Abstract
A loop-mediated isothermal amplification (LAMP) method for rapid detection of various Staphylococcus strains and associated antibiotic resistance determinant had been developed and evaluated in this study. Six primers, including outer primers, inner primers and loop primers, were specially designed for recognizing eight distinct sequences on three targets: 16SrRNA, femA and mecA.. Forty-one reference strains, including various species of gram-negative and -positive isolates, were included in this study to evaluate and optimize LAMP assays. The optimal reaction condition was found to be 65 °C for 45 min, with detection limits at 100 fg DNA/tube and 10 CFU/reaction for 16S rRNA, 100 fg DNA/tube and 10 CFU/reaction for femA, 1 pg DNA/tube and 100 CFU/reaction for mecA, respectively. Application of LAMP assays were performed on 118 various types of Staphylococcus isolates, the detection rate of LAMP assays for the 16SrRNA, femA and mecA was 100% (118/118), 98.5% (64/65) and 94.3% (66/70), and the negative predictive value (NPV) was 100%, 98.1% and 92.3% respectively; with a 100% positive predictive value (PPV) for all three targets. In conclusion, LAMP assays were demonstrated to be useful and powerful tools for rapid detection of various Staphylococcus strains, and undoubtedly, the rapidness, technical simplicity, and cost-effectiveness of LAMP assays will demonstrate broad application for bacteriological detection of food-borne Methicillin-resistant Staphylococcus (MRS) isolates.
Keywords: Loop-mediated isothermal amplification, Rapid detection, Food-borne Staphylococci, MRSA, MRCNS
1. Introduction
Food-borne infections and diseases still remain one of the greatest concerns in public health and food safety, which are caused by a large variety of pathogens that contaminate food and food products. Food-borne pathogens cause 14 million illnesses, 60,000 hospitalizations, and 1,800 deaths annually; staphylococcal food poisoning serves as one of the most economically important food-borne diseases and is a major issue for public health programs worldwide (Alarcon et al, 2006; Shimizu et al, 2000). During 1983–1997, the annual number of staphylococcal food poisoning cases in the USA had been estimated to be 185,000, with 1,750 hospitalizations and 2 deaths, totaling a cost of 1.5 billion dollars. From 1993 to 1998, 926 outbreaks were reported in 15 European Union (EU) countries (Smyth et al, 2006). In Japan, according to the Ministry of Health and Welfare of Japan, during a period of 20 years (1980–1999), a total of 2,525 outbreaks of staphylococcal food poisoning were reported, which involved 59,964 persons, resulting in three deaths (Alarcon et al, 2006). In China, raw meat, milk and dairy products, frozen products and cooked foods have been found as major food types contaminated by Staphylococcus aureus, taking up 38%, 20%, 16% and 14%, respectively. For many regions in China, S. aureus was recovered from more than 15% of food samples (data from various local reports), and in some special outbreak cases, the identification rate approached 90%. However, despite the lack of comprehensive surveillance and investigation, the prevalence and occurrence of staphylococcal food poisoning varied considerably among different regions and areas, this discrepancy may be explained by different local eating habits and food product usage. In addition, staphylococcal strain difference may also contribute to the variation. Some S. aureus strains are capable of producing enterotoxins (SEs) after ingestion which may cause intoxication as vomiting and diarrhoea. Several coagulase negative staphylococcal (CNS) species other than S. aureus reportedly produce SEs, including S. epidermis, S. haemolyticus, etc. (Bautista et al, 1988; Becker et al, 2001; Jay, 1992). Though S. aureus contamination can be readily avoided by heat treatment of food, its ability to grow in a wide range of temperatures (7 to 48.5°C), pH (4.2 to 9.3) and sodium chloride concentrations (up to 15% NaCl) facilitates the contamination and transmission of this organism to various types of foods (Alarcon et al, 2006; Bergdoll, 1983; Schmitt et al, 1990; Shimizu et al, 2000).
Indiscriminate use of existing antibiotics contributes to proliferation of antibiotic resistance and poses a dilemma for the treatment of several bacterial infections, including therapy for individuals with food poisoning. Antibiotic resistance in microbes still remains one of the global major concerns in public health, with methicillin-resistant staphylococci (MRS) strains representing one important group, commonly reffered to as “Super Bugs” (Xu et al, 2011). Since their first discovery in 1961, MRS (including methicillin-resistant S. aureus, MRSA; and methicillin-resistant coagulase-negative staphylococci, MRCNS) has become one group of the most prevalent pathogens causing nosocomial infections throughout the world (Alarcon et al, 2006; Shimizu, 2000; Smyth et al, 2006; Xu et al, 2008a, b). MRS strains show resistance to nearly all β-lactam antibiotics and commonly multiple other drugs due to the mecA and other resistance genes carried by a mobile genetic element, designated staphylococcal cassette chromosome mec (SCCmec).
First appearing as almost exclusive nosocomial pathogens and now emerging in community facilities in the past two decades, MRS strains were reported with low incidence in food poisoning. However, since the first transmission of MRSA by food contamination (Kluytmans et al, 1995), MRS had been identified from contamination of various food samples such as milk, pork, chicken, veal, beef, turkey and lamb meat (Andreoletti et al, 2008; De Boer et al, 2009; Kwon et al, 2005), as well as in food production animals such as cattle, chickens, pigs and cows (Khanna et al, 2008; Lee, 2006; Van der Haeghen et al, 2010). With the first report of a MRSA-mediated gastrointestinal illness outbreak (Jones et al, 2002), MRS strains have been considered a major contributor to both health-care associated and food-borne illnesses. Carriage of MRS strains in a wide variety of food and food production animals may not be limited solely to food hazard, but also poses a significant occupational risk for the industrial staff, such as handlers, asymptomatic carriers, and uncolonized individuals.
The concentration of S. aureus necessary to cause food poisoning ranges from 106 to 108 CFU/g in food samples, and for sensitive persons even 105 CFU/g of staphylococcal bacteria are capable of producing enough SEs (around 1 μg) to generate symptoms (Alarcon et al, 2006; Johnson et al, 1990). Though most official regulations strictly require the absence of S. aureus for ready-to-eat foods and a low level of tolerance of 102–103 CFU/g for raw products, examples of low level contamination by S. aureus have still been allowed in a large variety of foodstuffs in many countries, with an instance of 103 CFU/g in raw milk cheeses in France (Alarcon et al, 2006; Le Loir et al, 2003; Shimizu et al, 2000). Routine culture-based diagnostic detection and identification procedure for potentially pathogenic Staphylococcus strains includes: enrichment and enumeration in liquid media, subsequent recovery and isolation of colonies on selective culture broth such as Baird-Parker agar for 24–48 h at 37°C, followed by DNase or coagulase assays for suspicious colonies and further confirmation by biochemical tests. However, the lengthy recovery time to identify microbes at the species level (6 days), false negative results due to bacterial starvation and physical stress, as well as insufficient sensitivity of around 102 CFU/g for solid foodstuffs and 10 CFU/g for liquid samples, have raised concerns for these conventional methodologies (Alarcon et al, 2006). During the past decades, a number of polymerase chain reaction (PCR) and real-time quantitative PCR (RQ-PCR) based assays have been employed and proposed for rapid detection of food-borne pathogens (Alarcon et al, 2006; Brakstad et al, 1992; Hein et al, 2001; Palomares et al, 2003; Saruta et al, 1997; Straub et al, 1999; Shimizu et al, 2000; Wilson et al, 1991). However, disadvantages for PCR (time consumption for post determination, high risk of cross contamination and low detection limit levels) and real-time PCR (requirement for trained personnel, operating space, expensive equipment and reagents) posed significant obstacles for their broad application.
Increased awareness for the risk and hazard of food-borne MRS strains and demands for tests capable of early, cost-effective, timely, and sensitive detection of staphylococci and associated antibiotic resistance determinants; the global transport of food and food products has made these tests an urgent necessity. Recently, a novel nucleic acid amplification method, designated loop-mediated isothermal amplification (LAMP), had been well established and documented (Mori et al, 2001; Nagamine et al, 2001; Notomi et al, 2000). This method relies on an auto-cycling strand displacement DNA synthesis performed by the Bst DNA polymerase large fragment, with four or six primers recognizing 6–8 distinct regions of the target gene and generating the loop-mediated amplification under isothermal conditions between 60–65°C (Fig. 1). Amplicons are mixtures of many different sizes of stem-loop DNAs containing several inverted repeats of the target sequence and cauliflower-like structures with multiple loops (Wang et al, 2008; Zhao et al, 2010a). LAMP may serve as a potentially valuable tool for rapid diagnosis of food-borne pathogens. This study aimed at developing and evaluating simple and rapid testing methods based on LAMP assays for differentiation of various MRS strains and related antibiotic resistance determinant, and applying these assays to detection of a large scale of staphylococci strains from various samples.
Fig. 1.
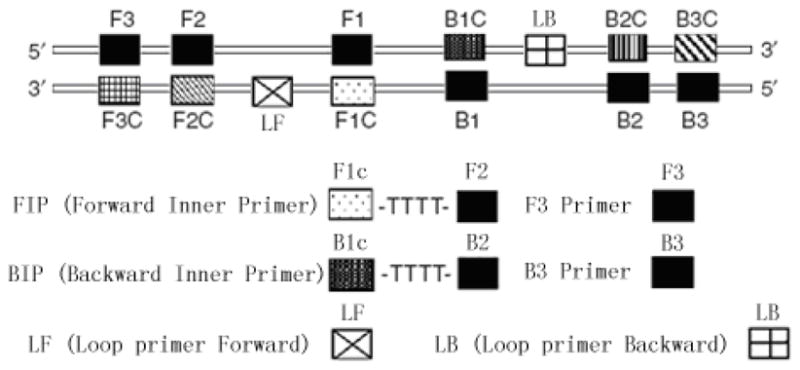
Schematic diagram of primers used in the LAMP assay.
2. Materials and methods
2.1. Bacterial strains
Forty-one reference strains, including various species of gram-positive and -negative isolates, were included in this study to evaluate and optimize the specificity and sensitivity of LAMP assays (Table 1). Optimized LAMP assays were performed on a total of 118 various types of Staphylococcus isolates, including 40 MRSA, 25 methicillin-sensitive Staphylococcus aureus (MSSA), 30 MRCNS and 23 methicillin-sensitive coagulase negative staphylococci (MSCNS) strains. LAMP results were comparatively validated by standard PCR amplification assays. These strains were isolated from various clinical samples, which had been preliminarily identified in the Lab of Clinical Microbiology, Zhongshan Supervision Testing Institute of Quality & Metrology.
Table 1.
Reference strains used in this study
| Reference strains | No. of samples | LAMP Assaysa
|
||
|---|---|---|---|---|
| 16s rRNA | femA | mecA | ||
| Gram-positive organisms | ||||
| Methicillin-resistant S. aureus 10442, COL, N315, 85/2082, CA05, JCSC 1978, JCSC 4469, MR108, M03-68, WIS | 10 | + | + | + |
| Methicillin-sensitive S. aureus ATCC 25923, ATCC 29213 | 2 | + | + | − |
| Methicillin-resistant coagulase-negative staphylococci: | ||||
| S. epidermidis ATCC 29887, ATCC 700586, 042219 | ||||
| S. haemolyticus 022407, 042403 | 8 | + | − | + |
| S. hominis 032315, 042306 | ||||
| S. warneri 012501 | ||||
| Methicillin-sensitive coagulase-negative staphylococci: | ||||
| S. epidermidis ATCC 12228, 012217 | ||||
| S. haemolyticus 022408 | 5 | + | − | − |
| S. hominis 012307 | ||||
| S. warneri 012502 | ||||
| Enterococcus faecalis GH152, GH299 | 2 | − | − | − |
| Stretococcus pyogenes GH126 | 1 | − | − | − |
| Stretococcus mitis GH185 | 1 | − | − | − |
| Stretococcus pnuemoniae GH165 | 1 | − | − | − |
| Stretococcus hemolyticus GH177 | 1 | − | − | − |
| Gram-negative organisms | ||||
| Escherichia coli ATCC 25922, C600 | 2 | − | − | − |
| Psuedomonas aeruginosa ATCC 27853 | 1 | − | − | − |
| Salmonella typhimurium ATCC 14028 | 1 | − | − | − |
| Salmonella choleraesuis ATCC 13312 | 1 | − | − | − |
| Klebsiella pneumoniae ATCC 13883 | 1 | − | − | − |
| Vibrio cholerae SK10 | 1 | − | − | − |
| Vibrio parahaemolyticus ATCC 17802 | 1 | − | − | − |
| Enterobacter cloacae ATCC 23355 | 1 | − | − | − |
| Acinetobacter baumannii GH31 | 1 | − | − | − |
| Total | 41 | |||
+, positive; −, negative.
2.2. Primer design
Three targets were selected to differentiate MRSA MSSA MRCNS MSCNS and non-Staphylococci strains. The protocol was designed to (i) detect any staphylococcal species to the exclusion of other bacterial pathogens using as an internal control, with primers corresponding to Staphylococcus-specific regions of the 16S rRNA genes; (ii) distinguish between S. aureus and CNS strains based on amplification of the S. aureus specific femA gene and (iii) provide an indication of the likelihood that the staphylococci present in the specimen are resistant to methicillin based on amplification of the mecA gene. For each target gene, a set of inner primers (including forward and backward inner primers), outer primers (including F3 and B3) and loop primers (including LF and LB, to accelerate reaction) were specially designed for LAMP reaction to target 8 distinct regions (Table 2). Forward inner primer (FIP)/backward inner primer (BIP) consisted of the complementary sequence of F1 (F1c)/B1 (B1c), a T–T–T–T linker and F2/B2; and outer primers F3 and B3 located outside of the F2 and B2 regions, respectively. Loop primers LF and LB were located between F2 and F1 or B1 and B2, which were designed to anneal at the loop structure of the amplicons and accelerate and enhance the sensitivity (Mori & Notomi, 2009; Nagamine et al, 2002). The primers had been designed using PrimerExplorer (PrimerExplorer, Eiken Chemical Co. Ltd.).
Table 2.
List of oligonucleotide primer sequences
| Target | Sequence (5′ to 3′) | Size | Position |
|---|---|---|---|
| 16s rRNA | |||
| F3 | CGTGGGGATCAAACAGGATT | 20 | 778–797 |
| B3 | CATGCTCCACCGCTTGTG | 18 | 943–960 |
| FIP | TAGCTGCAGCACTAAGGGGC-CCACGCCGTAAACGATGAG | 39 | 813–831, 854–873 |
| BIP | ACGCATTAAGCACTCCGCCT-GGGTCCCCGTCAATTCCT | 38 | 874–893, 924–941 |
| LF | GGAAACCCCCTAACACT | 17 | 837–853 |
| LB | GGGGAGTACGACCGCAAGGT | 20 | 894–913 |
| femA | |||
| F3 | ATGCTGGTGGTACATCAA | 18 | 1022–1039 |
| B3 | TGGTTTAATAAAGTCACCAACAT | 23 | 1217–1239 |
| FIP | GGTCAATGCCATGATTTAATGCATA-GCATTCCGTCATTTTGCC | 43 | 1042–1059, 1093–1117 |
| BIP | CAGAAGATGCTGAAGATGCTGG-TCAATAATTTCAGCATTGTAACC | 45 | 1151–1172, 1192–1214 |
| LF | AATCATTTCCCATTGCACT | 22 | 1068–1089 |
| LB | TGTAGTTAAATTCAA | 15 | 1173–1187 |
| mecA | |||
| F3 | AAGATGGCAAAGATATTCAACT | 22 | 956–977 |
| B3 | AGGTTCTTTTTTATCTTCGGTTA | 23 | 1148–1170 |
| FIP | GTGGATAGCAGTACCTGAGCC-TTGATGCTAAAGTTCAAAAGAGT | 44 | 983–1005, 1033–1053 |
| BIP | CCTCAAACAGGTGAATTATTAGCAC-CTTCGTTACTCATGCCATAC | 45 | 1054–1078, 1116–1135 |
| LF | TAATCATTTTTCATGTTG | 18 | 1014–1031 |
| LB | TGTAAGCACACCTTCATATGACGT | 24 | 1080–1103 |
2.3. Establishment of Lamp assays
Forty-one reference strains were used to develop and evaluate the specificity and sensitivity of LAMP assays. Cultural conditions and DNA extraction of Gram-positive and –negative strains were performed as described previously (Xu et al, 2007; Xu et al, 2008a; Xu et al, 2009; Xu et al, 2010). The detection limits of LAMP and PCR assays were ascertained by both minimal CFU of bacteria and template DNA amount. In brief, overnight cultures and template DNA from MRSA strain 85/2082 were serially diluted 10-fold with sterile water, ranging from 102 to 108 CFU/ml and 10−14 to 10−7 g DNA, respectively. A negative control was performed using sterile water instead of culture or DNA template. LAMP assays were carried out in a total of 25 μl reaction mixture containing 1.6 μM (each) of the primers FIP and BIP, 0.2 μM (each) of the primers F3 and B3, 0.8 μM (each) of primers LF and LB, 1.6 mM of deoxynucleoside triphosphates, 6 mM MgSO4, 1 M betain (Sigma, St. Louis, MO, USA), 1 X thermopol buffer (New England Biolabs, Ipswich, MA, USA), and the specified amounts of target genomic DNA. The reaction was heated at 95°C for 3 min, then chilled on ice, 1 μl (8 U) of Bst DNA polymerase (New England Biolabs, Ipswich, MA, USA) was added, after incubation at 65°C for 45 min, the reaction was terminated by heating at 80°C for 2 min. PCR amplification was carried out in a 50 μl reaction volume, using the two outer primers F3 and B3. The thermal profile for PCR was 94°C for 5 min, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min and a final extension cycle at 72°C for 7 min. The amplified products (5 μl/well) were analyzed by gel electrophoresis in 2% agarose gels and stained with ethidium bromide for 10 min. For LAMP assays, the lowest bands from amplicons were purified using the QIA quick PCR purification kit (QIAGEN, Hilden, Germany) and cloned into the pMD18-T vector (TaKaRa, Shiga, Japan), followed by sequencing performed by an ABI PRISM 310 genetic analyzer (PE Biosystems, Foster City, CA, USA).
2.4. Application of LAMP assays on clinical isolates
One hundred and eighteen clinical staphylococci isolates were subjected to detection by LAMP and PCR assays as aforementioned. Template DNA was prepared through a rapid procedure as described. Overnight Luria-Bertani (LB) broth cultures were diluted 10-fold in 10 mM Tris-HCl (pH 8.0) containing 1 mM EDTA and the suspension was boiled for 15 min and kept on ice. After centrifugation at 12,000 g for 2 min, the resulting supernatant was used as templates for LAMP and PCR assays. Heating and isothermal amplification were performed on a heating block and water bath. Positive LAMP reactions were measured by several qualitative criteria. White magnesium pyrophosphate precipitates, generated during the strand displacement auto-cycling reaction, could be seen at the bottom of microfuge tubes. In addition, staining with a 1/10 dilution of SYBR Green I allowed for the visualization of a positive reaction both colorimetrically by the naked eye as well as imaging under an ultraviolet (UV) light source; PCR amplicons were evaluated by electrophoresis as mentioned above. These experiments were replicated to ensure reproducibility..
3. Results
3.1. Optimization of the conditions of LAMP assays
In order to determine the optimal conditions of LAMP, DNA from MRSA 85/2082 was used as target template. The specific amplification generated many ladder-like pattern bands on agarose gel due to its characteristic secondary structure, with sizes ranging from 183 bp for 16S rRNA, 218 bp for femA and 215 bp for mecA respectively. Sequences of the bottom bands were identical to those PCR amplified with F3 and B3. No significant difference was observed when LAMP assays were performed under isothermal condition between 60°C and 65°C. However, the LAMP product amplified at 65°C showed higher levels of resolution of DNA when compared to other temperatures (data not shown), which was consistent with previous studies (Wang et al, 2008; Zhao et al, 2011). Reaction lengths of LAMP assays were varied between 15 min, 30 min, 45 min, 60 min, 75 min, 90 min, 105 min and 120 min, under 65°C, with 10 ng template DNA. Without loop primers, amplification products could not be observed until 90 min. With loop primers, the amplification was initially detected at 30 min, and reached maximal detection levels at 45 min. LAMP assays were performed with omission of one or two of the primers, under 65°C for 45 min. However, no amplification could be detected in the absence of each of FIP, BIP, F3 or B3 primers. Since each of the primers plays an indispensable role in auto-cycling strand displacement reaction by forming the loop out structure, LAMP assays were only successful in the existence of both inner and outer primers.
3.2. Sensitivity and specificity of LAMP assays
The sensitivity of LAMP and PCR assays were ascertained by both minimal CFU of bacteria and template DNA amount. The detection limits of LAMP assays were found to be 100 fg DNA/tube and 10 CFU/reaction (LAMP was positive for sample containing 104 CFU/ml, with 1 μl was included in the reaction system) for 16S rRNA, 100 fg DNA/tube and 10 CFU/reaction (LAMP was positive for sample containing 104 CFU/ml, with 1 μl was included in the reaction system) for femA, 1 pg DNA/tube and 100 CFU/reaction (LAMP was positive for sample containing 105 CFU/ml, with 1 μl was included in the reaction system) for mecA. PCR sensitivity was determined to be 100 pg DNA/tube and 103 CFU/reaction for 16S rRNA, femA and mecA, indicating that LAMP was at least 10-fold more sensitive than PCR (Fig. 2 & 3). To determine specificity of the primers, LAMP assays were also subjected to 31 gram-positive and 10 gram-negative isolates. No false positive amplification was observed, indicating the high specificity of the established LAMP assays (data not shown).
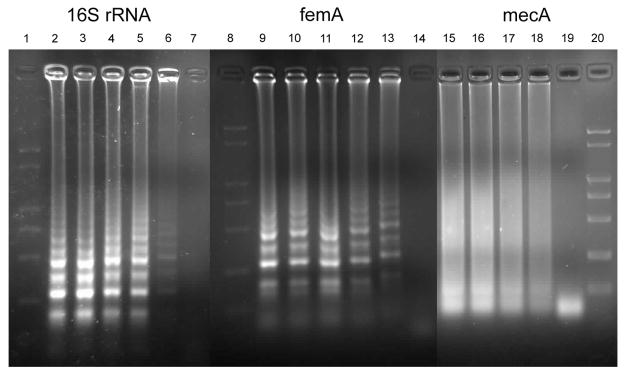
Fig. 2.
Sensitivity of LAMP assays for detection of MRSA 85/2082: lane 1–7, 8–14, 15–20 referring to LAMP assays of 16S rRNA, mecA and femA respectively. Lane 1, 8, 20: DNA Marker; lane 2, 9, 15: 1 ng template DNA; lane 3, 10, 16: 100 pg template DNA; lane 4, 11, 17: 10 pg DNA; lane 5, 12, 18: 1 pg DNA; lane 6, 13, 19: 100 fg DNA; lane 7, 14: 10 fg DNA.
Fig. 3.
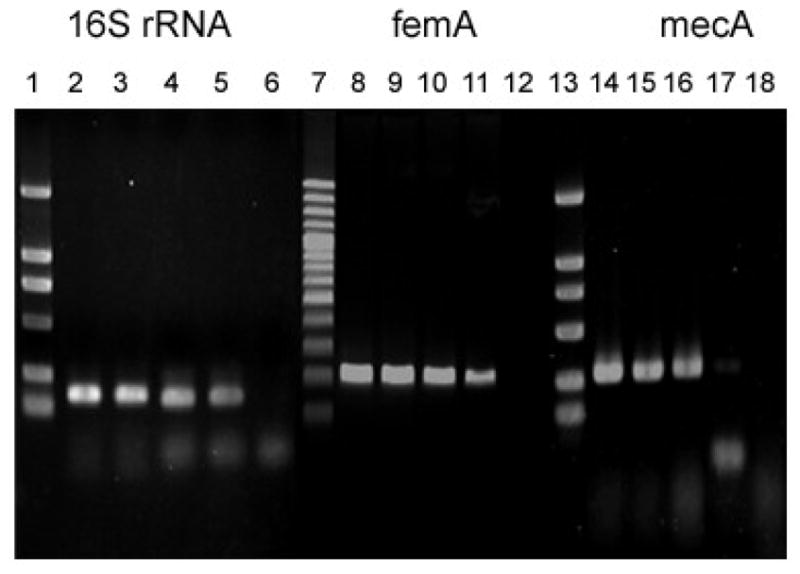
Sensitivity of PCR assays for detection of MRSA 85/2082: lane 1–6, 7–12, 13–18 referring to PCR assays of 16S rRNA, mecA and femA respectively. Lane 1, 7, 13: DNA Marker; lane 2, 8, 14: 100 ng template DNA; lane 3, 9, 15: 10 ng template DNA; lane 4, 10, 16: 1 ng DNA; lane 5, 11, 17: 100 pg DNA; lane 6, 12, 18: 10 pg DNA.
3.3. Application of LAMP assays on clinical isolates
The established LAMP assays were applied to detect 118 various types of Staphylococcus isolates using simple equipment and observed directly by naked eye and under UV light (Fig. 4–7). Of these strains, 118, 64 and 66 strains were detected to be positive for 16SrRNA, femA and mecA by LAMP respectively, where 113, 60 and 61 were detected by PCR (Table 3). The detection rate of LAMP assays for the 16SrRNA, femA and mecA was 100% (118/118), 98.5% (64/65) and 94.3% (66/70), versus 95.8% (113/118), 92.3% (60/65) and 87.1% (61/70) for PCR assays, respectively. The negative predictive value (NPV) of LAMP assays for femA and mecA was 98.1% and 92.3%, with 91.4% and 84.2% for PCR, respectively. The positive predictive value (PPV) was detected to be 100% for both LAMP and PCR, with no false positive had been observed. In comparison with conventional PCR methods, LAMP yielded better detection rate and NPV, while high PPV were obtained by both assays.
Fig. 4.
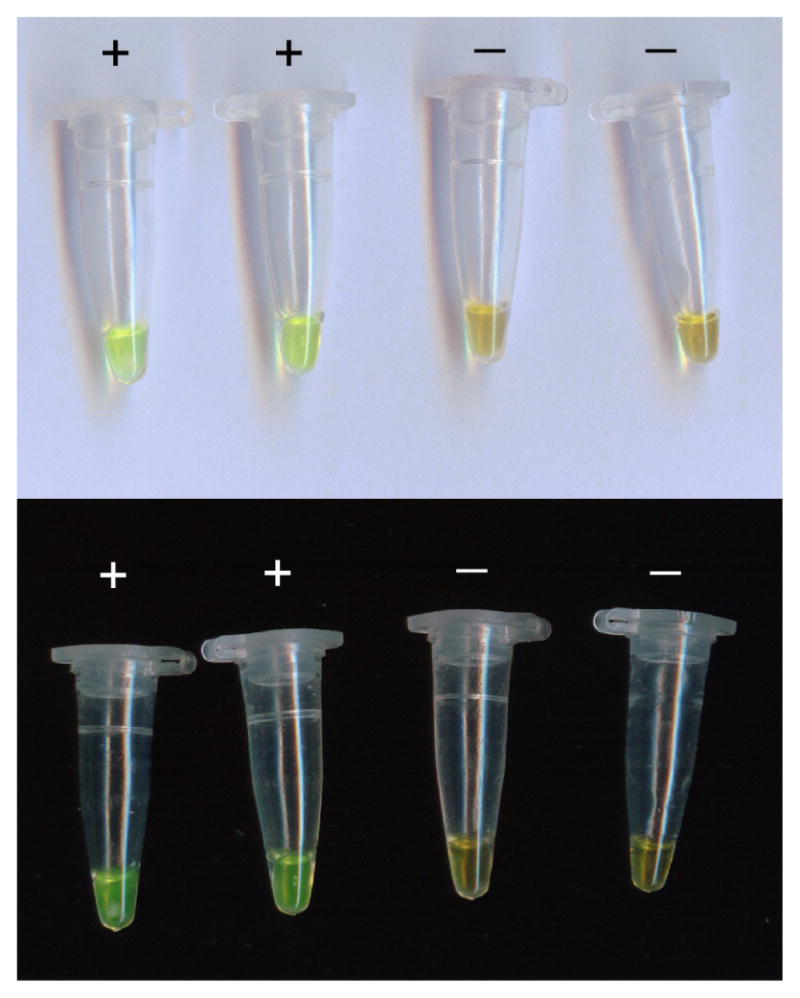
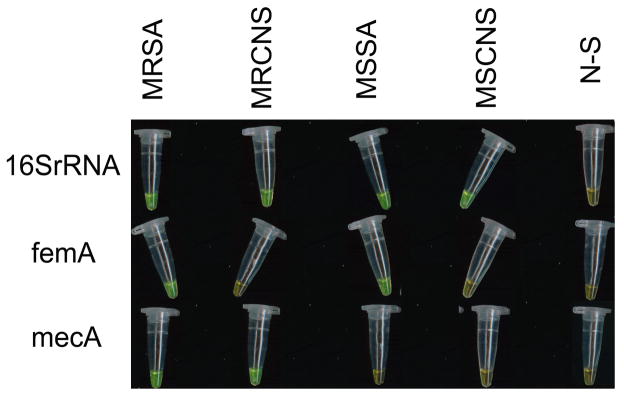
Amplification of LAMP products dyed with SYBR Green I were visually detected by oexamining color changes with the naked eye: green indicates a positive result and orange indicates a negative result.
Fig. 7.
Results determination through observation at the color change by naked eye when LAMP assays were employed to detect various Staphylococcus strains.
Table 3.
Results of application of LAMP assays on clinical MRSA isolates.
| Clinical isolates | LAMP VS PCR | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| MRSA | MSSA | MRCNS | MSCNS | Detection rate | False positive | PPV | NPV | |
| No. of isolates | 40 | 25 | 30 | 23 | ||||
| 16S rRNA | 40/38 | 25/25 | 30/28 | 23/22 | 100%/95.8% | 0/0 | 100%/100% | −/− |
| femA | 40/37 | 24/23 | 0/0 | 0/0 | 98.5%/92.3% | 0/0 | 100%/100% | 98.1%//91.4% |
| mecA | 38/35 | 0/0 | 28/26 | 0/0 | 94.3%/87.1% | 0/0 | 100%/100% | 92.3%//84.2% |
−, not applicable.
4. Discussion
Staphylococcus are currently widespread food-borne pathogens throughout the world and have prompted a heightened interest and concern for the low level detection of these food-borne pathogens as well as its related antibiotic resistance determinant. Thus, rapid and accurate detection approaches are needed for evaluation of foodstuffs to reduce risk of the associated food poisoning caused by infection and contamination with food-borne pathogens. In evaluating detection methodologies for ecologic and epidemiological purposes, a series of attributes should be considered and assessed, including specificity, sensitivity, simplicity, expense and time. As a novel nucleic acid amplification method, LAMP has been known as a rapid, specific, sensitive, cost-effective and easy-operating alternative for diagnosis of food-borne pathogens. Until recently, a number of isothermal amplification techniques, such as nucleic acid sequence-based amplification (NASBA) and the self-sustained sequence reaction (SSR), were reported to be less specific due to their low reaction stringency (40°C). Furthermore, these technologies require either a precision instrument for amplification or an elaborate method for detection of the amplified products.. With inner and outer primers recognizing six distinct regions, higher specificity was achieved with LAMP as compared to more conventional PCR-based methods. In the present study, high specificity had been illustrated by no false positive observations for reference strains and 100% PPV during application. Due to its powerful amplification efficiency, LAMP had been characterized by high sensitivity and low detection limits. In this present study, sensitivities of LAMP assays were found to be 10, 10 and 100 CFU/reaction for 16S rRNA, femA and mecA, showing significant advantages compared with previous LAMP detection on Staphylococcus (Misawa et al, 2007), in which 1000 and 100 copied were required for detection of spa and mecA. Despite a frequently reported loss in sensitivity when applied to artificially inoculated food (Alarcon et al, 2006; Brakstad et al, 1992), the detection limits of regular PCR and RQ-PCR ranged from 103–105 CFU/ml and 10-102 CFU/ml, respectively (Alarcon et al, 2006; Brakstad et al, 1992; Hein et al, 2001; Ramesh et al, 2002; Shimizu et al, 2000). In the current study, detection limits were found to be 10–102 CFU/reaction, demonstrating 10–1000 times more sensitive than conventional PCR, which may acceptably fulfill the requirement of low level detection of pathogenic MRS strains. Aside from stringency, a major concern for the food industry is rapidness of testing protocols. The total detection time, including DNA preparation, LAMP reaction and results determination, was approximately 70 min, while conventional PCR methods require nearly 2 hours for amplification reactions alone. Moreover, in the preliminary Staphylococcus LAMP detection (Misawa et al, 2007), 2 hours was needed for incubation, with template preparation and result determination excluded. Additionally, routine PCR and RQ-PCR require comparatively strict template DNA preparation, otherwise sensitivity could be compromised by PCR inhibitors present in biological samples (Horisaka et al, 2004). Since LAMP exhibits robustness in the presence of inhibitory substances, it may be less affected by various components than PCR (Tomita et al, 2008). Therefore, LAMP-based methodologies are advantageous due to the rapid and relatively simple template DNA purification process (Kaneko et al, 2007; Mori & Notomi, 2009). In previous studies, no amplification or low sensitivity (as low as 1.7 × 105 CFU/ml) for PCR assays had been reported when template DNA was not purified, which had been raised up to 27–180 CFU/ml following 2 hour incubation (Alarcon et al, 2006; Pitcher et al, 1989). However, the simple template preparation process in this study takes only 20 min and had been previously proved applicable in various food samples in preliminary studies (Zhao et al, 2010a, b, c; Zhao et al, 2011); therefore the current LAMP assays should be applicable for pathogen detection in real food samples. As aforementioned, laborious demand, expensive reagents and equipment restrict the broad application of RQ-PCR to clinical routine laboratories. However, with the reaction performed under isothermal conditions without a thermal cycler, only simple equipment like a heat block and water baths were needed for the operation of LAMP assays at low expense. As comparison, real-time turbidimeter are commonly required in previous publications on LAMP detection (Misawa et al, 2007). Therefore, simplicity and clarity of the endpoint result determination makes the LAMP assay a simple, rapid, and cost-effective alternative to current rapid detection methodologies for the detection of food-borne pathogens…
5. Conclusion
In conclusion, LAMP assays were demonstrated to be useful and powerful tools for the rapid detection of various Staphylococcus strains, with advantages on the extension and flexibility in application to either separate detection of staphylococci, S. aureus and methicillin-resistance or combined use for differentiation of MRSA, MSSA, MRCNS, MSCNS and non-staphylococci. This technology may serve as a valuable tool for the rapid diagnosis of food-borne pathogens in both commercial and clinical fields, especially for resource-limited laboratories in rural regions (Mori & Notomi, 2009). Undoubtedly, the rapidness, easiness and cost-effectiveness of LAMP assays will aid in the broad application of bacteriological detection of food-borne MRS isolates. Nevertheless, implementation of LAMP assays to routine clinical laboratory diagnoses requires accumulation of data on food samples and the validation and connection with current procedures (Alarcon et al, 2006). Further investigation should focus on the application on artificially or naturally contaminated food samples and comparative sensitivity and specificity with current culture- or PCR-based approaches.
Fig. 5.
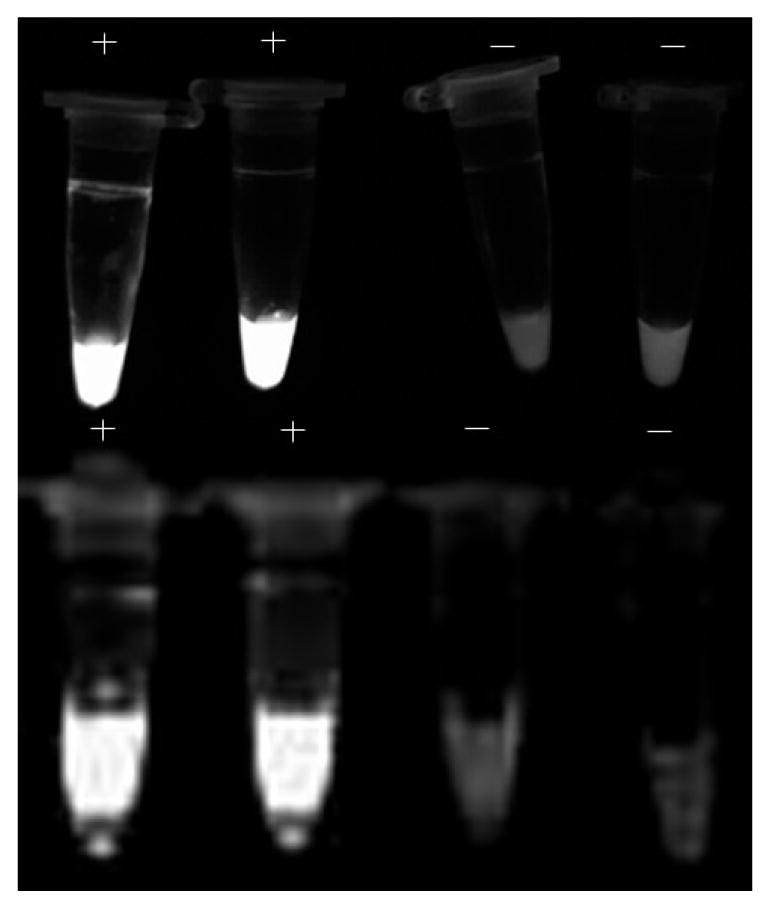
Amplification of LAMP products dyed with SYBR Green I were visually detected by fluorescence under UV light: intensely bright ones are determined to be positive and those lacking appreciable fluorescence are deemed negative.
Fig. 6.
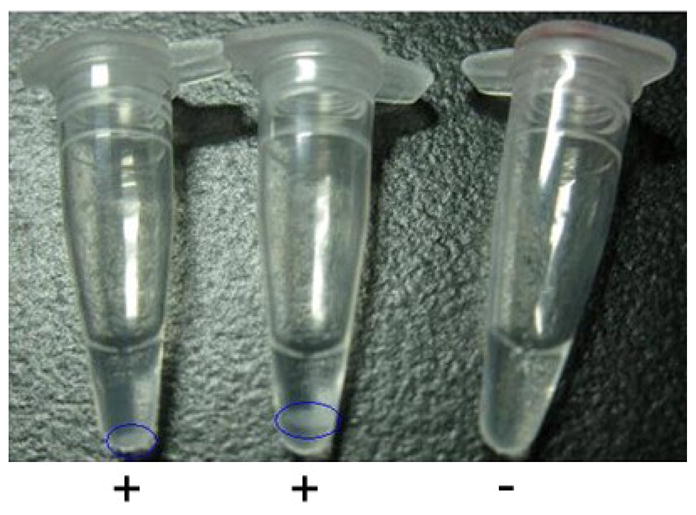
Amplification of LAMP products were visually determined by visually detecting turbidity derived from the white precipitate of magnesium pyrophosphate generated by the strand displacement auto-cycling reaction.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health grant (R01 AI69568-01A2), Science Foundation, Ministry of Education of China (706046), National Natural Science Foundation of China (20877028), National Science Foundation for Distinguished Young Scholars of China (31000781), State Scholarship Fund of China Scholarship Council (2008615044) and the Doctorate Foundation of South China University of Technology (2009–2010).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreoletti O, Budka H, Buncic S, Colin P, Collins JD, De Koeijer A, Griffin J, Havelaar A, Hope J, Klein G, Kruse H, Magnino S, López AM, McLauchlin J, Nguyen-Thé C, Noeckler K, Noerrung B, Maradona MP, Roberts T, Vagsholm I, Vanopdenbosch E. Foodborne antimicrobial resistance as a biological hazard. The EFSA Journal. 2008;765:2–87. [Google Scholar]
- Alarcon B, Vicedo B, Aznar R. PCR-based procedures for detection and quantification of Staphylococcus aureus and their application in food. Journal of Applied Microbiology. 2006;100:352–364. doi: 10.1111/j.1365-2672.2005.02768.x. [DOI] [PubMed] [Google Scholar]
- Bautista L, Gaya P, Medina M, Nunez M. A quantitative study of enterotoxin production by sheep milk staphylococci. Applied and Environmental Microbiology. 1988;54:566–569. doi: 10.1128/aem.54.2.566-569.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Keller B, Von Eiff C, Bruck M, Lubritz G, Etienne J, Peters G. Enterotoxigenic Potential of Staphylococcus intermedius. Applied and Environmental Microbiology. 2001;67:5551–5557. doi: 10.1128/AEM.67.12.5551-5557.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergdoll MS. Staphylococci and Staphylococcal Infections. London: Academic Press; 1983. Enterotoxins; pp. 559–598. [Google Scholar]
- Brakstad OG, Aasbakk K, Maeland JA. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. Journal of Clinical Microbiology. 1992;30:1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer E, Zwartkruis-Nahuis JTM, Wit B, Huijsdens XW, De Neeling AJ, Bosch T, Van Oosterom RAA, Vila A, Heuvelink AE. Prevalence of methicillin-resistant Staphylococcus aureus in meat. International Journal of Food Microbiology. 2009;134:52–56. doi: 10.1016/j.ijfoodmicro.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Hein I, Lehner A, Rieck P, Klein K, Brandl E, Wagner M. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Applied and Environmental Microbiology. 2001;67:3122–3126. doi: 10.1128/AEM.67.7.3122-3126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horisaka T, Fujita K, Iwata T. Sensitive and specific detection of Yersinia pseudotuberculosis by loop-mediated isothermal amplification. Journal of Clinical Microbiology. 2004;42:5349–5352. doi: 10.1128/JCM.42.11.5349-5352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay JM. Microbiological food safety. Critical Reviews in Food Science and Nutrition. 1992;31:1549–7852. doi: 10.1080/10408399209527567. [DOI] [PubMed] [Google Scholar]
- Jones TF, Kellum ME, Porter SS, Bell M, Schaffner W. An outbreak of community-acquired foodborne illness caused by methicillin-resistant. Staphylococcus aureus Emerging Infectious Diseases. 2002;8:82–84. doi: 10.3201/eid0801.010174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EA, Nelson JH, Johnson M. Microbiological safety of cheese made from heat-treated milk. Journal of Food Protection. 1990;53:519–540. doi: 10.4315/0362-028X-53.6.519. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Kawana T, Fukushima E, Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. Journal of Biochemical and Biophysical Methods. 2007;70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Khanna T, Friendship R, Dewey C, Weese JS. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Veterinary Microbiology. 2008;128:298–303. doi: 10.1016/j.vetmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Kluytmans J, Van Leeuwen W, Goessens W, Hollis R, Messer S, Herwaldt L, Bruining H, Heck M, Rost J, Van Leeuwen N, Van Belkum A, Vervrugh H. Food-initiated outbreak of methicillin-resistant Staphylococcus aureus analyzed by Pheno- and Genotyping. Journal of Clinical Microbiology. 33:1121–1128. doi: 10.1128/jcm.33.5.1121-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon NH, Park KT, Moon JS, Jung WK, Kim SH, Kim JM, Hong SK, Koo HC, Joo YS, Park YH. Staphylococcal cassette chromosome mec (SCCmec) characterization and molecular analysis for methicillin-resistant Staphylococcus aureus and novel SCCmec subtype IVg isolated from bovine milk in Korea. Journal of Antimicrobial Chemotherapy. 2005;56:624–632. doi: 10.1093/jac/dki306. [DOI] [PubMed] [Google Scholar]
- Le Loir Y, Baron F, Gautier M. Staphylococcus aureus and food poisoning. Genetic Molecular Research. 2003;2:63–76. [PubMed] [Google Scholar]
- Lee JH. Occurrence of methicillin-resistant Staphylococcus aureus strains from cattle and chicken, and analyses of their mecA, mecR1 and mecI genes. Veterinary Microbiology. 2006;114:155–159. doi: 10.1016/j.vetmic.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Misawa Y, Yoshida A, Saito R, Yoshida H, Okuzumi K, Ito N, Okada M, Moriya K, Koike K. Application of loop-mediated isothermal amplification technique to rapid and direction detection of methicillin-resistant Staphylococcus aureus (MRSA) in blood cultures. Journal of Infection Chemotherapy. 2007;13:134–140. doi: 10.1007/s10156-007-0508-9. [DOI] [PubMed] [Google Scholar]
- Mori Y, Nagamine K, Tomita N. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochemical and Biophisical Research Communications. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. Journal of Infection Chemotherapy. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine K, Watanabe K, Ohtsuka K. Loop-mediated isothermal amplification reaction using a nondenatured template. Clinical Chemistry. 2001;47:1742–1743. [PubMed] [Google Scholar]
- Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Molecular and Cellular Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomares C, Torres MJ, Torres A, Aznar J, Palomares JC. Rapid identification of Staphylococcus aureus from blood culture specimens using real-time fluorescence PCR. Diagnostic Microbiology and Infectious Diseases. 2003;45:183–189. doi: 10.1016/s0732-8893(02)00542-4. [DOI] [PubMed] [Google Scholar]
- Pitcher DG, Saunders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocynate. Letters in Applied Microbiology. 1989;8:151–156. [Google Scholar]
- Ramesh A, Padmapriya BP, Chandrashekar A, Varadaraj MC. Application of a convenient DNA extraction method and multiplex PCR for the direct detection of Staphylococcus aureus and Yersinia enterocolitica in milk samples. Molecular and Cellular Probes. 2002;16:307–314. doi: 10.1006/mcpr.2002.0428. [DOI] [PubMed] [Google Scholar]
- Saruta K, Matsunaga T, Kono M, Hoshina S, Ikawa S, Sakai O, Machida K. Rapid identification and typing of Staphylococcus aureus by nested PCR amplified ribosomal DNA spacer region. FEMS Microbiology Letters. 1997;146:271–278. doi: 10.1111/j.1574-6968.1997.tb10204.x. [DOI] [PubMed] [Google Scholar]
- Schmitt M, Schuler-Schmid U, Scmidt-Lorenz W. Temperature limits of growth, TNase, and enterotoxin production of Staphylococcus aureus strains isolated from foods. International Journal of Food Microbiology. 1990;11:1–19. doi: 10.1016/0168-1605(90)90036-5. [DOI] [PubMed] [Google Scholar]
- Shimizu A, Fujita M, Igarashi H, Takagi M, Nagase N, Sasaki A, Kawano J. Characterization of Staphylococcus aureus coagulase type VII isolates from staphylococcal food poisoning outbreaks (1980–1995) in Tokyo, Japan, by pulsed-field gel electrophoresis. Journal of Clinical Microbiology. 2000;38:3746–3749. doi: 10.1128/jcm.38.10.3746-3749.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DS, Kennedy J, Jane T, Miajlovic H, Bolton D, Smyth CJ. Staphylococcus aureus isolates from Irish domestic refrigerators possess novel enterotoxin and enterotoxin-like genes and are clonal in nature. Journal of Food Protection. 2006;69:508–515. doi: 10.4315/0362-028x-69.3.508. [DOI] [PubMed] [Google Scholar]
- Straub JA, Hertel C, Hammes WP. A 23S rDNA-targeted polymerase chain reaction-based system for detection of Staphylococcus aureus in meat starter cultures and dairy products. Journal of Food Protection. 1999;62:1150–1156. doi: 10.4315/0362-028x-62.10.1150. [DOI] [PubMed] [Google Scholar]
- Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nature Protocols. 2008;3:877–892. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Van der Haeghen W, Cerpentier T, Adriaensen C, Vicca J, Hermans K, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Veterinary Microbiology. 2010;144:166–171. doi: 10.1016/j.vetmic.2009.12.044. [DOI] [PubMed] [Google Scholar]
- Wang L, Shi L, Alam MJ, Geng Y, Li L. Specific and rapid detection of food-borne Salmonella by loop-mediated isothermal amplification method. Food Research International. 2008;41:69–74. [Google Scholar]
- Wilson IG, Cooper JE, Gilmour A. Detection of enterotoxigenic Staphylococcus aureus in dried skimmed milk: use of the polymerase chain reaction for amplification and detection of staphylococcal enterotoxin genes entB and entC1 and the thermonuclease gene nuc. Applied and Environmental Microbiology. 1991;57:1793–1798. doi: 10.1128/aem.57.6.1793-1798.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Shi L, Zhang C, Zhang L, Li X, Cao Y, Li L, Yamasaki S. Nosocomial infection caused by class 1 integron-carrying Staphylococcus aureus in a hospital in South China. Clinical Microbiology and Infection. 2007;13:980–984. doi: 10.1111/j.1469-0691.2007.01782.x. [DOI] [PubMed] [Google Scholar]
- Xu Z, Shi L, Alam MJ, Li L, Yamasaki S. Integron-bearing methicillin-resistant coagulase-negative staphylococci in South China, 2001–2004. FEMS Microbiology Letters. 2008a;278:223–230. doi: 10.1111/j.1574-6968.2007.00994.x. [DOI] [PubMed] [Google Scholar]
- Xu Z, Li L, Alam MJ, Yamasaki S, Shi L. First confirmation of integron-bearing methicillin-resistant Staphylococcus aureus. Current Microbiology. 2008b;57:264–268. doi: 10.1007/s00284-008-9187-8. [DOI] [PubMed] [Google Scholar]
- Xu Z, Li L, Shirtliff ME, Alam MJ, Yamasaki S, Shi L. Occurrence and characteristics of class 1 and 2 integrons in Pseudomonas aeruginosa isolates from patients in Southern China. Journal of Clinical Microbiology. 2009;47:230–234. doi: 10.1128/JCM.02027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Li L, Shirtliff ME, Peters BM, Peng Y, Alam MJ, Yamasaki S, Shi L. First report of class 2 integron in clinical Enterococcus faecalis and class 1 integron in Enterococcus faecium in South China. Diagnostic Microbiology and Infectious Diseases. 2010;68:315–317. doi: 10.1016/j.diagmicrobio.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Xu Z, Li L, Shirtliff ME, Peters BM, Li B, Peng Y, Alam MJ, Yamasaki S, Shi L. Resistance class 1 integron in clinical methicillin-resistant Staphylococcus aureus strains in Southern China, 2001–2006. Clinical Microbiology and Infection. 2011 doi: 10.1111/j.1469-0691.2010.03379.x. [DOI] [PubMed] [Google Scholar]
- Zhao X, Li Y, Wang L, You L, Xu Z, Li L, He X, Liu Y, Wang J, Yang L. Development and application of a loop-mediated isothermal amplification method on rapid detection Escherichia coli O157 strains from food samples. Molecular Biology Reports. 2010a;37:2183–2188. doi: 10.1007/s11033-009-9700-6. [DOI] [PubMed] [Google Scholar]
- Zhao X, Wang L, Li Y, Xu Z, Li L, He X, Liu Y, Wang J, Yang L. Development and application of a loop-mediated isothermal amplification method on rapid detection of Pseudomonas aeruginosa strains. World Journal of Microbiology and Biotechnology. 2011 doi: 10.1007/s11274-010-0429-0. [DOI] [Google Scholar]
- Zhao X, Wang L, Chu J, Li Y, Li Y, Xu Z, Li L, Shirtliff ME, He X, Liu Y, Wang J, Yang L. Rapid detection of Vibrio parahaemolyticus strains and virulent factors by loop-mediated isothermal amplification assays. Food Science and Biotechnology. 2010b;19:1191–1197. [Google Scholar]
- Zhao X, Wang L, Chu J, Li Y, Li Y, Xu Z, Li L, Shirtliff ME, He X, Liu Y, Wang J, Yang L. Development and application of a rapid and simple loop-mediated isothermal amplification method for food-borne Salmonella detection. Food Science and Biotechnology. 2010c;19:1655–1659. [Google Scholar]