Abstract
Effective targeted cancer therapeutic development depends upon distinguishing disease-associated ‘driver’ mutations, which have causative roles in malignancy pathogenesis, from ‘passenger’ mutations, which are dispensable for cancer initiation and maintenance. Translational studies of clinically active targeted therapeutics can definitively discriminate driver from passenger lesions and provide valuable insights into human cancer biology. Activating internal tandem duplication (ITD) mutations in FLT3 (FLT3-ITD) are detected in approximately 20% of acute myeloid leukaemia (AML) patients and are associated with a poor prognosis1. Abundant scientific2 and clinical evidence1,3, including the lack of convincing clinical activity of early FLT3 inhibitors4,5, suggests that FLT3-ITD probably represents a passenger lesion. Here we report point mutations at three residues within the kinase domain of FLT3-ITD that confer substantial in vitro resistance to AC220 (quizartinib), an active investigational inhibitor of FLT3, KIT, PDGFRA, PDGFRB and RET6,7; evolution of AC220-resistant substitutions at two of these amino acid positions was observed in eight of eight FLT3-ITD-positive AML patients with acquired resistance to AC220. Our findings demonstrate that FLT3-ITD can represent a driver lesion and valid therapeutic target in human AML. AC220-resistant FLT3 kinase domain mutants represent high-value targets for future FLT3 inhibitor development efforts.
Perhaps the most compelling evidence so far that FLT3-ITD could representa driver mutation inAML was the identificationof a secondary FLT3 kinase domain mutation that conferred moderate resistance to the multikinase inhibitor PKC412 in a single FLT3-ITD+ patient who relapsed after an initial bone marrow response8. Although the broad-spectrum kinase inhibitor sorafenib has anecdotally achieved bone marrow remissions in FLT3-ITD+ AML patients9, whether its mechanism of action is mediated through inhibition of FLT3 or a distinct kinase is unclear. Indeed, two patients who relapsed after an initial response to sorafenib had no detectable FLT3 kinase domain mutations at the time of resistance10.
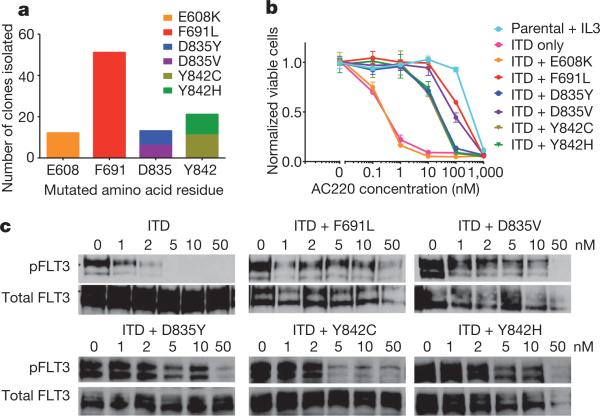
A recent interim analysis of 53 relapsed/refractory FLT3-ITD+ AML patients evaluable for efficacy in a multinational phase II trial of AC220 monotherapy documented a composite complete remission (<5% bone marrow blasts) rate of 45% (frequently associated with incomplete recovery of peripheral blood counts)7. We sought to use the clinical activity of AC220 as a tool to define FLT3-ITD as a driver or passenger mutation in human AML. Using a previously validated in vitro saturation mutagenesis assay11, we identified AC220 resistance-conferring mutations at four residues in the kinase domain of FLT3-ITD (Fig. 1a). Mutations at three of these amino acid positions conferred high degrees of in vitro AC220 resistance as demonstrated in proliferation (Fig. 1b) and cell-based biochemical assays (Fig. 1c). These residues consist of the ‘gatekeeper’ residue (F691) and two residues within the activation loop (D835, Y842). For unclear reasons, the E608K substitution did not confer substantial AC220 resistance and was not further characterized.
Figure 1. Mutation screen of FLT3-ITD reveals secondary kinase domain mutations that cause varying degrees of resistance to AC220.
a, Numbers of independent AC220-resistant Ba/F3 FLT3-ITD subpopulations with amino acid substitution at the indicated residue obtained from a saturation mutagenesis assay (n = 97 clones). b, Normalized cell viability of Ba/F3 populations stably expressing FLT3-ITD mutant isoforms after 48 h in various concentrations of AC220 (error bars represent standard deviations of triplicates from the same experiment). c, Western blot analysis using anti-phospho-FLT3 (pFLT3) or anti-FLT3 antibody performed on lysates from IL-3-independent Ba/F3 populations expressing the FLT3-ITD mutant isoforms indicated. Cells were exposed to AC220 at the indicated concentrations for 90 min.
We next assessed the presence of drug-resistant kinase domain mutations in FLT3-ITD in eight paired pre-treatment and relapse samples obtained from FLT3-ITD+ AML patients who initially achieved morphological reduction of bone marrow blasts to ≤5% with AC220 monotherapy, but subsequently relapsed despite continued AC220 treatment. In every case, subcloning and sequencing12 of individual FLT3-ITD alleles revealed mutations at the time of relapse (Table 1) that were not detected pre-treatment (Supplementary Table 1). Mutations were confined to two of the three critical residues identified in our in vitro screen. The activation loop mutation D835Y was detected in three cases, D835V in two, and the gatekeeper mutation F691L was identified in three. Additionally, one novel activation loop mutation, D835F, was identified in a single patient. This mutation confers substantial in vitro resistance to AC220 (Supplementary Fig. 1) and cross-resistance to sorafenib (data not shown), and was probably not detected in our saturation mutagenesis screen because its creation requires a two-nucleotide substitution. One patient (1011-007) seemed to have evolved polyclonal resistance, with both F691L and D835V mutations detected on separate FLT3-ITD sequences. Collectively, these findings suggest that clinical response and relapse in each of these eight patients is mechanistically mediated through modulation of FLT3-ITD kinase activity.
Table 1.
Summary of FLT3 kinase domain mutations in patients relapsed on AC220
| Subject number | Sex | Age (years) | Prior therapy | Karyotype at enrolment | Karyotype at relapse | Blasts in relapse sample (%) | New mutation at relapse | ITD+ clones with mutation | Weeks on study |
|---|---|---|---|---|---|---|---|---|---|
| 1009-003 | F | 75 | 7+3 | 45~54,XX,+3, + 6, + 7, +8,+13,+14,+21, + 22[cp15]/46,XX[5] | 52,XX,+3,+6,+7,+8, + 10,+12,+13[cp7]/46, XX[14] | 90 | D835F | 6/15 | 12 |
| 1011-006 | M | 70 | 7+3, low-dose cytarabine | Normal | ND | 10 | D835Y | 4/15 | 8 |
| 1011-007 | F | 56 | 7+3, HAM | Normal | 46,XX,del(11) (p?13p?15) [12]/46,XX[9] | 80 | F691L D835V |
4/24 5/24 |
11 |
| 1005-004 | F | 60 | Cytarabine and mitoxantrone | Normal | Normal | 92 | F691L | 9/22 | 19 |
| 1005-006 | M | 43 | 7+3, MEC, allogeneic stem cell transplant | 6,XY,t(1;15)(p22;q15) | ND | 59 | D835Y | 8/17 | 6 |
| 1005-007 | F | 59 | 7+3, HDAC | Normal | ND | 39 | D835V | 9/21 | 23 |
| 1005-009 | M | 68 | Cytarabine and mitoxantrone | Normal | ND | 58 | D835Y | 8/14 | 19 |
| 1005-010 | M | 52 | 7+3, HDAC, mitoxantrone and etoposide | 46,XY,t(4;12)(q26;p11.2), t(8;14)(q13;q11.2) | ND | 22 | F691L | 6/18 | 20 |
All patients achieved morphological bone marrow blasts of ≤5% at best response. 7+3, low-dose cytarabine for 7 days plus 3 daysanthracycline; HAM, high-dose cytarabine plus mitoxantrone; HDAC, high-dose cytarabine; MEC, mitoxantrone, etoposide, cytarabine. ND, not done.
To assess more precisely for resistance-conferring mutations at relapse, we used a recently described single molecule real-time (SMRT; Pacific Biosciences) sequencing platform, which can provide sequencing reads of sufficient length to enable focused interrogation of FLT3-ITD alleles (Supplementary Fig. 2)13. With this assay, hundreds of reads (range 19–930) spanning the ITD region and kinase domain with an average read length of greater than 1 kilobase (kb) (Supplementary Table 2) were reliably obtained from individual patient samples. Attention was focused on the amino acid codons identified in the in vitro screen for AC220 resistance-conferring mutations. SMRT sequencing confirmed the presence of resistance-conferring kinase domain mutations in FLT3-ITD at relapse in all eight patient samples (Table 2). Consistent with the results obtained by subcloning and sequencing, mutations at E608 and Y842 were not detected. The frequency of individual alternative codon substitutions within FLT3-ITD ranged from as low as 2.7% (D835F in patient 1005-006) to 50.6% (D835Y in patient 1005-009). The presence of polyclonal resistance was confirmed in patient 1011-007, and noted in an additional three cases: 1009-003, 1005-006 and 1005-007 (Table 2 and Supplementary Fig. 3). In general, mutations were detected on distinct molecules, although in the case of 1011-007, a subset of FLT3-ITD molecules with F691L also harboured a D835V mutation (5/21 observations; 23.8% of FLT3-ITD(F691L) alleles; data not shown). Analysis of three normal control samples revealed base substitutions at these residues at a very low frequency (Table 2 and Supplementary Table 3). The evolution of polyclonal resistance due to secondary kinase domain mutations in FLT3-ITD in four of eight relapsed patients is supportive of a central dependence upon FLT3-ITD signalling in the leukaemic clone of a subset of AML patients, and indicative of profound selective pressure exerted upon this clone by AC220. Additionally, these findings reveal the genetic complexity of drug-resistant disease that may evolve in cancer patients on clinically active therapy.
Table 2.
Third-generation sequencing identifies polyclonal FLT3 kinase domain mutations
| Pre-treatment |
Relapse |
Normal control no. 1 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Subject number | Mutation | Native codon | Alternative codon | Observed alternative codon frequency in ITD+ sequences (%) | Total number of ITD+ sequences sampled | Observed alternative codon frequency in ITD+ sequences (%) | Total number of ITD+ sequences sampled | Observed alternative codon frequency (%) | Total number of sequences sampled |
| 1009-003 | D835Y | GAT | TAT | 0.21 | 482 | 8.4 | 332 | 0.00 | 768 |
| D835V | GAT | GTT | 0.00 | 482 | 3.3 | 332 | 0.13 | 768 | |
| D835F | GAT | TTT | 0.00 | 482 | 10.2 | 332 | 0.00 | 768 | |
| 1011-006 | D835Y | GAT | TAT | 0.00 | 196 | 41.0 | 402 | 0.00 | 768 |
| 1011-007 | F691L | TTT | TTG | 0.18 | 561 | 6.2 | 341 | 0.22 | 450 |
| D835Y | GAT | TAT | 0.00 | 930 | 3.0 | 436 | 0.00 | 768 | |
| D835V | GAT | GTT | 0.43 | 930 | 29.6 | 436 | 0.13 | 768 | |
| 1005-004 | F691L | TTT | TTG | 0.00 | 496 | 29.6 | 513 | 0.22 | 450 |
| 1005-006 | D835Y | GAT | TAT | 0.00 | 171 | 39.5 | 261 | 0.00 | 768 |
| D835F | GAT | TTT | 0.00 | 171 | 2.7 | 261 | 0.00 | 768 | |
| 1005-007 | D835Y | GAT | TAT | 0.00 | 57 | 4.0 | 378 | 0.00 | 768 |
| D835V | GAT | GTT | 0.00 | 57 | 47.4 | 378 | 0.13 | 768 | |
| 1005-009 | D835Y | GAT | TAT | 0.00 | 19 | 50.6 | 445 | 0.00 | 768 |
| 1005-010 | F691L | TTT | TTG | 0.00 | 387 | 25.3 | 150 | 0.22 | 450 |
All P values <1 × 10-5 for alternative codon frequencies at relapse compared to a representative normal control sample (no. 1 refers to one of three normal control samples analysed).
All mutations identified at relapse were detected at a frequency significantly higher than that observed in a normal control, and although relapse occurred relatively rapidly in some patients, mutations were not convincingly detectable before treatment. The aggregate frequency of all mutations at relapse in individual patients ranged from approximately 20–50% in all cases, which is consistent with leukaemic blasts homozygous for FLT3-ITD and containing one drug-resistant allele per cell, although the presence of a heterogeneous blast population with only a subset of drug-resistant FLT3-ITD+ cells expressing kinase domain mutations cannot be excluded.
The five substitutions that conferred a high degree of resistance to AC220 in vitro were cross-resistant to sorafenib in cell-based growth and biochemical assays (Supplementary Fig. 4). The degree of relative resistance to sorafenib associated with these mutants was generally similar to that observed with AC220 (Supplementary Table 4).
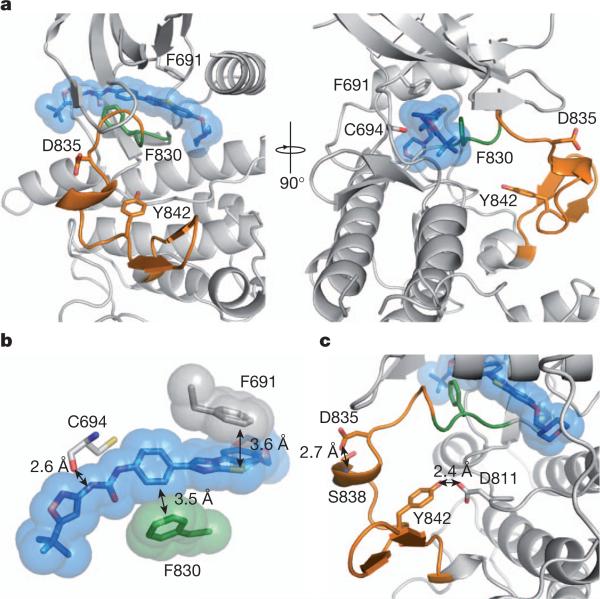
To understand the structural effects of AC220-resistance conferring mutations, we modelled the binding of AC220 to FLT3 (Fig. 2a). The crystal structure of the FLT3 kinase domain has been determined previously in an inactive conformation14 that resembles the inactive conformations of ABL15, KIT16 and insulin receptor tyrosine kinase17, with the activation loop folded back onto the ATP-binding cleft (loop-in conformation), thereby preventing substrate loading. The Asp-Phe-Gly (DFG) motif at the amino-terminal end of the activation loop adopts the DFG-out conformation, in which the Asp side chain, which normally coordinates a magnesium ion, is removed from the active site. The activation of FLT3 would require flipping of the DFG motif and reorganization of the activation loop, as observed in ABL18 and insulin receptor kinase19. Previously published binding data suggest that AC220 is a type II kinase inhibitor that preferentially binds to the inactive, DFG-out kinase conformation20. Our molecular docking analysis supports a model whereby AC220 interacts favourably with the DFG-out, inactive conformation. In the docked AC220–FLT3 model, the AC220 amide group is 2.6Å from the carbonyl group of C694, consistent with the formation of a hydrogen bond. The phenol ring of AC220 forms a perpendicular aromatic–aromatic interaction21 with F830 in the DFG motif (Fig. 2b). This interaction would not be possible in the DFG-in, active kinase conformation. The gatekeeper residue F691 forms a π–π stacking contact with the benzo-imidazol-thiazol moiety of AC220, which may further stabilize the complex. Substitutions at F691 with non-aromatic residues such as leucine may not compensate for the π–π stacking interaction.
Figure 2. Modelling of FLT3–AC220 interactions.
a, Docking model of the AC220-bound FLT3 kinase domain: AC220 (blue); activation loop (orange) and DFG motif (green); amino acid residues that confer AC220 resistance when mutated (F691, D835 and Y842) and that interact with AC220 (F691, C694 and F830) in sticks. b, Surface and stick presentation of AC220 and AC220-interacting residues on FLT3: the carbonyl oxygen of C694 interacts with one of the AC220 amide groups; F691 forms a π–π stacking interaction with AC220; F830 interacts with AC220 through a perpendicular aromatic–aromatic interaction. c, Structure of the activation loop: residues D835,Y842 and interacting residues on FLT3 depicted in sticks.
Residues D835 and Y842 stabilize the inactive conformation of the activation loop by forming hydrogen bonds with the main-chain amide group of S838 and the side chain of D811, respectively (Fig. 2c). Thus, replacement of either residue might destabilize this particular inactive conformation of the activation loop, which would then be expected to hinder the binding of AC220. In further support of this model, a binding study of AC220 and sorafenib, a crystallographically proven type II inhibitor22, revealed that the binding affinity of both inhibitors to the FLT3 D835V mutant is substantially reduced compared to native FLT3, both in the presence and absence of the juxtamembrane domain containing the ITD (Supplementary Fig. 5 and Supplementary Table 5).
Other potential explanations for the mechanism of resistance conferred by AC220-resistant mutants include increased kinase activity and differential activation of downstream effectors. Western blot analysis of cells expressing AC220-resistant FLT3-ITD-mutant isoforms revealed increased FLT3 autophosphorylation of D835 mutant iso-forms, but no discernable difference in phosphorylation of downstream targets, including the direct FLT3-ITD target STAT5A/B23, and no difference in cellular proliferation (Supplementary Fig. 6). Overall, these data support a primarily structural mechanism for AC220 resistance mediated by mutations at F691, D835 and Y842, although further studies are necessary for definitive confirmation. We speculate that the ability to retain inhibitory activity against activation loop substitutions at D835 and Y842 will require a type I FLT3 kinase inhibitor capable of effectively binding to the active, DFG-in conformation of the kinase.
Substitutions at gatekeeper residues such as FLT3-ITD(F691) are well-documented causes of resistance to kinase inhibitors12,24. Analogues of the FLT3-ITD(D835V) activation loop mutation have proven problematic for a number of kinase inhibitors: KIT(D816V), an activating mutation that is commonly detected in systemic mastocytosis, confers a high degree of resistance to imatinib and other KIT inhibitors25. Our data, although derived from a small cohort of patients that will need to be validated in larger studies, suggest that substitutions at F691 and D835 in FLT3-ITD will pose substantial barriers to disease control in AML patients treated with either AC220 or sorafenib, and therefore represent high-value targets for novel FLT3 inhibitor development efforts.
Compelling data suggest that activating FLT3 mutations are acquired relatively late during leukaemogenesis in a pre-established clone1,3, and alone are insufficient to cause acute leukaemia in pre-clinical models2. Recent evidence suggests that the molecular heterogeneity of individual leukaemias can be substantial, and can occur in both branching and linear fashions early during leukaemogenesis, including at the leukaemia-initiating or ‘leukaemic stem’ cell level26. Collectively, our data are consistent with acquisition of FLT3-ITD and drug-resistant FLT3 kinase domain mutations in a leukaemia-initiating cell population, although formal transplantation studies in mice are needed to address this question definitively. Our findings validate FLT3-ITD as a therapeutic target in human AML, and suggest that FLT3-ITD is capable of conferring a state of ‘oncogene addiction’, whereby cellular survival pathways associated with normal or precancerous cells can become hijacked, leading to a state of reliance upon key signalling molecules that can be exploited therapeutically. This work supports the exploration of therapeutic strategies targeting select activating mutations in other signalling molecules that are believed to be acquired relatively late in disease evolution, such as JAK2 (ref. 27) or RAS3, with agents capable of achieving clinically meaningful target inhibition. Further studies will be required to identify mechanisms of drug resistance that may circumvent reliance on activated FLT3 by activation of downstream or parallel pathways, as has been described with other kinase inhibitors28. To that end, translational studies using detailed molecular analyses of primary samples obtained from AML patients treated with clinically effective targeted therapeutics promise to further inform mechanisms of drug resistance, strategies for future drug development, and models of disease evolution.
METHODS SUMMARY
MSCVpuroFLT3-ITD plasmid DNA was mutagenized and used to generated AC220-resistant Ba/F3 clones as previously described11. The FLT3 kinase domain was sequenced from PCR-amplified genomic DNA isolated from AC220-resistant clones. Identified drug-resistant mutations were re-engineered into MSCVpuroFLT3-ITD using site-directed mutagenesis and Ba/F3 cell lines were created as detailed in Methods. Cell viability in the presence and absence of drug was assessed using trypan blue exclusion. FLT3 phosphorylation status was determined by western blot analysis of whole cell lysates prepared after 90 min of drug exposure from Ba/F3 cells stably expressing FLT3 mutant isoforms and from transfected 293T cells in the absence of drug. The FLT3 kinase domain was PCR amplified from cDNA derived from blood or bone marrow samples from patients enrolled on the exploratory portion of the phase II trial of AC220 in AML (http://clinicaltrials.gov/ct2/show/NCT00989261; identifier NCT00989261) and from normal control bone marrow or mobilized peripheral blood stem cells. PCR products were cloned into Escherichia coli and individual clones were sequenced using Sanger sequencing. Alternatively, SMRTBell libraries13 were prepared as per the manufacturer's instructions and sequenced on a Pacific Biosciences RS instrument. For details of the computational sequencing analysis, please see Methods. Molecular docking of FLT3-ITD to AC220 was performed using Autodock 4.2 package. Inhibitor binding constants were measured using an active-site-dependent competition binding assay as previously descibed20. For further details of methods, please see Methods.
METHODS
DNA constructs, mutagenesis and resistance screen
FLT3-ITD cDNA cloned from the MV4;11 cell line (ITD: residues 591–601) into the HpaI site of the pMSCVpuro retroviral vector (Clontech) was a gift from Ambit Biosciences and was used as a template for mutagenesis. We used a modified strategy for random mutagenesis previously described11. Briefly, 1 μg of MSCV puroFLT3-ITD was used to transform the DNA-repair-deficient Escherichia coli strain XL-1 Red (Stratagene) and plated on 20 ampicillin-agar bacterial plates. After incubation for 36 h, colonies were collected by scraping, and plasmid DNA was purified by using a plasmid MAXI kit (Qiagen). Subsequently, mutagenized FLT3-ITD plasmid stock and Ecopack packaging plasmid were cotransfected into 293T cells grown in DMEM (Invitrogen) containing 10% fetal calf serum (FCS; Omega Scientific) using Lipofectamine 2000 (Invitrogen) as per the manufacturer's protocol. Viral supernatants were collected at 48 h, purified using a 0.44 μm vacuum filter, and used to infect Ba/F3 cells at a 1:100 to 1:300 dilution of viral supernatant to fresh RPMI 1640 (Invitrogen) supplemented with 10% FCS. Alternatively, viral supernatant was aliquoted and frozen. Thawed supernatant was used to infect Ba/F3 cells at a 1:50 dilution. Viral supernatant was diluted with the goal of minimizing multiplicity of infection. For infection, 1–2 × 106 Ba/F3 cells were resuspended in 3 ml of the diluted viral stock supplemented with recombinant mouse IL-3 (Invitrogen), and 4 μg ml–1 polybrene, plated in each well of a 12-well tissue culture dish and centrifuged at 1,500g in a Beckman Coulter Allegra 6KR centrifuge with a microplate carrier for 90 min at 34 °C. Centrifuged cells were subsequently transferred to a 37 °C incubator overnight. Infected Ba/F3 cells were washed twice with media to remove IL-3 and plated in 3 ml of RPMI medium 1640 at 5 × 105 cells per well of a 6-well dish supplemented with 20% FCS and 1.2% Bacto-agar with 20 nM AC220 (a gift from Ambit Biosciences). After 10–21 days, visible colonies were plucked from agar and expanded in the presence of drug (20 nM AC220).
Sequencing and alignments
Expanded colonies were harvested 7–14 days after isolation from agar, and whole genomic DNA was isolated using the QIAamp kit (Qiagen). The FLT3 kinase domain was amplified by PCR from whole genomic DNA by using TopTaq DNA polymerase (Qiagen). The primers TK1F (5′- TGCTGTGCATACAATTCCCTTGGC-3′) and TK2R (5′- TCTCTGCTGAAAGGTCGCCTGTTT-3′) were used for kinase domain amplification and subsequent bidirectional sequencing was performed using these primers in addition to TK1R (5′- AGTCCTCCTCTTCTTCCAGCCTTT-3′) and TK2F (5′- GAGAGGCACTCATGTCAGAACTCA-3′). Alignments to the native FLT3-ITD sequence were performed using Sequencher software (Gene Codes Corporation).
Generation of mutants
Mutants isolated in the screen were engineered into pMSCVpuroFLT3-ITD by using the QuikChange mutagenesis kit (Stratagene). In all cases, individual point mutants were confirmed by sequence analysis.
Cell viability and proliferation assays
Stable Ba/F3 lines were generated by using retroviral spinfection with the appropriate mutated plasmid as outlined above, with the exception of the exclusion of polybrene. At 48 h post-infection, puromycin was added to infected cells at a concentration of 4 mg μl–1. Cells were selected in the presence of puromycin for 7–10 days and subsequently IL-3 was washed twice from the cells with media and cells were selected in RPMI medium 1640 plus 10% FCS in the absence of IL-3. Exponentially growing Ba/F3 cells (5 × 104) were plated in each well of a 24-well dish with 1 ml of RPMI 1640 plus 10% FCS containing the appropriate concentration of drug as indicated in triplicate. Cells were allowed to expand for 2 days and were counted by using a Vi-cell XR automated cell viability analyser (Beckman Coulter). The mean number of viable cells at varying concentrations of drug was normalized to the median number of viable cells in the no-drug sample for each mutant. Error bars represent the standard deviation. Numerical IC50 values were generated using nonlinear best-fit regression analysis using Prism 5 software (GraphPad).
For proliferation assays, on day 0, parental Ba/F3 cells and Ba/F3 cells stably expressing FLT3-ITD mutant isoforms were plated in triplicate with 1 mL of RPMI 1640 plus 10% FCS at a density of 5 × 104 cells per well in each well of a 24-well dish. Cells were allowed to expand and were counted by using a Vi-cell XR automated cell viability analyser (Beckman Coulter) on days 2, 3, 4, 5, 6 and 7. To maintain exponential growth of cells, 0.5 ml of cells from each well were used for counting on each day and 0.25 ml of cells from the remaining volume were transferred to a new well with 0.75 ml of fresh RPMI plus 10% FCS (including 2 ng ml–1 of IL-3 for parental Ba/F3 cells). Extrapolated cell counts were calculated from the measured count on each day using the appropriate dilution factor (1× on day 2, 4× on day 3, 16× on day 4, and so on). The number of viable cells at each time point was normalized to the starting number of cells for each cell line on day 0 and the mean normalized cell count on each day was calculated. Error bars represent the standard deviation.
Immunoblotting
Exponentially growing Ba/F3 cells stably expressing each mutation along with a native FLT3-ITD control were plated in RPMI medium 1640 plus 10% FCS supplemented with kinase inhibitor at the indicated concentration. After a 90-min incubation, the cells were washed in phosphate buffered saline (PBS) and lysed in Cell Extraction Buffer (Invitrogen) supplemented with protease and phosphatase inhibitors. The lysate was clarified by centrifugation and quantified by BCA assay (Thermo Scientific). Protein was subjected to sodium dodecylsulphate polyacrylamide electrophoresis and transferred to nitrocellulose membranes. Immunoblotting was performed using anti-phospho-FLT3, anti-phospho-STAT5, anti-STAT5, anti-phospho-ERK, anti-ERK, anti-phospho-S6, anti-S6, anti-GAPDH (Cell Signaling) and anti-FLT3 S18 antibody (Santa Cruz Biotechnology). Alternatively, 293T cells were plated in 6-cm plates and transfected with MSCVpuroFLT3-ITD plasmid containing FLT3 mutations of interest using Lipofectamine 2000 (Invitrogen) as per the manufacturer's protocol. After 48 h, cells were washed with PBS, collected, lysed and subjected to western blot analysis as described above.
Competition binding assays
Inhibitor binding constants were measured by using active site-dependent competition binding assays essentially as previously described20. In brief, FLT3 protein isoforms were labelled with a chimaeric double-stranded DNA tag containing the NFkB binding site (50-GGGAATTCCC-30) fused to an amplicon for qPCR readout, which was added directly to the expression extracts. Binding reactions were assembled by combining DNA-tagged kinase extract, affinity beads loaded with a kinase inhibitor probe molecule, and test compound in 13 binding buffer (PBS, 0.05% Tween 20, 10 mM DTT, 0.1% BSA, 2 mg ml–1 sonicated salmon sperm DNA). Extracts were used directly in binding assays without any enzyme purification steps at a ≥10,000-fold overall stock dilution (final DNA-tagged enzyme concentration <0.1 nM). Assays were incubated for 1 h at room temperature (23 °C), which was sufficient to establish equilibrium. Subsequent washing, elution, and qPCR readout steps were as described20.
Patients and FLT3 kinase domain sequencing analysis
Eight cases of acquired resistance to AC220 were analysed. Patients were enrolled on the exploratory cohort of the phase II clinical trial of AC220 in relapsed or refractory AML at the University of California, San Francisco (UCSF), University of Pennsylvania or Johns Hopkins University. Details of the clinical trials and results are reported elsewhere7. All patients were FLT3-ITD-positive at enrolment. Samples were collected pre-treatment and at the time of disease progression. Only patients who had achieved morphological clearance of bone marrow blasts to ≤5% at best response and subsequently relapsed with an increase in peripheral blood or bone marrow blasts are included in this analysis. The patients in this analysis included all the patients meeting the above criteria at the three participating institutions. All patients gave informed consent according to the Declaration of Helsinki to participate both in the clinical trials and for collection of samples. All research involving human subjects was approved by the relevant Institutional Review Board at each individual participating institution (UCSF, University of Pennsylvania or Johns Hopkins).
For sequencing, frozen Ficoll-purified mononuclear cells obtained from blood or bone marrow were lysed in Trizol (Invitrogen) and RNA was isolated according to the manufacturer's protocol. cDNA was synthesized using Superscript II (Invitrogen) as per the manufacturer's protocol. The FLT3 kinase domain and adjacent juxtamembrane domain were PCR amplified from cDNA using primers TK1F and TK2R as above. PCR products were cloned using TOPO TA cloning (Invitrogen) and transformed into competent E. coli. Individual colonies were plucked, expanded in liquid culture overnight and plasmid DNA for sequencing was isolated using the QIAprep Spin Miniprep kit (Qiagen). Each colony was considered representative of a single mRNA. To minimize contamination from PCR artefact, we sequenced at least 10 and up to 24 FLT3-ITD-containing clones from each sample and required that mutations be found in >15% of clones. The primers TK1F, TK1R, TK2F and TK2R were used for bidirectional sequencing as above. Alignments with native FLT3 sequence were performed using Sequencher software (Gene Codes Corporation).
Sample preparation and SMRT sequencing
PCR product containing the FLT3 kinase domain was generated from patient cDNA as described above using high fidelity DNA polymerase.Weprepared PCR products for Pacific Biosciences sequencing using standard commercial kits and reagents (http://www.pacificbiosciences.com/products/consumables/reagents) following the manufacturer's instructions. PCR products input amounts ranged from 0.3–3 μg, and we prepared SMRTBell libraries13 on the full PCR products without any fragmentation. We sequenced all samples on a Pacific Biosciences RS instrument and recorded sequence for 75 min.
Computational analysis of FLT3 mutations
We obtained samples from three healthy individuals with no cancer history (two bone marrow, one mobilized peripheral blood stem cells), isolated RNA, made cDNA, amplified the FLT3 kinase domain, and sequenced following a protocol identical to that used on the AML samples. We used the sequence from Normal Control no. 1 as a control for all process steps between sample acquisition and sequencing. Data from the remaining two normal controls were compared to the Normal Control no. 1 and revealed no significant differences (Supplementary Table 3). We use the circular consensus sequencing (CCS) mode to obtain high accuracy reads for the ~1.4 kb amplicon with PacBio RS. The CCS mode generates reads by combining multiple independent single-pass sequencing reads for individual molecules to correct raw errors and generate a better accuracy consensus (see Supplementary Fig. 2). We report only the CCS reads where the same molecule is sequenced at three or more times, that is, raw read length >4.2 kb for the 1.4 kb amplicon. With the CCS reads, we obtained the sequence of the ~1.4 kb amplicon containing the FLT3 juxtamembrane and kinase domains with up to about 98% to 99% accuracy (see alignment identity for ITD– samples in Supplementary Table 2). For each CCS read, we used tandem repeats finder (TRF)29 to identify the ITD sequence. To determine unambiguously whether a read was ITD– or ITD+ consistently, we used only the CCS reads that included at least the region from the 50-bp 5′-end upstream to the 50-bp 39-end downstream sequence of the ITD region in the analysis. This allowed us to determine the number of sequences containing the ITD more accurately despite a small percentage of insertion and deletion errors in the CCS reads. Two distinct peaks allowed us to identify ITD– versus ITD+ CCS reads unambiguously. We found that each sample had only one major ITD as expected, although in some cases the majority ITD differed at relapse compared to pre-treatment. We then passed the ITD+ population of the CCS reads to the next stage for codon mutation analysis. A list of the number of total CCS reads identified is listed in Supplementary Table 2. We identified ~200–1,300 CCS reads spanning the whole region between the ITD region and the furthest codon of interest (Y842) for codon analysis per sample.
For codon mutation analysis, we restricted our analysis to the 608, 691, 835 and 842 codons from reference sequence NM_004119 (Homo sapiens FLT3 mRNA) and then took the frequency of sequences obtained for each of these codons in the PCR amplicon of healthy Normal Control no. 1 and compared that to the frequency of sequences in each AML patient sample. A local quality filter that required exact matching of the codons before and after the codon of interest was used for filtering out low quality codon calls that might be due to sequencing errors. We used the observed frequencies from the control sample for calculating the significance of the observed mutation in the AML patient samples. The P value was calculated by comparing the numbers of native codons observed and the alternative codon between the control sample and the AML patient sample with Fisher's exact test on the contingency table30,31. Owing to the potential statistical bias that could arise if the number of observed mutations was small in some cases, or if sequencing error frequenciesdifferedbetween mutantand reference codon sequences, we onlyreport the mutations using a conservative significance threshold of P < 1 × 10–5. We used a simulation todetermine the sensitivityofthisanalysistodetecta true mutation ata given codon position. Sensitivity in this analysis was determined as a function of three parameters: the number of errors observed in the control sequence, the total number of times that position is sequenced in the control, and the number of times that position is sequenced in the patient sample. For the simulation, we conservatively assumed that all alternative codons seen in the control are actual errors. With this simulation, we estimated that this analysis allows us to detect variants in the patient sample above 3% with high confidence if we get more than 300 observations of the codon of interest. To refine further our search for mutations underlying relapse in these patients, we considered only those mutations that were in cis to an ITD, as defined on being on the same single DNA molecule sequence read. These mutations at both baseline and relapse are listed in Table 2.
Molecular docking
Molecular docking was performed using Autodock 4.2 package32. The FLT-ITD structure (residue 587–947) was prepared from the Protein Data Bank accession 1RJB14. All bound waters were removed from the protein. The structure was then added for hydrogens, and partial atomic charges were assigned using AutoDockTools (ADT)32. Residues K644, F830, F691 and E661 were selected as flexible residues. To define the flexible residues, we first analysed the crystal structure of imatinib-bound inactive KIT kinase domain. The structure of FLT3 is quite similar to that of KIT, so this comparison helps us identify potential ligand interacting residues in FLT3. In the KIT structure, residue L595, K623, E640, L644, T670, Y672, L799 and F811 are close to the ligand. We thus define the corresponding residues in the FLT3 kinase domain (Y693, F830, F691, K644, E661, L818, L616, M665, V624) as flexible. Our docking studies revealed that the conformations of Y693, L818, L616, M665 and V624 in the docking solutions are largely identical to their conformations in the crystal structure, and so we did not consider these five residues to be flexible in the final calculations.
The coordinates of AC220 were generated using the Dundee PROGRD2 server33, and its initial conformation was energy minimized by the GROMACS force field. The Gasteiger charges were then assigned to the ligand using ADT. Seven torsion bonds were defined as rotable during the docking procedure. The ligand was put into the kinase ATP-binding pocket and manually aligned to avoid atom clashes. A three-dimensional grid box (dimensions: 60 × 30 × 60 unit in number of grid points, grid spacing: 0.375Å) centred at the ligand defining the search space was then created by AutoGrid4.2 (ref. 32). Two hundred runs of Larmarckian Genetic Algorithm were performed to optimize the ligand–protein interactions. The solutions were clustered according to the root mean standard deviation values, and ranked by the binding free energy. Two general poses are observed. The top-ranked pose has an average energy of –10.32 kcal mol–1 (the lowest energy for this pose is –10.93 kcal mol–1). The second-ranked pose, which is flipped by 180° with respect to the top-ranked position, has 133 solutions (63%) with an average energy of –5.82 kcal mol–1 (the lowest energy for this pose is –6.98 kcal mol–1). Given the gap in the calculated energy, we only picked the lowest one for the purely illustrative purposes of this analysis.
Supplementary Material
Acknowledgements
We thank K. Lin for technical assistance. This work was funded by grants from the Leukemia and Lymphoma Society (to C.C.S. and N.P.S.), the Doris Duke Charitable Foundation (to N.P.S.), NCI Leukemia SPORE P50 CA100632-06 (to M.J.L.), NCI R01 CA12886 (to M.J.L.) and the NIH T-32 Molecular Mechanisms of Cancer (to C.C.S.). C.C.S. would like to acknowledge the EHA/ASH Translational Research Training Institute. N.P.S. would like to thank Art and Alison Kern and the Edward S. Ageno family for their support.
Footnotes
Author Contributions C.C.S., Q.W., C.-S.C., K.J.T., A.K., E.E.S. and J.K. designed experiments, performed research, analysed data and wrote the manuscript. N.P.S. designed experiments, analysed data and wrote the manuscript. L.E.D., S.W., J.P.H. and S.S. performed experiments and reviewed the manuscript. P.P.Z. was involved in study design and reviewed the manuscript. A.E.P. and M.J.L. provided reagents, performed research and reviewed the manuscript.
Author Information SMRT sequencing data is deposited online at http://www.ncbi.nlm.nih.gov/sra under accession number SRA050226.1. Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Thiede C, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 2.Lee BH, et al. FLT3 internal tandem duplication mutations induce myeloproliferative or lymphoid disease in a transgenic mouse model. Oncogene. 2005;24:7882–7892. doi: 10.1038/sj.onc.1208933. [DOI] [PubMed] [Google Scholar]
- 3.Shih LY, et al. Acquisition of FLT3 or N-ras mutations is frequently associated with progression of myelodysplastic syndrome to acute myeloid leukemia. Leukemia. 2004;18:466–475. doi: 10.1038/sj.leu.2403274. [DOI] [PubMed] [Google Scholar]
- 4.Knapper S, et al. A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first-line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood. 2006;108:3262–3270. doi: 10.1182/blood-2006-04-015560. [DOI] [PubMed] [Google Scholar]
- 5.Fischer T, et al. Phase IIB trial of oral Midostaurin (PKC412), the FMS-like tyrosine kinase 3 receptor (FLT3) and multi-targeted kinase inhibitor, in patients with acute myeloid leukemia and high-risk myelodysplastic syndrome with either wild-type or mutated FLT3. J. Clin. Oncol. 2010;28:4339–4345. doi: 10.1200/JCO.2010.28.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarrinkar PP, et al. AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML). Blood. 2009;114:2984–2992. doi: 10.1182/blood-2009-05-222034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortes J, et al. 16th Congress of the European Hematology Association; Haematologica. 2011. [Google Scholar]
- 8.Heidel F, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107:293–300. doi: 10.1182/blood-2005-06-2469. [DOI] [PubMed] [Google Scholar]
- 9.Metzelder S, et al. Compassionate use of sorafenib in FLT3-ITD-positive acute myeloid leukemia: sustained regression before and after allogeneic stem cell transplantation. Blood. 2009;113:6567–6571. doi: 10.1182/blood-2009-03-208298. [DOI] [PubMed] [Google Scholar]
- 10.Scholl S, et al. Secondary resistance to sorafenib in two patients with acute myeloid leukemia (AML) harboring FLT3-ITD mutations. Ann. Hematol. 2011;90:473–475. doi: 10.1007/s00277-010-1027-9. [DOI] [PubMed] [Google Scholar]
- 11.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 12.Shah NP, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 13.Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 2010;38:e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith J, et al. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol. Cell. 2004;13:169–178. doi: 10.1016/s1097-2765(03)00505-7. [DOI] [PubMed] [Google Scholar]
- 15.Levinson NM, et al. A Src-like inactive conformation in the Abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mol CD, et al. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J. Biol. Chem. 2004;279:31655–31663. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- 17.Hubbard SR, Wei L, Ellis L, Hendrickson WA. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature. 1994;372:746–754. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- 18.Mol CD, et al. Structure of a c-Kit product complex reveals the basis for kinase transactivation. J. Biol. Chem. 2003;278:31461–31464. doi: 10.1074/jbc.C300186200. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wodicka LM, et al. Activation state-dependent binding of small molecule kinase inhibitors: structural insights from biochemistry. Chem. Biol. 2010;17:1241–1249. doi: 10.1016/j.chembiol.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 22.Wan PT, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 23.Choudhary C, et al. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- 24.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbie DA, Deangelo DJ. Systemic mastocytosis: current classification and novel therapeutic options. Clin. Adv. Hematol. Oncol. 2006;4:768–775. [PubMed] [Google Scholar]
- 26.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 27.Kralovics R, et al. Acquisition of the V617F mutation of JAK2 is a late genetic event in a subset of patients with myeloproliferative disorders. Blood. 2006;108:1377–1380. doi: 10.1182/blood-2005-11-009605. [DOI] [PubMed] [Google Scholar]
- 28.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yates F. Tests of significance for 2 × 2 contingency tables. J. R. Stat. Soc. A. 1984;147:426–463. [Google Scholar]
- 31.Barnard GA. Must clinical trials be large? The interpretation of p-values and the combination of test results. Stat. Med. 1990;9:601–614. doi: 10.1002/sim.4780090606. [DOI] [PubMed] [Google Scholar]
- 32.Morris GM, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schüttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.