Abstract
The stomatogastric nervous system (STNS) of the American lobster Homarus americanus serves as a useful model for studies of neuromodulatory substances such as peptides and their roles in the generation of rhythmic behaviors. As a central component of the STNS, the stomatogastric ganglion (STG) is rich in neuropeptides and contains well-defined networks of neurons, serving as an excellent model system to study the effect of neuropeptides on the maturation of neural circuits. Here, we utilize multiple mass spectrometry (MS)-based techniques to study the neuropeptide content and abundance in the STG tissue as related to the developmental stage of the animal. Capillary electrophoresis (CE)-MS was employed to unambiguously identify low abundance neuropeptide complements, which were not fully addressed using previous methods. In total, 35 neuropeptides from 7 different families were detected in the tissue samples. Notably, 10 neuropeptides have been reported for the first time in this study. In addition, we utilized a relative quantitation method to compare neuropeptidomic expression at different developmental stages and observed sequential appearance of several neuropeptides. Multiple isoforms within the same peptide family tend to show similar trends of changes in relative abundance during development. We also determined that the relative abundances of tachykinin peptides increase as the lobster grows, suggesting that the maturation of circuit output may be influenced by the change of neuromodulatory input into the STG. Collectively, this study expands our knowledge about neuropeptides in the crustacean STNS and provides useful information about neuropeptide expression in the maturation process.
Keywords: Homarus americanus, matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry (MALDI TOF/TOF MS), capillary electrophoresis (CE), neuropeptide, stomatogastric ganglion (STG), stomatogastric nervous system (STNS), development
The crustacean stomatogastric nervous system (STNS) has been a premier model system for the understanding of the generation of rhythmic motor patterns in circuit analysis for decades.1,2 This model preparation contains relatively large neurons that are easily identifiable, with the capability to produce motor patterns even after being removed from the animal. The neuronal circuits are extensively modulated by many substances, ranging from amine neurotransmitters to a large array of neuropeptides.1,3−6 These features of the STNS make it an attractive preparation to investigate the underlying mechanisms of central pattern generators and other circuits.6,7
Among various crustacean species, the American lobster Homarus americanus has become a particularly useful model system for the study of neural development. The lobster starts its life as an embryo, followed by 3 larval stages before the metamorphosis into the postlarval stage (stage IV). Subsequent juvenile stages follow for several years before the lobster becomes an adult. Previous studies showed that the embryonic network could produce a single rhythmic pattern,8 but in a mature network (after mid larval stages), two distinct rhythms, the pyloric rhythm and the gastric mill rhythm, could be produced.8−10 This difference in functional output could be attributed to the immaturity of the synaptic connections, the neuromodulatory environment, or the membrane properties of the embryonic network. It has been reported that single embryonic networks could generate adult-like outputs upon manipulation, indicating that the adult STG network backbone is already established by midembryonic development.11−13 In addition, the embryonic motor pattern could also be produced from adult preparation when certain neuromodulators were applied.13 Therefore, it is tempting to hypothesize that the immature output of the embryonic network could be largely attributed to the neuromodulatory environment and that the neuropeptide complement changes as a function of the neural network development.
To study the neuropeptides in the STNS, extensive research has been carried out with a variety of approaches. Several studies using immunocytochemistry indicated that neuropeptides are acquired sequentially in the STNS network.14−17 Some neuromodulators are present in the STNS as early as midembryonic development, but others do not appear until mid larval stages. Marder and co-workers used an electrophysiological approach to demonstrate that receptors for many neuromodulators appear and function before the motor patterns of the STNS are mature.14,18,19 These previous studies suggest the sequential recruitment of neuropeptides as a function of development. However, given the large number of isoforms of several peptide families, it remains unknown whether different individual members of a given peptide family appear at different developmental stages. To address this question, a methodology with the capability to detect multiple peptides simultaneously is needed.
Mass spectrometry (MS) has evolved as a powerful tool for peptide analysis due to its high sensitivity, chemical specificity, speed, and capability for analyzing highly complex mixtures. The last several years have witnessed explosive discovery and elucidation of neuropeptides in the crustacean nervous system using MS-based approaches.17,20−32 With the advent of electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI), it became possible to detect biological species with molecular specificity in very limited samples such as single organs or single cells.33,34 Several sample preparation methods employing either direct tissue analysis or pooled tissue extraction have proven to be effective for neuropeptide analysis.25,35 We have previously identified a total of 102 peptides in the nervous system and neuroendocrine organs of Homarus americanus using a combination of direct tissue and pooled extract analysis.36,37 Furthermore, both methods have been employed to compare the neuropeptide complement of the STG and brain in the adult and embryonic lobsters.38 However, previous peptidomic studies mainly focused on relatively high abundance peptides in the adult nervous system, and the comparative peptide analysis in adult and embryonic preparations only examined two conditions. The characterization of lower abundance peptides with relative quantitation across multiple developmental stages has not been addressed yet.
Capillary electrophoresis (CE) provides a powerful tool to alleviate extreme chemical complexity of tissue extracts by overcoming analyte suppression and the limited dynamic range of MS analysis. We demonstrated that the off-line coupling of CE to MALDI MS analysis enabled global analysis of neuropeptides with enhanced peptide detection using even a 10-fold less sample amount as compared to direct MALDI MS analysis.39,40 The current study expands upon previous investigations on neuropeptide identification by characterizing low abundance neuropeptides with the incorporation of CE separation prior to MS analysis to improve peptidome coverage. Overall, 35 neuropeptides from seven different families were detected in the STG, including 10 neuropeptides reported for the first time in this study. A more complete neuropeptide list would provide a useful guide for further functional studies that would lead to improved mechanistic insight into the regulatory role of these signaling peptides in neural network development. In addition, the neuropeptidomic expression patterns at different developmental stages were compared and sequential appearance of neuropeptides was also highlighted. Collectively, this work employed analytical tools to generate a more complete STG peptidomic profile and presented a relative quantitation method to survey STG peptide expression at multiple developmental stages. These results will lead to an improved understanding of neuropeptide regulation in the maturation process at the network level.
Results and Discussion
The stomatogastric ganglion (STG) connects to the central nervous system (CNS) via a single input nerve (the stomatogatric nerve, stn) and a single output nerve to control motor movements of the stomach. Despite size differences, the general morphology of the STG and the number of neurons within the STG remain the same throughout developmental stages, thus yielding similar peptide extraction efficiencies. The discovery and identification of neuropeptides in the STG of H. americanus via mass spectrometry (MS) has been described in several previous reports.17,36,38,41 However, due to the chemical complexity and small tissue size, the majority of previous studies used only accurate mass matching method for the identification of relatively high-abundance neuropeptides. The identification and assignment of neuropeptides in low abundance were proven to be difficult. Here, we utilized a combination of direct tissue analysis, tissue extraction profiling, sequence-specific tandem MS fragmentation, and CE separation of tissue extract to unambiguously assign several low-intensity peaks in the mass spectra. Furthermore, the quantification study mapped the neuropeptide changes at different stages and demonstrated similar trends of isoform level changes within one family. This integrated methodology enabled a more in-depth characterization of neuropeptidome in developing H. americanus STG.
Neuropeptide Assignment and Identification by MALDI TOF/TOF
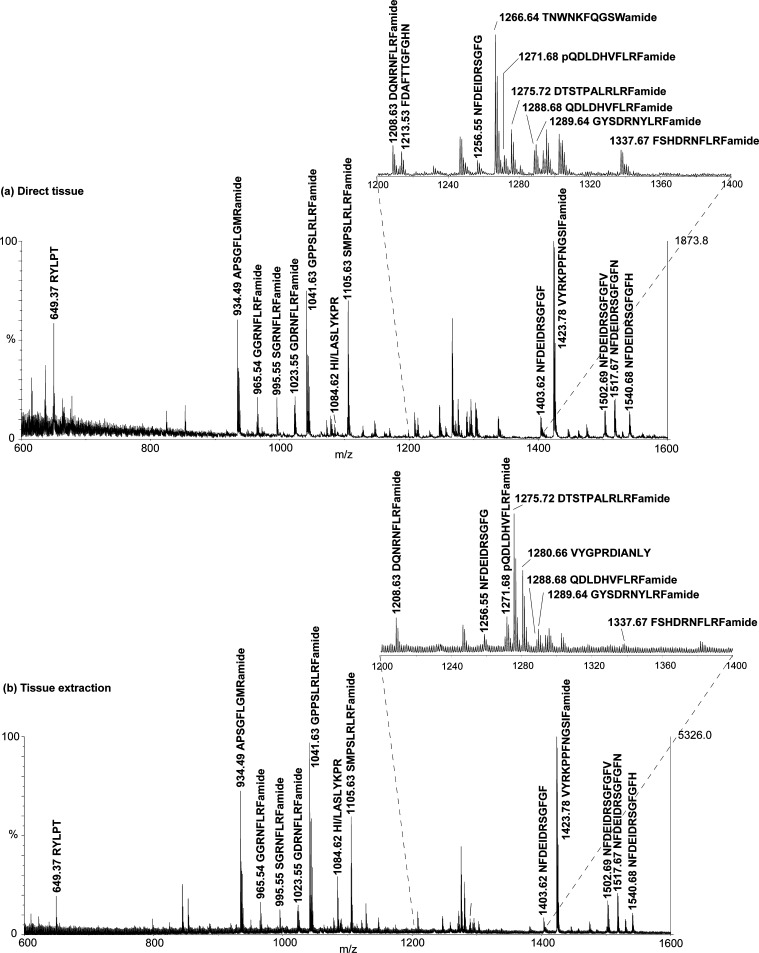
Figure 1 shows representative mass spectra of the adult STG acquired using direct tissue and tissue extraction analyses. Tissue extraction of 15 STGs (Figure 1b) produces a cleaner spectrum, presumably due to a combination of larger peptide amounts and a Ziptip cleanup procedure, whereas direct tissue analysis of the STG produces more noise in the low mass range (Figure 1a), possibly due to the limited sample amount (1 single tissue) and incomplete salt removal. However, the direct tissue method is superior to tissue extraction, considering its speed and simplicity of sample preparation. It only requires mashing of the sample to expose neuropeptides to the surface and then applying acidic dihydroxybenzoic acid (DHB) to extract and crystallize the analytes. Both methods generated spectra with similar peptide patterns, including relative abundances, indicating relatively stable neuropeptide expression levels and the feasibility of both methods (Figure 1 and Figure S1 (Supporting Information)). The peaks were first assigned based on accurate mass measurements by matching with calculated masses from previous reports36,38 and an in-house H. americanus neuropeptide database. The mass measurement accuracy for assignment was set at 30 ppm.
Figure 1.
Neuropeptides detected in (a) single adult STG by direct tissue analysis and (b) tissue extract of 15 pooled adult STGs using MALDI TOF/TOF MS. Peak assignment is based on accurate mass measurement and CID confirmation. While peptide profiles are very similar, minor differences are noted due to the removal of salts and additional sample preparation steps in tissue extraction. Unlabeled peaks are unidentified peaks. The right axis in (a) shows the absolute intensity of the spectrum. Mass spectra with entire mass range analyzed (m/z 600–2000) are provided in Supporting Information, Figure S1.
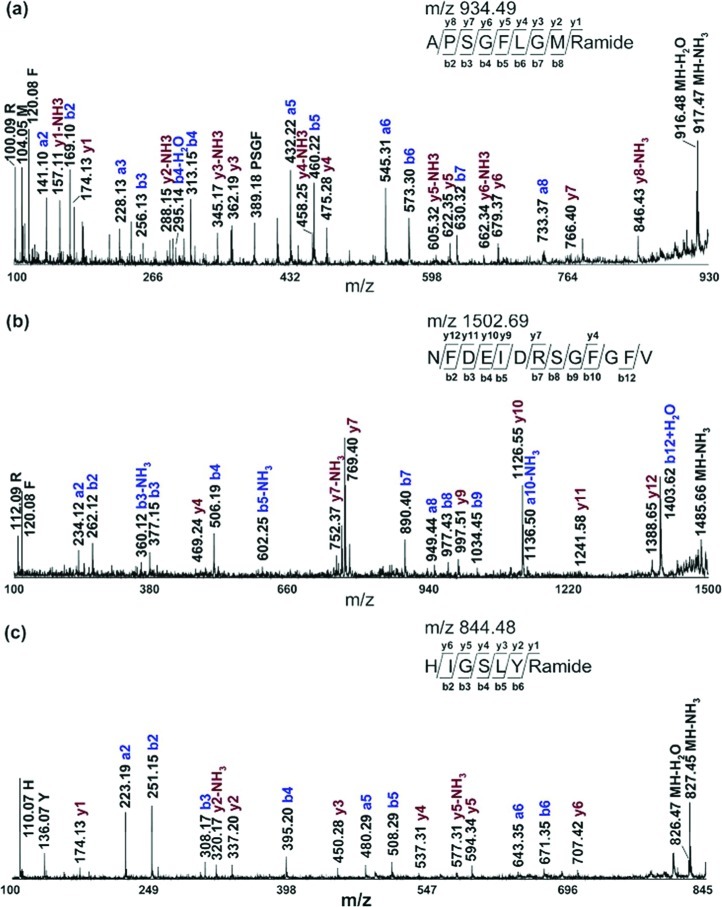
Most of the peptide identities were also confirmed by collisional-induced dissociation (CID) fragmentation performed directly on tissue or tissue extract, as indicated in Table 1. Figure 2a shows a representative tandem mass spectrum for Cancer borealis tachykinin related peptide (CabTRP) Ia APSGFLGMRamide (m/z 934.49). As seen, an almost complete peptide sequence can be obtained, providing high confidence for neuropeptide identification. Figure 2b and c shows MS/MS spectra of lower abundance peptide peaks, an orcokinin peptide NFDEIDRSGFGFV (m/z 1502.69), and a peptide not belonging to any known family, HIGSLYRamide (m/z 844.48). The majority of sequence-specific fragment ions are observed, offering solid confirmation for peptide peak assignment.
Table 1. Neuropeptides Detected in the STG of Adult American Lobster Homarus americanusa.
| [M + H]+ |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| peptide family | sequence | theoretical | observed | error (ppm) | MS/MS | CE fractions | ESTs identified | accession no. | BLAST score | E-value |
| AST-A type | RQYAFGLamide | 853.47 | 853.47 | –3.3 | 25 | – | ||||
| PRNYAFGLamide | 936.51 | 936.5 | –7.66 | xb | 22, 23 | – | ||||
| PRDYAFGLamide | 937.49 | 937.5 | 8.82 | xb | 20, 21 | – | ||||
| AST-B type | TNWNKFQGSWamide | 1266.64 | 1266.63 | –3.97 | x | 22, 23 | – | |||
| STNWSSLRSAWamide | 1293.63 | 1293.64 | 5.13 | 24, 25, 26 | – | |||||
| FaRPs | GGRNFLRFamide | 965.54 | 965.54 | –4.15 | x | 33, 34 | – | |||
| SGRNFLRFamide | 995.55 | 995.52 | –30.89 | 32, 33 | – | |||||
| GNRNFLRFamide | 1022.56 | 1022.57 | 5.57 | 33 | – | |||||
| GDRNFLRFamide | 1023.55 | 1023.56 | 7.24 | x | 30 | – | ||||
| GPPSLRLRFamide | 1041.63 | 1041.63 | 0.4 | x | 31, 32, 33, 34 | – | ||||
| APSKNFLRFamide | 1078.62 | 1078.62 | –4.04 | x | 31, 32 | – | ||||
| GAHKNYLRFamide | 1104.61 | 1104.61 | –4.29 | 30, 31 | – | |||||
| SMPSLRLRFamide | 1105.63 | 1105.63 | –3.34 | 31, 32, 33, 34 | – | |||||
| DQNRNFLRFamide | 1208.63 | 1208.62 | –9.86 | 25, 26, 27 | – | |||||
| pQDLDHVFLRFamide | 1271.68 | 1271.64 | –28.66 | x | 17 | + | FD699285 | 163 | 6E-10 | |
| DTSTPALRLRFamide | 1275.72 | 1275.71 | –5.76 | x | 22, 23, 24, 25 | – | ||||
| QDLDHVFLRFamide | 1288.68 | 1288.64 | –29.92 | x | 27 | + | FD699285 | 163 | 6E-10 | |
| GYSDRNYLRFamide | 1289.64 | 1289.63 | –6.79 | 28 | – | |||||
| FSHDRNFLRFamide | 1337.67 | 1337.69 | 11.44 | N/A | – | |||||
| orcokinin | FDAFTTGFGHN | 1213.53 | 1213.54 | 12.13 | x | N/A | + | DV774522c | 171 | 5E-07 |
| VYGPRDIANLY | 1280.66 | 1280.66 | 2.73 | x | 16, 17 | + | DV774081c | 198 | 5E-07 | |
| SSEDMDRLGFGFN | 1474.63 | 1474.65 | 10.19 | x | N/A | + | FC071459c | 197 | 5E-07 | |
| NFDEIDRSGFGFV | 1502.69 | 1502.69 | 0.99 | x | 16, 17 | + | DV772231c | 143 | 6E-07 | |
| NFDEIDRSGFGFN | 1517.67 | 1517.66 | –4.26 | x | 16 | + | DV771438c | 170 | 6E-07 | |
| NFDEIDRSGFGFH | 1540.68 | 1540.68 | –1.66 | x | 17, 18 | + | DV774848c | 197 | 6E-07 | |
| NFDEIDRSGFG | 1256.55 | 1256.57 | 13.09 | N/A | – | |||||
| NFDEIDRSGFA | 1270.57 | 1270.6 | 23.61 | 17 | – | |||||
| NFDEIDRSGFGF | 1403.62 | 1403.63 | 7.3 | x | 16 | – | ||||
| proctolin | RYLPT | 649.37 | 649.37 | –1.35 | x | 22, 23, 24, 25 | – | |||
| SIFamide | VYRKPPFNGSIFamide | 1423.78 | 1423.78 | 1.91 | x | 27, 28, 29, 30 | + | CN952058 | 235 | 1E-18 |
| tachykinin | APSGFLGMRamide | 934.49 | 934.49 | 1.34 | x | 23, 24, 25, 26, 27 | + | EX567979 | 373 | 3E-34 |
| APSGFLGM(O)Ramide | 950.49 | 950.49 | 5.1 | 23, 24, 25, 26 | + | EX567979 | 373 | 3E-34 | ||
| TPSGFLGMRamide | 964.5 | 964.51 | 11.5 | 23, 24, 25, 26 | – | |||||
| others | HI/LASLYKPR | 1084.62 | 1084.62 | 2.12 | x | 32, 33 | – | |||
| HIGSLYRamide | 844.48 | 844.48 | –2.49 | x | 27, 28 | - | ||||
AST, allatostatin; FaRPs, FMRFamide-related peptides. Others are peptides that do not fall into any known peptide families. Previously known peptides are shown in normal font; peptides previously reported in other species or other organs of the lobster, but observed in the STG for the first time, are shown in bold. Neuropeptides with similar sequences have similar retention time. CE elution order of the neuropeptides in lobster STG under negative mode was pGlu modified > orcokinin > AST-A > AST-B > proctolin > tachykinin > high mass (m/z > 1200) FaRPs > SIFamide > low mass (m/z < 1200) FaRPs. Notably, the overlaps of isoforms in different peptide families are expected.
Tandem MS was performed together with the isotopic peak of CabTRP Ia.
FDAFTTGFGHN, VYGPRDIANLY, SSEDMDRLGFGFN, NFDEIDRSGFGFV, NFDEIDRSGFGFN, and NFDEIDRSGFGFH can all be found in these six ESTs.
Figure 2.
Collisional induced dissociation (CID) mass spectra of neuropeptides from the adult STG of the lobster H. americanus acquired on a MALDI TOF/TOF mass spectrometer. (a) CabTRP Ia, APSGFLGMRamide (m/z 934.49); (b) orcokinin, NFDEIDRSGFGFV (m/z 1502.69); (c) other peptide, HIGSLYRamide (m/z 844.48). All precursor ions are singly charged. The sequence-specific b- and y-type fragment ions and immonium ions are labeled.
Comparisons with Previous Studies on Adult STG by MS
The MALDI TOF/TOF analysis indicated that Val1-SIFamide (m/z 1423.78 VYRKPPFNGSIFamide) was the highest peak in the mass spectra, which is in accordance with a previous study of adult STG by MALDI FTMS.41 It appears to be the sole isoform in the SIFamide family in the lobster, H. americanus. Immunocytochemical and physiological studies have demonstrated its widespread distribution and local neuromodulatory role throughout the STNS.41 Nonetheless, another previous analysis of the adult lobster STG using high pressure MALDI FTMS revealed that [Asn13]orcokinin (m/z 1517.67 NFDEIDRSGFGFN) is the most abundant peak in a majority of the mass spectra.38 To address this difference, we further tested the adult STG on high pressure MALDI FTMS and observed a result very similar to that of MALDI TOF/TOF with Val1-SIFamide being the highest peak (Figure S2, Supporting Information). This discrepancy might be due to the different batches of animals, different environmental conditions, or different parameters of the instrument. In crustaceans, SIFamide might play a role in processing or transmitting tactile, olfactory, and visual stimuli.42 The highest signal response in MALDI MS measurement also might be due to the presence of multiple basic residues in its sequence, and therefore, further investigations would be necessary to establish the absolute concentrations of these peptides.
CE-MALDI for Improved Neuropeptide Characterization
In addition to mass matching and tandem mass fragmentation, a microscale separation technique, CE was also incorporated prior to MS detection. With CE, interfering components could be eliminated, and closely spaced peaks in isotopic cluster peaks could be identified with greater confidence due to another dimension of information.
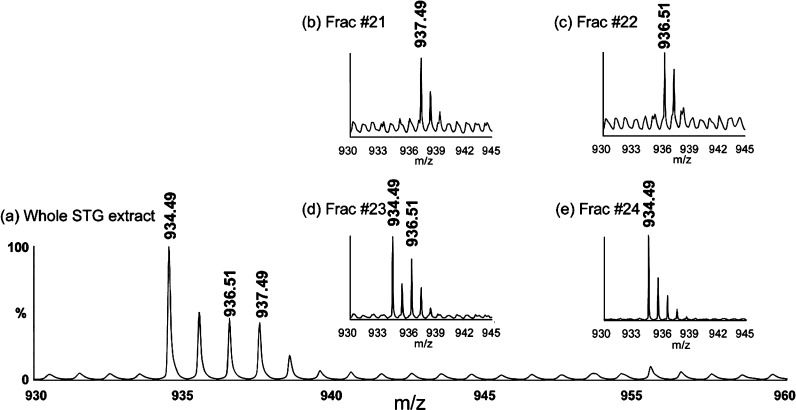
In both direct tissue and extract analysis of the STG, we could observe the peak of CabTRP Ia (m/z 934.49) (Figure 3a). Tandem MS results support the putative identity of this peak. However, it is observed that the isotopic peaks m/z 936.5 and m/z 937.5 are much higher than the theoretical isotopes for m/z 934.49. This suggests the existence of contaminating species which overlap with CabTRP Ia. Interestingly, two A-type allatostatins PRNYAFGLamide (m/z 936.51) and PRDYAFGLamide (m/z 937.49) were recently sequenced from the pericardial organs of H. americanus by nanoLC ESI QTOF MS/MS.37 To test whether they exist in the STG, CE separation was employed to resolve these peaks. Figure 3a is a zoom-in spectrum of the STG extract, showing CabTRP Ia and its isotope envelope. After CE separation of the STG extract, different peaks are found eluted into different fractions. Although the peak intensities are not high enough to perform MS/MS, we do observe individual peaks that could very possibly be PRNYAFGLamide (m/z 936.51) and PRDYAFGLamide (m/z 937.49) based on the accurate mass matching (<10 ppm) and their expected relative migration time in CE.
Figure 3.
MALDI TOF/TOF mass spectra of (a) the adult STG extract and (b–e) CE separation of three peaks APSGFLGMRamide (m/z 934.49), PRNYAFGLamide (m/z 936.51), and PRDYAFGLamide (m/z 937.49). (a) Two peaks m/z 936.51 and m/z 937.49 overlap with the isotopic cluster envelope of peak m/z 934.49 in the tissue extract profiling. After CE fractionation, these three peaks are separated, with (b) m/z 937.49 present only in fraction #21, (c) m/z 936.51 in fraction #22, (d) both m/z 934.49 and m/z 936.51 in fraction #23, and (e) m/z 934.49 only in fraction #24. The appearances of isolated peaks at m/z 936.51 and m/z 937.49 in distinct fractions confirm the existence of these two peptides.
In addition to resolving overlapped peaks, CE separation also enhances the peptidome coverage and confidence of peak assignment in the MS detection. In the spectra of the tissue extract of adult lobsters, we observe two peaks (m/z ∼950.5 and ∼964.5) with low abundances (Figure S3a, Supporting Information). It is not easy to assign these peaks, especially when the peak cluster of m/z 964.5 is dominated by a peptide from the FaRP family GGRNFLRFamide (m/z 965.54).23 These two tachykinin peptides APSGFLGM(O)Ramide (m/z 950.49) and TPSGFLGMRamide (m/z 964.5) were shown to be present in the brain of the American lobster, but neither peptide was reported in the STG previously. Our study thus represents the first report about the existence of these two tachykinins in the STG based on similar CE migration patterns. As shown in Figure S3 (Supporting Information), peaks at m/z 950.5 and 964.5 are better resolved after coeluting with the other tachykinin neuropeptide CabTRP Ia (m/z 934.49) with a much improved intensity. All tachykinins are eluted much earlier than the peak m/z 965.54 from the FaRP family, so the ambiguity is eliminated. More detailed mapping of peptides and their CE migration time is shown in Table 1.
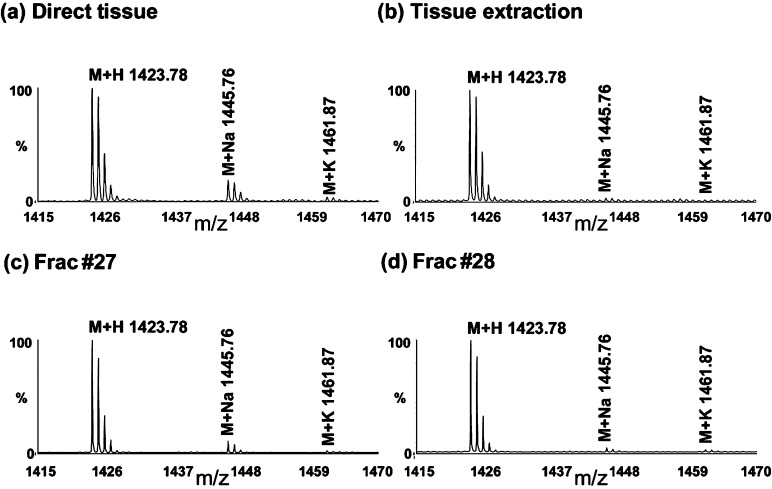
Another example of utilizing CE to assist peptide identification is to assign sodium and potassium adduct peaks based on CE profiles. In direct MS profiling experiments, we detect both Val1-SIFamide VYRKPPFNGSIFamide (m/z 1423.78) and two additional peaks at m/z 1445.8 and 1461.8 in the STG of lobster. Notably, the relative intensities of these two peaks are higher in the direct tissue analysis than tissue extract detection (Figure 4a and b), with the first method bearing more salts than the second one. After CE separation, we observe that the peaks m/z 1445.8 and 1461.8 always appear together with Val1-SIFamide (Figure 4c and d), as in fractions #27 and #28, suggesting that these two peaks are likely Na+ and K+ adducts. These results are also in agreement with the accurate mass measurement result (<10 ppm). Because of the presence of these two adduct peaks, the sum of ion abundance of all three peaks are used for quantification.
Figure 4.
MALDI TOF/TOF mass spectra of the adult STG. (a,b) Direct tissue profiling and extraction profiling of Val1-SIFamide VYRKPPFNGSIFamide (m/z 1423.78), its sodium adduct peak (m/z 1445.76), and potassium adduct peak (m/z 1461.87), with higher salt adduct intensities in direct tissue analysis. (c,d) After CE separation, the peaks with larger masses are coeluted with m/z 1423.78 in fractions #27 and #28, confirming their identities of salt adducts of Val1-SIFamide.
Database Searches of the Peptides Detected by Mass Spectrometry
While many orcokinin peptide isoforms have been identified using mass spectrometry, only some of them match with the expressed sequence tags (ESTs) from mRNA/cDNA libraries (Table 1). This is due to the lack of sequence information of preproorcokinin for this specific species. Via BLAST analysis, six orcokinin ESTs in American lobster H. americanus are identified. It is noted that 6 out of 9 detected neuropeptides (FDAFTTGFGHN, VYGPRDIANLY, SSEDMDRLGFGFN, NFDEIDRSGFGFV, NFDEIDRSGFGFN, and NFDEIDRSGFGFH) are predicted to be the mature peptides from the processing of the prohormone of orcokinin.43 The other three peptides (NFDEIDRSGFG, NFDEIDRSGFA, and NFDEIDRSGFGF) are the truncated forms of the full-length isoforms and have been detected in previous studies.17,43 NFDEIDRSGFG (m/z 1256.55) and NFDEIDRSGFGF (m/z 1403.62) are unlikely to be the fragments of the full-length isoforms (e.g., m/z 1502.69 NFDEIDRSGFGFV) because they can be detected using direct tissue analysis, a method that only requires seconds for sample preparation (Figure 1), thus greatly reducing the possibility of degradation and sample handling artifacts. In addition, we look into the MS/MS spectrum of NFDEIDRSGFGFV (m/z 1502.69) (Figure 2b) and determine that the peak m/z 1256.55 is not one of its fragments, removing the possibility that this peak is generated from m/z 1502.69 during MS analysis. The above results confirm that the truncated orcokinins come from the tissue sample themselves, not from sample preparation or detection.
Combining in silico searching of the publicly accessible ESTs for unannotated transcripts with the predicted processing of the encoded precursor proteins enables the identification of TPSGFLGMRamide in American lobster H. americanus(44) and HIGSLYRamide in green crab Carcinus maenas.45 These two peptides are not in the EST database yet but are predicted to be encoded by the open reading frame (ORF) of the full-length cDNA (accession no. EU408472 for TPSGFLGMRamide and DV111329 for HIGSLYRamide). The initial discoveries of these two peptides by MS prompt follow-up studies using database searches. These additional analyses help solve the ambiguity between Ile and Leu, which cannot be readily differentiated by MS. Given the rapid pace at which new crustacean ESTs are being added to searchable databases and the fast improvements in MS detection, we expect to identify more novel neuropeptides in the near future.
Neuropeptide Profiling at Multiple Stages and MS-Based Relative Quantification Methodology
Besides identifying neuropeptides, it is also of particular interest to compare the neuropeptide abundances at different stages of development. We first tested the sample preparation and animal variation by comparing the mass spectra of multiple animals at one stage (Figure S4, Supporting Information). The results showed sufficient consistency in direct tissue STG mass spectra at the same stage and pronounced differences for several neuropeptides in different developmental stages. Then, we systematically studied the time course of appearance and the abundances of neuromodulators in the STG at each developmental stage. Our results indicated that the maturation of neuromodulatory inputs could play a role in the final functional maturation of the output of the neuronal circuit. The results were compared with previous developmental studies of H. americanus using immunocytochemical techniques.
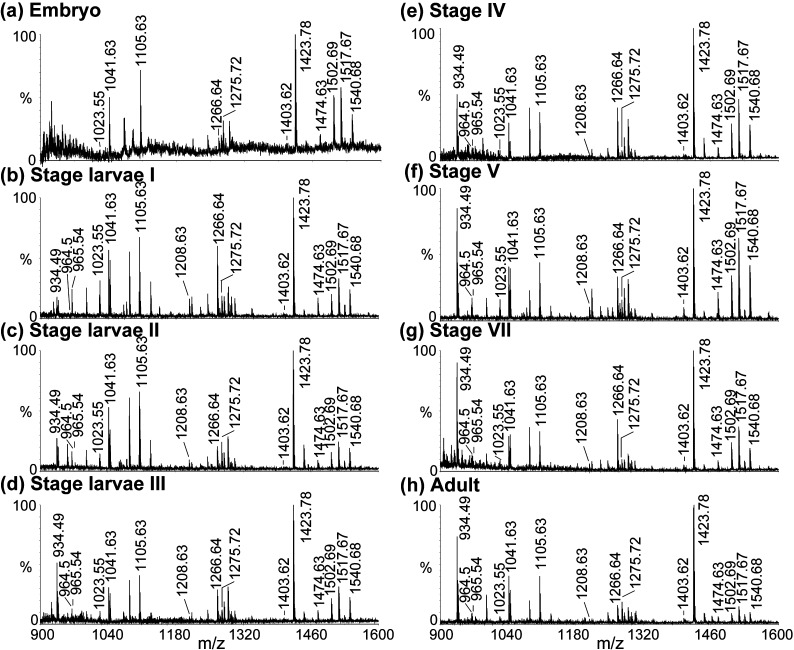
Figure 5 shows comparisons of direct tissue analysis of the STG at multiple developmental stages, from embryo through larval (I, II, III, and IV) and juvenile (V and VII) stages and into adult. The extremely tiny size of embryonic STG causes relatively higher noise background in the spectrum, but the peptide peaks are still clearly seen. Neuropeptide profiles are similar throughout various stages with SIFamide, orcokinin, and RFamide family in relatively high abundances. To better understand the neuropeptide changes at different stages, quantification of neuropeptides at each stage is investigated. Bucknall et al. reported utilizing MALDI TOF MS as a tool for quantifying a diverse array of biomolecules in liquid state.46 In this method, the peak heights showed good correlation with the molecule concentration through validation. However, due to the tiny size of each STG tissue in our experiments, especially in early developmental stages (<50 μm in embryo), we were unable to collect enough tissue samples to produce extracts for quantification. Another report involved the use of standard addition method for quantifying signaling peptides directly in tissue samples.47 Following this, Dickinson et al. used mass spectral peak intensities of peptides from tissue extracts to quantify relative abundances of peptides in the sinus glands and commissural ganglia of Homarus americanus.43
Figure 5.
Comparison of direct tissue spectra of STG at multiple stages using MALDI TOF/TOF: (a) embryo; (b) stage larvae I; (c) stage larvae II; (d) stage larvae III; (e) stage IV; (f) stage V; (g) stage VII; and (h) adult. The masses for known neuropeptides are highlighted in the spectra. The three larval stages last ∼15–30 days. The first 10 stages last about a year. The lobster can reach adult stage within ∼5 years, after 25 molts.
We adapted a similar strategy by using the relative abundance of each peak in the spectrum to represent the relative quantity of each neuropeptide because the relative proportion of each neuropeptide in the ensemble of neuropeptide complement at each stage is more relevant. As several stages are compared in parallel and only the relative ratios are utilized for quantitation, the errors brought by different ionization efficiencies of distinct peptide sequences are greatly reduced and compensated. Moreover, by using relative ratios, we avoid the incorporation of peptide standard which could be difficult to perform on the STG samples due to the large dynamic range of various peptides associated with distinct tissue sizes at different developmental stages. Supporting Information Table S1 provides the normalized average amplitude and standard deviation for neuropeptide measurements in the STG from multiple stages.
There are two types of signal suppression effects observed in MALDI.48 One is related to the contaminants in the sample. The quick washing step prior to STG deposition removes the majority of the salts to significantly enhance the intensity and resolution of peptide detection. The other type of suppression stems from physiochemical properties of the matrix and sample, like the matrix sample composition. We found that if we applied the same concentration of matrix (100 mg/mL) to early stage samples as the adult ones, the signals were substantially impaired due to the high signal suppression from DHB (Figure S5, Supporting Information). Therefore, the concentration of matrix was lowered (20 mg/mL) in early stages in order to obtain better signal, presumably due to the overall low-levels of endogenous peptides in early developmental stages. Notably, the various concentrations of DHB do not alter the relative intensities of the resulting peaks.
Quantification of Tachykinin and SIFamide
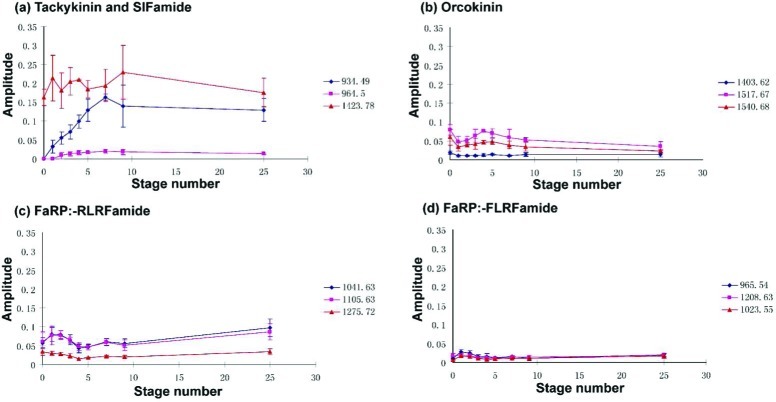
Figure 6a (blue trace) provides a trend showing the relative abundance of CabTRP Ia APSGFLGMRamide (m/z 934.49) (tachykinin peptide) in the STG of the lobster. As shown in Figure 5 and Table S1 (Supporting Information), although the majority of the neuropeptides are already present in the embryo, CabTRP Ia is rarely seen at this early stage. It starts to appear in the stage I larvae (Figure 6a blue trace, Table S1), and the abundance increases up to about 5-fold until stage V (from 3% to 15%), at which point the concentration levels off (p = 3.88 × 10–19, ANOVA). We also observe a similar trend for another tachykinin-related peptide TPSGFLGMRamide m/z 964.5 (Figure 6a pink trace), even though its relative abundance is lower. In BLAST analysis of H. americanus, translation of one EST (EX567979) encodes APSGFLGMR but not TPSGFLGMR. Recently, Christie et al. presented data identifying a TRP-encoding full-length cDNA (accession no. EU408472) whose open reading frame could encode TPSGFLGMR (m/z 964.5) in the American lobster Homarus americanus.44 This cDNA encodes seven tachykinin neuropeptides, with six of them being APSGFLGMRamide (m/z 934.49) and one of them being TPSGFLGMRamide (m/z 964.5). Interestingly, in our MS analyses, the relative ratio of m/z 934.49 is about 6–8 times higher than that of m/z 964.5 (Figure 6a and Table S1, Supporting Information). The findings are also in agreement with previous immunocytochemical studies indicating that tachykinin-like staining is not obvious until larval stage II.16 MS is more sensitive than immunocytochemistry considering that low abundance CabTRP Ia is already detectable in larval stage I (Figure 5b and Table S1, Supporting Information), even though it is not abundant enough to show positive immunoreactivity.
Figure 6.
Quantitative comparisons of the neuropeptide abundances from multiple families at different developmental stages of lobsters. (a) Tachykinin (m/z 934.49 and 964.5) and SIFamide (m/z 1423.78); (b) orcokinin; (c) FaRP-RLRFamide subfamily; (d) FaRP-FLRFamide subfamily. Amplitude is calculated using the intensity of a certain peak divided by the total neuropeptide peak intensities in the spectrum. Stage 0 represents embryonic stage, and stage 25 is assumed as adult (n = 5). Embryonic STG is ∼50 μm in length and ∼50 μm in width; larvae II STG is ∼120 μm in length and ∼80 μm in width; larvae IV STG is ∼160 μm in length and ∼100 μm in width; adult STG is ∼800 μm in length and ∼400 μm in width.
Previous studies showed that in Homarus, the different rhythmic motor patterns generated by the adult STNS are segregated progressively from a single embryonic network during the course of the postlarval development.8,9 The progressive increases in the frequency and regularity of the rhythms coincide well with the gradually increased amount of tachykinin neuropeptides detected by MS. It is possible that besides descending neuromodulatory input, many other factors like synaptic connections and/or cellular properties of the network itself could also influence output maturation. For instance, Rehm et al. compared the effects of CabTRP1a on intact and isolated STGs of the lobster.19 It was noted that CabTRP exerts similar modulatory effects on motor patterns in embryos and adults when neuromodulatory input remains intact,49 indicating that the neuronal network has been established before the mature output can be produced. However, in an isolated STG, the modulatory effect of CabTRP is dramatically different in embryonic and adult preparations, demonstrating the immaturity of the responses to modulators. Although fine-tuning or subtle changes in network itself might be needed before the output of the circuit becomes fully matured, the complete establishment of neuromodulatory input may play a more crucial role in network maturation. The gradual increment in the abundance of the tachykinin peptide family helps to generate mature functional output, suggesting its potential roles of regulation in maturation process.
Compared to the tachykinin-related peptides, the SIFamide neuropeptide Val1-SIFamide (m/z 1423.78) exhibits relatively stable expression levels throughout developmental stages (Figure 6a red trace, Table S1, Supporting Information). One EST (CN952058, Table 1) encodes the putative SIFamide precursor. It was previously shown that bath application of the Val1-SIFamide peptide to the STG results in an increase of both the amplitude and duration of the neuronal activity controlling the pyloric motor pattern. The threshold concentration for this effect is between 10–7 and 10–8 M, suggesting a local mode of the delivery of this peptide rather than hormonal. Immunocytochemical results revealed few developmental differences in the Val1-SIFamide distribution.19 As mentioned before, SIFamide is suggested to be involved in processing tactile, olfactory, and visual stimuli, which can be acquired in early stages, and it is not surprising that this peptide displays a relatively stable ratio throughout the lobster life cycle.
Quantification of Orcokinins and FaRPs
In database searches, six ESTs (Table 1) encoding orcokinin precursors in American lobster H. americanus have been identified. The trends for relative abundance changes of three orcokinin peptides (m/z 1403.62, 1517.67 and 1540.68) are shown in Figure 6b. The amplitudes of peptides in this family show similar patterns, with minor fluctuation at multiple stages, and each orcokinin peptide isoform represents less than 8% of total neuropeptide complement. As one of the largest peptide families, orcokinins were first identified in crayfish50 and have been detected in the pericardial organs of the lobster.37 The whole mount staining of STGs at multiple developmental stages demonstrated that the neuropil of the STG of an E57 embryo was already densely immunoreactive for orcokinin.17 Our result confirms the study with highly abundant peaks shown in the embryonic stage (Figure 5a). The early appearance of this peptide family in relatively high abundance suggests that this peptide may participate in basic functions acquired early in development. Functional study of the orcokinin peptides revealed that they can decrease the burst of the adult pyloric circuit.43 Also, when [Asn13]-orcokinin was tested for biological activity, no increase in the hindgut motility was observed as in other species.43 This finding suggests the need to further explore possible unique functionality of orcokinin peptides or target tissue for these peptides in Homarus americanus.
FaRPs represent an even larger and more diverse peptide group, which can be found in both vertebrates and invertebrates.51 The RFamide peptide sequences are highly conserved throughout evolution, and these peptides can affect essential functions such as pain, food intake, and cardiovascular regulation. Because of the incompletion of the current ESTs, only two RFamide peptide-encoding transcripts are identified by transcriptome mining. FaRPs like GPPSLRLRFamide (m/z 1041.63) and SMPSLRLRFamide (m/z 1105. 63) are present in high abundance at very early stages as shown in Figure 5a. This is in accord with the previous immunocytochemical study showing that FaRPs are present in embryos E50. There are several subfamilies with different C-terminal motifs, including -RLRFamide, -FLRFamide, and -YLRFamide. Minor differences in ratios are observed for different RFamides, but most individual isoforms from the same subfamily exhibit similar trends. For example, the patterns for GPPSLRLRFamide (m/z 1041.63), SMPSLRLRFamide (m/z 1105.63), and DTSTPALRLRFamide (m/z 1275.72) are shown in Figure 6c. Another subfamily GGRNFLRFamide (m/z 965.54), GDRNFLRFamide (m/z 1023.55), and DQNRNFLRFamide (m/z 1208.63) exhibit similar trends as well (Figure 6d). Most of the -FLRFamide and -RLRFamide subfamily peptides are low abundance peptides, as shown by the low percentage in amplitude, and there is no obvious change of abundance with development.
Compared to traditional immunocytochemistry offering cellular or subcellular spatial localization of peptides, mass spectrometry detection is limited by its spatial resolution due to hardware limitation such as laser beam size or sample preparation via matrix crystallization. However, MS offers a chemical information-rich technique that can provide a more comprehensive view of all isoforms involved in neural development. This can address the shortcomings of the immunocytochemical approach which suffers from the difficulty to differentiate individual isoforms in a family. A more in-depth investigation is needed to fully understand the functional consequences of expression level changes in orcokinin and various RFamide isoforms during the course of neuronal network development. The similar trends of isoform patterns within a family might suggest that the peptides in the family are expressed and processed simultaneously from a single neuropeptide gene and that their release and actions also occur concurrently in the development and maturation of a neural network. These MS-based peptidomic analyses are complementary to previous anatomical mapping of neuromodulators via immunocytochemistry while expanding our previous knowledge by providing more detailed and quantitative descriptions of individual peptide players involved in development.
Methods
Animals and Dissection
Early stage lobsters were obtained from New England Aquarium (Boston, MA, USA), and adult lobsters were ordered from the Maine Lobster Direct Web site (http://www.mainelobsterdirect.com). All animals were kept in a circulating artificial seawater tank at 10–15 °C. Ages of embryos were estimated using an eye index scale.52 Embryos used in this study were at a stage of 85%–100% of embryonic developments (E85–E100). The ages of hatched lobsters were divided into distinct life-history phases, including larvae, juvenile, and adult. Larval lobsters include the first three stages (I, II, and III, where stages are distinguished by molts), while postlarvae and juveniles are stage IV and beyond, respectively. The stage of each lobster was determined based on its carapace length or the total length in the lobster growth chart.
To dissect embryonic lobsters, embryos were removed from egg cases and pinned with the dorsal side up onto the Petri dish. The carapace was cut into half from the posterior side, and the yolk on both sides was removed. The stomach was clearly seen, with the STG on top of it. STG was cut apart from the stomatogastric nerve, rinsed, and transferred onto the MALDI plate. The dissections of lobsters at larval and early juvenile stages followed similar procedures with embryos except that no egg case removal was needed. The dissection of adult lobsters was described previously.17,53 The raising and dissection of lobsters has been approved by the Animal Care Facility at the University of Wisconsin—Madison. All handling was performed in accordance with the university guidelines.
Direct Tissue Analysis
Upon dissection and desheathing, the STG was briefly rinsed by acidified methanol [90% methanol (Fisher Scientific, Pittsburgh, PA, USA), 9% glacial acetic acid (Fisher Scientific), and 1% deionized water]. Tissue was then desalted in 10 mg/mL 2, 5-dihydroxybenzoic acid (DHB; ICN Biomedical Inc., Costa Mesa, CA, USA) prepared in deionized water. The tissue was placed on the MALDI plate and mashed before the matrix (DHB) was applied. For STGs from adults and juveniles, 0.3 μL of DHB [100 mg/mL in 50:50 water/purge-trap grade methanol (Fisher Scientific), v/v] was deposited onto the tissue. For smaller size STGs from larvae and embryos, 2–3 STGs were placed onto one spot and mixed with DHB (20 mg/mL) to allow for crystallization prior to MALDI MS detection. Caution was exercised to make sure that the dissections throughout development were consistent. Specifically, two short pieces of stomatogastric nerves were kept on the STG during dissection to maintain tissue integrity.
Tissue Extraction
For tissue extraction, 15 STGs from adult lobsters were pooled and homogenized in a 0.1-mL tissue grinder (Wheaton Inc., Millville, NJ, USA) with acidified methanol. The homogenate was transferred to a 0.6-mL microtube and centrifuged at 16,100g for 10 min in an Eppendorf 5415 D microcentrifuge (Brinkmann Instruments Inc., Westbury, NY, USA). The resulting supernatant was reserved, and the pellet was re-extracted twice using the same method. Supernatants from three extractions were combined and then subjected to a Savant SC 110 SpeedVac concentrator (Thermo Electron Corporation, West Palm Beach, FL, USA) to dry down. Subsequently, the resulting STG extract was resuspended in 10 μL of aqueous 0.1% formic acid (v/v) and then desalted by ZipTipC18 pipet tips (Millipore Corporation, Billerica, MA, USA).
CE Separation of STG Extract
The CE setup was previously described elsewhere.39 Capillary (50 μm i.d. × 360 μm o.d.) with a total length of 67 cm was purchased from Polymicro Technologies (Phoenix, AZ, USA). The CE was off-line coupled to MALDI TOF/TOF by a cellulose acetate membrane-coated porous joint. Prior to use, a new capillary was treated with a combination of solvents in the following order: (1) 75:25, NaOH (1.0 M)/MeOH (v/v), (2) deionized water, (3) 0.1 M NaOH, (4) air, (5) deionized water again, and (6) running buffer under 0.5 psi in sequence for 5 min in each step, followed by electrophoretic equilibration with the separation buffer (H3PO4/NaH2PO4, pH 2.8) for 10 min. Acidic phosphate buffer promotes better ionization in the positive mode of MALDI. The pH of 2.8 is within the buffer capacity range and is compatible with neuropeptide analysis.
Sample injection was performed under pressure at 0.5 psi for 3 s (∼50 nL) followed by the initiation of electroosmotic flow (EOF). The separation was run under negative mode (−15 kV), so the EOF countered the pressure initiated capillary siphoning flow, which could further enhance the resolution. The fractions were deposited, every 60 s, onto nanoliter volumes of α-cyano-4-hydroxy-cinnamic acid [CHCA (Sigma-Aldrich, St. Louis, MO, USA)] matrix spots which were already predeposited onto the MALDI plate. CHCA matrix was 5 mg/mL CHCA in 50% water and 50% acetonitrile (v/v) solutions with addition of 0.1% formic acid.
MALDI TOF/TOF
A model 4800 MALDI TOF/TOF analyzer (Applied Biosystems, Framingham, MA) equipped with a 200 Hz, 355 nm Nd:YAG laser was used for both direct peptide profiling and tandem MS fragmentation analysis. Acquisitions were performed in positive ion reflectron mode. Instrument parameters were set using the 4000 Series Explorer software (Applied Biosystems). Mass spectra were obtained by averaging 1000 laser shots covering mass range m/z 500 to 2000 with a majority of the neuropeptides being detected. Accurate mass measurement within 30 ppm was used to identify peptides. MS/MS was achieved by 2 kV CID using air as collision gas. The peptide sequences were predicted with the help of de novo Explorer software (Applied Biosystems) and manually verified.
Comparison of Neuropeptide Profiles of the STG from Different Stages of Lobsters
Quantification was conducted for direct tissue analysis of STG. Peptide peak identifications were made by using mass matching with calculated peptide masses based on previous literature and in house-developed tissue and species-specific neuropeptide database. Mass spectra obtained from lobsters’ STGs (n = 5) of each stage were compared for commonly detected peaks.
In the STG, the relative peak intensities were used to represent the quantities of neuropeptides. The relative abundance of each peak in the overall neuropeptide profiles at multiple stages was calculated and compared. The calculation was done using the monoisotopic peaks of the assigned neuropeptides ranging from m/z 900–1600. The amplitude of each monoisotopic peak for neuropeptide signal was extracted using Explorer software from each spectrum acquired under MS mode and divided by the total intensity of neuropeptide peaks in the spectrum. This processing helped to reduce the variation caused by the tissue size or shot-to-shot variation. For two peaks with around 1 Da mass difference, the isotopic peak intensity of the lower mass one would be subtracted from that of the higher mass one for corrected peak amplitude. Each STG was assayed in duplicate, and the average was used to represent the peak intensity measurement for each neuropeptide in every animal. If a neuropeptide was absent in one spectrum, a data point equal to noise intensity was used for calculation. One-way ANOVA was used to compare the differences across all developmental stages (α = 0.05).
Database Searches
Database searches were conducted using methods described in recent publications.54 Specifically, the online program tblastn (National Center for Biotechnology Information [NCBI], Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST/) was used to mine for ESTs encoding putative H. americanus peptide precursors. For all searches, the default settings of the program were used, with the exception that the database searched was set to nonhuman, nonmouse ESTs (i.e., EST_others) and was restricted to H. americanus transcripts (i.e., taxid: 6706). For each of the neuropeptide-encoding transcripts identified, the BLAST score and BLAST-generated E-value for significant alignment are provided in Table 1.
Acknowledgments
We thank Dr. Kristina Rehm from Professor Eve Marder’s lab at Brandeis University for teaching the Li Laboratory the lobster dissection. We also thank Yuzhuo Zhang (Li Laboratory) for assistance with the MALDI TOF/TOF instrument. We also thank the University of Wisconsin-Biotechnology Center Mass Spectrometry Facility and Drs. Amy Harms and Michael Sussman for access to the MALDI TOF/TOF instrument. The Massachusetts Division of Marine Fisheries is thanked for complimentary shipment of lobster embryos, larvae, and juvenile animals. We thank the anonymous reviewers for their constructive suggestions. Tyler Greer (Li Laboratory) is thanked for his editorial assistance.
Glossary
Abbreviations
- STNS
stomatogastric nervous system
- MS
mass spectrometry
- CE
capillary electrophoresis
- STG
stomatogastric ganglion
- MALDI TOF/TOF
matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry
- FaRPs
FMRFamide-related peptides
- CabTRP
Cancer borealis tachykinin related peptide
Supporting Information Available
The entire mass range analysis of STG (Figure S1), high pressure MALDI FTMS direct tissue analysis of STG (Figure S2), mass spectral analysis of CE separation and identification of three tachykinin peptides (Figure S3), demonstration of spectral variability which was caused by sample preparation and animal individual variation (Figure S4), comparisons of different concentrations of DHB for analyzing the STG of early stage lobster (Figure S5), and Table S1. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
X.J. and L.L. designed the research; X.J., R.C., and J.W. performed the research; A.M. and M.T. assisted with sample collection; X.J. analyzed the data; X.J. and L.L. wrote the article.
National Institutes of Health through grant 1R01DK071801 and the Wisconsin Alumni Research Foundation. L.L. acknowledges a Vilas Associate Award and an H. I. Romnes Faculty Fellowship.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Marder E.; Bucher D. (2007) Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu. Rev. Physiol. 69, 291–316. [DOI] [PubMed] [Google Scholar]
- Maynard D. M. (1972) Simpler networks. Ann. N.Y. Acad. Sci. 193, 59–72. [DOI] [PubMed] [Google Scholar]
- Marder E. (1976) Cholinergic motor neurones in the stomatogastric system of the lobster. J. Physiol. 257, 63–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E.; Eisen J. S. (1984) Transmitter identification of pyloric neurons: electrically coupled neurons use different transmitters. J. Neurophysiol. 51, 1345–1361. [DOI] [PubMed] [Google Scholar]
- Katz P. S.; Eigg M. H.; Harris-Warrick R. M. (1989) Serotonergic/cholinergic muscle receptor cells in the crab stomatogastric nervous system. I. Identification and characterization of the gastropyloric receptor cells. J. Neurophysiol. 62, 558–570. [DOI] [PubMed] [Google Scholar]
- Nusbaum M. P.; Beenhakker M. P. (2002) A small-systems approach to motor pattern generation. Nature 417, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiebe P. (2001) Neuropeptides are ubiquitous chemical mediators: Using the stomatogastric nervous system as a model system. J. Exp. Biol. 204, 2035–2048. [DOI] [PubMed] [Google Scholar]
- Casasnovas B.; Meyrand P. (1995) Functional differentiation of adult neural circuits from a single embryonic network. J. Neurosci. 15, 5703–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards K. S.; Miller W. L.; Marder E. (1999) Maturation of lobster stomatogastric ganglion rhythmic activity. J. Neurophysiol. 82, 2006–2009. [DOI] [PubMed] [Google Scholar]
- Bucher D.; Taylor A. L.; Marder E. (2006) Central pattern generating neurons simultaneously express fast and slow rhythmic activities in the stomatogastric ganglion. J. Neurophysiol. 95, 3617–3632. [DOI] [PubMed] [Google Scholar]
- Fenelon V. S.; Le Feuvre Y.; Meyrand P. (2004) Phylogenetic, ontogenetic and adult adaptive plasticity of rhythmic neural networks: a common neuromodulatory mechanism?. J. Comp. Physiol., A 190, 691–705. [DOI] [PubMed] [Google Scholar]
- Le Feuvre Y.; Fenelon V. S.; Meyrand P. (2001) Ontogeny of modulatory inputs to motor networks: early established projection and progressive neurotransmitter acquisition. J. Neurosci. 21, 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Feuvre Y.; Fenelon V. S.; Meyrand P. (1999) Central inputs mask multiple adult neural networks within a single embryonic network. Nature 402, 660–664. [DOI] [PubMed] [Google Scholar]
- Pulver S. R.; Thirumalai V.; Richards K. S.; Marder E. (2003) Dopamine and histamine in the developing stomatogastric system of the lobster Homarus americanus. J. Comp. Neurol. 462, 400–414. [DOI] [PubMed] [Google Scholar]
- Kilman V.; Fenelon V. S.; Richards K. S.; Thirumalai V.; Meyrand P.; Marder E. (1999) Sequential developmental acquisition of cotransmitters in identified sensory neurons of the stomatogastric nervous system of the lobsters, Homarus americanus and Homarus gammarus. J. Comp. Neurol. 408, 318–334. [DOI] [PubMed] [Google Scholar]
- Fenelon V. S.; Kilman V.; Meyrand P.; Marder E. (1999) Sequential developmental acquisition of neuromodulatory inputs to a central pattern-generating network. J. Comp. Neurol. 408, 335–351. [DOI] [PubMed] [Google Scholar]
- Li L.; Pulver S. R.; Kelley W. P.; Thirumalai V.; Sweedler J. V.; Marder E. (2002) Orcokinin peptides in developing and adult crustacean stomatogastric nervous systems and pericardial organs. J. Comp. Neurol. 444, 227–244. [DOI] [PubMed] [Google Scholar]
- Richards K. S.; Marder E. (2000) The actions of crustacean cardioactive peptide on adult and developing stomatogastric ganglion motor patterns. J. Neurobiol. 44, 31–44. [PubMed] [Google Scholar]
- Rehm K. J.; Deeg K. E.; Marder E. (2008) Developmental regulation of neuromodulator function in the stomatogastric ganglion of the lobster Homarus americanus. J. Neurosci. 28, 9828–9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggerman G.; Verleyen P.; Clynen E.; Huybrechts J.; De Loof A.; Schoofs L. (2004) Peptidomics. J. Chromatogr., B 803, 3–16. [DOI] [PubMed] [Google Scholar]
- Bulau P.; Meisen I.; Schmitz T.; Keller R.; Peter-Katalinic J. (2004) Identification of neuropeptides from the sinus gland of the crayfish Orconectes limosus using nanoscale on-line liquid chromatography tandem mass spectrometry. Mol. Cell Proteomics 3, 558–564. [DOI] [PubMed] [Google Scholar]
- Fu Q.; Christie A. E.; Li L. (2005) Mass spectrometric characterization of crustacean hyperglycemic hormone precursor-related peptides (CPRPs) from the sinus gland of the crab Cancer productus. Peptides 26, 2137–2150. [DOI] [PubMed] [Google Scholar]
- Fu Q.; Kutz K. K.; Schmidt J. J.; Hsu Y. W.; Messinger D. I.; Cain S. D.; de la Iglesia H. O.; Christie A. E.; Li L. (2005) Hormone complement of the Cancer productus sinus gland and pericardial organ: an anatomical and mass spectrometric investigation. J. Comp. Neurol. 493, 607–626. [DOI] [PubMed] [Google Scholar]
- Huybrechts J.; Nusbaum M. P.; Bosch L. V.; Baggerman G.; De Loof A.; Schoofs L. (2003) Neuropeptidomic analysis of the brain and thoracic ganglion from the Jonah crab Cancer borealis. Biochem. Biophys. Res. Commun. 308, 535–544. [DOI] [PubMed] [Google Scholar]
- Li L.; Kelley W. P.; Billimoria C. P.; Christie A. E.; Pulver S. R.; Sweedler J. V.; Marder E. (2003) Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab Cancer borealis. J. Neurochem. 87, 642–656. [DOI] [PubMed] [Google Scholar]
- Messinger D. I.; Kutz K. K.; Le T.; Verley D. R.; Hsu Y. W.; Ngo C. T.; Cain S. D.; Birmingham J. T.; Li L.; Christie A. E. (2005) Identification and characterization of a tachykinin-containing neuroendocrine organ in the commissural ganglion of the crab Cancer productus. J. Exp. Biol. 208, 3303–3319. [DOI] [PubMed] [Google Scholar]
- Behrens H. L.; Chen R.; Li L. (2008) Combining microdialysis, NanoLC-MS, and MALDI-TOF/TOF to detect neuropeptides secreted in the crab Cancer borealis. Anal. Chem. 80, 6949–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiebe P.; Dreger M.; Meseke M.; Evers J. F.; Hucho F. (2002) Identification of orcokinins in single neurons in the stomatogastric nervous system of the crayfish Cherax destructor. J. Comp. Neurol. 444, 245–259. [DOI] [PubMed] [Google Scholar]
- Skiebe P.; Dreger M.; Borner J.; Meseke M.; Weckwerth W. (2003) Immunocytochemical and molecular data guide peptide identification by mass spectrometry: orcokinin and orcomyotropin-related peptides in the stomatogastric nervous system of several crustacean species. Cell. Mol. Biol. 49, 851–871. [PubMed] [Google Scholar]
- Torfs P.; Baggerman G.; Meeusen T.; Nieto J.; Nachman R. J.; Calderon J.; De Loof A.; Schoofs L. (2002) Isolation, identification, and synthesis of a disulfated sulfakinin from the central nervous system of an arthropods the white shrimp Litopenaeus vannamei. Biochem. Biophys. Res. Commun. 299, 312–320. [DOI] [PubMed] [Google Scholar]
- Yasuda A.; Yasuda-Kamatani Y.; Nozaki M.; Nakajima T. (2004) Identification of GYRKPPFNGSIFamide (crustacean-SIFamide) in the crayfish Procambarus clarkii by topological mass spectrometry analysis. Gen. Comp. Endrocrinol. 135, 391–400. [DOI] [PubMed] [Google Scholar]
- Yasuda-Kamatani Y.; Yasuda A. (2000) Identification of orcokinin gene-related peptides in the brain of the crayfish Procambarus clarkii by the combination of MALDI-TOF and on-line capillary HPLC/Q-Tof mass spectrometries and molecular cloning. Gen. Comp. Endrocrinol. 118, 161–172. [DOI] [PubMed] [Google Scholar]
- Li L.; Garden R. W.; Sweedler J. V. (2000) Single-cell MALDI: a new tool for direct peptide profiling. Trends Biotechnol. 18, 151–160. [DOI] [PubMed] [Google Scholar]
- Li L.; Romanova E. V.; Rubakhin S. S.; Alexeeva V.; Weiss K. R.; Vilim F. S.; Sweedler J. V. (2000) Peptide profiling of cells with multiple gene products: combining immunochemistry and MALDI mass spectrometry with on-plate microextraction. Anal. Chem. 72, 3867–3874. [DOI] [PubMed] [Google Scholar]
- Kutz K. K.; Schmidt J. J.; Li L. (2004) In situ tissue analysis of neuropeptides by MALDI FTMS in-cell accumulation. Anal. Chem. 76, 5630–5640. [DOI] [PubMed] [Google Scholar]
- Ma M.; Chen R.; Sousa G. L.; Bors E. K.; Kwiatkowski M. A.; Goiney C. C.; Goy M. F.; Christie A. E.; Li L. (2008) Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the American lobster Homarus americanus. Gen. Comp. Endrocrinol. 156, 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.; Jiang X.; Conaway M. C.; Mohtashemi I.; Hui L.; Viner R.; Li L. (2010) Mass spectral analysis of neuropeptide expression and distribution in the nervous system of the lobster Homarus americanus. J. Proteome Res. 9, 818–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cape S. S.; Rehm K. J.; Ma M.; Marder E.; Li L. (2008) Mass spectral comparison of the neuropeptide complement of the stomatogastric ganglion and brain in the adult and embryonic lobster Homarus americanus. J. Neurochem. 105, 690–702. [DOI] [PubMed] [Google Scholar]
- Wang J.; Ma M.; Chen R.; Li L. (2008) Enhanced neuropeptide profiling via capillary electrophoresis off-line coupled with MALDI FTMS. Anal. Chem. 80, 6168–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Jiang X.; Sturm R. M.; Li L. (2009) Combining tissue extraction and off-line capillary electrophoresis matrix-assisted laser desorption/ionization Fourier transform mass spectrometry for neuropeptide analysis in individual neuronal organs using 2,5-dihydroxybenzoic acid as a multi-functional agent. J. Chromatogr., A 1216, 8283–8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie A. E.; Stemmler E. A.; Peguero B.; Messinger D. I.; Provencher H. L.; Scheerlinck P.; Hsu Y. W.; Guiney M. E.; de la Iglesia H. O.; Dickinson P. S. (2006) Identification, physiological actions, and distribution of VYRKPPFNGSIFamide (Val1)-SIFamide) in the stomatogastric nervous system of the American lobster Homarus americanus. J. Comp. Neurol. 496, 406–421. [DOI] [PubMed] [Google Scholar]
- Verleyen P.; Huybrechts J.; Schoofs L. (2009) SIFamide illustrates the rapid evolution in Arthropod neuropeptide research. Gen. Comp. Endrocrinol. 162, 27–35. [DOI] [PubMed] [Google Scholar]
- Dickinson P. S.; Stemmler E. A.; Barton E. E.; Cashman C. R.; Gardner N. P.; Rus S.; Brennan H. R.; McClintock T. S.; Christie A. E. (2009) Molecular, mass spectral, and physiological analyses of orcokinins and orcokinin precursor-related peptides in the lobster Homarus americanus and the crayfish Procambarus clarkii. Peptides 30, 297–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie A. E.; Cashman C. R.; Stevens J. S.; Smith C. M.; Beale K. M.; Stemmler E. A.; Greenwood S. J.; Towle D. W.; Dickinson P. S. (2008) Identification and cardiotropic actions of brain/gut-derived tachykinin-related peptides (TRPs) from the American lobster Homarus americanus. Peptides 29, 1909–1918. [DOI] [PubMed] [Google Scholar]
- Christie A. E.; Cashman C. R.; Brennan H. R.; Ma M.; Sousa G. L.; Li L.; Stemmler E. A.; Dickinson P. S. (2008) Identification of putative crustacean neuropeptides using in silico analyses of publicly accessible expressed sequence tags. Gen. Comp. Endrocrinol. 156, 246–264. [DOI] [PubMed] [Google Scholar]
- Bucknall M.; Fung K. Y.; Duncan M. W. (2002) Practical quantitative biomedical applications of MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 13, 1015–1027. [DOI] [PubMed] [Google Scholar]
- Rubakhin S. S.; Sweedler J. V. (2008) Quantitative measurements of cell-cell signaling peptides with single-cell MALDI MS. Anal. Chem. 80, 7128–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Z.; Fitzgerald M. C. (2001) A solid sample preparation method that reduces signal suppression effects in the MALDI analysis of peptides. Anal. Chem. 73, 625–631. [DOI] [PubMed] [Google Scholar]
- Rehm K. J.; Taylor A. L.; Pulver S. R.; Marder E. (2008) Spectral analyses reveal the presence of adult-like activity in the embryonic stomatogastric motor patterns of the lobster Homarus americanus. J. Neurophysiol. 99, 3104–3122. [DOI] [PubMed] [Google Scholar]
- Stangier J.; Hilbich C.; Burdzik S.; Keller R. (1992) Orcokinin: a novel myotropic peptide from the nervous system of the crayfish Orconectes limosus. Peptides 13, 859–864. [DOI] [PubMed] [Google Scholar]
- Zajac J. M.; Mollereau C. (2006) RFamide peptides. Introduction. Peptides 27, 941–942. [DOI] [PubMed] [Google Scholar]
- Helluy S. M.; Beltz B. S. (1991) Embryonic development of the American lobster (Homarus americanus): Quantitative staging and characterization of an embryonic molt cycle. Biol. Bull. 180, 355–371. [DOI] [PubMed] [Google Scholar]
- Thirumalai V.; Marder E. (2002) Colocalized neuropeptides activate a central pattern generator by acting on different circuit targets. J. Neurosci. 22, 1874–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M.; Gard A. L.; Xiang F.; Wang J.; Davoodian N.; Lenz P. H.; Malecha S. R.; Christie A. E.; Li L. (2010) Combining in silico transcriptome mining and biological mass spectrometry for neuropeptide discovery in the Pacific white shrimp Litopenaeus vannamei. Peptides 31, 27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.