythmic genes in keratinocytes, we established a circadian in vitro model system using primary neonatal human epidermal keratinocytes (NHEKs). To this end, we used a luciferase reporter for Bmal1 promoter activity stably integrated into NHEK cells. Serum shock combined with temperature cycles led to persistent circadian rhythms in Bmal1 promoter activity (Fig. 2B). As reported for human adult low calcium temperature (HaCaT) keratinocytes earlier (20) circadian rhythms showed strong dampening after release into constant conditions. Of note, circadian period in HaCaT keratinocytes was temperature compensated (Fig. S5A) similar to other cellular clocks (21). Next, we tested whether canonical clock genes and clock-controlled genes identified from our microarray analysis can be entrained to temperature cycles. To this end, we harvested synchronized NHEKs in regular 2 h intervals. Similar to Bmal1 promoter activity Bmal1 mRNA abundance was highly rhythmic with a peak at the beginning of the warm phase during entrainment and persistent (albeit damped) oscillation in free running conditions (Fig. 2C). Other clock and clock-controlled genes also showed significant circadian rhythms (Fig. 2C and Fig. S5 B and C). For instance, Klf9 gene expression [which is circadian in vivo (Fig. 2A)] was highly rhythmic during temperature entrainment but not unambiguously in constant conditions (Fig. 2C) suggesting that Klf9 is at least in part driven by systemic cues (such as temperature or others). Together, our NHEK model system demonstrates a cell autonomous circadian clock in primary keratinocytes consistent with our in vivo results.
Klf9 Is a Cortisol-Dependent Marker for Epidermal Differentiation.
Among the clock-controlled genes in epidermis a large subset of transcription factors was found that possibly mediate timed transcription of downstream genes thereby mediating circadian control of cellular functions.
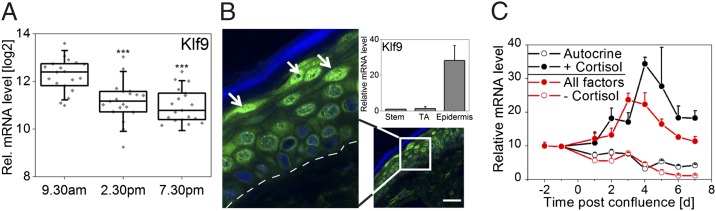
In particular, Klf9 raised our interest as it has been implicated in differentiation and proliferation of epithelial tissues (22), both essential processes in epidermal homeostasis. We found robust diurnal oscillations of Klf9 mRNA in epidermal suction blister, as well as punch-biopsy samples in vivo (Figs. 2A and 3A) with a peak around noon. Of note, Klf9 mRNA is also circadian in mouse liver (http://bioinf.itmat.upenn.edu/circa) and peritoneal macrophages (8). We found that (i) an E-box downstream of the Klf9 transcription start site could be mildly activated by CLOCK/BMAL1 (Fig. S5E); and (ii) BMAL1 binds to this and a second further upstream E-box in a daytime-dependent manner (Fig. S5E), suggesting that although Klf9 mRNA does not exhibit robust oscillations in vitro, the keratinocyte clock might contribute to Klf9 oscillation in vivo. Notably, Klf9 knockdown did not significantly affect circadian rhythms in U2OS cells (Fig. S5D) indicating that Klf9 functions as clock output gene rather than being part of the core clock machinery.
Fig. 3.
Klf9 is an epidermal differentiation marker. (A) Log2 Klf9 mRNA levels (background-corrected signal intensity) in epidermis of 19 subjects is shown in a box plot. Box, 25–75 percentile; vertical bar, median value; error bars, SDs. Individual signal intensities are shown in gray rectangles. ***P < 0.001 (t test). (B) KLF9 protein levels in human skin section determined by immunofluorescence staining. KLF9 immunoreactivity is shown in green and nuclei are stained with DAPI (blue). The stratum corneum layer (outermost epidermal layer) is indicated in blue, and the basal membrane is indicated as a white dashed line. Arrowheads show strong staining in differentiated cells in upper epidermal layers (stratum granulosum). (Scale bar: 50 μm.) (Inset) Klf9 mRNA levels in validated cell fractions (23) of stem cells, transient amplifying keratinocytes (TA Cell), and fully differentiated epidermis (Epidermis) and given relative to the stem cell fraction (four subjects; mean ± SD). (C) Keratinocytes were differentiated by cultivating cells into postconfluence using different culture conditions: full growth medium ± cortisol (All factors and -Cortisol, respectively) or autocrine growth conditions ± cortisol (Autocrine and +Cortisol, respectively). Klf9 mRNA levels of two experiments (mean ± maximum or minimum) are shown relative to minimum expression.
To test whether Klf9 is differentially expressed during epidermal differentiation in vivo we compared validated cell fractions (23) of epidermal stem cells, transit amplifying (TA) keratinocytes (i.e., cell fraction with high proliferative capacity) and fully differentiated non proliferating epidermis. Klf9 mRNA levels were highly elevated in differentiated epidermis compared with stem cells and TA cells (Fig. 3B). In addition, KLF9 protein was strongly expressed in nuclei of suprabasal epidermal layers (Fig. 3B) whereas only weak KLF9 immunoreactivity was observed in the basal cell layer comprising epidermal stem cells and TA cells. To confirm these findings in vitro Klf9 mRNA levels were monitored in cultured keratinocytes during confluence induced differentiation (24). As Klf9 expression is influenced by serum factors and cortisol in other tissues (25, 26) Klf9 expression was assessed in growth conditions with and without growth factors and cortisol. We found strong induction of Klf9 expression during keratinocyte differentiation in a cortisol dependent manner. When cells were cultured in growth factor supplemented medium Klf9 gene expression gradually increased after cells reached confluence (Fig. 3C). Peak expression was observed 4 d postconfluence, correlating with induction of the early differentiation marker Keratin10 (Krt10) and the differentiation associated cell cycle inhibitor p21 (compare Fig. 3C and Fig. S6 A and B). In contrast, in autocrine culture conditions lacking growth factors no differentiation dependent induction of Klf9 was detected, which was not due to impaired differentiation in general as differentiation markers showed similar (albeit not identical) induction kinetics (Fig. S6 A–C). Importantly, supplementing autocrine medium with cortisol restored Klf9 induction during keratinocyte differentiation. Accordingly, removing cortisol from the growth factor mix led to similar abrogation of Klf9 induction as removing all growth factors (Fig. 3C). This shows that cortisol is a necessary and sufficient factor in the differentiation dependent induction of Klf9 expression. Notably, cortisol sensitivity during epidermal differentiation was specific for Klf9, whereas other Klf genes (as well as clock genes) did not - or only weakly - respond to cortisol (Fig. S6D). To monitor Klf9 induction kinetics autocrine keratinocyte cultures were treated with cortisol or the synthetic glucocorticoid dexamethasone (dex). Both cortisol and dex induced Klf9 expression in a time dependent manner with strong (>two-fold) induction occurring already 3 h after treatment (Fig. S6E). This suggests that Klf9 is a fast responding gene in keratinocytes similar to observations in vertebrate tissues (25). In line with this, we found a significant correlation between cortisol levels in suction-blister fluid and Klf9 transcript abundance in suction-blister epidermis throughout the day (Fig. S6F). Thus, Klf9 oscillation in vivo seems to be mainly driven by rhythmic systemic cortisol levels.
Klf9 Regulates Proliferation in Primary Keratinocytes.
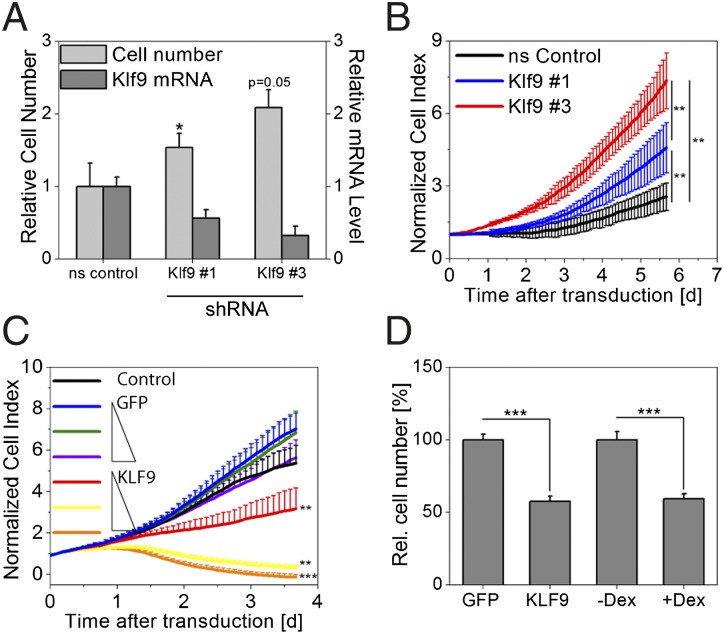
Keratinocyte differentiation is associated with cell cycle withdrawal, and forced Klf9 expression has been reported to cause growth arrest in mouse cancer stem cells (27). Together with the differentiation dependent induction of Klf9 expression reported here, it is conceivable that Klf9 exerts a similar role in human keratinocytes. To test this, we down-regulated Klf9 mRNA using stably integrated microRNA-adapted short hairpin RNA (shRNAmir) constructs and subsequently assessed keratinocyte proliferation. Both flow cytometry as well as real-time proliferation studies revealed a significant increase in cell proliferation in Klf9-deficient cells. This effect was concentration dependent with lower Klf9 levels resulting in a stronger increase in cell growth (Fig. 4 A and B). Ectopic, stable KLF9 overexpression in primary keratinocytes (Fig. S7 A and B) induced the opposite effect: a concentration-dependent decrease in cell growth was observed in cells overexpressing KLF9 whereas no effect was observed in GFP expressing control cells (Fig. 4 C and D). Interestingly, dex treatment of keratinocyte cultures resulting in Klf9 induction led to a similar attenuation of proliferation (Fig. 4D) confirming results reported earlier (28). Together these data suggest that cortisol dependent Klf9 expression may modulate proliferation of differentiating keratinocytes.
Fig. 4.
Klf9 attenuates keratinocyte proliferation. (A) Klf9 transcript was down-regulated in primary keratinocytes using stably integrated shRNAmir constructs (#1 and #3), and cell numbers were assessed 5 d after transduction. Klf9 mRNA levels and cell numbers are given relative to cells transduced with a nonsilencing (ns control) vector (mean ± SD of three experiments; *P < 0.05, t test). (B) Cells were treated similar to A, and cell growth was assessed using real-time impedance measurements (mean value of six wells ± SD; representative plot of three experiments is shown). (C) Primary keratinocytes were transduced with increasing (1×, 2×, and 4×) viral titers carrying GFP or KLF9 expression constructs or left untreated (Control). Cell growth was subsequently analyzed in real-time impedance measurements (mean value of six wells ± SD; representative plot of three experiments is shown). **P < 0.01; ***P < 0.001 (Mann–Whitney U test). (D) Cells were treated similar to C, and a different set of cells was treated with dex. Cell number was analyzed by cell counting 4 d after transduction/treatment [1× virus; mean ± SEM of 9 wells (virus) or 24 wells (dex treatment) of two pooled independent experiments; ***P < 0.01, t test).
Is KLF9 function rhythmic in the epidermis? To address this question, we identified putative KLF9 target genes via transcriptome microarray analysis of KLF9 over-expressing keratinocytes. Within the set of genes down-regulated upon KLF9 overexpression, we found significantly more genes, whose expression is also daytime-dependent in vivo, indicating that KLF9-mediated transcriptional regulation indeed conveys circadian rhythmicity to KLF9 target genes (Fig. S7D). Supporting the hypothesis that (at least a subset) of these genes are direct targets of KLF9 we found a significant enrichment of KLF9 binding sites in the corresponding promoter regions (Fig. S7D). Interestingly, gene ontology analysis of putative KLF9 target genes points to biological processes such as ‘epidermal development’ and “pigment biosynthetic process” (Table S1D). In fact, putative KLF9 target genes include proliferation/differentiation markers [such as Ell3 (29)], as well as genes implicated in glucocorticoid signaling [such as Fkbp5 (30)]. These putative KLF9 targets also showed significant rhythmic expression in the epidermis, with a phase predicted from KLF9 being a repressor of these genes (Fig. S7C), and have putative KLF9 binding motifs in their promoters (Fig. S7E).
In summary, these data suggest that circadian Klf9 gene expression translates into rhythmic repression of putative target genes, including proliferation markers.
Discussion
Human Epidermis Harbors a Molecular Circadian Clock.
Accumulating evidence points to a central role of the circadian system for cellular and organ homeostasis. Although circadian rhythms have been detected in virtually all tested tissues in mice, studies in humans addressing the molecular mechanisms of circadian rhythms have been hindered mostly by accessibility of sample material. To our knowledge, no genome-wide transcriptional profiling for daytime-dependent variations in gene expression has been reported in a human organ so far. In this study, we present an easy yet robust tool to study circadian gene expression in human epidermis. Collecting samples during the day, we identified about 300 genes with significant daytime-dependent expression. Validation experiments using more time points, as well as computer simulations, show that our three-time-point sampling strategy is a very good alternative to sampling over the whole 24-h cycle in humans. Clock gene transcripts with robust diurnal variations in epidermis samples in vivo also showed circadian expression in keratinocytes in vitro. Other rhythmic genes such as Klf9 seem to require systemic signals for strong oscillation. Thus, we believe our suction-blister approach is feasible to identify daytime-dependent-regulated genes in human epidermis in vivo. By collecting suction-blister fluid together with epidermis samples, it is possible to directly correlate systemic rhythms (such as cortisol) to circadian gene expression.
Skin forms the natural barrier between body and environment and, as such, is strongly exposed to diurnal changes in environmental conditions; however, studies of the circadian systems of the skin are surprisingly rare. Circadian gene expression has been shown in mouse skin, and some skin functions (e.g., barrier recovery, sebum secretion, pH) show diurnal variations (13). Only one study has reported circadian gene expression of (only two) clock genes in vivo, however, without differentiating between dermal and epidermal expression (31). Our approach allows studying genome wide circadian gene expression specifically in the human epidermis. This is crucial because physiology of epidermal and dermal compartments vary considerably. Functional grouping of rhythmic transcripts in the epidermis suggested circadian control of various pathways ranging from cell cycle control to metabolic processes. Future studies may address the question whether and how circadian gene expression is translated into rhythmic skin function.
Klf9 Is an Epidermal Circadian Transcription Factor Regulating Keratinocyte Proliferation.
The epidermis undergoes a constant process of self-renewal in which epidermal stem cells gradually commit to a complex program of terminal differentiation (32). A key element in this process is precise spatial and temporal control of keratinocyte proliferation and cell cycle withdrawal (33). Disruption of this tightly regulated homeostatic process leads to severe skin diseases such as psoriasis, dermatitis, and nonmelanoma skin cancer (34). The spatiotemporal organization of keratinocyte cell cycle events, however, is still not completely understood. This study offers molecular insights into a role of the circadian system in controlling keratinocyte proliferation. We identified Klf9 as a circadian transcription factor in human epidermis that seems predominantly driven by rhythmic systemic cortisol and possibly also temperature cycles. Klf9 expression is highly sensitive to glucocorticoids and shows diurnal expression patterns in vivo with a similar phase to systemic cortisol rhythms in suction-blister fluid. However, we also observed Klf9 oscillations in vitro during temperature entrainment with a similar phase to E-box driven genes. Because we found that CLOCK/BMAL1 can activate E-boxes in the Klf9 gene and BMAL1 binds to these E-boxes in a daytime-dependent manner, it is conceivable that the local keratinocyte clock also contributes to Klf9 oscillation in vivo.
Klf9 has been associated with differentiation and proliferation in mouse uterus epithelial tissue (35). However, a role for Klf9 in epidermal homeostasis has not yet been reported in mice nor in humans. Here, we find highly elevated Klf9 mRNA and protein levels late in the multistep differentiation program of human epidermis. We validated these findings using a confluence induced differentiation model in primary keratinocyte in vitro. Interestingly, however, differentiation associated induction of Klf9 was strongly dependent on glucocorticoids. Previously, it has been shown that topical or systemic administration of glucocorticoids as well as stress induced increase in systemic cortisol levels causes decreased keratinocyte proliferation in human and mouse epidermis (36, 37). In line with this, we observed a strongly attenuated keratinocytes proliferation upon either forced Klf9 expression or glucocorticoid treatment, whereas depletion of Klf9 led to hyperproliferation. Moreover, putative Klf9 target genes (including proliferation/differentiation markers) also show rhythmic expression in the epidermis. It remains to be elucidated in future studies to what extent the glucocorticoid phenotype is Klf9-dependent. In fact, it is conceivable that glucocorticoid-dependent mechanisms independent of Klf9 contribute to the inhibition of keratinocyte proliferation. However, our data support the hypothesis that Klf9 might be involved in glucocorticoid-dependent modulation of the complex program of differentiation-associated cell cycle exit in epidermal keratinocytes. It is possible that this effect is daytime-dependent because our data suggest a rhythmic function of Klf9 in the epidermis; however, whether this is dependent on the local keratinocyte clock or systemic time cues remains to be investigated. Such a daytime-dependent mechanism might be important for chronotherapeutic applications because glucocorticoids are commonly used for treatment of various skin diseases such as dermatitis and psoriasis. However, long-term glucocorticoid treatment results in severe adverse effects, particularly in the skin (38). Optimizing glucocorticoid administration for treating dermatological diseases according to daytime might ultimately improve efficiency and reduce side effects, as recently shown for rheumatoid arthritis (39).
Future studies could address the precise role of Klf9 in keratinocyte proliferation control and its possible involvement in skin cancer. This might open avenues for chronotherapeutic approaches in treating skin diseases, as well as help understanding the consequences of disrupted circadian rhythms (e.g., by shift work) for epidermal homeostasis.
Materials and Methods
Epidermal suction blister samples were collected according to the recommendations of the Declaration of Helsinki as well as applicable laws for a nondrug study. All donors provided written, informed consent. Suction-blister samples were obtained at the study center of Beiersdorf AG and approved by the Beiersdorf AG Legal Review Board. Ten male and 10 female volunteers aged 28.35 ± 4.36 y (mean ± SD) participated in the study. Individual chronotypes were assessed using the Munich Chronotype Questionnaire (MCTQ) (40). During the study equicaloric food was provided, and blood glucose levels were determined at 0730 hours, 1000 hours, 1230 hours, 1500 hours, and 1730 hours using the Accu Check system (Roche Diagnostics). Saliva samples were obtained at similar times and cortisol levels were determined by Demeditec Diagnostics. Suction-blister procedure was started at 0700 hours, 1200 hours, and 1700 hours, as described previously (41). Briefly, epidermis samples were detached by applying a negative pressure of 180–320 mmHg for ∼2.5 h. Consequently, suction blisters were harvested at 0930 hours, 1430 hours, and 1930 hours, respectively. Suction-blister fluid was removed and stored at −20 °C, and suction-blister epidermis was snap frozen in liquid nitrogen and stored at −80 °C. Cortisol/cortisone levels in suction-blister fluid were determined by ultrapressure liquid chromatography–tandem mass spectrometry as described previously (42).
See SI Material and Methods for a full description of methods.
Supplementary Material
Acknowledgments
We thank Janosch Hildebrand for providing validated keratinocyte cell fractions; Astrid Grudziecki, Maike Mette-Thaben, and Ronny Kaufmann for technical assistance; Judit Meyer-Kovac for critical reading of the manuscript; Jürgen Ripperger for technical advice on ChIP experiments; and Michael Brunner for providing anti-BMAL1 antibody. F.S. is a doctoral student of the Charité Universitätsmedizin Berlin. This study was supported by Deutsche Forschungsgemeinschaft Grants SFB 618/A4 and SPP 1395 InKomBio (to A.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE36014 and GSE35635).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118641109/-/DCSupplemental.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 3.Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- 4.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schibler U. Circadian time keeping: The daily ups and downs of genes, cells, and organisms. Prog Brain Res. 2006;153:271–282. doi: 10.1016/S0079-6123(06)53016-X. [DOI] [PubMed] [Google Scholar]
- 6.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 7.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller M, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 10.Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: From experimental genetics to human disease. Circ Res. 2010;106:447–462. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 13.Geyfman M, Andersen B. How the skin can tell time. J Invest Dermatol. 2009;129:1063–1066. doi: 10.1038/jid.2008.384. [DOI] [PubMed] [Google Scholar]
- 14.Kiessling S, Eichele G, Oster H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J Clin Invest. 2010;120:2600–2609. doi: 10.1172/JCI41192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 16.Zieker D, et al. Circadian expression of clock- and tumor suppressor genes in human oral mucosa. Cell Physiol Biochem. 2010;26:155–166. doi: 10.1159/000320547. [DOI] [PubMed] [Google Scholar]
- 17.Kusanagi H, et al. Expression profiles of 10 circadian clock genes in human peripheral blood mononuclear cells. Neurosci Res. 2008;61:136–142. doi: 10.1016/j.neures.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 18.James FO, Boivin DB, Charbonneau S, Bélanger V, Cermakian N. Expression of clock genes in human peripheral blood mononuclear cells throughout the sleep/wake and circadian cycles. Chronobiol Int. 2007;24:1009–1034. doi: 10.1080/07420520701800736. [DOI] [PubMed] [Google Scholar]
- 19.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spörl F, et al. A circadian clock in HaCaT keratinocytes. J Invest Dermatol. 2011;131:338–348. doi: 10.1038/jid.2010.315. [DOI] [PubMed] [Google Scholar]
- 21.Izumo M, Johnson CH, Yamazaki S. Circadian gene expression in mammalian fibroblasts revealed by real-time luminescence reporting: Temperature compensation and damping. Proc Natl Acad Sci USA. 2003;100:16089–16094. doi: 10.1073/pnas.2536313100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simmen FA, et al. Dysregulation of intestinal crypt cell proliferation and villus cell migration in mice lacking Kruppel-like factor 9. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1757–G1769. doi: 10.1152/ajpgi.00013.2007. [DOI] [PubMed] [Google Scholar]
- 23.Hildebrand J, et al. A comprehensive analysis of microRNA expression during human keratinocyte differentiation in vitro and in vivo. J Invest Dermatol. 2011;131:20–29. doi: 10.1038/jid.2010.268. [DOI] [PubMed] [Google Scholar]
- 24.Minner F, Herphelin F, Poumay Y. Study of epidermal differentiation in human keratinocytes cultured in autocrine conditions. Methods Mol Biol. 2010;585:71–82. doi: 10.1007/978-1-60761-380-0_6. [DOI] [PubMed] [Google Scholar]
- 25.Bonett RM, Hu F, Bagamasbad P, Denver RJ. Stressor and glucocorticoid-dependent induction of the immediate early gene kruppel-like factor 9: Implications for neural development and plasticity. Endocrinology. 2009;150:1757–1765. doi: 10.1210/en.2008-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang XL, Simmen FA, Michel FJ, Simmen RC. Increased expression of the Zn-finger transcription factor BTEB1 in human endometrial cells is correlated with distinct cell phenotype, gene expression patterns, and proliferative responsiveness to serum and TGF-beta1. Mol Cell Endocrinol. 2001;181:81–96. doi: 10.1016/s0303-7207(01)00536-6. [DOI] [PubMed] [Google Scholar]
- 27.Ying M, et al. Krüppel-like family of transcription factor 9, a differentiation-associated transcription factor, suppresses Notch1 signaling and inhibits glioblastoma-initiating stem cells. Stem Cells. 2011;29:20–31. doi: 10.1002/stem.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stojadinovic O, et al. Novel genomic effects of glucocorticoids in epidermal keratinocytes: Inhibition of apoptosis, interferon-gamma pathway, and wound healing along with promotion of terminal differentiation. J Biol Chem. 2007;282:4021–4034. doi: 10.1074/jbc.M606262200. [DOI] [PubMed] [Google Scholar]
- 29.Johnstone RW, et al. Functional analysis of the leukemia protein ELL: Evidence for a role in the regulation of cell growth and survival. Mol Cell Biol. 2001;21:1672–1681. doi: 10.1128/MCB.21.5.1672-1681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies TH, Ning YM, Sánchez ER. A new first step in activation of steroid receptors: Hormone-induced switching of FKBP51 and FKBP52 immunophilins. J Biol Chem. 2002;277:4597–4600. doi: 10.1074/jbc.C100531200. [DOI] [PubMed] [Google Scholar]
- 31.Bjarnason GA, et al. Circadian expression of clock genes in human oral mucosa and skin: Association with specific cell-cycle phases. Am J Pathol. 2001;158:1793–1801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat Rev Genet. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- 33.Okuyama R, LeFort K, Dotto GP. A dynamic model of keratinocyte stem cell renewal and differentiation: Role of the p21WAF1/Cip1 and Notch1 signaling pathways. J Investig Dermatol Symp Proc. 2004;9:248–252. doi: 10.1111/j.1087-0024.2004.09308.x. [DOI] [PubMed] [Google Scholar]
- 34.Kamstrup M, Faurschou A, Gniadecki R, Wulf HC. Epidermal stem cells - role in normal, wounded and pathological psoriatic and cancer skin. Curr Stem Cell Res Ther. 2008;3:146–150. doi: 10.2174/157488808784223087. [DOI] [PubMed] [Google Scholar]
- 35.Simmen RC, et al. Subfertility, uterine hypoplasia, and partial progesterone resistance in mice lacking the Kruppel-like factor 9/basic transcription element-binding protein-1 (Bteb1) gene. J Biol Chem. 2004;279:29286–29294. doi: 10.1074/jbc.M403139200. [DOI] [PubMed] [Google Scholar]
- 36.Choi EH, et al. Glucocorticoid blockade reverses psychological stress-induced abnormalities in epidermal structure and function. Am J Physiol Regul Integr Comp Physiol. 2006;291:R1657–R1662. doi: 10.1152/ajpregu.00010.2006. [DOI] [PubMed] [Google Scholar]
- 37.du Vivier A, Phillips H, Hehir M. Applications of glucocorticosteroids. The effects of twice-daily vs once-every-other-day applications on mouse epidermal cell DNA synthesis. Arch Dermatol. 1982;118:305–308. doi: 10.1001/archderm.118.5.305. [DOI] [PubMed] [Google Scholar]
- 38.Hengge UR, Ruzicka T, Schwartz RA, Cork MJ. Adverse effects of topical glucocorticosteroids. J Am Acad Dermatol. 2006;54:1–15. doi: 10.1016/j.jaad.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Buttgereit F, et al. Targeting pathophysiological rhythms: Prednisone chronotherapy shows sustained efficacy in rheumatoid arthritis. Ann Rheum Dis. 2010;69:1275–1280. doi: 10.1136/ard.2009.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zavada A, Gordijn MC, Beersma DG, Daan S, Roenneberg T. Comparison of the Munich Chronotype Questionnaire with the Horne-Ostberg’s Morningness-Eveningness Score. Chronobiol Int. 2005;22:267–278. doi: 10.1081/cbi-200053536. [DOI] [PubMed] [Google Scholar]
- 41.Südel KM, et al. Tight control of matrix metalloproteinase-1 activity in human skin. Photochem Photobiol. 2003;78:355–360. doi: 10.1562/0031-8655(2003)078<0355:tcomma>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 42.Kulle AE, Riepe FG, Melchior D, Hiort O, Holterhus PM. A novel ultrapressure liquid chromatography tandem mass spectrometry method for the simultaneous determination of androstenedione, testosterone, and dihydrotestosterone in pediatric blood samples: Age- and sex-specific reference data. J Clin Endocrinol Metab. 2010;95:2399–2409. doi: 10.1210/jc.2009-1670. [DOI] [PubMed] [Google Scholar]
- 43.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21:350–361. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.