Background: Phosphoinositide 3-kinase lipid signals exert important biological effects through proteins with specific recognition domains.
Results: We identify a novel such protein domain in IQGAP proteins and define its crystal structure and phosphoinositide binding preferences.
Conclusion: This domain is a distinct cellular phosphatidylinositol 3,4,5-trisphosphate sensor, characteristic of select IQGAP proteins.
Significance: These observations open a new and unexpected window on phosphoinositide 3-kinase signaling networks.
Keywords: Cell signaling, Crystal Structure, PI 3-Kinase (PI3K), Protein Domains, Receptors, C2 Domain, IQGAP, PH Domain, PtdInsP3, aPI Domain
Abstract
Class I phosphoinositide (PI) 3-kinases act through effector proteins whose 3-PI selectivity is mediated by a limited repertoire of structurally defined, lipid recognition domains. We describe here the lipid preferences and crystal structure of a new class of PI binding modules exemplified by select IQGAPs (IQ motif containing GTPase-activating proteins) known to coordinate cellular signaling events and cytoskeletal dynamics. This module is defined by a C-terminal 105–107 amino acid region of which IQGAP1 and -2, but not IQGAP3, binds preferentially to phosphatidylinositol 3,4,5-trisphosphate (PtdInsP3). The binding affinity for PtdInsP3, together with other, secondary target-recognition characteristics, are comparable with those of the pleckstrin homology domain of cytohesin-3 (general receptor for phosphoinositides 1), an established PtdInsP3 effector protein. Importantly, the IQGAP1 C-terminal domain and the cytohesin-3 pleckstrin homology domain, each tagged with enhanced green fluorescent protein, were both re-localized from the cytosol to the cell periphery following the activation of PI 3-kinase in Swiss 3T3 fibroblasts, consistent with their common, selective recognition of endogenous 3-PI(s). The crystal structure of the C-terminal IQGAP2 PI binding module reveals unexpected topological similarity to an integral fold of C2 domains, including a putative basic binding pocket. We propose that this module integrates select IQGAP proteins with PI 3-kinase signaling and constitutes a novel, atypical phosphoinositide binding domain that may represent the first of a larger group, each perhaps structurally unique but collectively dissimilar from the known PI recognition modules.
Introduction
Class I phosphoinositide (PI)4 3-kinases play a central role in the regulation of key cellular processes including cell growth, division, motility and metabolism, communicating to the cell interior the information encoded by diverse extracellular cues (1, 2). These receptor-regulated enzymes achieve this primarily through their ability to catalyze the 3-phosphorylation of their PtdIns(4,5)P2 substrate at the inner face of the plasma membrane to produce the lipid second messenger, PtdInsP3 (3–5). This pivotal signal is removed by the 3-phosphatase PTEN (phosphatase and tensin homologue deleted on chromosome 10) (6) or modified sequentially by the regulated action of 5- and 4-phosphatase enzymes to produce PtdIns(3,4)P2 and PtdIns3P, which may act as additional intracellular signals (7–9). The biological significance of PI 3-kinase signaling is emphasized by the diverse disease conditions associated with the dysregulation of these primary events and those further downstream in this pathway (10, 11). The cellular effects of 3-phosphoinositides are mediated by the ability of these lipids to modify the localization and/or activity of an array of effector proteins with appropriate recognition domains selective for one or, possibly more 3-PI signals (12, 13). The precise cellular consequences of PI 3-kinase activation are thus dependent on the balance of 3-PI concentrations and on both the repertoire and inositol headgroup preferences of the specific effector proteins expressed.
Among the wider set of established protein modules several, including pleckstrin homology (PH), phox homology (PX), and FYVE domains in particular, represent important 3-PI binding units and their recognition and characterization has greatly facilitated the understanding of PI 3-kinase signaling (12, 14). However, numerous proteins, including many of the enzymes involved in the maintenance of cellular phosphoinositide and inositol phosphate concentrations, lack such domains but nevertheless, recognize their cognate phosphoinositol targets with a degree of selectivity and affinity tuned appropriately to their biological functions. This suggests that additional protein modules capable of selective 3-PI binding but distinct from those already established may yet await recognition. On this premise, we recently described a proteomic screen for novel 3-PI effector proteins that, in addition to numerous other candidates, identified IQGAP1 as a potential PtdInsP3-responsive protein (15). This ∼190-kDa putative tumor promoter expresses several protein interaction modules that facilitate its cellular role, integrating multiple signaling inputs with cytoskeletal re-organization (16–19). Intriguingly, however, IQGAP1 lacks known lipid binding domains yet localizes similarly to PtdInsP3 in response to stimuli that activate PI 3-kinase and regulates the activity of the small guanosine triphosphatase (GTPase) Rac1, which is also responsive to inputs from PI 3-kinase signaling (16–22). Thus, the potential, direct interaction between IQGAP1 and 3-PIs offers the opportunity not only to establish new functional connections to the wider PI 3-kinase signaling network but also, perhaps, to identify novel 3-PI protein recognition domains.
Here we show that a module at the extreme C terminus of select IQGAP family members accounts for the binding of IQGAP1 to 3-PIs. A combination of in vitro and cellular approaches demonstrates that this domain from IQGAP1 and -2 but not IQGAP3, binds selectively to PtdInsP3 with an affinity and other lipid binding characteristics comparable with those displayed by the PH domain of GRP1 (general receptor for phosphoinositides or cytohesin-3), an established PtdInsP3 effector protein. By solving the crystal structure of the IQGAP2 C-terminal domain, we demonstrate that this domain represents a protein module distinct from those currently recognized to bind phosphoinositides but with features reminiscent of some C2 domains. These results thus suggest that this novel, atypical phosphoinositide (aPI) binding domain may serve to integrate PI 3-kinase signaling with the activity of select IQGAP family members. Most pertinently, the identification of this aPI binding unit raises the exciting prospect that it may reflect the first of a wider set of such protein modules, not all necessarily similar but collectively identifiable by their distinction from the existing, canonical groups of PI binding domains.
EXPERIMENTAL PROCEDURES
Materials
Cells were from the European Collection of Animal Cell Cultures. The glutathione S-transferase (GST)-tagged PH domains of general receptor for phosphoinositides 1 (GRP1) and phospholipase Cδ (PLCδ), were obtained from the Division of Signal Transduction Therapy, University of Dundee. Insulin-like growth factor-1 (IGF-1) was from Sigma. All synthetic and/or tagged inositol phosphates and phosphoinositides were from Cell Signals (Columbus, OH) or Echelon. Streptavidin-dextrose sensor chips for surface plasmon resonance (SPR) were from GE Healthcare. Protease inhibitor mixture and benzonase were from Sigma. Talon resin was from Clontech. Other reagents were from the sources indicated or as defined previously (15, 23, 24).
Cell Culture
Swiss 3T3 fibroblasts were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal calf serum and an additional 2 mm glutamine.
Molecular Cloning of IQGAP1 and the aPI Domain
The C-terminal region of IQGAP1 (GST-IQGAP1(718–1657)) was cloned as described previously (15). The cDNA of GST-IQGAP1(718–1657) was used as a template for PCR amplification of the aPI binding domain from IQGAP1 (amino acids 1551–1657) using the following primers: sense, 5′-TCAACGGATCCAAGCTTATGAAAGGAAAGAAAAGC-3′ and antisense, 5′-CGAAGCGGCCGCAGATCTTTACTTCCCGTAGAAC-3′. The resulting PCR product was subcloned into pGEX-6P-1 following BamHI/NotI digestion. The cDNAs encoding the aPI domains from IQGAP2 and IQGAP3 (amino acids 1470–1575 and 1527–1631, respectively) were obtained by minigene synthesis and purchased from Dundee Cell Products (Dundee, UK). An enhanced green fluorescent protein (EGFP) construct for aPIIQGAP1 was generated by subcloning from the aPIIQGAP1-GST vector (BglII/SmaI-EGFPC1 and BamHI/BsaAI-aPIIQGAP1-GST). The sequence of all constructs was confirmed by The Sequencing Service, University of Dundee. For crystallography, an N-terminal His-tagged construct of IQGAP2 residues 1476–1571 was incorporated into pET28-mhl (GI: 134105571).
Cellular Translocation Studies
Cells were seeded at 15,000/cm2 onto pre-sterilized coverslips in 6-well plates in 1.5 ml of appropriate culture medium. After 16 h, the cells were transfected with 1 μg of cDNA encoding the EGFP constructs indicated using FuGENE (Promega) according to the manufacturer's instructions. Following the expression of proteins over 24 h, the cells were serum starved for 2 h, then further incubated as indicated, washed twice with 1.5 ml of ice-cold phosphate-buffered saline (PBS, pH 7.4, 10 mm Na2HPO4, 1.8 mm KH2PO4, 2.7 mm KCl, 137 mm NaCl), and fixed with 1.5 ml of 2% (v/v) paraformaldehyde in PBS at room temperature for 10 min. The fixed cells were washed twice with 1.5 ml of PBS and once with 1.5 ml of Tris-buffered saline (20 mm Tris, pH 7.5, 150 mm NaCl), then incubated at room temperature for 5 min in 1.5 ml of TBS supplemented with 0.1% (v/v) Nonidet P-40. The fixed cells were then stained with DAPI (4′,6-diamidino-2-phenylindole, 1 μg/ml) in 1.5 ml of TBS for 15 min. Following a final wash with 1.5 ml of TBS, the coverslips were mounted with Mowiol 4-88 (Calbiochem) and 2.5% (w/v) diazabicyclo[2.2.2]octane (DABCO) and the cells were imaged using a DeltaVision Spectris microscope.
Expression of Recombinant Proteins for Biochemical Studies
The pGEX-6P-1 constructs encoding GST-aPI domains were transformed into BL21 Escherichia coli and cultures were grown for 16 h at 37 °C in 50 ml of Luria Bertani (LB) broth supplemented with 100 μm ampicillin. The cultures were then diluted into 500 ml of LB supplemented similarly and subsequently allowed to reach an A600 of ∼0.6 before being incubated further at 18 °C for 16 h following the addition of isopropyl β-d-thiogalactoside (50 μm). The bacterial cells were pelleted by centrifugation at 6000 × g and 4 °C for 30 min then re-suspended in 25 ml of lysis buffer (50 mm Tris, pH 7.5, 50 mm sodium fluoride, 1 mm EDTA, 1 mm EGTA, 5 mm sodium pyrophosphate, 10 mm glycerol 2-phosphate, 0.5% (w/v) Triton X-100, supplemented with fresh 0.1% (v/v) 2-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride, 0.1 mm benzamidine, and 1 mm sodium vanadate) prior to a single round of freeze-thawing and sonication. Following a second round of similar centrifugations, the supernatants were loaded onto columns (1–3 ml) of glutathione-Sepharose pre-equilibrated with lysis buffer at 4 °C. The columns were washed with 10–30 ml of buffer (50 mm Tris-HCl, 150 mm NaCl, pH 7.2) supplemented with an additional 350 mm NaCl and then with 10–30 ml of buffer alone. The proteins were eluted with 5–15 ml of buffer supplemented 15 mm glutathione and the eluate was dialyzed against 5 liters of buffer supplemented with 1 mm dithiothreitol and 20% (v/v) glycerol at 4 °C for 12 h. Aliquots (100 μl) of the proteins were frozen in a dry ice/methanol bath and stored at −80 °C prior to analysis.
Expression of Recombinant Protein for Structural Studies
The His-tagged C-terminal fragment of IQGAP2 was expressed in a BL21 E. coli strain harboring the pRARE2 plasmid by inoculating 100 ml of overnight culture grown in LB broth into 2 liters of Terrific Broth medium in the presence of 50 μg/ml of kanamycin and 25 μg/ml of chloramphenicol at 37 °C. When the A600 reached ∼3.0, the temperature of the medium was lowered to 15 °C and the culture was induced with 0.5 mm isopropyl β-d-thiogalactoside. The cells were allowed to grow overnight then harvested, flash frozen in liquid nitrogen, and stored at −80 °C. The frozen cells from 2 liters of TB culture were thawed and re-suspended in 150 ml of extraction buffer (20 mm HEPES, pH 7.5, 500 mm NaCl, 5% (v/v) glycerol, 2 mm 2-mercaptoethanol, 5 mm imidazole) supplemented with fresh 0.5% (v/v) CHAPS, protease inhibitor mixture, and 3 μl of benzonase (250units/μl), then lysed using a micro-fluidizer at 15,000 p.s.i. The lysate was centrifuged at 22,500 × g for 45 min and the supernatants were mixed with 5 ml of a 50% (w/v) slurry of Talon beads, then incubated at 4 °C on a rotary shaker for 1 h. The mixture was then centrifuged at 850 × g for 5 min and the supernatant was discarded. The beads were then washed successively with buffer (20 mm HEPES, pH 7.5, 500 mm NaCl, 5% (v/v) glycerol, 2 mm 2-mercaptoethanol) containing 30 and 75 mm imidazole, and finally eluted with the same buffer containing 300 mm imidazole. The eluate was collected and further purified on a Superdex-75 gel filtration column pre-equilibrated with 20 mm HEPES, pH 7.5, 500 mm NaCl, and 1 mm tris(2-carboxymethyl)phosphine. Fractions containing the protein were collected and concentrated using an Amicon Ultra-15 centrifugal filter. The purity of the preparation by SDS-PAGE was found to be greater than 95%.
Crystallization, Data Collection, and Structure Solution
Crystals were grown from 30% (w/v) PEG5000 monomethyl ether, 0.2 m (NH4)2SO4, 0.1 m MES buffer, pH 6.5, with 1:100 chymotrypsin (w/w) (25) in a sitting drop setup. The cryoprotectant used was paratone-N. A hexaiodoplatinate(IV) derivative was obtained by soaking in an ammonium sulfate-based precipitant formula. Data were collected at the GM/CA CAT, beam line 23ID, Advanced Photon Source synchrotron and processed using DENZO (26) (Table 1). Using HKL2MAP/SHELX (27) six Pt sites were identified and used to generate initial phases to 2.0-Å resolution. A partial model, covering two molecules in the asymmetric unit, was built with warpNtrace (28), the phases of which were combined with the amplitudes of the 1.5-Å native data set. The resulting maps were of excellent quality and warpNtrace was able to automatically build 180 of 192 possible residues. This model was then completed by iterative model building in Coot (29) and refinement with REFMAC5 (30) to R/Rfree = 21.0/23.9.
TABLE 1.
Data collection and refinement statistics
Data in parentheses refer to the highest resolution shell.
| IQGAP2 native | IQGAP2 Pt-derivative SAD | |
|---|---|---|
| Wavelength (Å) | 0.97946 | 1.0720 |
| Resolution (Å) | 50.0-1.50 (1.53-1.50) | 50.00-2.00 (2.03-2.00) |
| Space group | P21 | P21 |
| Unit cell (Å,o) | ||
| a | 45.1 | 45.7 |
| b | 47.2 | 46.7 |
| c | 46.6 | 48.1 |
| β | 95.5 | 98.0 |
| Reflections | ||
| Observed | 110,112 (3,413) | 58,756 (1,990) |
| Unique | 30,938 (1,365) | 13,539 (622) |
| Redundancy | 3.6 (2.5) | 4.3 (3.2) |
| Rmerge | 0.033 (0.230) | 0.093 (0.261) |
| I/σI | 39.2 (4.1) | 32.6 (5.3) |
| Completeness (%) | 98.7 (88.3) | 98.9 (91.6) |
| Rcryst (%) | 21.0 | |
| Rfree (%) | 23.9 | |
| Total number of atoms | ||
| Protein | 1,518 (188 residues) | |
| Water | 156 | |
| 〈B〉 Protein (Å2) | 8.7 | |
| 〈B〉 Water (Å2) | 20.9 | |
| Root mean square deviations from ideal geometry | ||
| Bond lengths (Å) | 0.011 | |
| Bond angles (°) | 1.30 | |
Mass Spectrometry
To allow the lipid binding capability reported to be attributed to the stated proteins, the purity of each was verified by SDS-PAGE following prior reduction and alkylation as described previously (15). The protein bands were visualized with colloidal blue (Invitrogen) and the predominant bands (see “Results”) were subjected to tryptic digestion and the fragments were reconstituted in 1% (v/v) formic acid in water and analyzed by the Fingerprints Proteomics Facility, University of Dundee, on a LTQ-Orbitrap XL mass spectrometer system (ThermoScientific) as described previously (15).
Measurement of Protein-Lipid Binding
Protein-lipid interactions were measured by SPR or by time-resolved fluorescence energy transfer (TR-FRET) as described previously but with minor modifications (15, 24). Briefly, SPR was performed using a Biacore 3000 biosensor (GE Healthcare) and streptavidin sensor chips were coated with biotinylated lipids presented in a four flow-cell format that allowed measurement at 25 °C of protein binding to a reference surface (no lipid) to be compared simultaneously with that to test surfaces loaded with PtdIns(3,4)P2, PtdIns(4,5)P2, or PtdInsP3. To increase the sensitivity and prolong the lifespan of the streptavidin chips, these were primed, prior to lipid loading, with three 60-s pulses of 5 mm NaOH delivered at a flow rate of 5 μl/min with three running buffer (10 mm HEPES, pH 7.2, 250 mm NaCl, 0.1 mm EDTA, 0.1 mm EGTA) washes at the same flow rate between consecutive priming cycles. The lipids (100 μm), diluted in running buffer, were loaded at a flow rate of 5 μl/min for 1 min or to achieve a comparable loading of each lipid corresponding to ∼200–250 response units. This is equivalent to a lipid mass of ∼200–300 fmol or a flow cell concentration of ∼10 μm, assuming the estimate from the manufacturer that 1 response unit corresponds to 1 pg/mm2 of bound analyte. However, the tetravalent capacity of streptavidin together with the potential steric hindrance associated with densely tethered ligands may reduce the effective mass of immobilized lipid. The sensor surfaces were then blocked by washing with running buffer supplemented with essentially fatty acid-free bovine serum albumin (BSA) (0.2 mg/ml) at the same flow rate before application under identical conditions of the test proteins at successively increasing concentrations as indicated. After each injection, the streptavidin chips were regenerated using a 30-s pulse of 5 mm NaOH followed by three successive washes with BSA supplemented running buffer as described for chip priming. For some experiments, 0.02% (w/v) cholic acid was included in all buffers but did not alter qualitatively the results obtained.
Protein lipid interactions were also measured using a TR-FRET assay essentially as described previously (24). In association experiments, a stock detector mixture containing APC (18.6 nm), europium chelate anti-GST antibody (228 nm), and the GST fusion of the required binding protein (100 nm) was prepared giving each component twice the final required concentration. Aliquots (25 μl) of this mixture were added to wells of a 96-well lumitrac white plate (Greiner) containing 25 μl of the required biotinylated lipids at the concentrations indicated. For dissociation experiments, a detector complex was formed similarly and the required biotinylated ligand added to the final concentrations indicated in the same manner. In both types of experiments, generation of a FRET complex was detected by reading in an LJL Analyst instrument in TR-FRET mode as described previously (24).
RESULTS
The C-terminal aPI Binding Domain of IQGAP1 Confers 3-PI Binding Capability
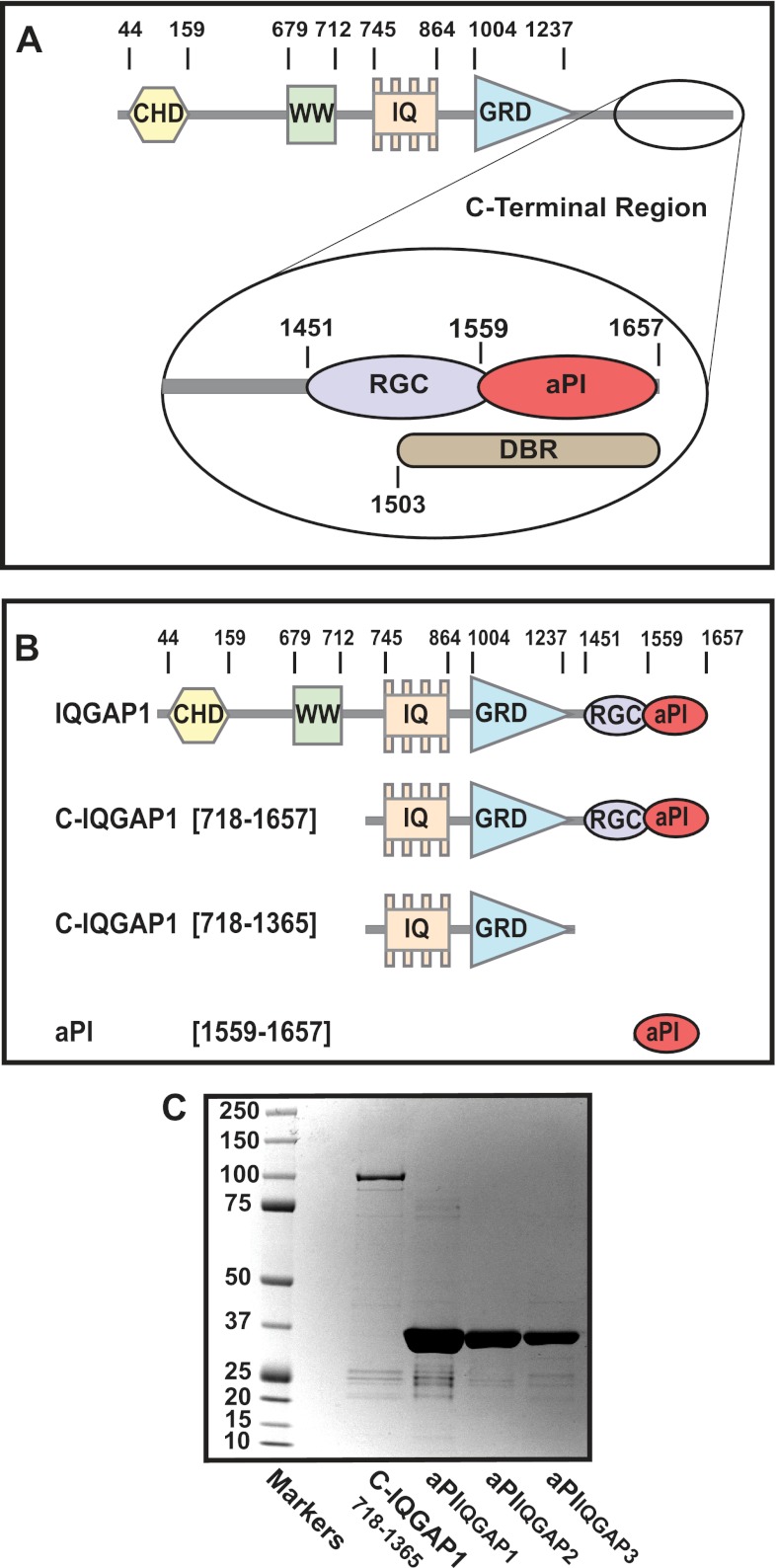
In a recent screen for novel 3-PI effector proteins we identified IQGAP1 and showed that the C-terminal half of this protein binds these lipids selectively (15). The present study aims to define the 3-PI binding module within this protein, to investigate its functional conservation across IQGAP family members, and to characterize its 3-PI lipid preference. To address these issues, a series of GST-tagged constructs reflecting sequential truncations of the C-terminal section of IQGAP1 (C-IQGAP1(718–1657)) examined previously for 3-PI binding (15) was developed and their purity verified as illustrated in Fig. 1. The full-length IQGAP1 protein has multiple domains (16) of which several are retained within C-IQGAP1(718–1657) including, the calmodulin binding IQ motifs, the RasGAP-related domain, and a further conserved module, corresponding to amino acids 1451–1581, known as the RasGAP C terminus (Interpro, IPR000593, and Pfam, PF03836, both as at February 2012) (31). In addition, an earlier study (32) reported that a region overlapping the latter and extending from amino acid 1503 to the extreme C terminus accounts for the interaction of IQGAP1 with the Dia1 protein and termed this the Dia1 binding region. In preliminary experiments the 3-PI binding capability of each of the constructs illustrated was examined by SPR, using a method established previously (15). The results (not shown, but see Figs. 2 and 3) revealed that beyond IQGAP1(718–1657), lipid binding was unique to the C-terminal IQGAP1(1551–1657) fragment (aPIIQGAP1), which we have termed the aPI binding domain on the basis of the observations described below.
FIGURE 1.
The architecture of the IQGAP1 protein shows the location of the aPI binding module. A, the domain architecture of IQGAP1. The linear sequence of the established, functional domains within IQGAP1 is shown. These include the calponin homology domain (CHD), the proline-rich WW domain (WW), the repeat IQ motifs (IQ), and the RasGAP-related domain (GRD). The expanded view of the C terminus shows the RasGAP C terminus (RGC), Dia1-binding region (DBR), and the atypical PI binding module (aPI). B, the truncated IQGAP1 constructs examined for 3-PI binding. The central panel shows a schematic representation relative to full-length IQGAP1 of the three GST-tagged, truncated IQGAP1 constructs, C-IQGAP1(718–1657), C-IQGAP1(718–1365), and C-IQGAP1(1559–1657) (aPIIQGAP1) examined for 3-PI binding by surface plasmon resonance. C, the purity of IQGAP1 constructs. The purity of each construct determined by SDS-PAGE and colloidal blue staining is indicated. The results shown are representative of at least two separate preparations of each construct. The identity of the bands indicated was confirmed by mass spectrometry as described under ”Experimental Procedures.“
FIGURE 2.
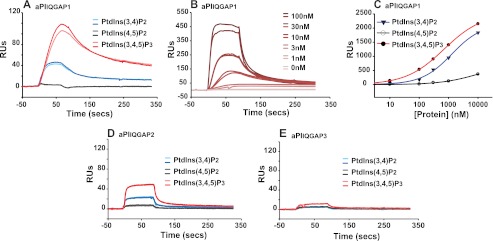
The aPI module of IQGAP1 and -2 but not IQGAP3 binds 3-phosphoinositides. A, aPIIQGAP1 binds 3-PIs. The binding of aPIIQGAP1 (30 nm) to biotinylated PtdInsP3, PtdIns(3,4)P2, and PtdIns(4,5)P2, each tethered to a streptavidin-coated surface, was compared by SPR relative to that measured in the absence of lipid. The association of protein with the lipid surface was monitored over a 90-s period of protein injection during which binding approached an approximate steady-state. The corresponding dissociation of protein when further application of aPIIQGAP1 was ceased is also shown. B, the protein concentration dependence of aPIIQGAP1 binding to PtdInsP3. The binding of aPIIQGAP1 (0–100 nm) to biotinylated PtdInsP3 was determined by SPR as indicated for panel A. C, aPIIQGAP1 binds selectively to PtdInsP3. The comparative protein concentration dependence of aPIIQGAP1 binding to biotinylated PtdInsP3, PtdIns(3,4)P2, and PtdIns(4,5)P2 was measured by SPR under conditions where protein association versus dissociation approximated to a steady-state as described for panels A and B and fitted using the sigmoidal, four-parameter logistic function afforded by SigmaPlot: y = yo + a/1 + (x/xo)b (where y = the measured binding, yo = the nonspecific binding, a = max − min binding, x = ligand concentration, xo = ligand concentration giving half-maximal response (binding or displacement) and b = slope). No constraints were applied on the basis of assumptions of binding stoichiometry or other parameters. D and E, aPIIQGAP2 but not aPIIQGAP3 binds 3-PIs. The binding of aPIIQGAP2 (D) or aPIIQGAP3 (E) to biotinylated PtdInsP3, PtdIns(3,4)P2, and PtdIns(4,5)P2 was measured as described for panel A. For all panels the results from individual experiments are representative of a minimum of at least three experiments using two or more batches of recombinant protein. A, B, D, and E show sequential traces for duplicate samples. Note that quantitative comparisons across panels should be avoided because of protein/chip deterioration with aging.
FIGURE 3.
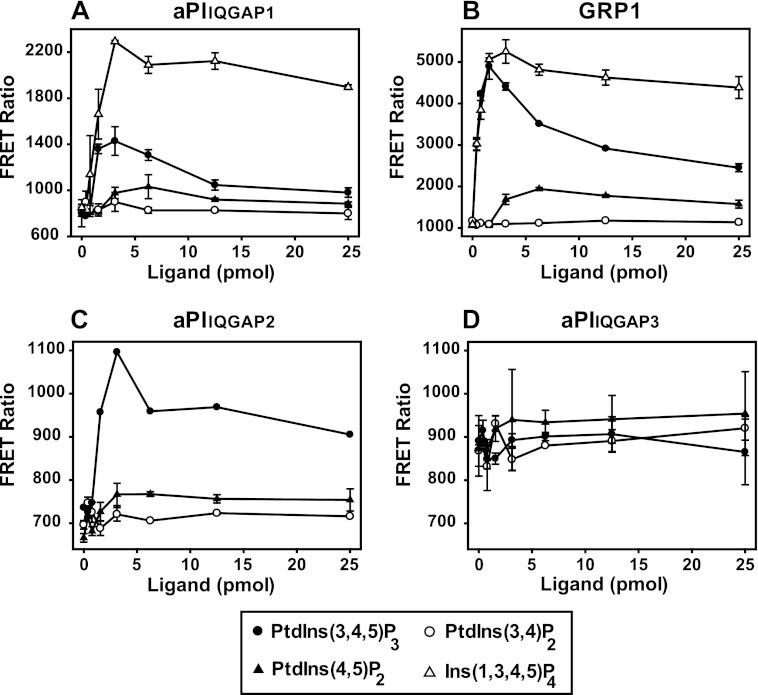
The aPI binding module and PH domain of GRP1 share PtdInsP3 recognition characteristics. The ligand concentration dependence with which equivalent concentrations (50 nm) of aPIIQGAP1 (A), aPIIQGAP2 (C), aPIIQGAP3 (D), and the PH domain of GRP1 (B) establish a FRET complex with biotinylated PtdInsP3 or biotinylated Ins(1,3,4,5)P4 (A and B) is illustrated. The results show that for aPIIQGAP1, PHGRP1, and aPIIQGAP2 (A–C) an optimal FRET signal was achieved with ∼2.5 pmol of PtdInsP3 or with 3.0 pmol of Ins(1,3,4,5)P4 for aPIIQGAP1 and PHGRP1 (A and B) and that lipid but not inositol headgroup in excess of the capacity of the streptavidin-coated APC FRET-acceptor yielded unproductive protein-ligand binding that secondarily reduced the maximal signal. The data shown are the mean ± the range of duplicate determinations in a single experiment representative of several which gave similar results.
The aPI Binding Modules of IQGAP1 and -2 but Not IQGAP3 Bind PtdInsP3 Selectively
The inositol phospholipid binding preference of this minimal aPI protein module from IQGAP1 and the corresponding units from the other IQGAP family members, IQGAP2 and -3, was examined further by SPR as shown in Fig. 2. Consistent with our previous observations (15) and in support of the results reported above, Fig. 2A shows that aPIIQGAP1 bound to PtdInsP3 and PtdIns(3,4)P2, both lipid products generated following the activation of class I PI 3-kinases, but did not bind PtdIns(4,5)P2, the substrate of these enzymes. Thus, Fig. 2A demonstrates the rapid association of aPIIQGAP1 with biotinylated PtdInsP3 or PtdIns(3,4)P2 but not with PtdIns(4,5)P2 immobilized on a steptavidin-coated surface and the corresponding dissociation of the bound aPIIQGAP1 when further protein application was ceased. Fig. 2B shows similar results but which demonstrate importantly that the binding of this aPIIQGAP1 module to PtdInsP3 was dependent on protein concentration. This feature is further examined in Fig. 2C, which compares the aPIIQGAP1 binding to each PtdInsP3, PtdIns(3,4)P2, or PtdIns(4,5)P2, determined from the limit of the association phase when binding was at or close to steady-state, across a range of protein concentrations. These results show that maximal and half-maximal binding to PtdInsP3 occurred, respectively, at ≥10 and ∼0.5 μm protein. The binding to other lipids required sufficiently higher protein concentrations that certainty of saturation binding could not be achieved. Importantly, aPIIQGAP1 also recognized the inositol 1,3,4,5-tetrakisphosphate (Ins(1,3,4,5)P4) headgroup of PtdInsP3 because protein binding to surface-tethered PtdInsP3 was reduced to ∼50% by an equal molar amount of this water-soluble ligand (data not shown, but see Figs. 3 and 5). Collectively, these observations suggest a significant preference of the aPIIQGAP1 module for phosphoinositides with a particular headgroup configuration over other closely related lipids of similar structure and charge. Fig. 2D demonstrates that the corresponding region of IQGAP2 also bound to 3-PIs and likewise expressed selectivity for PtdInsP3. By contrast, Fig. 2E establishes clearly that the similar, but not identical, C-terminal unit of IQGAP3 showed little measurable lipid binding under comparable conditions. These results are thus consistent with the notion that the 3-PI interaction of IQGAP1 and -2 is a characteristic conferred specifically by the aPI binding domain but that strict sequence and/or structural constraints govern this property as might be anticipated for an important, functional protein module.
FIGURE 5.
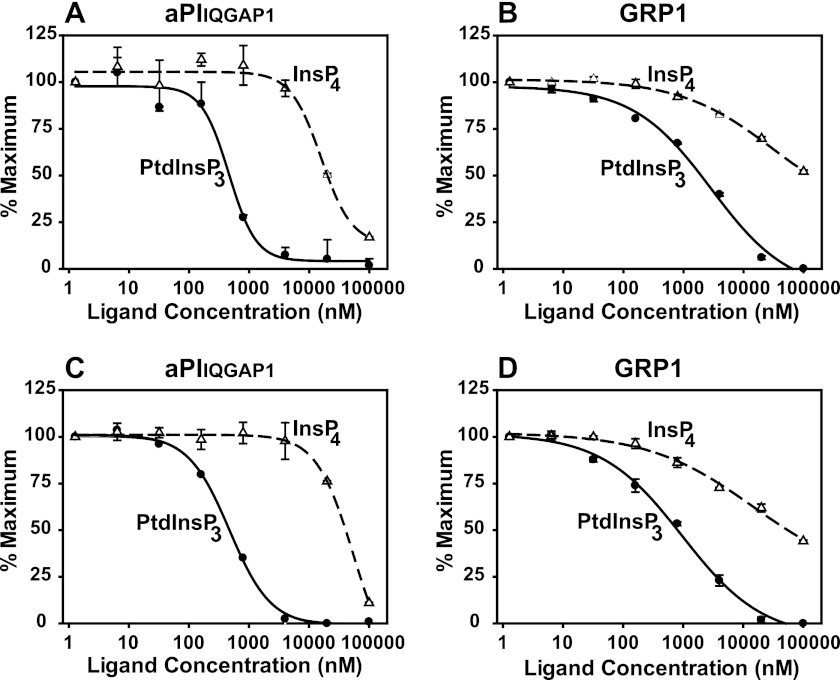
The aPIIQGAP1 and PH domain of GRP1 share a preference for PtdInsP3 over Ins(1,3,4,5)P4. The concentration dependence with which aPIIQGAP1 (A and C) and the PH domain of GRP1 (B and D) bind to PtdInsP3 (closed circles) or Ins(1,3,4,5)P4 (open triangles) is compared using a competition TR-FRET assay. Binding was measured by the reduction in the optimal FRET signal resulting from the introduction the free ligand indicated to FRET complexes pre-established with biotinylated PtdInsP3 (2.5 pmol, A and B) or biotinylated Ins(1,3,4,5)P4 (3.0 pmol; C and D). The data indicate the mean ± the range of duplicate measurements from a single experiment representative of two that gave similar results. Curves were fitted as described in the legend for Fig. 2C.
As the characteristics of lipid-protein interactions are rarely addressed adequately by a single technique (13), the analysis of phosphoinositide binding by the same aPIIQGAP modules was complemented using a TR-FRET method (24) that facilitates the ready comparison of protein binding to multiple lipid ligands. In this approach a europium-coupled anti-GST antibody bound at high affinity to a GST-tagged lipid binding module establishes a FRET signal on interaction of the latter with an allophycocyanin (APC) acceptor coated with streptavidin and pre-bound to a specific, biotinylated lipid target molecule. This strategy allows either the direct measurement of the FRET signal generated by equilibrium binding of the test protein to its lipid target (Fig. 3) or the secondary diminution of this resulting from the inclusion of free, competing ligands (Fig. 4 and 5). For technical and practical reasons the second, competition format is more flexible and provides a more reliable index of the lipid selectivity profiles of individual protein modules (24). In Fig. 3, however, the first, more direct, of these approaches is used to compare the ability of the aPI domain from each IQGAP1, -2, and -3 to bind phosphoinositides. The results presented confirm the earlier SPR studies showing that this module from IQGAP1 and -2 bound to PtdInsP3, whereas that from IQGAP3 did not bind this or the other PIs tested. In contrast to the SPR analysis, however, these results suggest a different secondary preference of the aPI binding domain from both IQGAP1 and -2, neither of which generated significant FRET in response to PtdIns(3,4)P2 (or PtdIns3P) but gave limited signals through interaction with PtdIns(4,5)P2. At present the cause of this difference is unclear but additional results (see Fig. 6) suggest that the binding to PtdIns(4,5)P2 is unlikely to be relevant in an authentic cellular context. Fig. 3 further demonstrates another important feature of this FRET approach, shown particularly by the binding of aPIIQGAP1 or the PH domain of GRP1 to PtdInsP3 where the FRET signal diminishes at high concentrations of biotinylated lipid that exceed the capacity of the APC acceptor. This occurs because the maximal FRET signal is limited not only by the concentrations of both the donor and acceptor protein components and the affinity of the test protein for its lipid target but also by the lipid loading capacity of the steptavidin-APC. Thus, excess biotinylated ligand yields unproductive binding that undermines the optimal FRET signal. Significantly, however, Fig. 3 also shows that, in contrast to PtdInsP3, biotinylated Ins(1,3,4,5)P4 in excess of that required for an optimal FRET signal resulted in comparatively little unproductive binding of aPIIQGAP1. This suggests that Ins(1,3,4,5)P4 tethered to the APC surface is recognized by the aPI-binding domain with a higher affinity than that in free solution. Similar results were obtained for the PH domain of GRP1. These results are important because they imply that features in addition to inositol headgroup preference contribute to lipid binding by both the aPI and PH domains (see also Fig. 5).
FIGURE 4.
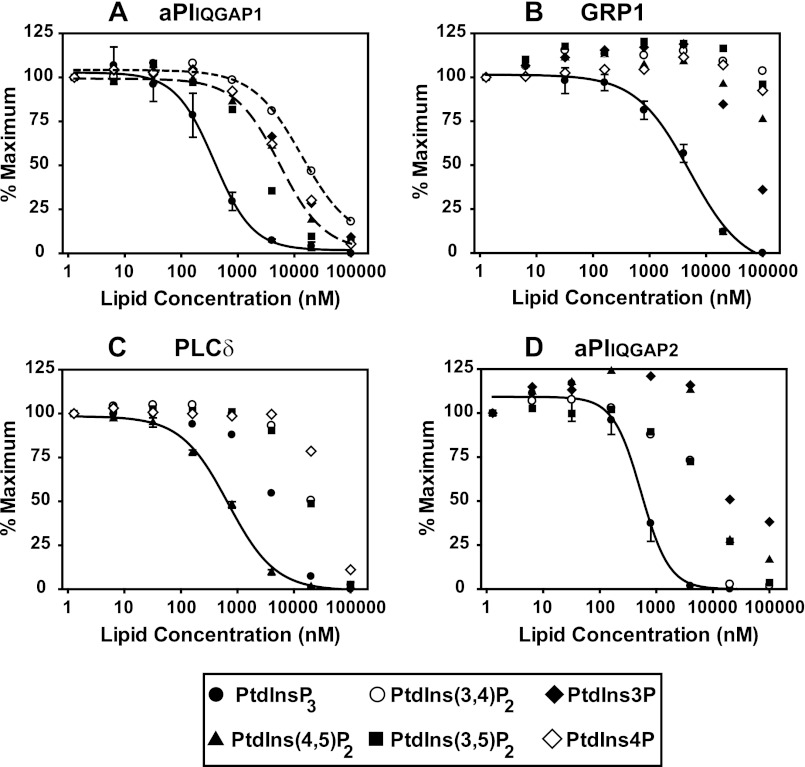
The phosphoinositide selectivity profile of aPIIQGAP1/2 is comparable with that of established PI-binding domains. The concentration dependence with which aPIIQGAP1 (A), aPIIQGAP2 (D), the PH domain of GRP1 (B), the PH domain of PLCδ (C) bound to phosphoinositides including, PtdInsP3, PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2, PtdIns4P, and PtdIns3P is compared. Lipid binding to each protein module (50 nm) was determined by TR-FRET using a competition assay in which ligand binding was measured by the reduction of an optimal FRET signal following the introduction of the indicated, free lipid ligand to the FRET complex pre-established with biotinylated PtdInsP3 (2.5 pmol, A, B, and D) or biotinylated PtdIns(4,5)P2 (2.5 pmol, C). The data indicate the mean ± the range of duplicate measurements made in one experiment representative of three that gave comparable results. Where shown, curves were fitted as described in the legend to Fig. 2C.
FIGURE 6.
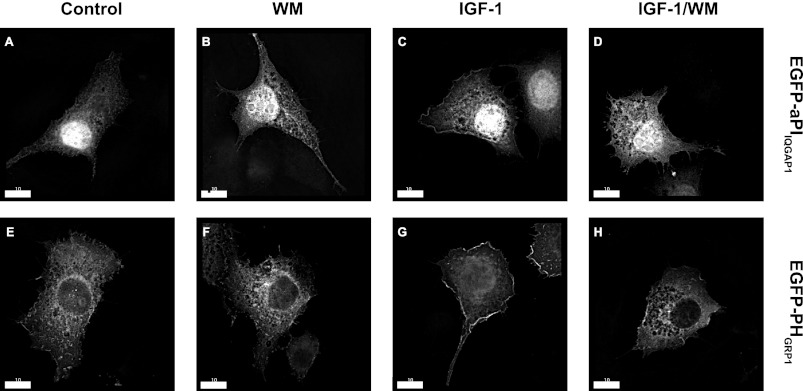
The aPIIQGAP1 and PH domain of GRP1 show similar recognition of cellular 3-PIs. EGFP-tagged protein constructs of aPIIQGAP1 (EGFP-aPIIQGAP1; A–D) and the PH domain of GRP1 (EGFP-PHGRP1; E–H) were expressed separately in Swiss 3T3 fibroblasts. The cells were then incubated for 15 min at 37 °C with vehicle (A and C; E and G) or 100 nm wortmannin (B and D; F and H) prior to a similar incubation for a further 5 min without (A and B; E and F) or with 30 nm IGF-1 (C and D; G and H). The cells were then fixed and the cellular distribution of the EGFP-tagged probes was examined. The results shown are from a single experiment and are typical of those obtained reproducibly in 6 separate experiments. Similar numbers of cells were examined per coverslip for EGFP-aPIIQGAP1 and EGFP-PHGRP1 and the proportion showing plasma membrane localization in response to IGF-1 was ∼70 and ∼80%, respectively. Note that the EGFP-PHGRP1, but not the EGFP-aPIIQGAP1, probe incorporated a nuclear export signal to facilitate its preferential cytosolic localization in resting cells. See also supplemental Fig. S1.
In further experiments the FRET parameters were optimized to configure a competition format of the assay, which allowed the comparative binding character of the aPI from IQGAP1 and -2 to be contrasted with that of established PI recognition domains as described in Fig. 4. Figs. 4, A–D, thus show the separate binding preferences of the PH domains of GRP1 (Fig. 4B) and phospholipase Cδ (PLCδ, Fig. 4C) and aPIIQGAP1 and aPIIQGAP2 (Fig. 4, A and D) for a series of competing PIs. This was measured in each case by the reduction of an optimal FRET signal, generated for the aPI domains using biotinylated PtdInsP3 on the basis of the results shown above and for the PH domains using the biotinylated lipid consistent with the established, respective PtdInsP3 and PtdIns(4,5)P2 selectivity of GRP1 and PLCδ (33–35). This approach provides a reliable index of the rank order of lipid selectivity for each separate protein module and yields a measure of the apparent affinity of these modules for individual lipid targets. It must be emphasized, however, that as measurements of affinity are made indirectly, in competition with an APC-tethered ligand target for which the true affinity of individual protein modules is not established or necessarily identical, the displacement values (IC50 values) obtained may reflect underestimates of actual affinity for some proteins. These results reveal several important features. First, they confirm the anticipated ability of this FRET approach to reproduce the PI selectivity profiles expected for well characterized lipid binding modules. Thus, Fig. 4, B and C, show clearly that the PH domain of GRP1 bound with high selectivity and apparent affinity to PtdInsP3 and the PH domain of PLCδ bound similarly to PtdIns(4,5)P2, both results are consistent with the wide use of these protein modules as cellular probes for these particular target lipids (35). In support of our earlier results (15), Fig. 4A shows that the aPI from IQGAP1 bound to PtdInsP3 with an apparent affinity for this lipid similar to that of GRP1. Thus, the half-maximal reduction (IC50) in the optimal FRET signal, reflecting displacement of biotinylated PtdInsP3 from the FRET complex by the same untagged lipid, occurred, respectively, for aPIIQGAP1 and the PH domain of GRP1 at ∼0.5 and 2–3 μm. This sensitivity of aPIIQGAP1 for PtdInsP3 is consistent both with that measured by SPR and the capability of this module to sense cellular fluctuations in the concentrations of this lipid (see below). By contrast to the PH domain of GRP1, however, the aPI module from IQGAP1 appeared less selective in its lipid preference, also expressing low micromolar apparent affinity for PtdIns(3,5)P2 but successively lower sensitivity in rank order for PtdIns(4,5)P2, PtdIns4P, PtdIns3P, and PtdIns(3,4)P2. The limited binding to the last of these lipids in particular is intriguing because it is in direct contrast to the results obtained by SPR but is consistent with the low cellular preference of this aPI module for PIs other than PtdInsP3 (see Fig. 6). Most importantly, however, these results establish directly, in vitro at least, the ability of the aPI domain of IQGAP1 to recognize inositol phospholipids and to bind PtdInsP3 in particular with an apparent affinity comparable with that of protein domains known to execute their cellular functions via interaction with this lipid. Fig. 4D moreover confirms that these characteristics with respect to PI binding are shared by the similar aPI domain from IQGAP2, consistent with the view that this reflects a retained protein module of conserved function.
As water-soluble inositol phosphates, many corresponding to the polar headgroups of membrane phosphoinositides, also occur as significant cellular metabolites (36), the ability of the aPIIQGAP1 to recognize these molecules was also examined by TR-FRET. Fig. 5 compares quantitatively the binding of aPIIQGAP1 and the PH domain of GRP1 to PtdInsP3 and to the corresponding Ins(1,3,4,5)P4 headgroup of this lipid. Fig. 5, B and D, show, respectively, that either biotinylated PtdInsP3 or biotinylated Ins(1,3,4,5)P4 could be used to establish effective FRET complexes for the PH domain of GRP1. Fig. 5, A and C, establish the same feature for the aPI domain from IQGAP1. Most importantly, however, these results demonstrate that when competing, free ligand was introduced, the FRET signal generated via either biotinylated molecule was reduced preferentially by PtdInsP3 for both protein modules. Interestingly, where tested, excess biotinylated Ins(1,3,4,5)P4 behaved similarly to the same untagged ligand in these experiments (data not shown), suggesting that the biotin tag itself has little, direct influence on binding. Hence, whereas the aPI domain of IQGAP1 and the PH domain of GRP1 were both able to bind to Ins(1,3,4,5)P4 when this was presented tethered to the surface of the APC FRET acceptor, both proteins bound this molecule in free solution much less efficiently than either recognized free PtdInsP3. Additional experiments (data not shown) similar to those illustrated in Fig. 5 demonstrated further that aPIIQGAP1 and the PH domain of GRP1 also both bound a series of other inositol phosphates if at all, only with very much lower apparent affinity than either expressed for PtdInsP3. These included, respectively, the Ins(1,4,5)P3 and Ins(1,3,4)P3 headgroups of PtdIns(4,5)P2 and PtdIns(3,4)P2, together with Ins(3,4,5)P3 and the Ins(1,3,5,6)P4 enantiomer of Ins(1,3,4,5)P4. The low apparent affinity displayed for these latter two molecules, however, precluded an estimate of the inositol phosphate headgroup stereoselectivity of either protein. Most pertinently, these results reveal that the aPI domain of IQGAP1 expresses a high apparent affinity and strong preference for PtdInsP3 over other like molecules, a requisite feature for a PtdInsP3-reponsive protein, but also imply that it shares other, perhaps surface recognition, characteristics that appear common to similar lipid binding domains.
aPIIQGAP1 Recognizes Cellular PtdInsP3 Selectively
The ability of aPIIQGAP1 to recognize PtdInsP3 in an authentic membrane environment was examined using an EGFP-tagged construct expressed in Swiss 3T3 fibroblasts as illustrated in Fig. 6. Fig. 6, A–D, compares the localization of EGFP-aPIIQGAP1 in resting cells or those exposed to IGF-1, which stimulates an increase in the concentrations of PtdInsP3 and PtdIns(3,4)P2 through the wortmannin-sensitive activation of class I PI 3-kinases in these cells (37, 38). Fig. 6, E–H, shows comparable results for the EGFP-tagged PH domain of GRP1 (EGFP-PHGRP1). In control cells EGFP-PHGRP1 was cytosolic, whereas EGFP-aPIIQGAP1 was found in the cytosol and in the nucleus. The latter was unexpected, given the established, predominant distribution of full-length IQGAP1 between the cytoplasm and the cell periphery (39). However, as a similar dual localization of EGFP-PHGRP1 is also observed unless a nuclear export signal is incorporated into this probe as employed here (38) and is not altered in resting cells by wortmannin in either case, this likely reflects technically rather than physiologically relevant issues. Importantly, however, in resting cells little or no EGFP-aPIIQGAP1 was localized in a manner consistent with its membrane association, suggesting that this probe does not recognize cellular PtdIns(4,5)P2 or other PIs/phospholipids whose concentrations are of a comparable order under basal conditions. By contrast, however, comparison of Fig. 6, A and C, shows that stimulation with IGF-1 caused an increase in the concentration of EGFP-aPIIQGAP1 at the cell periphery. This response was blocked by the prior treatment of cells with wortmannin (Fig. 6D) and was qualitatively similar to the parallel, peripheral re-localization of EGFP-PHGRP1 that is accounted for in these cells by an IGF-1-induced increase of ∼10-fold in PtdInsP3 concentrations (37, 38). Its re-localization thus suggests a similar responsiveness of EGFP-aPIIQGAP1 to modulation of 3-PI concentrations mediated via the activation of class I PI 3-kinase. Furthermore, this result most probably reflects binding of EGFP-aPIIQGAP1 to PtdInsP3 because IGF-1 stimulation is not reported to modify the cellular concentrations of PtdIns(3,5)P2, whereas the treatment of other cell types under conditions that dramatically elevate PtdIns(3,4)P2 but not PtdInsP3 concentrations (40) had relatively little impact on EGFP-aPIIQGAP1 localization (data not shown). In conjunction with our in vitro binding analyses, these results thus identify aPIIQGAP1 as a highly selective PtdInsP3-responsive protein module with a binding preference and affinity for this lipid target akin to that of other well characterized domains of documented physiological importance despite its notably dissimilar structure.
aPI Domain from IQGAP1 and -2 Is Structurally Distinct 3-PI Binding Protein Module
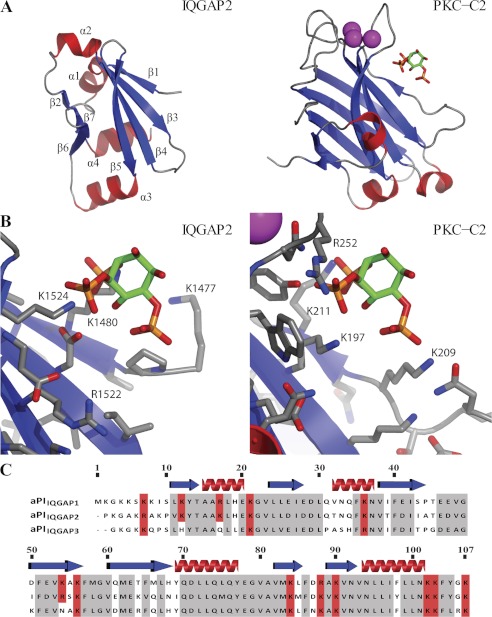
Each of the known classes of lipid binding domain comprises a particular sequence of secondary structural units folded to achieve a characteristic tertiary structure (12, 41–43). This structure usually involves an arrangement of basic side chains to interact with the negatively charged lipid headgroup. The sequence of the IQGAP proteins does not display any similarity to that of FYVE, PX, C2, or PH domains. To understand how the IQGAP2 C-terminal fold would compare with established phosphoinositide-interacting domains, we determined the first crystal structure of this family of proteins. The IQGAP2 C-terminal domain was cloned, produced recombinantly in E. coli, and purified using immobilized metal ion affinity chromatography. The protein was crystallized from PEG solutions and the structure solved by single-wavelength anomalous dispersion with the help of a platinum derivative (Table 1). The 1.5-Å native structure reveals an α/β-fold, consisting of a β-sandwich comprising a small, three-stranded β-sheet (β2, β6, and β7) and a larger four-stranded β-sheet (β1, β3, β4, and β5) separated on three sides by four, short α-helices (Fig. 7A). Searches with DALI (44) or SSM (45) initially failed to reveal any structurally similar proteins from the PDB database. Strikingly, however, when the topology connectivity restraint was removed, the SSM server identified distant structural similarity to C2 domains. For example, the C2 domain of PKCα (PDB code 3GPE and Ref. 46) superposes with an root mean square deviation of 2.8 Å for 55 equivalent Cα atoms (Fig. 7A). The IQGAP2 C-terminal domain is much smaller (94 versus 137 residues) and lacks the loops that bind the calcium ions that are important for PtdSer binding in the C2 domains (47, 48). Most of the largest 5-stranded β-sheet in the C2 domain is replaced by a considerably smaller 3-stranded β-sheet (β2, β6, and β7) in IQGAP2 (Fig. 7A, PDB code 4EZA). Strikingly, the larger β-sheet of the IQGAP2 domain still forms a twisted, concave surface reminiscent of the C2 domains, which represents the phosphoinositide-binding site in the C2 domain of PKCα (46, 49) (Fig. 7B). Indeed there are structural equivalents in IQGAP2 for the lysine/arginine residues that bind the phosphoinositide in the PKCα C2 domain (Fig. 7A), which suggests that this may also represent the site of polyphosphoinositide recognition in IQGAP2. These structural insights reveal that the aPI domain is distinct from established lipid binding scaffolds and propose a putative binding pocket for polyphosphoinositides. Interestingly, sequence alignment of the IQGAP family shows that the lysine/arginine residues identified as potentially involved in polyphosphoinositide recognition are conserved in IQGAP1 and -2 (Lys-1562/Lys-1604 and Lys-1480/Arg-1522, respectively) but not in IQGAP3 (Fig. 7C), implying that these may have a primary role in ligand recognition.
FIGURE 7.
The three-dimensional structure of the IQGAP2 C-terminal or aPIIQGAP2 domain. A and B, comparison of the IQGAP2 C-terminal domain structure (PDB code 4EZA) to that of the C2 domain of protein kinase Cα (PDB code 3GPE and (46)) in complex with the headgroup of PtdIns(4,5)P2. Structures are shown with the same viewing matrix after superposition. Secondary structure is shown with red helices and blue strands. Calcium ions are shown as magenta spheres. The PtdIns(4,5)P2 headgroup is shown as sticks. The position of the PtdIns(4,5)P2 headgroup in the protein kinase Cα C2 domain complex structure is also shown together with the IQGAP2 C-terminal domain in panel B to facilitate comparison of interacting side chains. C, a sequence alignment of the aPI domains from the IQGAP protein family (aPIIQGAP1, 1551–1657; aPIIQGAP2, 1470–1575; aPIIQGAP3, 1527–1631) reveals a high level of homology shown in gray. Basic amino acids conserved in aPIIQGAP1 and aPIIQGAP2 are shown in red. The absence of two of these, numbered 12 and 54 (corresponding to Lys-1562 and Lys-1604 in aPIIQGAP1 and Lys-1480 and Arg-1522 in aPIIQGAP2) from aPIIQGAP3, coupled to their location in the proposed binding pocket shown in panel B, suggests a possible role in ligand recognition.
DISCUSSION
The cellular consequences of PI 3-kinase activation are mediated predominantly by the expression of appropriate effector proteins incorporating modules selective for one or more phosphoinositides (1, 2, 5, 12, 14). Many proteins, however, are responsive to fluctuations in the cellular concentrations of their phosphoinositol partners but lack these recognized, targeting domains. Recently, we described a screen for novel 3-PI effector proteins that avoided bias toward candidates expressing known PI recognition modules and identified IQGAP1, a 190-kDa putative tumor promoter (16–18) with several modular domains but none known to bind 3-PIs. Preliminary evidence suggested that the C-terminal half of IQGAP1 can bind select 3-PIs, suggesting this may express an aPI binding domain (15). The present study defines the novel, protein module responsible for lipid interaction and establishes its PI binding and structural characteristics.
The lipid binding capability of IQGAP1 was found to reside within the extreme C-terminal 107 amino acids (C-IQGAP1(1559–1667) or aPIIQGAP1). This unit is distinct from the IQ motifs and RasGAP-related domains responsible, respectively, for mediating the interaction of IQGAP1 with calmodulin and the small GTPases Rac1 and CDC42 (16–18) and separate from the RasGAP C terminus, which extends between residues 1451 and 1581 (Interpro, IPR000593, and Pfam, PF03836) (31). Alternatively, aPIIQGAP1 overlaps significantly with the region known to bind Dia1 (32), suggesting that 3-PI and protein binding may be mutually exclusive with competition leading to potentially important, biological consequences. The corresponding C-terminal region from IQGAP2 also bound 3-PIs but that from IQGAP3, which differs only modestly (see below), bound only weakly if at all, suggesting that lipid binding is a conserved feature but subject to the strict constraints that are the hallmark of a functional protein domain.
Complementary cellular and in vitro approaches were used to compare the PI binding properties of aPIIQGAP1 with those expressed by the PH domains of GRP1 and PLCδ, established intracellular receptors for PtdInsP3 and PtdIns(4,5)P2 (33–35), respectively. Analyses using both SPR and a TR-FRET approach demonstrated unequivocally the ability of aPIIQGAP1 and of aPIIQGAP2 to bind PtdInsP3. The micromolar apparent affinity of aPIIQGAP1 for this lipid was comparable with that measured similarly for the PH domain of GRP1 and is consistent with that requisite for proteins responsive to the estimated cellular range of PtdInsP3 concentrations (3). The relative selectivity of aPIIQGAP1 and aPIIQGAP2 for PtdInsP3 over other PIs, however, was lower than that displayed by either PH domain and varied somewhat between in vitro methods, both features that may reflect the difficulties inherent in the adequate presentation of lipid molecules out with the physicochemical context of an authentic membrane bilayer (13). Compelling support for this view emerged from the cellular expression of EGFP-tagged aPIIQGAP1 and EGFP-PHGRP1, which revealed that both proteins localized to the cell periphery consistent with their association with plasma-membrane lipids only following the activation of PI 3-kinase. The behavior of EGFP-PHGRP1 in this respect is consistent with that reported previously and is accounted for by its ability to faithfully reflect changes in cellular PtdInsP3 concentrations (38). Collectively our results support the conclusion that aPIIQGAP1 is comparably responsive to the same PtdInsP3 lipid target. Pertinently, our TR-FRET studies suggest that aPIIQGAP1 recognizes Ins(1,3,4,5)P4, the headgroup of this lipid and an important, independent cellular inositol metabolite (50), only with a much lower sensitivity. Intriguingly, however, both aPIIQGAP1 and the PH domain of GRP1 recognize Ins(1,3,4,5)P4 preferentially when this molecule is presented as a surface-tethered ligand rather than in free solution. At present the reason for this is uncertain, although it may reflect surface or other secondary, target-recognition features of these proteins that are not yet understood. Nevertheless, the shared expression of this property by aPIIQGAP1 and an established PtdInsP3 binding domain together with their common lipid preference emphasizes the similar functional character of these protein modules.
Despite this, it is clear that IQGAP1 and -2 do not express modular protein units of the types known currently to recognize PIs. Typically, the separate classes (PH, PX etc.) of these modules are defined individually by a characteristic three-dimensional frame assembled from an established and ordered group of particular secondary structure units (12, 14, 35). Held within this frame, the precise spatial presentation of a limited set of amino acids, usually including several basic residues, confers the individual headgroup preference that specifies the recognition of a particular phosphoinositide target(s). Hydrophobic and basic residues peripheral to the binding pocket, however, may also contribute to membrane interaction (42). The crystal structure of the IQGAP2 C-terminal domain presented here demonstrates that neither the linear sequence and number of secondary structure units nor their spatial arrangement conforms to those which define the established 3-PI binding modules. It is apparent that these features of IQGAP proteins differ also from those characteristic of certain C2 domains that bind PIs (46, 51). The crystal structure for the IQGAP2 C-terminal domain reveals a novel fold with distant topological similarity to the C2 domain fold including a pocket lined with basic residues. We propose that this structural frame represents a new, atypical PI binding protein module whose target specificity is conferred primarily by the sequence and orientation of particular basic residues within the surface groove. In support of this view, a comparative alignment of IQGAP family members reveals potentially key amino acids, including Lys-1562/Lys-1604 and Lys-1480/Arg-1522 that are conserved in parallel with the PtdInsP3 binding capability between IQGAP1 and -2 but not IQGAP3. Further studies will be required to confirm the active participation of these in lipid binding. Remarkably, however, comparison of the aPI binding modules from their parent IQGAP proteins reveals near complete identity across species from frog through rodents to humans, showing up to 95% identical sequence (see supplemental Fig. S2), providing compelling evidence that the aPI binding domain is a functionally important and persistently conserved module.
As the aPI binding domain of select IQGAP family members confers PtdInsP3 recognition and responsiveness, this capability must be integrated with other molecular interactions that contribute to the biological role of these proteins. Intact IQGAP proteins are large, multimodular units that integrate upstream signals from diverse cell-surface receptors to modulate cytoskeletal dynamics and other downstream events through their direct interaction with key binding partners. Among these, the inclusion of actin, E-cadherin, catenins, calmodulin, and components of the ERK pathway, allow IQGAP1 to facilitate coordinated regulation of cell motility/adhesion and proliferation (16–18). In contrast to the isolated aPI binding module therefore, it is unlikely that the cellular localization of full-length IQGAP proteins is mediated exclusively via the binding of the aPI domain to PtdInsP3. Nevertheless, the shared capacity of IQGAP1 and PI 3-kinase to regulate the activity of Rac1, together with the common localization of IQGAP1 and PtdInsP3 at the leading edge of migrating cells (16–18, 21, 22) and other recent observations (31) argue persuasively for the integrative coupling of IQGAP1 and 3-PI signaling. Through its ability to bind directly to PtdInsP3 with an affinity poised in the appropriate cellular range, the aPI binding domain may provide the requisite means by which this coupling is achieved. Thus, mechanisms might be envisaged in which the recognition of PtdInsP3 by the aPI binding module acts upstream as a component of a coincidence detection system enabling flexible responses to parallel signaling inputs or acts downstream to favor selective binding to protein partners that promote discrete cellular outcomes. In either case, the capability conferred by the aPI binding domain is likely fundamental to the biological function of IQGAP proteins. Its recognition reflects a significant, new connection to both the IQGAP and PI 3-kinase signaling networks and may help clarify mutually the molecular mechanisms underlying the role of these complex systems in healthy and disease states.
In brief, we have identified and characterized a novel PtdInsP3-binding protein module distinct from those shown previously to recognize phosphoinositides. This is exemplified by select members of the IQGAP protein family and is likely integral to the function of these proteins, although its wider distribution across the proteome remains to be established. The description of this module supports the premise that a larger group of aPI binding protein domains may exist, each perhaps structurally unique but collectively distinct from those classified previously. This prospect adds a novel dimension to the current concept of PI 3-kinase signaling.
Supplementary Material
Acknowledgments
We are grateful to the Division of Signal Transduction Therapy, University of Dundee, for the generous provision of proteins, the Fingerprints Proteomics Facility, University of Dundee, for the mass spectrometric analysis of protein samples, and the Sequencing Service, University of Dundee, for the verification of cDNA sequences. GM/CA CAT has been funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the Advanced Photon Source was supported by the United States Department of Energy, Basic Energy Sciences, Office of Science, under contract DE-AC02-06CH11357. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institute, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.
This work was supported in part by Medical Research Council, United Kingdom Programme Grant G0801865 (to M. J. D., A. G., M. S., N. R. L., C. P. D., and I. H. B.).

This article contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (code 4EZA) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PI
- phosphoinositide
- APC
- allophycocyanin
- aPI binding domain
- atypical phosphoinositide binding domain
- C2
- second conserved region of PKC
- EGFP
- enhanced green fluorescent protein
- FYVE
- Fab-1, YOTB, Vac1, and EEA1
- GAP
- GTPase activating protein
- GRP1
- general receptor for phosphoinositides
- Ins(1,3,4,5)P4
- inositol 1,3,4,5-tetrakisphosphate
- Ins(1,3,5,6)P4
- inositol 1,3,5,6-tetrakisphosphate
- Ins(1,4,5)P3
- inositol 1,4,5-trisphosphate
- Ins(1,3,4)P3
- inositol 1,3,4-trisphosphate
- Ins(3,4,5)P3
- inositol 3,4,5-trisphosphate
- IQGAP
- IQ motif containing GTPase activating protein
- PH
- pleckstrin homology
- PLCδ
- phospholipase Cδ
- PtdIns
- phosphatidylinositol
- PtdIns3P
- phosphatidylinositol 3-phosphate
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PtdIns(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PtdInsP3
- phosphatidylinositol 3,4,5-trisphosphate
- PTEN
- phosphatase and tensin homologue deleted on chromosome 10
- PX
- phagocytic oxidase (phox) homology
- SPR
- surface plasmon resonance
- TR-FRET
- time-resolved fluorescence resonance energy transfer.
REFERENCES
- 1. Cantley L. C. (2002) The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 2. Di Paolo G., De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 3. Stephens L. R., Jackson T. R., Hawkins P. T. (1993) Agonist-stimulated synthesis of phosphatidylinositol(3,4,5)-trisphosphate. A new intracellular signalling system? Biochim. Biophys. Acta 1179, 27–75 [DOI] [PubMed] [Google Scholar]
- 4. Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. (2001) Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70, 535–602 [DOI] [PubMed] [Google Scholar]
- 5. Hawkins P. T., Anderson K. E., Davidson K., Stephens L. R. (2006) Signaling through class I PI3Ks in mammalian cells. Biochem. Soc. Trans. 34, 647–662 [DOI] [PubMed] [Google Scholar]
- 6. Leslie N. R., Downes C. P. (2002) PTEN. The down side of PI 3-kinase signaling. Cell Signal. 14, 285–295 [DOI] [PubMed] [Google Scholar]
- 7. Majerus P. W., Kisseleva M. V., Norris F. A. (1999) The role of phosphatases in inositol signaling reactions. J. Biol. Chem. 274, 10669–10672 [DOI] [PubMed] [Google Scholar]
- 8. Downes C. P., Leslie N. R., Batty I. H., van der Kaay J. (2007) Metabolic switching of PI3K-dependent lipid signals. Biochem. Soc. Trans. 35, 188–192 [DOI] [PubMed] [Google Scholar]
- 9. Stenmark H., Aasland R. (1999) FYVE-finger proteins. Effectors of an inositol lipid. J. Cell Sci. 112, 4175–4183 [DOI] [PubMed] [Google Scholar]
- 10. Marone R., Cmiljanovic V., Giese B., Wymann M. P. (2008) Targeting phosphoinositide 3-kinase. Moving towards therapy. Biochim. Biophys. Acta 1784, 159–185 [DOI] [PubMed] [Google Scholar]
- 11. Kok K., Geering B., Vanhaesebroeck B. (2009) Regulation of phosphoinositide 3-kinase expression in health and disease. Trends Biochem. Sci. 34, 115–127 [DOI] [PubMed] [Google Scholar]
- 12. Lemmon M. A. (2003) Phosphoinositide recognition domains. Traffic 4, 201–213 [DOI] [PubMed] [Google Scholar]
- 13. Narayan K., Lemmon M. A. (2006) Determining selectivity of phosphoinositide-binding domains. Methods 39, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 15. Dixon M. J., Gray A., Boisvert F. M., Agacan M., Morrice N. A., Gourlay R., Leslie N. R., Downes C. P., Batty I. H. (2011) A screen for novel phospho-inositide 3-kinase effector proteins. Mol. Cell Proteomics 10, M110.003178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brown M. D., Sacks D. B. (2006) IQGAP1 in cellular signaling. Bridging the GAP. Trends Cell Biol. 16, 242–249 [DOI] [PubMed] [Google Scholar]
- 17. White C. D., Brown M. D., Sacks D. B. (2009) IQGAPs in cancer. A family of scaffold proteins underlying tumorigenesis. FEBS Lett. 583, 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noritake J., Watanabe T., Sato K., Wang S., Kaibuchi K. (2005) IQGAP1, a key regulator of adhesion and migration. J. Cell Sci. 118, 2085–2092 [DOI] [PubMed] [Google Scholar]
- 19. Brandt D. T., Grosse R. (2007) Get to grips. Steering local actin dynamics with IQGAPs. EMBO Rep. 8, 1019–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tirnauer J. S. (2004) A new cytoskeletal connection for APC. Linked to actin through IQGAP. Dev. Cell 7, 778–780 [DOI] [PubMed] [Google Scholar]
- 21. Cain R. J., Ridley A. J. (2009) Phosphoinositide 3-kinases in cell migration. Biol. Cell 101, 13–29 [DOI] [PubMed] [Google Scholar]
- 22. Stephens L., Milne L., Hawkins P. (2008) Moving towards a better understanding of chemotaxis. Curr. Biol. 18, R485–494 [DOI] [PubMed] [Google Scholar]
- 23. Batty I. H., Downes C. P. (1994) The inhibition of phosphoinositide synthesis and muscarinic receptor-mediated phospholipase C activity by Li+ as secondary, selective, consequences of inositol depletion in 1321N1 cells. Biochem. J. 297, 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gray A., Olsson H., Batty I. H., Priganica L., Peter Downes C. (2003) Nonradioactive methods for the assay of phosphoinositide 3-kinases and phosphoinositide phosphatases and selective detection of signaling lipids in cell and tissue extracts. Anal. Biochem. 313, 234–245 [DOI] [PubMed] [Google Scholar]
- 25. Dong A., Xu X., Edwards A. M., Midwest Center for Structural Genomics, Structural Genomics Consortium, Chang C., Chruszcz M., Cuff M., Cymborowski M., Di Leo R., Egorova O., Evdokimova E., Filippova E., Gu J., Guthrie J., Ignatchenko A., Joachimiak A., Klostermann N., Kim Y., Korniyenko Y., Minor W., Que Q., Savchenko A., Skarina T., Tan K., Yakunin A., Yee A., Yim V., Zhang R., Zheng H., Akutsu M., Arrowsmith C., Avvakumov G. V., Bochkarev A., Dahlgren L. G., Dhe-Paganon S., Dimov S., Dombrovski L., Finerty P., Jr., Flodin S., Flores A., Gräslund S., Hammerström M., Herman M. D., Hong B. S., Hui R., Johansson I., Liu Y., Nilsson M., Nedyalkova L., Nordlund P., Nyman T., Min J., Ouyang H., Park H. W., Qi C., Rabeh W., Shen L., Shen Y., Sukumard D., Tempel W., Tong Y., Tresagues L., Vedadi M., Walker J. R., Weigelt J., Welin M., Wu H., Xiao T., Zeng H., Zhu H. (2007) In situ proteolysis for protein crystallization and structure determination. Nat. Methods 4, 1019–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27. Pape T., Schneider T. R. (2004) HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 28. Perrakis A., Morris R., Lamzin V. S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 31. White C. D., Erdemir H. H., Sacks D. B. (2012) IQGAP1 and its binding proteins control diverse biological functions. Cell Signal. 24, 826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brandt D. T., Marion S., Griffiths G., Watanabe T., Kaibuchi K., Grosse R. (2007) Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J. Cell Biol. 178, 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klarlund J. K., Guilherme A., Holik J. J., Virbasius J. V., Chawla A., Czech M. P. (1997) Signaling by phosphoinositide 3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science 275, 1927–1930 [DOI] [PubMed] [Google Scholar]
- 34. Garcia P., Gupta R., Shah S., Morris A. J., Rudge S. A., Scarlata S., Petrova V., McLaughlin S., Rebecchi M. J. (1995) The pleckstrin homology domain of phospholipase C-delta 1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry 34, 16228–16234 [DOI] [PubMed] [Google Scholar]
- 35. Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., Lemmon M. A. (2000) Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373–384 [DOI] [PubMed] [Google Scholar]
- 36. Irvine R. F., Schell M. J. (2001) Back in the water. The return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2, 327–338 [DOI] [PubMed] [Google Scholar]
- 37. Van der Kaay J., Beck M., Gray A., Downes C. P. (1999) Distinct phosphatidylinositol 3-kinase lipid products accumulate upon oxidative and osmotic stress and lead to different cellular responses. J. Biol. Chem. 274, 35963–35968 [DOI] [PubMed] [Google Scholar]
- 38. Gray A., Van Der Kaay J., Downes C. P. (1999) The pleckstrin homology domains of protein kinase B and GRP1 (general receptor for phosphoinositides-1) are sensitive and selective probes for the cellular detection of phosphatidylinositol 3,4-bisphosphate and/or phosphatidylinositol 3,4,5-trisphosphate in vivo. Biochem. J. 344, 929–936 [PMC free article] [PubMed] [Google Scholar]
- 39. Li Z., Kim S. H., Higgins J. M., Brenner M. B., Sacks D. B. (1999) IQGAP1 and calmodulin modulate E-cadherin function. J. Biol. Chem. 274, 37885–37892 [DOI] [PubMed] [Google Scholar]
- 40. Batty I. H., van der Kaay J., Gray A., Telfer J. F., Dixon M. J., Downes C. P. (2007) The control of phosphatidylinositol 3,4-bisphosphate concentrations by activation of the Src homology 2 domain containing inositol polyphosphate 5-phosphatase 2, SHIP2. Biochem. J. 407, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DiNitto J. P., Cronin T. C., Lambright D. G. (2003) Membrane recognition and targeting by lipid-binding domains. Sci. STKE 2003, re16. [DOI] [PubMed] [Google Scholar]
- 42. Kutateladze T. G. (2010) Translation of the phosphoinositide code by PI effectors. Nat. Chem. Biol. 6, 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moravcevic K., Oxley C. L., Lemmon M. A. (2012) Conditional peripheral membrane proteins. Facing up to limited specificity. Structure 20, 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Holm L., Sander C. (1995) Dali, a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 45. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 46. Guerrero-Valero M., Ferrer-Orta C., Querol-Audí J., Marin-Vicente C., Fita I., Gómez-Fernández J. C., Verdaguer N., Corbalán-García S. (2009) Structural and mechanistic insights into the association of PKCα-C2 domain to PtdIns(4,5)P2. Proc. Natl. Acad. Sci. U.S.A. 106, 6603–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sutton R. B., Davletov B. A., Berghuis A. M., Südhof T. C., Sprang S. R. (1995) Structure of the first C2 domain of synaptotagmin I. A novel Ca2+/phospholipid-binding fold. Cell 80, 929–938 [DOI] [PubMed] [Google Scholar]
- 48. Verdaguer N., Corbalan-Garcia S., Ochoa W. F., Fita I., Gómez-Fernández J. C. (1999) Ca2+ bridges the C2 membrane-binding domain of protein kinase Cα directly to phosphatidylserine. EMBO J. 18, 6329–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Corbalán-García S., García-García J., Rodríguez-Alfaro J. A., Gómez-Fernández J. C. (2003) A new phosphatidylinositol 4,5-bisphosphate-binding site located in the C2 domain of protein kinase Cα. J. Biol. Chem. 278, 4972–4980 [DOI] [PubMed] [Google Scholar]
- 50. Batty I. R., Nahorski S. R., Irvine R. F. (1985) Rapid formation of inositol 1,3,4,5-tetrakisphosphate following muscarinic receptor stimulation of rat cerebral cortical slices. Biochem. J. 232, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Corbalán-García S., Guerrero-Valero M., Marín-Vicente C., Gómez-Fernández J. C. (2007) The C2 domains of classical/conventional PKCs are specific PtdIns(4,5)P2-sensing domains. Biochem. Soc. Trans. 35, 1046–1048 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.