Creating protein capture reagents on a proteome-wide scale is a grand challenge in molecular medicine.[1] High quality reagents are critical for elucidating protein function, and developing diagnostic and therapeutic agents. Although antibodies remain the gold standard, their cumbersome production in animals is expensive, time consuming, and can fail for many targets.[2] Alternative technologies like single-chain antibodies and aptamers overcome some of these limitations, but still require many iterative rounds of selection and amplification to produce high quality binders.[3] Problems such as these have created a pressing need for next generation technologies that lend themselves as tools to explore the human proteome, but are more efficient and cost-effective to produce than existing reagents. Ideal protein capture reagents would be constructed with minimal effort from easy-to-assemble building blocks that are readily available and inexpensive.[4] The most promising reagents would function in assays that already exist for antibodies.
Bivalent reagents are attractive candidates for synthetic antibodies because the interactions they produce can be much stronger than their corresponding monovalent interactions.[5] This approach has been used to create many protein affinity reagents,[6] including a recent bivalent peptoid that targets vascular endothelial growth factor receptor 2 (VEGFR-2) in vitro and in cells.[7] Unfortunately, the discovery process used to create these reagents has been difficult to scale. Recently, we developed a new type of protein affinity reagent called a “DNA synbody” whose synthesis does not require in vitro selection or challenging synthetic chemistry.[8] Instead, DNA synbodies are produced by a process that we refer to as ligand interactions by nucleotide conjugates (LINC). LINC uses a short double-stranded DNA scaffold to determine the optimal separation distance and angular geometry needed to transform two ligands into a single high affinity protein capture reagent.[9]
In our original demonstration, we relied on peptide microarrays to identify ligands that recognize distinct sites on the surface of a desired protein target.[8] While this strategy provided a rapid method of peptide identification, ligands produced in this way are limited in their ability to be optimized for higher binding affinity.[10] Recognizing that many disease-associated proteins have peptides that are well documented and thoroughly characterized, we wondered whether such molecules could be used as a readily available source of chemical parts to create high quality DNA synbodies. Here, we demonstrate the feasibility of this strategy by using previously discovered peptides with known affinity to growth factor receptor-bound protein 2 (Grb2) to create an anti-Grb2 DNA synbody. We chose Grb2 because of its importance in growth factor-mediated cell signaling,[11] which is involved in numerous cellular responses including pathways that contribute to tumor growth and metastasis.[12]
Two peptides that recognize non-overlapping sites on the surface of human Grb2 were identified (Supporting Information, Figure 1). The SH2-binding peptide (ASpYVNVSA) contains a phospho-tyrosine (pY) residue that is essential for high affinity binding, and closely mimics the natural Grb2 binding partner, phosphorylated tyrosine kinase in the signal transduction pathway.[13a] This SH2-binding peptide is reported to have a dissociation constant (Kd) of 0.5 mM. The second peptide, YEVPPPVPPRRR, which selectively binds the N-terminal SH3 domain of Grb2 with a reported Kd of 5 mM, is a natural proline-rich ligand.[13b] The dissociation constants of bothGrb2-binding peptides were verified by surface plasmon resonance (SPR). The two peptides were found to bind Grb2 near their reported literature values; 0.4 ± 0.1 mM for the SH2-binding peptide and 7.5 ± 5.7 mM for the SH3-binding peptide.
Figure 1.
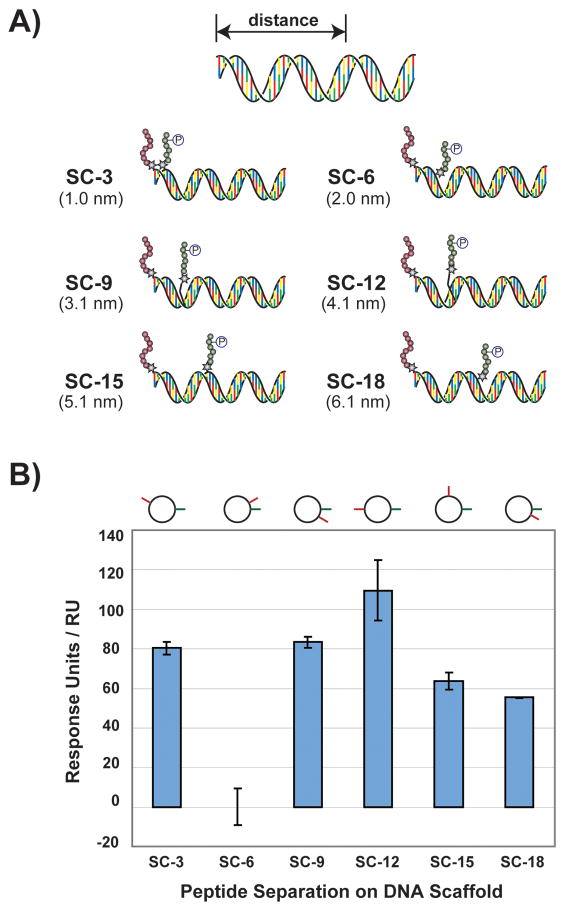
Design and affinity measurement of synbody constructs. A) Cartoon representation illustrating six synbody constructs (SCs) screened for affinity to Grb2. SCs were designed to spatially separate the SH3-binding peptide and the SH2-binding peptide by 3, 6, 9, 12, 15, and 18 base pairs (nanometer distance given in parenthesis). B) The SCs were assayed for relative binding affinity to Grb2 using surface plasmon resonance. The cartoon image above each bar indicates the relative position of each peptide with respect to the longitudinal axis of the dsDNA helix.
We began by creating a focused library of synbody constructs (SC) based on the two Grb2 peptides. SCs were assembled in both the forward and reverse orientations by separately conjugating one peptide to the 5′-end of the sense strand and attaching the other peptide to downstream positions on the antisense strand. Annealing the two strands together yielded a series of bivalent synbodies that sampled a distance of ~1–6 nanometers on the DNA scaffold (Figure 1a). The forward orientation was arbitrarily defined as the set of synbodies that contained the SH3-binding peptide on the sense strand and the SH2-binding peptide at variable positions on the antisense strand. By default, the reverse orientation contained the opposite set with the two peptides spaced at identical distances but on opposing strands of the DNA helix.
An SPR T-100 instrument equipped with auto-injection capability was used to rapidly screen each SC for affinity to recombinant Grb2. Measurements were made by flowing the SCs over a Grb2 sample that was immobilized on a CM5 biosensor chip. From this data, a clear trend emerged in which SCs assembled in the forward orientation produced higher relative binding responses than SCs produced in the reverse orientation. Close inspection of the synbodies constructed in the forward orientation revealed that synbody construct 12 (SC-12) with an estimated separation distance of 4.1 nm produced the highest binding response relative to the other five SCs assembled with that orientation (Figure 1b). This is an interesting arrangement for a bivalent affinity reagent because it positions the two peptides roughly 180° apart on the DNA helix. Other synbody constructs, such as SC-9 or SC-15, which narrow or widen the space between the two peptides (~3.1 and ~5.1 nm, respectively) have lower binding, indicating that these SCs are suboptimal relative to SC-12. SC-6 showed no detectable binding in three independent trials, suggesting that this configuration may produce an intramolecular peptide-peptide interaction that precludes Grb2 binding. All of the other SCs examined showed intermediate binding to Grb2, indicating that they bind Grb2 less efficiently than SC-12.
One recurring question that is often raised about synthetic antibodies is how well do these reagents compare to antibodies. Recognizing the importance that alternative affinity reagents could play in large-scale proteomics research.[1b] we decided to explore this question through a series of side-by-side assays that compare the binding properties of SC-12 to a typical commercial antibody. Using a standard ELISA-like assay, we found that Ab-4 and H-9, two randomly selected antibodies purchased from separate vendors bind Grb2 with Kd’s of 67 ± 31 and 45 ± 16 nM, respectively. In our experience, this level of binding is typical for a standard commercial antibody. SC-12 binds to Grb2 with a Kd of 6.9 ± 0.4 nM, which is 5–10-fold stronger than either commercial antibody (Figure 2). This result is particularly striking when considering the fact that DNA synbodies represent a class of affinity reagents that are structurally much simpler than any protein fold, especially one as complex as an immunoglobin.
Figure 2.
Comparison of the binding properties of SC-12 to a standard commercial antibody. Dissociation constants for A) synbody SC-12 and B) antibody H-9 bound to Grb2 were measured using an ELISA assay. SC-12 bound Grb2 with a Kd of 6.9 nM, while the commercial antibody H-9 bound Grb2 with a Kd of 45 nM. C–D) Dissociation constants were validated by surface plasmon resonance. SC-12 was found to have an on-rate that is 5 times faster than the antibody H-9, and both reagents have similar off-rates. This difference in binding kinetics results in tighter binding affinity for SC-12 when compared to H-9.
To further explore the binding properties of our Grb2 synbody, SPR was used to measure the kinetic profiles for SC-12 and H-9. The H-9 antibody was chosen for this experiment because it binds to Grb2 with higher affinity than the Ab-4 antibody. When Grb2 is passed over a surface containing SC-12, an immediate increase in binding response is observed and equilibrium is reached within 30 seconds (Figure 2c). This result is indicative of rapid binding between Grb2 and the immobilized SC-12 affinity reagent. In contrast, the same assay performed on the H-9 antibody showed a much slower rate of binding and equilibrium is not yet reached after 75 seconds. Analysis of the individual rate constants indicates that the on-rate (kon) of SC-12 is more than 5 times faster than the on-rate of H-9 (kon = 7.3 × 105 M−1s−1 and 1.3 × 105 M−1s−1, respectively). In contrast, SC-12 and H-9 have similar off-rates (koff = 0.0014 s−1 and 0.0021 s−1, respectively), indicating that once Grb2 is bound protein dissociation occurs relatively slowly. Calculation of the dissociation constants from the kinetic terms reveals that SC-12 has ~10-fold higher affinity for Grb2 than H-9 (Kd of 1.9 nM versus 16.5 nM, respectively), which is consistent with the binding results obtained from the ELISA assay. Overall, these results are very encouraging as high affinity binding and slow dissociation kinetics represent a hallmark of a high quality protein capture reagent.
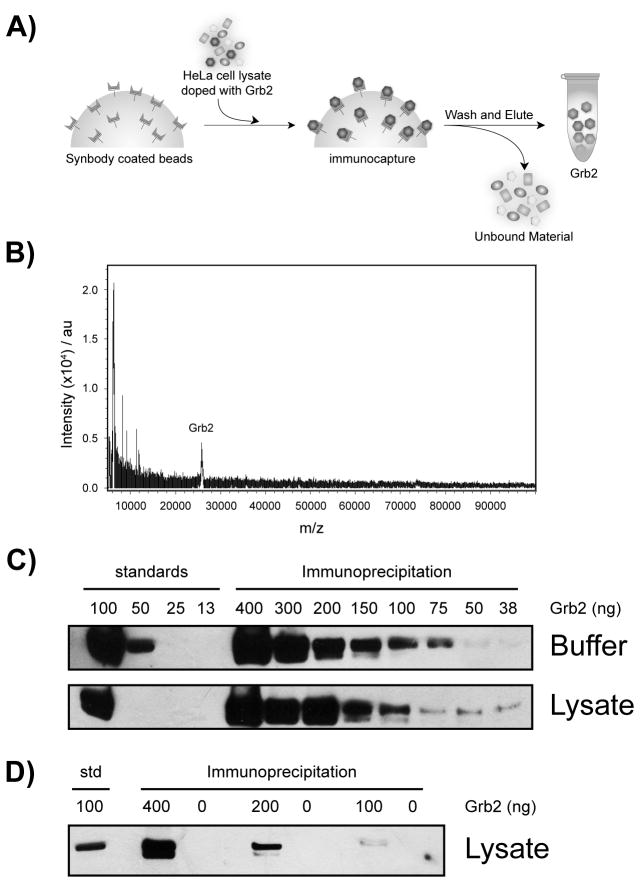
In addition to ligand binding affinity, protein specificity is an equally important parameter to consider when evaluating the quality of a protein affinity reagent. We therefore examined the ability of SC-12 to discriminate against other proteins commonly found in complex biological mixtures. Using a standard immunoprecipitation assay, SC-12 was challenged to isolate recombinant Grb2 protein spiked into a fresh solution of HeLa cell lysate (Figure 3a). SC-12 immobilized on streptavidin-coated magnetic beads was incubated with spiked HeLa cell lysate for 1 hour at 24°C, after which the beads were thoroughly washed, and the bound material was eluted with biotin and analyzed by MALDI-TOF mass spectrometry (Figure 3b). The resulting spectrum shows a clear peak at ~26 kDa that is identical in mass to our pure recombinant Grb2 protein standard. Since no other significant peaks were observed with a molecular mass above 15 kDa, we concluded that SC-12 functions with high specificity.
Figure 3.
Specificity of SC-12 in HeLa cell lystate. A) Synbody-coated magnetic beads specifically bind and remove Grb2 from Hela cell lysate. B) Mass spectrum of the eluate recovered from the pull-down assay reveals a single peak that is equivalent in mass to pure Grb2. C–D) Western blot analysis of the eluate from pull-down assays demonstrate efficient removal of Grb2 from buffer and lysate solutions. Bands observed in the western blot correspond to recombinant Grb2.
To validate the identity of the Grb2 antigen and determine the efficiency of protein capture, a second assay was performed in which immunoprecipitated Grb2 was analyzed by western blot analysis. Solutions of either PBS buffer or crude HeLa cell lysate were spiked with 40 to 400 ng of pure recombinant Grb2. Following a 1 hour incubation at 24°C, the beads were washed and the bound material was eluted with 0.1% SDS. All of the material present in the eluate was analyzed by western blot analysis. Comparison of the amount of Grb2 recovered from the buffered solutions with the HeLa cell lysate demonstrates that SC-12 functions with high efficiency since both gels yield similar amounts of protein and no bands other than Grb2 were observed in the gel (Figure 3c). By comparing the amount of protein present in each lane to recombinant Grb2 standards run on the same gel, we concluded that SC-12 can easily detect ~100 ng of Grb2 in either solution. To ensure that the bands observed in the western blot corresponded to the recombinant Grb2 protein, a third assay was performed on Hela cell lysate in which samples either contained or were missing the Grb2 spike (Figure 3d). Analysis of the resulting western blot demonstrates that Grb2 protein was only detected in solutions that contained the recombinant Grb2 spike, which is consistent with the ability of SC-12 to bind and remove Grb2 from a complex mixture.
In summary, we demonstrate that readily available peptides can be combined using the LINC technology to create DNA synbodies that recapitulate antibody function by capturing their cognate antigens in complex biological mixtures. We suggest that the simplicity of this process in combination with quality of the reagent produced make LINC an attractive strategy for accelerating the pace of affinity reagent production.
Experimental Section
Materials
DNA oligonucleotides were purchased from the Keck Facility at Yale University. Peptides were purchased from GenScript Corporation with 95% purity. Peptides were synthesized with a C-terminal Gly-Ser-Cys linker. Mouse monoclonal anti-Grb2 antibodies H9 and Ab-4 were purchased from Santa Cruz Biotechnology and Thermo Scientific, respectively.
Synbody Construction
All synbody constructs were designed and synthesized as previously described [8, 14]. In brief, the template strand (5′-d-CCC GAA ACA ACC GCG AGA GGC ACG CGC GTA GC-37prime;), which contains an amine modified deoxycytidine residue at position 1 (shown in bold) was separately conjugated to each peptide. A series of complementary strands (5′-d-GCT ACG CGC GTG CCT CTC GCG GTT GTT TCG GG-3′), which contain amine modified deoxynucleotides (shown in bold) at internal positions that are 3, 6, 9, 12, 15, and 18 nucleotides (from the 3′-end) were separately conjugated to both peptides. Peptide-oligonucleotide conjugates (POCs) were PAGE purified, isolated, and quantified by UV absorbance. POCs were annealed in 1x HBS-P buffer (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 0.005% v/v surfactant P20) supplemented with MgCl2 (5 mM) by heating to 55 °C for5 min and cooling on ice.
Synbody Distance Screen
Grb2 synbody constructs (SCs) were screened for relative binding affinity to Grb2 using a Biacore T-100 SPR instrument. Grb2 (0.025 mg/ml, 8,344 response units) was immobilized onto the surface of a CM5 chip (GE Healthcare) using the manufacturers recommended protocol. Synbody constructs (1 mM) were passed over the Grb2 chip in 1x HBS-P buffer at a flow rate of 30 ml/min. The binding responses were measured in triplicate, and all sensograms were corrected by background subtraction from the reference cell.
Affinity Determination
The affinity of SC-12 was measured by coating a microtiter 96-well plate with streptavidin protein (2 mg/ml in 0.1 M sodium bicarbonate, pH 9.8) overnight at 4 °C in a humidifier. The solution was removed and replaced with blocking buffer (100 ml, 2% BSA in 1x HBS-P, pH 7.4), and incubated for an additional 1 hour at 37°C in the humidifier. The solution was removed, and the plate was washed three times with 1x HBS-P and tapped dry. The biotin-modified Grb2 SC-12 (10 nM) was added to the plate in 1x HBS-P buffer supplemented with MgCl2 (5 mM) and incubated for 30 min at room temperature. Unbound synbody was removed from the well and a concentration series of Cy3-labeled Grb2 (100 ml) was incubated with the plate for 1 hour at 37 °C. The solution was removed and the plate was washed three times with 1x HBS-P. The plate was scanned using a SpectroMax plate reader for fluorescence (excitation at 550 nm and emission at 570 nm). Assays were conducted in duplicate and the data was corrected by background subtraction. The data was plotted and analyzed using GraphPad Prism software. The affinity of anti-Grb2 antibodies H-9 and Ab-4 were determined in the same manner with the exception that the antibodies were coated directly onto the 96-well plate.
Kinetic Characterization
Kinetic parameters of the SC-12 and the H-9 antibody were determined using a Biacore T-100 surface plasmon resonance instrument. Avidin (0.025 mg/ml, 15,000 response units) was immobilized onto a CM5 chip using standard amine coupling chemistry. SC-12 containing a biotin label at the 3′ end of template strand was added to the chip in 1x HBS-P buffer at a flow rate of 10 ml/min for 1,000 sec, which resulted in 1,400 response units immobilized to the chip. Grb2 protein (250 nM) was passed over the chip at a rate of 30 ml/min for a contact time of 80 sec, followed by a 300 sec dissociation time. The on-rate (kon), off-rate (koff) and the solution dissociation constant (Kd) were determined using a 1:1 binding model in the Biacore software package. The solution binding parameters of the H-9 antibody was determined in the same manner by immobilizing the antibody directly onto the surface of CM5 chip (1,402 response units).
Pull-Down Assay
The streptavidin coated magnetic beads (DynaBeads, Invitrogen) were washed three times with of 1x HBS-P buffer (30 ml) supplemented with MgCl2 (5 mM). The biotin containing Grb2 SC-12 (150 pmol) was incubated with the beads for 15 min at room temperature. The beads were washed, and then incubated with HeLa cell lysate (5 ml of A280 = 15.68 in 1x HBS-P) spiked with Grb2 (220 pmol) for 15 min at 4 °C. The beads were washed three times with 1x HBS-P buffer (30 ml), the bound proteins were eluted using excess biotin (1 mM). The elution was lyophilized down to 5 ml and 1 ml elution was mixed with 3 ml of matrix (sinapic acid saturated, 125 ml acetonitrile, 235 ml water, 16 ml trifluoroacetic acid). A negative control was conducted in a similar manner by incubating the magnetic beads with HeLa cell lysate spiked with Grb2 for 15 min at 4 °C. Mass spectrometry analysis of the pure Grb2 protein, elution from the pull-down assay, and the elution from the negative control were performed on a Bruker Autoflex III MALDI-TOF mass spectrometer operating in linear delayed-extraction mode with 19.00 kV full accelerating potential.
Immunoprecipitation
Biotin-conjugated SC-12 (20 μl, 1 μM) was incubated with streptavidin-coupled M-270 Dynabeads (20 μl, Invitrogen) at room temperature for 1 hour. Grb2 protein was prepared in 100 mM phosphate buffered saline (PBS). The synbody-coated beads coated were washed three times with PBS (20 μl) and incubated overnight with the desired amount of target protein at 4 °C. For the specificity detection, serial dilutions of protein were prepared in diluted soluble Hela cell lysate. (A280 = 1.4244) After binding, the beads were washed again three times with PBS (20 μl) and heated at 90 °C in SDS (0.1%, 15 μl) diluted in PBS and 4× LDS loading buffer (5 μl, Invitrogen) for 10 min. The samples were run on a 4–12% precast SDS-PAGE gel (Invitrogen) at 200V for 30 min. The proteins were verified by western blot detection using anti-Grb2 primary antibody (Santa Cruz biotechnology) and goat anti-mouse IgG HRP secondary antibody (Bethyl Laboratories).
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA126622-01) and the Science Foundation of Arizona (CAA 0265-08). We thank the National Cancer Institute for generously providing the recombinant Grb2 protein, B. Williams and B. Page for technical assistance with the synbodies, and L. Hartwell for helpful comments and suggestions on the manuscript.
References
- 1.a) Blow N. Nature. 2007;447:741–744. doi: 10.1038/447741a. [DOI] [PubMed] [Google Scholar]; b) Taussig MJ, Stoevesandt O, Borrebaeck C, Bradbury A, Cahill D, Cambillau C, de Daruvar A, Dubel S, Eichler J, Frank R, Gibson T, Gloriam D, Gold L, Herberg F, Hermjakob H, Hoheisel J, Joos T, Kallioniemi O, Koegll M, Konthur Z, Korn B, Kremmer E, Krobitsch S, Landegren U, van der Maarel S, McCafferty J, Muyldermans S, Nygren P, Palcy S, Pluckthun A, Polic B, Przybylski M, Saviranta P, Sawyer A, Sherman D, Skerra A, Templin M, Ueffing M, Uhlen M. Nat Methods. 2007;4:13–17. doi: 10.1038/nmeth0107-13. [DOI] [PubMed] [Google Scholar]; c) Uhlen M. Mol Cell Proteomics. 2007;6:1455–1456. [PubMed] [Google Scholar]; d) Uhlen M, Hober S. J Mol Recognit. 2008;22:57–64. doi: 10.1002/jmr.891. [DOI] [PubMed] [Google Scholar]; e) Haab BB, Paulovich AG, Anderson NL, Clark AM, Downing GJ, Hermjakob H, LaBaer J, Uhlen M. Mol Cell Proteomics. 2006;5:1996–2007. doi: 10.1074/mcp.T600020-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Chambers RS. Curr Opin Chem Biol. 2005;9:46–50. doi: 10.1016/j.cbpa.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Hey T, Fiedler E, Rudolph R, Fiedler M. Trends in Biotechnol. 2005;23:514–522. doi: 10.1016/j.tibtech.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Kodadek T. Nat Chem Biol. 2010;6:162–165. doi: 10.1038/nchembio.303. [DOI] [PubMed] [Google Scholar]
- 5.a) Mammen M, Choi SK, Whitesides GM. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; b) Kodadek T, Reddy MM, Olivios HJ, Bachhawat-Sikder K, Alluri PG. Acc Chem Res. 2004;37:711–718. doi: 10.1021/ar030145l. [DOI] [PubMed] [Google Scholar]
- 6.a) Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]; b) Schaffer L, Brissette RE, Spetzler JC, Pillutla RC, Ostergaard S, Lennick M, Brandt J, Fletcher PW, Danielsen DM, Hsiao KC, Anderson AS, Dedova O, Ribel U, Hoeg-Jensen T, Hansen PH, Blume AJ, Markussen J, Goldstein NI. Proc Natl Acad Sci USA. 2003;8:4435–4439. doi: 10.1073/pnas.0830026100. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kim Y, Cao Z, Tan W. Proc Natl Acad Sci USA. 2008;105:5664–5669. doi: 10.1073/pnas.0711803105. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Handl HL, Sankaranarayanan R, Josan JS, Vagner J, Mash EA, Gillies RJ, Hruby VJ. Bioconjugate Chem. 2007;18:1101–110. doi: 10.1021/bc0603642. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Muller J, Wulffen B, Potzsch B, Mayer G. ChemBioChem. 2007;8:2223–2226. doi: 10.1002/cbic.200700535. [DOI] [PubMed] [Google Scholar]
- 7.Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T. J Am Chem Soc. 2008;130:5744–5752. doi: 10.1021/ja711193x. [DOI] [PubMed] [Google Scholar]
- 8.Williams BAR, Diehnelt CW, Belcher P, Greving M, Woodbury NW, Johnston SA, Chaput JC. J Am Chem Soc. 2009;131:17233–17241. doi: 10.1021/ja9051735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ligand interactions by nucleotide conjugates (LINC) formerly known as combinatorial examination of ligands and linkers (CELL).
- 10.Zhang J, Williams BAR, Nilsson MT, Chaput JC. Chem Commun. 2010;46:7778–7780. doi: 10.1039/c0cc01475c. [DOI] [PubMed] [Google Scholar]
- 11.Simon JA, Schreiber SL. Chem Biol. 1995;2:53–60. doi: 10.1016/1074-5521(95)90080-2. [DOI] [PubMed] [Google Scholar]
- 12.Giubellino A, Burke TR, Bottaro DP. Expert Opin Ther Targets. 2008;12:1021–1033. doi: 10.1517/14728222.12.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Kessels HWHG, Ward AC, Schumacher TNM. Proc Natl Acad Sci USA s. 2002;99:8524–8529. doi: 10.1073/pnas.142224499. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nguyen JT, Turck CW, Cohen FE, Zuckermann RN, Lim WA. Science. 1998;282:2088–2092. doi: 10.1126/science.282.5396.2088. [DOI] [PubMed] [Google Scholar]
- 14.Williams BAR, Chaput JC. Current Protocols in Nucleic Acid Chemistry. 2010;4.41:1–20. doi: 10.1002/0471142700.nc0441s42. [DOI] [PMC free article] [PubMed] [Google Scholar]