Background: Stress granules (SG) have been implicated in the formation of pathological FUS and TDP-43 inclusions.
Results: SG recruitment of FUS and TDP-43 requires cytosolic mislocalization and their main RNA binding domain and glycine-rich domain.
Conclusion: FUS and TDP-43 have similar requirements for SG recruitment.
Significance: Understanding how FUS and TDP-43 are recruited to SG is critical for understanding FTLD/ALS pathology.
Keywords: Amyotrophic Lateral Sclerosis (Lou Gehrig's Disease), Neurodegeneration, Neurodegenerative Diseases, RNA-binding Protein, Stress Granule, Frontotemporal Lobar Degeneration (FTLD), Fused in Sarcoma (FUS), TAR DNA-binding Protein of 43 kDa (TDP-43)
Abstract
Cytoplasmic inclusions containing TAR DNA-binding protein of 43 kDa (TDP-43) or Fused in sarcoma (FUS) are a hallmark of amyotrophic lateral sclerosis (ALS) and several subtypes of frontotemporal lobar degeneration (FTLD). FUS-positive inclusions in FTLD and ALS patients are consistently co-labeled with stress granule (SG) marker proteins. Whether TDP-43 inclusions contain SG markers is currently still debated. We determined the requirements for SG recruitment of FUS and TDP-43 and found that cytoplasmic mislocalization is a common prerequisite for SG recruitment of FUS and TDP-43. For FUS, the arginine-glycine-glycine zinc finger domain, which is the protein's main RNA binding domain, is most important for SG recruitment, whereas the glycine-rich domain and RNA recognition motif (RRM) domain have a minor contribution and the glutamine-rich domain is dispensable. For TDP-43, both the RRM1 and the C-terminal glycine-rich domain are required for SG localization. ALS-associated point mutations located in the glycine-rich domain of TDP-43 do not affect SG recruitment. Interestingly, a 25-kDa C-terminal fragment of TDP-43, which is enriched in FTLD/ALS cortical inclusions but not spinal cord inclusions, fails to be recruited into SG. Consistently, inclusions in the cortex of FTLD patients, which are enriched for C-terminal fragments, are not co-labeled with the SG marker poly(A)-binding protein 1 (PABP-1), whereas inclusions in spinal cord, which contain full-length TDP-43, are frequently positive for this marker protein.
Introduction
Amyotrophic lateral sclerosis (ALS)3 and frontotemporal lobar degeneration (FTLD) are related neurodegenerative diseases in which the majority of cases are characterized by the pathological accumulation of the TAR DNA-binding protein 43 (TDP-43) or the Fused in sarcoma (FUS) protein (1). TDP-43 and FUS are DNA/RNA-binding proteins that are involved in transcriptional regulation, pre-mRNA splicing, microRNA processing, and mRNA transport (for review, see in Refs. 2 and 3). Although both proteins exert their function predominantly in the nucleus, pathological TDP-43 and FUS inclusions are mostly observed in the cytoplasm. Strikingly, inclusion-bearing cells often show a loss or reduction of nuclear TDP-43 or FUS staining (4–11). This has led to the hypothesis that loss of nuclear TDP-43 or FUS is a crucial step in disease progression.
Both proteins have multiple RNA binding domains as well as a protein interaction domain predicted to have prion-like properties. TDP-43 has two RNA recognition motif (RRM) domains, RRM1 and RRM2, with RRM1 being the predominant functional RNA binding domain (12). In addition, TDP-43 contains a C-terminal glycine-rich domain that mediates interactions with other heterogeneous nuclear ribonucleoproteins and is required for splicing regulation (13). This domain is highly aggregation-prone (14–18) and due to its amino acid composition has been suggested to have prion-like properties (19–22). FUS has multiple RNA binding domains with arginine-glycine-glycine (RGG) motifs, a RRM domain, and a zinc finger domain shown to mediate RNA binding (23, 24). In addition, FUS contains an N-terminal glutamine-rich domain that functions as a potent transcriptional activation domain (25) and was predicted to be a prion-like domain (19–21).
The relevance of TDP-43 and FUS in the pathogenesis of ALS and FTLD was strongly supported by the discovery of autosomal dominant mutations within TARDBP (the gene encoding TDP-43) and FUS in familial forms of ALS (6, 7, 26). So far, almost 40 different TARDBP mutations have been reported; most of them are missense mutations in the glycine-rich C-terminal domain. Although it has been claimed that TARDBP mutations increase aggregation tendency (14, 15, 27, 28), alter the protein cellular localization (29–31), or alter the protein half-life and interactions with other proteins (32), the pathogenic mechanism of these mutations is still unclear, as many inconsistencies among different studies have been reported. Pathogenic mutations in the FUS gene are mostly clustered in the C-terminal proline-tyrosine nuclear localization signal (PY-NLS) and impair Transportin-mediated nuclear import of FUS (33–36). Interestingly, mutations that show a very severe nuclear import defect, such as P525L, cause an unusually early disease onset and rapid disease progression (37–39), suggesting that impaired nuclear import of FUS is causally linked to the disease (33, 40). Even though it is still unclear how reduced nuclear import of FUS leads to neurodegeneration, it has been shown that blockade of Transportin-mediated nuclear import or FUS mutations leads to recruitment of FUS into stress granules (SG), implicating SG and reduced nuclear transport in disease pathogenesis (33, 34, 36, 40, 41). This is supported by the presence of SG markers in inclusions in ALS/FTLD-FUS patients (33, 42).
SG are cytosolic structures that form transiently upon exposure of cells to environmental stress, such as heat, viral infection, oxidative stress, or hypoxia (43). They arise from polysomes and store mRNAs encoding housekeeping proteins but exclude mRNAs encoding chaperones and enzymes involved in damage repair. In addition to mRNAs, SG contain many RNA-binding proteins, such as poly(A)-binding protein 1 (PABP-1) and T cell intracellular antigen 1 (TIA-1), which serve as specific markers for SG (44). In cultured cells, SG formation can be elicited with a variety of stress treatments, such as heat shock (42–44 °C), osmotic shock, UV irradiation, or substances that elicit mitochondrial and/or oxidative stress (44). SG have also been observed in vivo (41, 45–47), and SG marker proteins were found to label the pathological FUS inclusions in post mortem brains of ALS/FTLD patients (33, 42). Thus, it has been suggested that SG might be the precursors of the pathological FUS inclusions in ALS/FTLD-FUS patients (33).
How FUS is recruited to SG is currently unknown. Because FUS is an RNA-binding protein, it is conceivable that it is recruited into these structures via its associated mRNAs. Alternatively, protein-protein interactions might be involved in localization of FUS to SG. Interestingly, TIA-1 contains a prion-like glutamine-rich domain that has homology to the N-terminal glutamine-rich domain of FUS and promotes SG assembly by a prion-like aggregation mechanism (48, 49). Whether this domain of FUS is required for SG recruitment or aggregation is still unknown.
TDP-43 has also been described to be recruited to SG under various stress conditions (31, 50–55), and SG-associated proteins have been identified as TDP-43-interacting proteins (56). However, it is still controversial whether TDP-43 inclusions in human patients contain SG markers. Two studies found a lack of SG markers in TDP-43 inclusions of ALS/FTLD-TDP patients (33, 50), whereas two other studies reported co-labeling of TDP-43 inclusions with SG markers (31, 57). Furthermore, it is still not clear if or how TARDBP mutations affect SG recruitment. One cell culture study reported that TARDBP mutations increase the number of cells with TDP-43 inclusions in response to stress (31), whereas another group found that mutant (R361S) TDP-43 impairs SG formation (54), and a third study reported that overexpression of mutant (G348C) TDP-43 leads to larger SG (51).
To address how FUS and TDP-43 are recruited into SG, we mapped the domains required for SG recruitment of FUS and TDP-43. In addition, we analyzed the effect of various forms of ALS-associated TARDBP mutations on SG recruitment of TDP-43 and further investigated the presence of SG marker proteins in TDP-43 inclusions in ALS/FTLD-TDP cortex and spinal cord.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Human cervical carcinoma cells (HeLa) were cultured in Dulbecco's modified Eagle's medium with Glutamax (Invitrogen) supplemented with 10% (v/v) fetal calf serum (Invitrogen) and penicillin/streptomycin (PAA Laboratories). Transfection of HeLa cells was carried out with FuGENE 6 (Roche Applied Science) or Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Hippocampal neurons were isolated from embryonic day 18 rats as described previously (58). Neurons were plated at densities of 18,000 cells/cm2 in 6-cm tissue culture dishes containing poly-l-lysine (1 mg/ml; Sigma)-coated glass coverslips and Neurobasal medium supplemented with 2% B27 and 0.5 mm glutamine (all from Invitrogen). On day in vitro 7 (DIV 7), cultured neurons were transfected with FUS or TDP-43 constructs using Lipofectamine 2000 (Invitrogen) and were analyzed on DIV 9.
Stress and Inhibitor Treatment
Heat shock was performed by incubating cells for 1 h in a tissue culture incubator heated to 44 °C. For recovery experiments, cells were shifted back to 37 °C and incubated another 60 min. Where indicated, cycloheximide (Sigma) was added at a concentration of 20 μg/ml immediately before shifting cells to 44 °C. Clotrimazole (Sigma C6019) was dissolved in DMSO (20 mm stock) and was added to cells under serum-free conditions in Opti-MEM (Invitrogen) at a final concentration of 20 μm for 30 min. Sodium (meta)arsenite (Sigma S71287) was dissolved in water (100 mm stock) and added to cells at a final concentration of 0.5 mm for 30 min.
Antibodies
The following antibodies were used: β-actin-specific mouse monoclonal antibody clone AC-74 (Sigma); GFP-specific rabbit polyclonal antibody (BD Living Colors); HA-specific mouse monoclonal antibody HA.11 (Covance); horseradish peroxidase (HRP)-coupled rat monoclonal anti-HA antibody 3F10 (Roche Applied Science); myc-specific mouse monoclonal antibody 9E10 (sc-40, Santa Cruz); PABP-1-specific rabbit polyclonal antibody (Cell Signaling); TDP-43-specific rabbit polyclonal antibody TARDBP (Proteintech); polyclonal antibodies raised against amino acid residues 6–24 of TDP-43 (N-t TDP-43) and amino acid residues 394–414 of TDP-43 (C-t TDP-43) (59); phosphoserine 409/410-specific TDP-43 rat monoclonal antibody clone 1D3 (60); TIA-1-specific goat polyclonal antibody (C-20, Santa Cruz); α-tubulin-specific mouse monoclonal antibody clone B-5–1-2 (Sigma); βΙΙΙ-tubulin-specific rabbit polyclonal antibody clone Tuj1 (Sigma); V5-specific mouse monoclonal antibody (R960–25, Invitrogen). Secondary antibodies for immunoblotting were HRP-coupled goat anti-mouse or anti-rabbit IgGs (Promega). For immunofluorescence stainings, Alexa-488, Alexa-555, Alexa-594, and Alexa-647-conjugated donkey anti-mouse, anti-rabbit, anti-rat or anti-goat IgG (Invitrogen) were used.
cDNA Constructs and Primers
HA-FUS-WT, HA-FUS-P525L, and GFP-Bimax were described in Dormann et al. (33). For FUS deletion constructs, the individual domains of FUS were amplified by PCR, and PCR products were cloned into the pcDNA3.1/Hygro(−) vector (Invitrogen) via BamHI/XhoI restriction digest. TDP-WT-V5, myc-TDP-WT, and TDP-Δ1–173-V5 were described in Dormann et al. (61). For TDP-NLSmut, amino acids 82–84 of TDP-WT-V5 were mutated to alanine by QuikChange mutagenesis (Stratagene) as described by Winton et al. (62). ALS-associated point mutations (A315T, M337V, and G348C) were introduced into myc-TDP-WT or TDP-NLSmut-V5 by QuikChange mutagenesis (Stratagene). NLSmut-ΔC encoding amino acids 1–273 of human TDP-43 was amplified by PCR and after BamHI/XbaI restriction digest was cloned into the pcDNA6/V5-His vector (Invitrogen) that contained a stop codon between the V5 and the polyhistidine tag sequence. GFP-tagged constructs were generated by subcloning the respective sequences into pEGFP-C1 (Clontech). For all constructs, sequence integrity was verified by sequencing. Oligonucleotides sequences are available upon request.
Human Post Mortem Tissue
Histological analysis included five cases of FTLD-TDP (FTLD-TDP subtype A (n = 2), subtype B (n = 2), and subtype C (n = 1) according to Mackenzie et al. (63)) and four ALS cases with TDP-43 pathology.
Immunocytochemistry and Immunohistochemistry
For immunocytochemistry of HeLa cells, cells were fixed for 15 min in 4% paraformaldehyde in PBS, permeabilized for 5 min in 0.2% Triton X-100 with 50 mm NH4Cl, and subsequently blocked for 20–30 min in 5% donkey serum in PBSS (PBS with 0.1% saponin). Cells were stained with the indicated primary and secondary antibodies diluted in 5% donkey serum in PBSS for 30 min and washed 3–5 times in PBSS. To visualize nuclei, cells were stained with TO-PRO-3 iodide (Invitrogen, 1:500 in PBS) for 15 min and washed 3 times in PBS. Coverslips were mounted onto glass slides using ProLong Gold Antifade Reagent (Invitrogen).
For immunocytochemistry of hippocampal neurons, neurons were fixed with 4% paraformaldehyde on DIV 9, quenched in 50 mm ammonium chloride for 10 min, and permeabilized with 0.1% Triton X-100 for 3 min. After blocking with 2% fetal bovine serum (Invitrogen), 2% bovine serum albumin (Sigma), and 0.2% fish gelatin (Sigma) dissolved in PBS, neurons were incubated with respective primary and secondary antibodies diluted in 10% blocking solution. 4′-6-Diamidino-2-phenyl-indol (DAPI, Invitrogen) was used as a nuclear counterstain.
Immunohistochemistry on human post mortem material was performed on 5-μm-thick sections of formalin-fixed, paraffin-embedded sections from spinal cord or hippocampus with the N- and C-terminal TDP-43-specific antibodies and anti-PABP-1 using the NovoLink™ Polymer Detection kit (Novocastra) and developed with 3,3′-diaminobenzidine. Microwave antigen retrieval was performed for all stainings. Double-label immunofluorescence for PABP-1 and pTDP-43 was performed using Alexa-488- and -594-conjugated secondary antibodies. DAPI (Vector Laboratories) was used for nuclear counterstaining.
Image Acquisition and Quantification
Confocal images were obtained with an inverted laser scanning confocal microscope (Zeiss Axiovert 200 m) with a 63×/1.4 N.A. oil immersion lens using a pinhole diameter of 1 Airy unit in the red channel. Pictures were taken and analyzed with the LSM 510 confocal software (Zeiss). For HeLa cells, single confocal images were taken in the plane of the largest cytosolic area. For neurons, a series of images along the z axis was taken and projected into a single image using the maximal projection tool of the LSM 510 software. Immunofluorescence images of brain sections were obtained by wide-field fluorescence microscopy (BX61 Olympus with digital camera F-view, Olympus).
Nuclear and cytosolic localization was quantified with the LSM 510 colocalization tool as described in Dormann et al. (33). Stress granule localization was quantified with Image J as follows. Image identity was blinded, and FUS or TDP-43 (green)/TIA-1 (red) double positive cytoplasmic granules as well as the entire cell were manually encircled to measure fluorescence intensities of the green channel. After background subtraction, the percentage of FUS or TDP-43 in SG was calculated. For each condition, 10–20 cells were analyzed. Means across all cells and standard deviations were calculated.
Cell Lysates and Immunoblotting
Total cell lysates were prepared in ice cold radioimmune precipitation assay (RIPA) buffer freshly supplemented with complete EDTA-free protease inhibitor mixture (Roche Applied Science) for 15 min on ice. Lysates were sonicated in a bioruptor (Diagenode, 45 s on high), and protein concentration was determined by BCA protein assay (Pierce). Equal amounts of protein were separated by SDS-PAGE, transferred onto a PVDF membrane (Immobilon-P, Millipore), and analyzed by immunoblotting using the indicated antibodies. Bound antibodies were detected with the chemiluminescence detection reagents ECL (Amersham Biosciences) or Immobilon (Millipore).
RNA Binding Assay
RNA binding of FUS and TDP-43 domains was determined in an in vitro RNA binding assay according to Lerga et al. (24). Briefly, proteins were in vitro transcribed and translated using the TNT T7 Coupled Reticulocyte Lysate System (Promega) and labeled with 20 μCi of [35S]methionine (Amersham Biosciences). Strep-Tactin-Sepharose (IBA) was blocked with 200 μg/ml yeast tRNA (Roche Applied Science) and 0.125 mg/ml bovine serum albumin (BSA, New England Biolabs) in wash buffer (10 mm Tris-HCl, pH 7.4, 100 mm NaCl, 2.5 mm MgCl2, 0.5 mm DTT, 0.5 mm EGTA, 0.5% Triton X-100, 10% glycerol) for 30 min at 4 °C. Afterward, biotinylated RNA oligonucleotides (UG12, UGUGUGUGUGUGUGUGUGUGUGUG; GGUG, UUGUAUUUUGAGCUAGUUUGGUGAU; CCUC, UUGUAUUUUGAGCUAGUUUCCUCAU, all from Thermo Fisher Scientific) were bound to the preblocked Sepharose beads. Beads were rinsed twice with wash buffer and incubated with radiolabeled samples for 10 min at 4 °C in blocking buffer. Beads were washed 5 times in wash buffer and boiled 5 min in Laemmli buffer. Bound radiolabeled proteins were separated by SDS-PAGE and visualized by fluorography. 10% of the radiolabeled sample was directly used for SDS-PAGE and fluorography to visualize the input of radiolabeled protein.
RESULTS
RGG-Zinc Finger Domain of FUS Is Most Important for SG Recruitment, Whereas Glutamine-rich Domain Is Dispensable
We and others previously found that cytosolically mislocalized FUS is recruited to SG upon cellular stress (33, 34, 41). It is, however, still unknown whether cytosolic FUS is recruited into SG via its bound mRNAs or via protein-protein interactions, involving for example its TIA-1-related N-terminal domain or even both. To determine which domains are responsible for recruitment of FUS to SG, we expressed individual FUS domains in HeLa cells with an N-terminal HA tag and analyzed their SG recruitment in comparison to full-length FUS (see Fig. 1A for a schematic diagram). To this end we introduced the P525L mutation into the PY-NLS of the respective constructs, because this mutation causes cytosolic retention of FUS and allows its efficient recruitment to TIA-1-positive SG (33). In contrast, constructs carrying a wild-type (WT) PY-NLS were almost exclusively nuclear and, hence, were not recruited to SG upon cellular stress (supplemental Fig. S1).
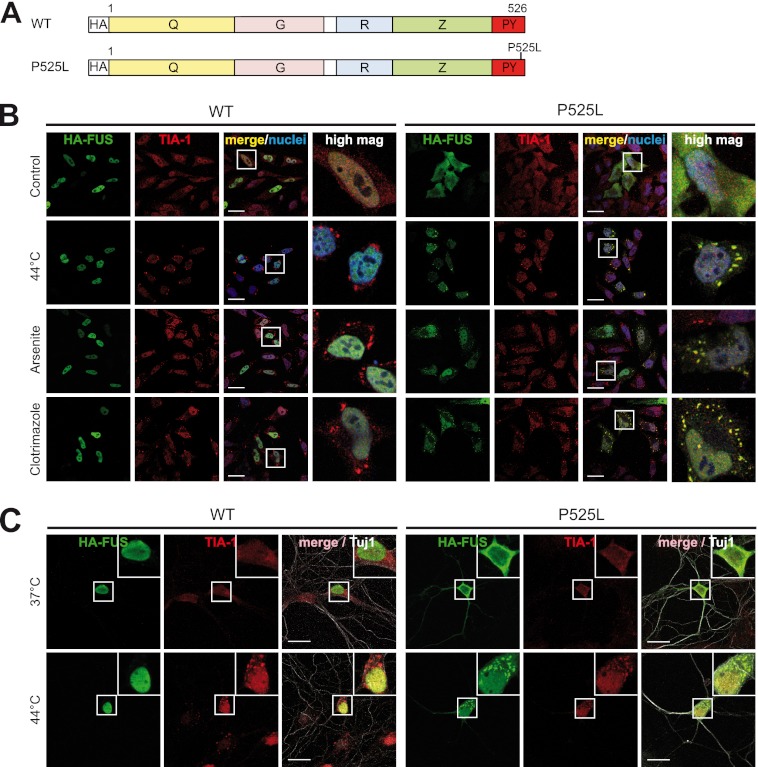
FIGURE 1.
Cytosolic FUS is recruited to SG upon treatment with various stressors. A, shown is a schematic diagram of FUS wild-type (WT) and P525L mutant used for transient transfection in HeLa cells. HA, HA epitope tag; Q, glutamine-rich domain; G, glycine-rich domain; R, RRM domain; Z, arginine-glycine-glycine (RGG) zinc finger domain. B, HeLa cells were transiently transfected with N-terminal-HA-tagged FUS-WT or FUS-P525L. 24 h after transfection cells were subjected to heat shock (44 °C for 1 h), sodium arsenite (0.5 mm for 30 min), or clotrimazole (20 μm for 30 min) or were left untreated (Control). Cells were fixed, stained with an HA-specific antibody (green), a TIA-1-specific antibody (red), and a nuclear counterstain (blue) and analyzed by confocal microscopy. Panels to the right show a higher magnification of the boxed region. Although FUS-WT remained nuclear, FUS-P525L was sequestered into SG under all stress conditions examined. Scale bars, 20 μm. C, primary rat hippocampal neurons were transiently transfected with HA-FUS-WT or P525L on DIV 7. 48 h after transfection, neurons were subjected to heat shock (44 °C) for 1 h or left untreated (37 °C). Neurons were fixed and stained with an HA-specific antibody (green), a TIA-1-specific antibody (red), and the neuronal marker antibody Tuj1 (white) to visualize neuronal morphology. FUS-P525L showed cytoplasmic mislocalization and was recruited to TIA-1-positive SG upon heat stress. Insets in the upper right corner show a higher magnification of the boxed region. Scale bars, 20 μm.
Initially, we investigated a variety of stress conditions such as heat shock (44 °C), oxidative stress caused by sodium arsenite treatment, and mitochondrial stress caused by clotrimazole treatment for their ability to induce SG formation. Consistent with our previous findings (33), FUS-WT was located in the nucleus and, therefore, was not recruited to SG upon cellular stress (Fig. 1B, upper panels). In contrast, FUS-P525L was efficiently recruited into SG under all stress conditions examined (Fig. 1B, lower panels). FUS-P525L-positive granules were bona fide stress granules, as they were co-localized with the SG marker TIA-1 and disassembled upon cycloheximide treatment or recovery from heat stress (supplemental Fig. S2A). SG recruitment of FUS-P525L was not a cell type-specific phenomenon, as it could also be observed in primary hippocampal neurons (Fig. 1C) and in SH-SY5Y neuroblastoma cells (supplemental Fig. S2B).
Using heat shock as stress condition, we next examined how well the individual domains of FUS are recruited to SG (see the schematic diagram in Fig. 2A). To obtain quantitative information, we measured the percentage of FUS protein localized in TIA-1-positive SG (see “Experimental Procedures” for details). In contrast to full-length FUS-P525L, the glutamine-rich domain (Q) remained diffusely distributed in the cytosol after heat shock, and no granular localization became evident even though TIA-1-positive SG formed in transfected cells (Fig. 2, B and C). Thus, despite its homology to the prion-like domain of TIA-1 (48, 49), the Q domain of FUS does not seem to be involved in SG recruitment. The glycine-rich domain (G) and the RRM domain (R) remained predominantly diffusely cytosolic, but small amounts were found in TIA-1-positive SG (Fig. 2, B and C). Finally, the C-terminal RGG-zinc finger domain (Z), which was rendered cytosolic by addition of the P525L mutation (ZP525L), showed more SG recruitment than all other domains examined. Because the HA-tagged Q and ZP525L domain showed very weak expression compared with the other constructs and could not be detected by Western blot (supplemental Fig. S3), we expressed these apparently unstable domains as GFP fusion proteins along with GFP-FUSP525L as a control. This yielded higher expression levels (supplemental Fig. S4A) and confirmed that the ZP525L domain shows SG association, whereas the Q domain does not (supplemental Fig. S4B).
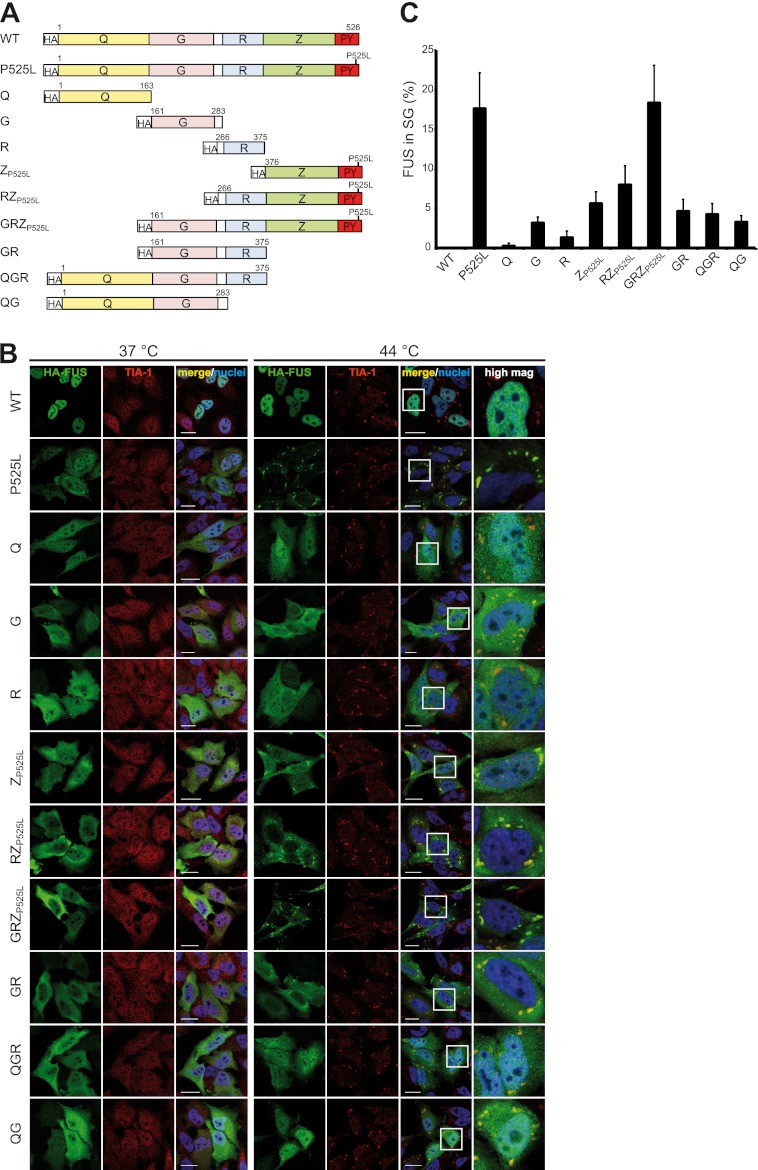
FIGURE 2.
The C-terminal RGG-zinc finger domain of FUS is the most important domain for SG recruitment. A, shown is a schematic diagram of different FUS constructs analyzed for SG recruitment. The P525L mutation was introduced into the PY-NLS to obtain proteins mislocalized in the cytosol. B, shown is immunocytochemistry of HeLa cells expressing the different FUS constructs shown in A. Before fixation, cells were subjected to heat shock (44 °C for 1 h) or left untreated (37 °C). Cells were stained with an HA-specific antibody (green), a TIA-1-specific antibody (red), and a nuclear counterstain (blue) and analyzed by confocal microscopy. Panels to the right show a higher magnification of the boxed region. The Z domain is most important for SG recruitment, whereas the Q domain is dispensable. The G and R domains also contribute to SG recruitment but to a lesser extent than Z. Scale bars = 20 μm. C, the percentage of FUS localized in TIA-1-positive SG was quantified using ImageJ. 10–20 cells were analyzed in a blinded manner, means across all cells were calculated, and S.D. are indicated by error bars. Note that the percentage of FUS-P525L in SG seems surprisingly low when looking at the corresponding confocal images in B. However, SG are very small compared with the remaining cellular volume, and therefore, FUS-P525L diffusely distributed in the cytosol and nucleus amounts to a significant percentage of the total protein (more than 80%).
Because our quantitative analysis revealed that none of the individual domains was recruited to SG to the same extent as the full-length protein (Fig. 2C), we analyzed combinations of the three domains (RZP525L, GRZP525L, and GR) and asked if this would enhance SG recruitment. Indeed, the combination of these domains showed an additive effect compared with the individual domains (Fig. 2, B and C), suggesting that all three domains contribute to SG recruitment. Finally, we analyzed combinations of the Q domain with other domains (QGR, QG), to exclude that the Q domain might have a different effect in the context of the other domains. However, QGR and QG did not differ in their SG localization from GR and G, respectively, demonstrating that the Q domain is indeed dispensable for SG recruitment. Consistently, the GRZP525L protein, which lacks the Q domain, was recruited to SG equally well as full-length FUS-P525L. Furthermore, the relatively weak SG recruitment efficiency of the QGR protein confirms that the C-terminal Z domain plays the most important role for SG association of FUS.
In summary, the RGG-zinc finger domain (Z) is the most important domain for SG recruitment of FUS. The glycine-rich domain (G) and to a minor extent the RRM domain (R) also contribute to SG recruitment, whereas the prion-like glutamine-rich domain (Q) is dispensable.
RGG-Zinc Finger Domain Is Main RNA Binding Domain of FUS
To explore if SG recruitment of the different FUS domains can be correlated with their ability to bind RNA, we examined their RNA binding capacity in an RNA binding assay. To this end we in vitro translated the same FUS constructs and performed a pulldown assay with biotinylated RNA oligonucleotides immobilized on streptavidin beads. Because FUS is known to preferentially bind to UG-rich sequences, specifically to oligonucleotides containing a GGUG motif (24), we first tested the ability of FUS-WT to bind to UG12 or a GGUG-containing oligonucleotide (UUGUAUUUUGAGCUAGUUUGGUGAU, named GGUG). The same oligonucleotide with a CCUC motif (named CCUC) was used as a negative control. Consistent with the previously reported finding that FUS binds to UG-rich sequences, FUS-WT was efficiently pulled down by UG12 and to a lesser extent by GGUG but not CCUC (Fig. 3A).
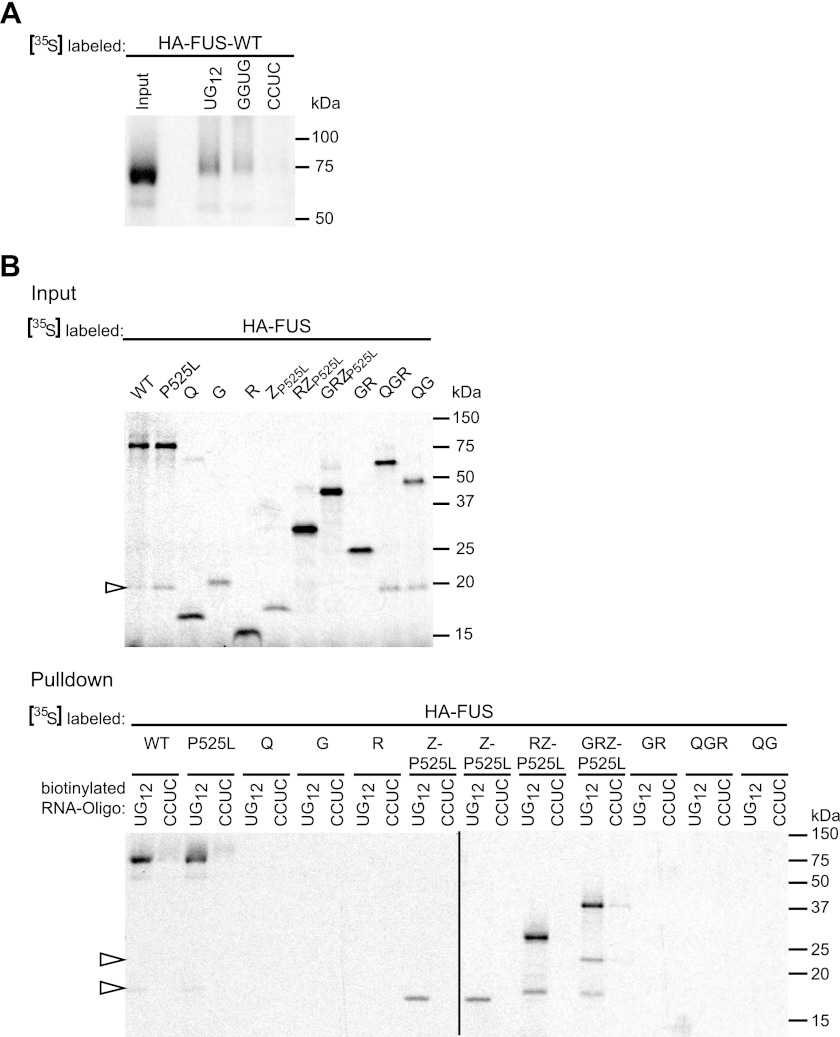
FIGURE 3.
The RGG-zinc finger domain of FUS binds to UG-rich RNA. A, FUS-WT was in vitro translated in the presence of [35S]methionine (left lane, input) and was analyzed for binding to different RNA oligonucleotides immobilized on streptavidin beads (right lanes, UG12, UGUGUGUGUGUGUGUGUGUGUGUG; GGUG, UUGUAUUUUGAGCUAGUUUGGUGAU; CCUC, UUGUAUUUUGAGCUAGUUUCCUCAU). FUS was pulled down most efficiently by UG12 and to a lesser extent by GGUG RNA. B, the indicated FUS constructs were in vitro translated in the presence of [35S]methionine (upper panel, Input). Biotinylated UG12 RNA immobilized on streptavidin beads was used to pull down radioactively labeled proteins (lower panel, Pulldown). CCUC RNA was used as a negative control. FUS-WT and P525L and all proteins comprising the ZP525L domain were specifically pulled down by UG12 RNA, whereas the other proteins did not show detectable RNA binding. Open arrowheads indicate degradation products.
Using UG12 as RNA bait, we next examined the RNA binding capacity of the individual FUS domains and combinations thereof (Fig. 3B). This demonstrated that only proteins containing the Z domain (ZP525L, RZP525L, and GRZP525L) showed efficient and selective binding to UG12 RNA (note that compared with ZP525L, RZP525L and GRZP525L showed stronger signals already in the input gel, and therefore, signals obtained in the pulldown assay cannot be compared directly). In contrast, the Q, G, and R domain and different combinations thereof (GR, QGR, QG) were not pulled down in our RNA binding assay, demonstrating that Q, G, and R show no or only weak binding to UG12 RNA. Thus, the C-terminal Z domain seems to be responsible for the preferential binding to UG-rich RNA. Interestingly, the domain with the highest RNA binding capacity (Z) was the one most important for SG recruitment (Fig. 2). This correlation suggests that FUS might be recruited to SG by virtue of its RNA binding capacity. The domains that contribute to SG recruitment to a lesser extent (G and R) showed no RNA binding capacity in our in vitro binding assay, suggesting that they might contribute to SG recruitment through other means, possibly protein-protein interactions.
Cytosolic Mislocalization Is Prerequisite for SG Recruitment of TDP-43
Similar to FUS, TDP-43 has been described to be localized in SG under various experimental conditions (31, 50–55). Because cytosolic mislocalization is a prerequisite for efficient SG recruitment of FUS (Fig. 1 and Refs 33 and 41), we speculated that this might also be the case for TDP-43. To test this hypothesis, we mutated three essential amino acids of the classical bipartite nuclear localization signal (62) and analyzed SG recruitment of this artificial NLS mutant (NLSmut, see Fig. 4A for a schematic diagram) in comparison to wild-type TDP-43 (TDP-WT) upon exposure to different stressors. TDP-WT was predominantly nuclear with and without stress and was not detectable in cytoplasmic granules (Fig. 4B, upper panels). In contrast, the partially cytosolic NLSmut protein was readily detectable in TIA-1-positive SG upon heat shock, clotrimazole treatment, and sodium arsenite treatment (Fig. 4B, lower panels). TDP-43-positive granules dissolved upon cycloheximide treatment or recovery from heat stress (supplemental Fig. S5A), demonstrating that they are indeed SG and not protein aggregates. Moreover, SG recruitment of NLSmut but not TDP-WT was observed in primary hippocampal neurons (Fig. 4C) and SH-SY5Y cells (supplemental Fig. S5B). Thus, in all cell types examined, only cytosolic but not nuclear TDP-43 is efficiently recruited to SG.
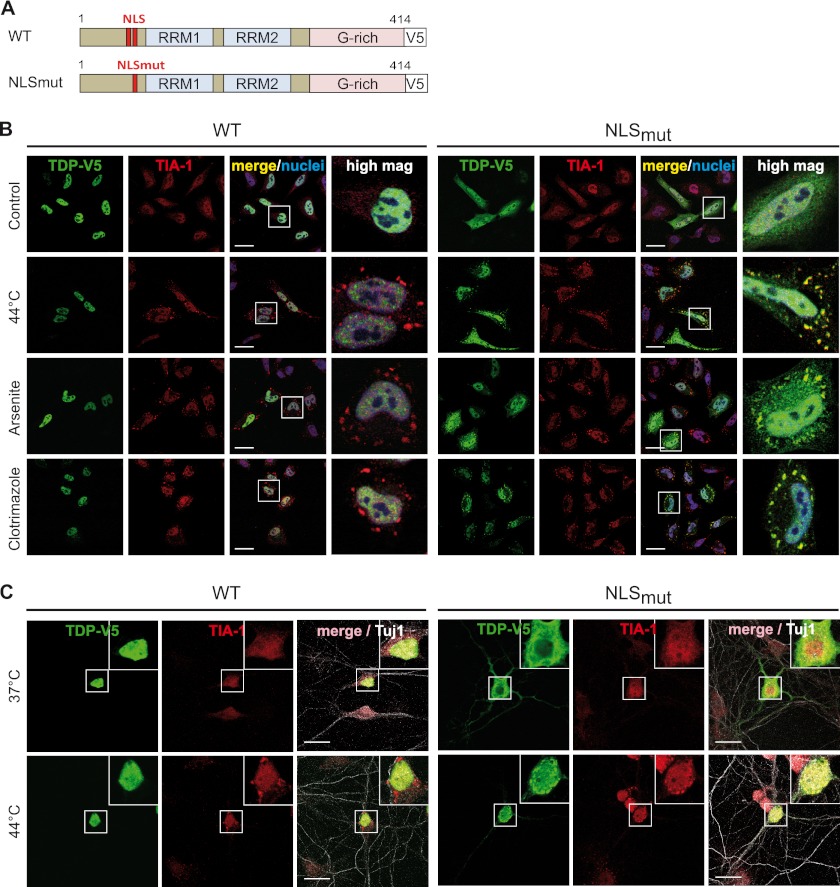
FIGURE 4.
Cytosolic mislocalization is a prerequisite for SG recruitment of TDP-43. A, shown is a schematic diagram of TDP-43 wild-type (WT) and NLS mutant (NLSmut). NLSmut, triple point mutation in the classical nuclear localization signal (K83A/R84A/K85A); G-rich, glycine-rich domain; V5, V5 epitope tag. B, C-terminal-V5-tagged TDP-WT or NLSmut were transiently transfected into HeLa cells and 24 h later were subjected to heat shock (44 °C for 1 h), sodium arsenite (0.5 mm for 30 min), or clotrimazole (20 μm for 30 min) treatment or were left untreated (Control). Cells were fixed, stained with a V5 (green) and TIA-1 (red)-specific antibody and a nuclear counterstain (blue), and analyzed by confocal microscopy. Panels to the right show a higher magnification of the boxed region. Although the cytosolic NLS mutant was sequestered into SG, TDP-WT remained nuclear under all stress conditions examined. Scale bars = 20 μm. C, primary rat hippocampal neurons were transiently transfected with V5-tagged TDP-WT or NLSmut. 48 h post-transfection, neurons were subjected to heat shock (44 °C) for 1 h or left untreated (37 °C). Neurons were fixed and stained with a V5-specific antibody (green), a TIA-1-specific antibody (red), and the neuronal marker antibody Tuj1 (white) to visualize neuronal morphology. NLSmut showed partial cytoplasmic mislocalization and was recruited to TIA-1-positive SG upon heat stress. Insets in the upper right corner show a higher magnification of the boxed region. Scale bars, 20 μm.
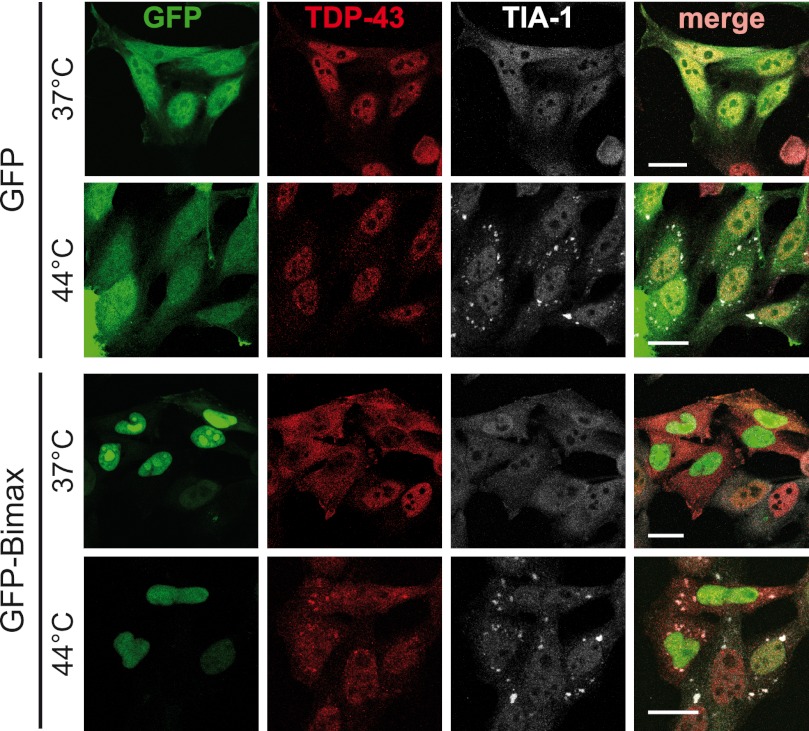
To corroborate this finding, we expressed an Importin α/β inhibitor peptide fused to GFP (GFP-Bimax) (64) in HeLa cells. Consistent with our previous findings (33), this caused endogenous TDP-43 to accumulate in the cytosol (Fig. 5). In line with the view that cytosolic mislocalization of TDP-43 is required for SG recruitment, endogenous TDP-43 was detectable in heat shock-induced SG only when its nuclear import was blocked by expression of the GFP-Bimax inhibitor (Fig. 5). In contrast, in control (GFP)-transfected cells, TDP-43 remained nuclear and was not detectable in SG upon heat shock. Together these findings demonstrate that similar to FUS, SG recruitment of TDP-43 requires at least a partial mislocalization of the nuclear protein to the cytosol.
FIGURE 5.
Endogenous TDP-43 is sequestered into heat shock-induced SG upon inhibition of Importin α/β-dependent nuclear import. HeLa cells were transfected with an Importin α/β-specific peptide inhibitor fused to GFP (GFP-Bimax) or GFP as a control (green). 24 h post-transfection cells were subjected to heat shock (44 °C for 1 h) or kept at control temperature (37 °C) before fixation. Cells were co-stained for endogenous TDP-43 (red) and TIA-1 (white) and analyzed by confocal microscopy. Expression of GFP-Bimax resulted in cytosolic mislocalization of endogenous TDP-43 and recruitment into SG upon heat shock. Under control conditions (GFP), TDP-43 was predominantly nuclear and did not colocalize with SG after heat shock. Scale bars = 20 μm.
ALS-associated TARDBP Mutations Do Not Affect Nuclear Localization or SG Recruitment
Despite extensive research over the last few years, the pathogenic mechanism of ALS-associated TARDBP mutations remains unclear. Some TARDBP mutations have been reported to cause cytosolic missorting of the protein (30, 31); however, this could not be confirmed in other studies (28, 32, 51). Furthermore, it is still not clear if and how TARDBP mutations affect SG recruitment, as controversial findings have been reported (31, 51, 54).
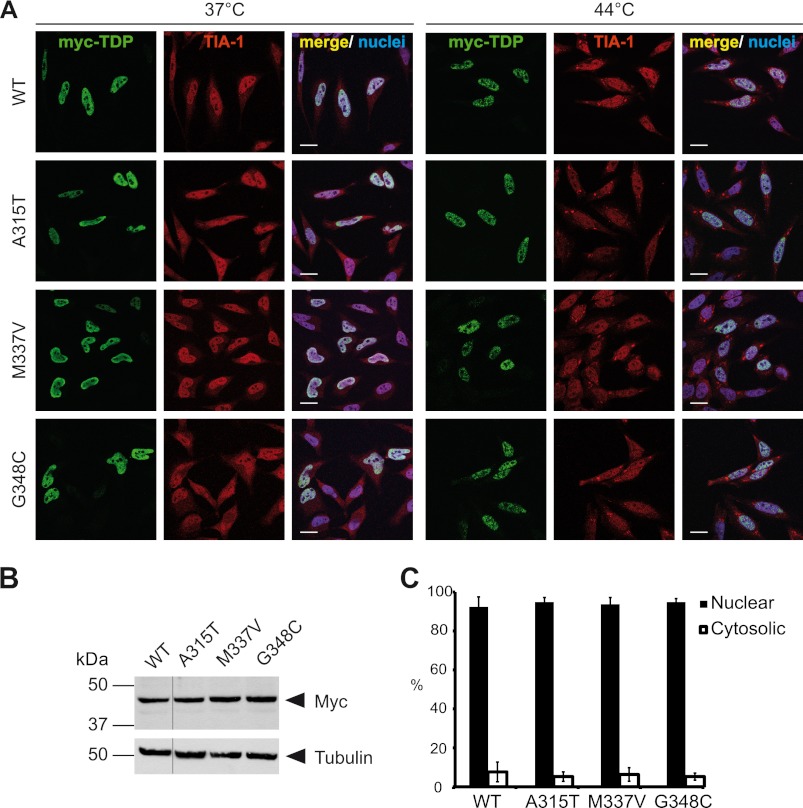
Because our data above imply that SG recruitment could be indicative of cytosolic mislocalization, we examined the localization of three well studied ALS-associated TARDBP mutations (A315T, M337V, and G348C) upon heat shock to see if these mutants would be preferentially detected in SG. However, none of the three examined ALS-associated point mutants showed detectable localization in TIA-1-positive SG upon heat shock (44 °C); instead, they were located entirely in the nucleus like TDP-WT (Fig. 6A; for expression levels see Fig. 6B). Consistently, a quantification of the amount of nuclear/cytosolic TDP-43 in cells cultured under normal culture conditions (37 °C) demonstrated that the three point mutants had an almost exclusive nuclear localization and did not differ from the WT protein (Fig. 6C). Thus, the examined ALS-associated TARDBP mutations (A315T, M337V, and G348C) do not cause cytoplasmic mislocalization or SG recruitment of TDP-43.
FIGURE 6.
ALS-associated TARDBP mutations do not alter the cellular localization of TDP-43. A, Myc-tagged wild-type TDP-WT or TDP-43 carrying the indicated ALS-associated point mutations (A315T, M337V, or G348C) were transiently transfected into HeLa cells. 24 h post-transfection cells were subjected to heat shock (44 °C for 1 h) or kept at control temperature (37 °C). Afterward cells were fixed, stained with a myc (green) and TIA-1 (red)-specific antibody and a nuclei counterstain (blue), and analyzed by confocal microscopy. Both TDP-WT and the ALS-associated point mutants were nuclear under control conditions and remained nuclear upon heat shock. Scale bars = 20 μm. B, shown are expression levels of TDP-43 constructs used in A. Total cell lysates were analyzed by immunoblotting with a myc-specific antibody (upper panel). Tubulin served as a loading control (lower panel). All lanes were from the same exposure of the same blot. C, quantification of nuclear and cytosolic fluorescence intensities of myc staining at 37 °C is shown. Error bars indicate S.D.
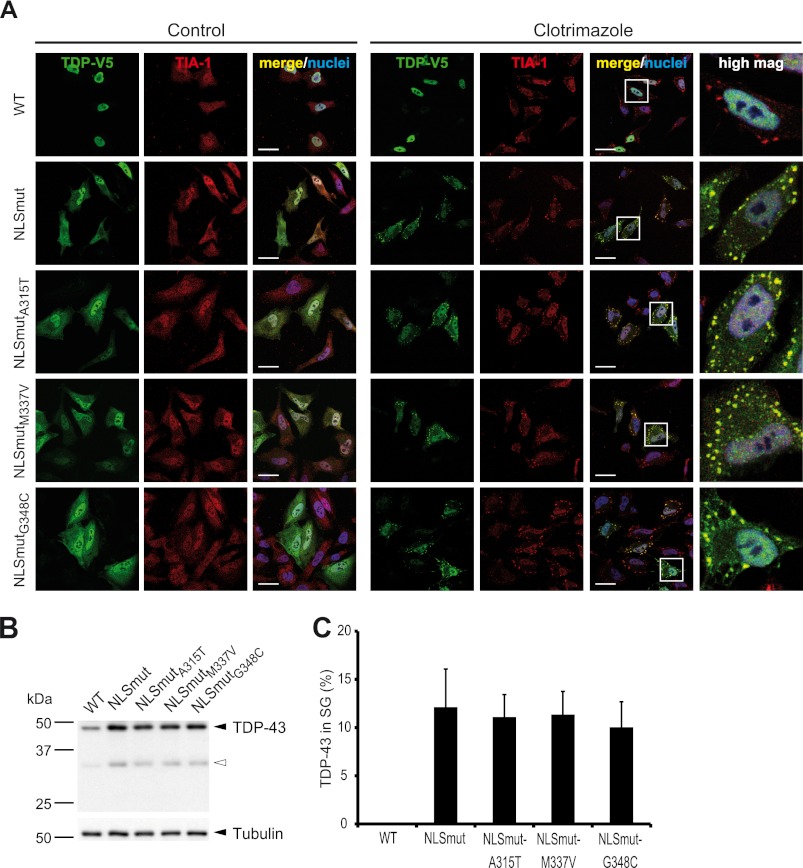
Because it is possible that TARDBP mutations may affect SG recruitment once the protein has accumulated in the cytosol, for example as a consequence of axonal injury (65) or reduced expression of nuclear import factors (66), we introduced the same ALS-associated TARDBP mutations into the NLS mutant of TDP-43 to see if the mutations would impair or enhance SG recruitment of cytosolic TDP-43. As expected, TDP-NLSmutA315T, NLSmutM337V, and NLSmutG348C showed the same cytosolic mislocalization as NLSmut (Fig. 7A, left panels) and similar expression levels (Fig. 7B). Upon cellular stress elicited by clotrimazole treatment, all mutants were readily detectable in TIA-1-positive SG (Fig. 7A, right panels). A quantitative analysis of SG recruitment showed that the ALS-associated TARDBP mutants were incorporated into SG to a similar degree as NLSmut (Fig. 7C). Taken together, the examined ALS-associated point mutations in the glycine-rich C-terminal domain of TDP-43 (A315T, M337V, and G348C) neither cause cytosolic mislocalization of nuclear TDP-43 nor do they affect SG recruitment of cytosolic TDP-43.
FIGURE 7.
ALS-associated TARDBP mutations do not affect SG recruitment of cytosolic TDP-43. ALS-associated point mutations (A315T, M337V, G348C) were introduced into the TDP-43 NLS mutant (NLSmutA315T, NLSmutM337V, NLSmutG348C), and the effect of mutations on SG recruitment was analyzed. A, HeLa cells transiently transfected with the indicated TDP-43 constructs were incubated with clotrimazole for 30 min or left untreated (Control). Cells were fixed, stained with a V5 (green) and TIA-1 (red)-specific antibody and a nuclear counterstain (blue), and analyzed by confocal microscopy. ALS-associated point mutations did not affect SG recruitment of cytosolic TDP-43. Scale bars = 20 μm. B, protein levels in total cell lysates were analyzed by immunoblotting with a V5-specific antibody (upper panel), and tubulin served as a loading control (lower panel). The black arrowhead indicates full-length TDP-43, and the white arrowhead indicates caspase-generated 35-kDa CTF frequently observed under transient transfection conditions (61, 75). C, the percentage of TDP-43 localized in TIA-1-positive SG was quantified using ImageJ. 15–20 cells were analyzed in a blinded manner, means across all cells were calculated, and S.D. are indicated by error bars.
TDP-43 Inclusions in Spinal Cord but Not in Cortex Contain SG Marker PABP-1
Even though TDP-43 can be recruited to SG under various experimental conditions (this study and Refs. 31, 50, 51, 53, and 56), it is still controversial whether TDP-43 inclusions in ALS/FTLD patients contain SG marker proteins. Two studies showed a lack of SG markers in TDP-43 inclusions (33, 50), whereas two other studies reported co-labeling of TDP-43 inclusions with SG markers (31, 57). We reasoned that these discrepancies might be due to the fact that TDP-43 inclusions differ in their TDP-43 species composition, with inclusions in the spinal cord of ALS and FTLD patients containing predominantly full-length TDP-43 and inclusions in the cortex and hippocampus of ALS and FTLD patients being highly enriched for C-terminal fragments (CTFs) of ∼25 kDa (59, 60).
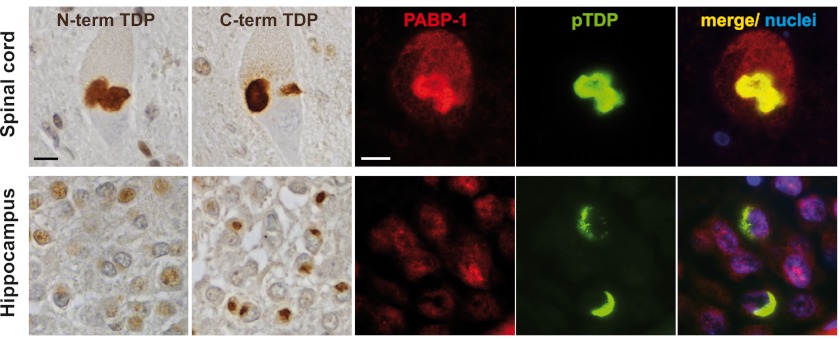
To see if differences in the TDP-43 species composition might account for the different results regarding co-labeling of TDP-43 inclusions with SG markers, we stained sections of spinal cord or cortex (hippocampus) from ALS-TDP and FTLD-TDP cases with N- and C-terminal TDP-43 antibodies as well as antibodies specific for the SG marker protein PABP-1. Inclusions in the spinal cord were consistently labeled with both N- and C-terminal TDP-43 antibodies, whereas inclusions in the cortex, including those in dentate granule neurons, were only labeled with the C-terminal antibody (Fig. 8), confirming previous results (59). Cortical TDP-43 inclusions were not immunoreactive for PABP-1, confirming our previous results (33). However, we revealed PABP-1 positivity in a subset (∼66%) of TDP-43-positive inclusions in the spinal cord, as demonstrated by double-label immunofluorescence (Fig. 8 and supplemental Fig. S6). Thus, TDP-43 inclusions in spinal cord and cortex show a differential co-labeling for the SG marker protein PABP-1.
FIGURE 8.
TDP-43 inclusions in spinal cord but not hippocampus are frequently co-labeled for the SG marker protein PABP-1. TDP-43 immunohistochemistry performed on formalin-fixed, paraffin-embedded tissue sections of spinal cord (upper panels) or hippocampus (lower panels) from FTLD-TDP and ALS-TDP cases is shown. Staining with N-terminal and C-terminal TDP-43-specific antibodies demonstrated labeling of neuronal cytoplasmic inclusions in motor neurons in the spinal cord with both antibodies (ALS case #1 shown), whereas inclusions in dentate granule neurons in the hippocampus were labeled only with the C-terminal antibody (FTLD-TDP case #1 shown). Double-label immunofluorescence stainings of the same cases showed co-labeling of phospho-TDP-43 positive inclusions (green) with the SG marker protein PABP-1 (red) in the spinal cord but not in cortical inclusions. Nuclei were stained with DAPI (blue). Scale bars = 10 μm.
25-kDa CTF of TDP-43 Is Not Recruited to SG
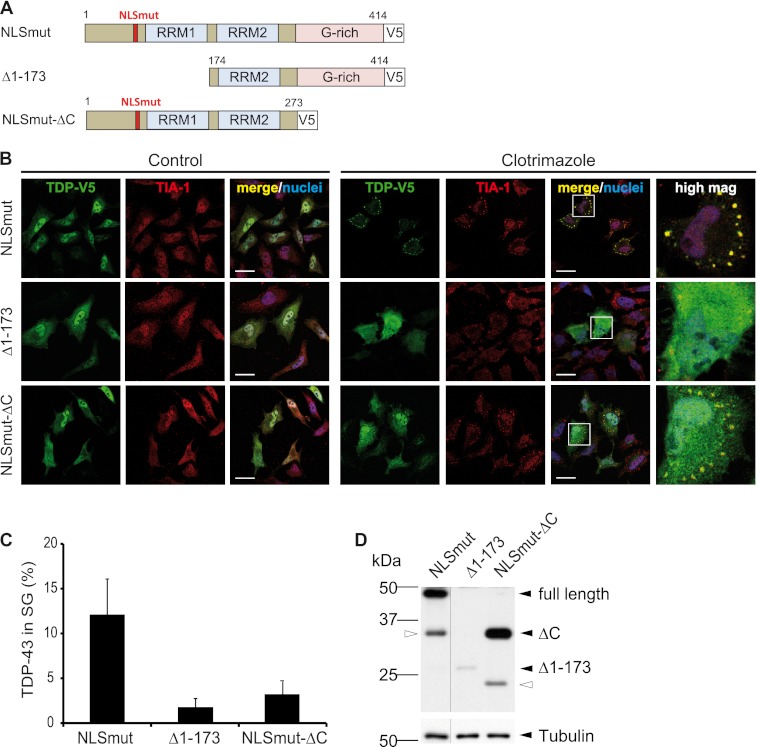
We speculated that the differences in SG marker co-labeling of TDP-43 inclusions in different tissues may be due to the presence of different TDP-43 species, with distinct abilities to be recruited to SG. Because inclusions in cortex are highly enriched in CTFs of TDP-43 (59, 60) and are negative for PABP-1 (Fig. 8), we wondered if CTFs may be unable to associate with SG. To test this hypothesis, we expressed a CTF of ∼25 kDa (TDPΔ1–173, see Fig. 9A for a schematic diagram) in HeLa cells and analyzed its recruitment to TIA-1-positive SG upon cellular stress. Like NLSmut, TDPΔ1–173 was partially localized in the cytosol under control conditions (Fig. 9B, left panels), as it lacks the N-terminal domain including the protein NLS (Fig. 9A). However, in contrast to NLSmut, TDPΔ1–173 remained diffusely distributed in the cytosol upon clotrimazole treatment and, consistent with our hypothesis, was very poorly incorporated into TIA-1-positive SG (Fig. 9, B, right panels, and C). Because TDPΔ1–173 showed very low expression levels compared with NLSmut (Fig. 9D), we repeated the experiment with GFP-tagged TDPΔ1–173 to exclude that the lack of SG association was simply due to low protein levels. Similar to V5-tagged TDPΔ1–173, the highly expressed GFP-tagged TDPΔ1–173 remained diffusely distributed upon cellular stress (supplemental Fig. S7, A and B). Thus, independent of expression levels, the 25-kDa CTF fails to associate with SG. This might explain why cortical TDP-43 inclusions, which are highly enriched in CTFs and contain little full-length TDP-43 (Fig. 8 and Refs. 59 and 60) are not co-labeled with SG marker proteins.
FIGURE 9.
25 kDa C-terminal fragment and a C-terminal deletion mutant of TDP-43 are poorly sequestered into SG. A, shown is a schematic diagram of TDP-43 deletion mutants analyzed for SG recruitment. The deletion mutant Δ1–173 was chosen to mimic the 25-kDa CTF found to be deposited in cortical regions of FTLD-TDP patients (4, 59). The C-terminal deletion mutant (NLSmutΔC) lacks the prion-like glycine-rich domain. B, the indicated TDP-43 constructs were transiently transfected in HeLa cells. Before fixation, cells were treated with clotrimazole (20 μm, 30 min) or left untreated (control). Subsequently, cells were stained with a V5 (green) and TIA-1 (red)-specific antibody and a nuclear counterstain (blue) and analyzed by confocal microscopy. Panels to the right show a higher magnification of the boxed region. In contrast to full-length TDP-NLSmut, both deletion mutants remained predominantly diffuse in the cytosol upon heat shock and were poorly recruited to SG. Scale bars = 20 μm. C, the percentage of TDP-43 localized in TIA-1-positive SG was quantified using ImageJ. 15–20 cells were analyzed in a blinded manner, means across all cells were calculated, and S.D. are indicated by error bars. D, protein levels in total cell lysates were analyzed by immunoblotting with a V5-specific antibody (upper panel); tubulin served as a loading control (lower panel). Black arrowheads indicate full-length TDP-NLSmut or the two deletion mutants, and white arrowheads indicate degradation products. All lanes were from the same exposure of the same blot.
C-terminal Glycine-rich Domain of TDP-43 Is Required for Efficient SG Recruitment
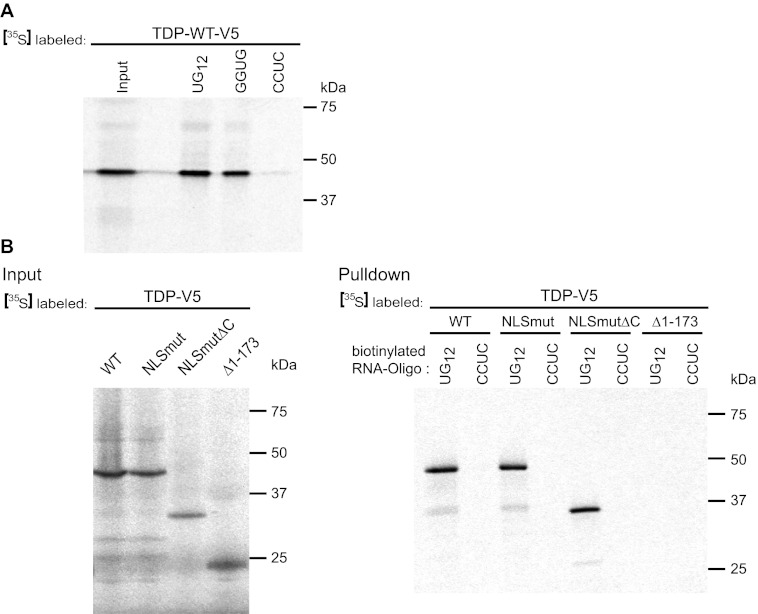
Surprisingly, not only deletion of amino acids 1–173 but also deletion of the C-terminal glycine-rich domain from TDP-43 NLSmut (NLSmut-ΔC, see the schematic diagram in Fig. 9A) led to a strong reduction in SG recruitment, as NLSmut-ΔC remained mostly diffusely distributed in the cytoplasm upon cellular stress (Fig. 9, B and C). Expression level differences could not account for this effect, as NLSmut-ΔC was at least as well expressed as NLSmut (Fig. 9D). To test whether reduced RNA binding may be responsible for reduced SG recruitment of NLSmut-ΔC, we performed an RNA binding assay using UG12 RNA as a TDP-43-specific target sequence (12). As expected, TDP-WT was efficiently pulled down by UG12 (Fig. 10A). Interestingly, TDP-WT bound equally well to GGUG but not the corresponding CCUC oligonucleotide, consistent with the recent finding that TDP-43 can bind to sequences other than UG repeats (67).
FIGURE 10.
The C-terminal deletion mutant of TDP-43 still binds to UG12 RNA. A, TDP-WT was in vitro translated in the presence of [35S]methionine (left lane, Input) and was analyzed for binding to different RNA oligonucleotides immobilized on streptavidin beads (right lanes, UG12, UGUGUGUGUGUGUGUGUGUGUGUG; GGUG, UUGUAUUUUGAGCUAGUUUGGUGAU; CCUC, UUGUAUUUUGAGCUAGUUUCCUCAU). TDP-43 bound to both UG12 and GGUG RNA but not to CCUC RNA). B, the indicated TDP-43 constructs were in vitro translated in the presence of [35S]methionine (upper panel, Input). Biotinylated UG12 RNA or CCUC control RNA were immobilized on streptavidin beads and were used to pull down radioactively labeled proteins (lower panel, Pulldown). TDP-WT and NLSmut as well as the C-terminal deletion mutant NLSmut-ΔC were specifically pulled down by UG12 RNA, whereas the Δ1–173 deletion mutant resembling the 25-kDa CTF did not bind to UG12 RNA.
Using UG12 as the RNA bait, we next compared the RNA binding capacity of full-length TDP-43 (WT and NLSmut) and the two deletion mutants NLSmutΔC and Δ1–173. The full-length proteins were specifically pulled down in our RNA binding assay, whereas TDP-Δ1–173, which lacks the protein main RNA recognition motif (RRM1) (12), failed to bind to UG12 RNA (Fig. 10B), which might explain why this fragment is not recruited to SG in FTLD/ALS patients (Fig. 8) and in cultured cells (Fig. 9). In contrast, NLSmut-ΔC bound to UG12 RNA as efficiently as full-length TDP-43 (Fig. 10B), demonstrating that the reduced SG recruitment capacity of this deletion mutant cannot be explained by reduced RNA binding. Thus, the glycine-rich domain seems to possess other, so far unknown features that are important for SG recruitment.
In summary, RNA binding of TDP-43 depends on the N-terminal RRM1 domain but not the C-terminal glycine-rich domain. Because both domains are required for SG recruitment of TDP-43, we suggest that RNA binding plus additional features encoded in the C-terminal glycine-rich domain such as protein-protein interactions might contribute to SG recruitment of TDP-43.
DISCUSSION
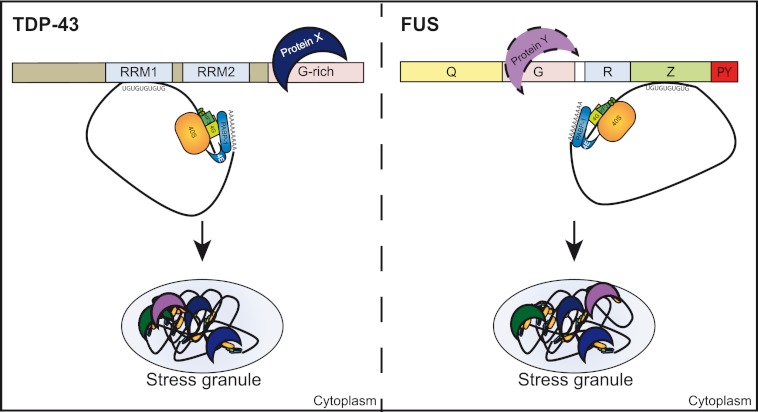
Two different mechanisms of how FUS and TDP-43 are recruited to SG can be envisaged. First, because both proteins have multiple RNA binding motifs (12, 24, 68), it is conceivable that they are recruited into SG via bound mRNAs. Second, protein-protein interactions with other SG-associated proteins could be involved. Our results suggest that RNA binding plays a crucial role for SG recruitment of both FUS and TDP-43, as deletion mutants lacking the principal RNA binding domains (Z domain of FUS and RRM1 domain of TDP-43) showed poor recruitment to SG. This correlation between RNA binding and SG recruitment suggests that FUS and TDP-43 might be recruited into SG through binding to UG-rich RNA sequences (see the model in Fig. 11), although we cannot exclude that protein-protein interactions mediated by the Z and RRM1 domain are involved as well.
FIGURE 11.
Model of SG recruitment of TDP-43 and FUS. Upon cellular stress, translation of mRNAs is arrested and translationally silent preinitiation complexes that contain mRNA, the small ribosomal subunit (40 S), early initiation factors (e.g. eIF3, eIF4A, eIF4G), and PABP-1 are packaged into SG. We suggest that recruitment of TDP-43 (left) or FUS (right) into SG involves both protein-RNA and protein-protein interactions. TDP-43 and FUS bind to UG-rich mRNA sequences via their main RNA binding domain (RRM1 in TDP-43 and RGG-zinc finger (Z) domain in FUS, respectively) and thus might be recruited into SG via their associated mRNAs. Because additional domains that did not show binding to UG-rich RNA in our RNA binding assay also contribute to SG recruitment of TDP-43 and FUS, we suggest that additional protein-protein interactions with proteins X and Y are involved in SG recruitment of TDP-43 and FUS.
In addition, domains that did not bind to UG-rich RNA in our in vitro assay (G and R domain of FUS and the C-terminal glycine-rich domain of TDP-43) seem to contribute to SG recruitment as well, as deletion of these domains impaired SG recruitment. Given their lack of RNA binding, these domains may contribute to SG recruitment by providing protein-protein interactions with other SG proteins (symbolized by protein X and Y in Fig. 11). However, we cannot exclude that these putative protein-interacting domains bind to RNA sequences not represented in our in vitro binding assay and that these protein-RNA interactions contribute to SG recruitment. Indeed, recent cross-linking and immunoprecipitation (CLIP) experiments have shown that FUS can bind to AU-rich stem loop structures (69) and TDP-43 can bind to sequences other than UG repeats (67). Which domain(s) of FUS and TDP-43 mediates binding to these alternative target sequences remains to be investigated.
Our finding that the C-terminal glycine-rich domain of TDP-43 (amino acid residues 274–414) is required for efficient SG recruitment is consistent with previously published data showing that residues 268–315 are necessary for localization of TDP-43 within SG (50, 51). Because the C-terminal domain has been reported to mediate protein-protein interactions, such as interactions with other heterogeneous nuclear ribonucleoproteins (13) including FUS (70), it seems likely that protein-protein interactions contribute to SG recruitment of TDP-43, although the protein(s) involved remains to be identified (protein X in Fig. 11). Freibaum et al. (56) identified numerous proteins involved in translation and SG-associated proteins as TDP-43-interacting proteins. Furthermore, TDP-43 and TIA-1 were found to interact in co-immunoprecipitation assays; however, this was only seen upon overexpression of both proteins (31). Nevertheless, TIA-1 and other SG-associated proteins are obvious candidates for proteins that recruit TDP-43 into SG via its C-terminal domain.
Although we did not observe myc- or V5-tagged TDP-WT or endogenous TDP-43 in SG after various stress treatments (Fig. 4–7), it is possible that very small amounts of TDP-43, undetectable by our antibodies, are present in SG under these conditions. That this might be the case is suggested by several reports describing at least small amounts of wild-type TDP-43 in SG upon various stress treatments (31, 50, 51, 53, 54, 56). Nevertheless, we could show that an artificial mutation in the TDP-43 NLS (NLSmut) or inhibition of Importin α/β-mediated nuclear import readily caused SG localization of TDP-43 upon cellular stress. We, therefore, suggest that cytosolic mislocalization is a prerequisite for recruitment of TDP-43 into SG. Cytosolic mislocalization may be artificially caused by high expression levels of TDP-WT and thus may allow unphysiological SG recruitment. Under physiological expression levels, we suggest that a nuclear import defect as the primary hit and cellular stress as the second hit are required for SG recruitment of TDP-43 (33, 40). In vivo, axonal injury (65) or reduced expression of nuclear transport factors, such as cellular apoptosis susceptibility protein (CAS) and Importin-α2 (66), might constitute such a primary hit leading to the cytoplasmic mislocalization of TDP-43 in ALS/FTLD-TDP patients.
What remains enigmatic is the cellular mechanism of ALS-associated TARDBP mutations. In contrast to ALS-associated FUS mutations, TARDBP mutations in the glycine-rich C-terminal domain (A315T, M337V, and G348C) did not affect nuclear localization in our study, consistent with previous reports (28, 32, 51). Whether ALS-associated TARDBP mutations affect SG formation was controversial. Two studies reported that TARDBP mutations increase the number or size of SG, suggesting a toxic gain-of-function mechanism (31, 51), whereas another group found that R361S is a loss-of-function mutation with regard to SG formation, leading to fewer SG (54). In our study the percentage of TDP-NLSmut in SG was not significantly altered by the presence of ALS-associated point mutations (A315T, M337V, and G348C) despite the importance of the C-terminal domain for SG recruitment. However, it remains possible that the dynamics of SG formation or dissolution are affected by ALS-associated TARDBP mutations, as recently suggested for R361S (54).
Another dispute that has remained unresolved is whether TDP-43 inclusions in FTLD and ALS patients contain SG marker proteins or not. Two studies reported a co-labeling of TDP-43 inclusions with SG markers (31, 57), whereas two other studies did not find evidence for SG marker co-labeling (33, 50). Our data suggest that SG marker co-labeling is dependent on the presence of full-length TDP-43, which is much more abundant in spinal cord inclusions than in cortical TDP-43 inclusions (this study and Refs. 59 and 60). We, therefore, suggest that the reported discrepancies could be due to the presence of different TDP-43 species in inclusions in different regions of the central nervous system. How CTFs are generated and why cortical TDP-43 inclusions are highly enriched in these fragments is still unclear. The absence of SG markers from CTF-containing inclusions suggests that these inclusions either arise independently of SG or that CTFs dissociate from SG upon proteolytic cleavage of full-length TDP-43, presumably due to its reduced RNA binding capacity. How exactly SG relate to CTF generation and TDP-43 inclusion formation remains to be investigated.
The presence of SG marker proteins and RNA in pathological FUS or TDP-43 inclusions has led to the hypothesis that these inclusions could arise from stress granules (31, 33, 42, 55). Indeed, various forms of stress have been implicated in the pathogenesis of ALS, including oxidative stress, mitochondrial dysfunction, damage to the vasculature, and inflammatory reactions (71–73). Even though it remains to be seen whether SG are actually pathogenic or protective, this model offers a plausible mechanism for how pathological aggregation of cytosolic FUS or TDP-43 might be triggered in response to cellular stress. Recruitment into SG most likely increases the local FUS or TDP-43 concentration, which might seed aggregation of these otherwise soluble proteins in a prion-like manner (74). Whether the prion-like domains of FUS and TDP-43 are secondarily involved in seeding aggregation remains to be seen. We note that so far cellular models were unable to recapitulate bona fide FUS or TDP-43 aggregation, as FUS- or TDP-43-containing SG are rapidly disassembled upon stress removal (Ref. 41 and this study). We, therefore, believe that additional hits or chronic stress might be needed for irreversible aggregation of SG-localized proteins (40).
Supplementary Material
Acknowledgments
We thank Claudia Abou-Ajram, Andrea Seibel, and Stephanie Kunath for technical assistance. We are grateful to the Hans and Ilse Breuer Foundation for the Confocal Microscope.
This work was supported by the Sonderforschungsbereich Molecular Mechanisms of Neurodegeneration (SFB 596), the Competence Network for Neurodegenerative Diseases (KNDD) of the Bundesministerium für Bildung und Forschung (BMBF), and an EMBO post-doctoral fellowship (to D. D.).

This article contains supplemental Figs. S1–S7.
- ALS
- amyotrophic lateral sclerosis
- TDP-43
- TAR DNA-binding protein of 43 kDa
- FUS
- Fused in sarcoma
- FTLD
- frontotemporal lobar degeneration
- SG
- stress granule
- RRM
- RNA recognition motif
- CTF
- C-terminal fragment
- PY-NLS
- proline-tyrosine nuclear localization signal
- PABP-1
- poly(A)-binding protein 1
- TIA-1
- T cell intracellular antigen 1
- DIV
- days in vitro.
REFERENCES
- 1. Mackenzie I. R., Rademakers R., Neumann M. (2010) TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 9, 995–1007 [DOI] [PubMed] [Google Scholar]
- 2. Buratti E., Baralle F. E. (2008) Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 13, 867–878 [DOI] [PubMed] [Google Scholar]
- 3. Lagier-Tourenne C., Cleveland D. W. (2009) Rethinking ALS: the FUS about TDP-43. Cell 136, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 5. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. (2006) TDP-43 is a component of ubiquitin-positive Tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 6. Kwiatkowski T. J., Jr., Bosco D. A., Leclerc A. L., Tamrazian E., Vanderburg C. R., Russ C., Davis A., Gilchrist J., Kasarskis E. J., Munsat T., Valdmanis P., Rouleau G. A., Hosler B. A., Cortelli P., de Jong P. J., Yoshinaga Y., Haines J. L., Pericak-Vance M. A., Yan J., Ticozzi N., Siddique T., McKenna-Yasek D., Sapp P. C., Horvitz H. R., Landers J. E., Brown R. H., Jr. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 [DOI] [PubMed] [Google Scholar]
- 7. Vance C., Rogelj B., Hortobágyi T., De Vos K. J., Nishimura A. L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., Ganesalingam J., Williams K. L., Tripathi V., Al-Saraj S., Al-Chalabi A., Leigh P. N., Blair I. P., Nicholson G., de Belleroche J., Gallo J. M., Miller C. C., Shaw C. E. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neumann M., Rademakers R., Roeber S., Baker M., Kretzschmar H. A., Mackenzie I. R. (2009) A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132, 2922–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lashley T., Rohrer J. D., Bandopadhyay R., Fry C., Ahmed Z., Isaacs A. M., Brelstaff J. H., Borroni B., Warren J. D., Troakes C., King A., Al-Saraj S., Newcombe J., Quinn N., Ostergaard K., Schrøder H. D., Bojsen-Møller M., Braendgaard H., Fox N. C., Rossor M. N., Lees A. J., Holton J. L., Revesz T. (2011) A comparative clinical, pathological, biochemical, and genetic study of fused in sarcoma proteinopathies. Brain 134, 2548–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mackenzie I. R., Ansorge O., Strong M., Bilbao J., Zinman L., Ang L. C., Baker M., Stewart H., Eisen A., Rademakers R., Neumann M. (2011) Pathological heterogeneity in amyotrophic lateral sclerosis with FUS mutations. Two distinct patterns correlating with disease severity and mutation. Acta Neuropathol. 122, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mackenzie I. R., Munoz D. G., Kusaka H., Yokota O., Ishihara K., Roeber S., Kretzschmar H. A., Cairns N. J., Neumann M. (2011) Distinct pathological subtypes of FTLD-FUS. Acta Neuropathol. 121, 207–218 [DOI] [PubMed] [Google Scholar]
- 12. Buratti E., Baralle F. E. (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276, 36337–36343 [DOI] [PubMed] [Google Scholar]
- 13. Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y. M., Baralle F. E. (2005) TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail. An important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 280, 37572–37584 [DOI] [PubMed] [Google Scholar]
- 14. Johnson B. S., Snead D., Lee J. J., McCaffery J. M., Shorter J., Gitler A. D. (2009) TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nonaka T., Kametani F., Arai T., Akiyama H., Hasegawa M. (2009) Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum. Mol. Genet. 18, 3353–3364 [DOI] [PubMed] [Google Scholar]
- 16. Furukawa Y., Kaneko K., Nukina N. (2011) Molecular properties of TAR DNA=binding protein-43 fragments are dependent upon its cleavage site. Biochim. Biophys. Acta 1812, 1577–1583 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y. J., Xu Y. F., Cook C., Gendron T. F., Roettges P., Link C. D., Lin W. L., Tong J., Castanedes-Casey M., Ash P., Gass J., Rangachari V., Buratti E., Baralle F., Golde T. E., Dickson D. W., Petrucelli L. (2009) Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang C., Tan W., Whittle C., Qiu L., Cao L., Akbarian S., Xu Z. (2010) The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PLoS One 5, e15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gitler A. D., Shorter J. (2011) RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion 5, 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Udan M., Baloh R. H. (2011) Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion 5, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cushman M., Johnson B. S., King O. D., Gitler A. D., Shorter J. (2010) Prion-like disorders. Blurring the divide between transmissibility and infectivity. J. Cell Sci. 123, 1191–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fuentealba R. A., Udan M., Bell S., Wegorzewska I., Shao J., Diamond M. I., Weihl C. C., Baloh R. H. (2010) Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J. Biol. Chem. 285, 26304–26314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iko Y., Kodama T. S., Kasai N., Oyama T., Morita E. H., Muto T., Okumura M., Fujii R., Takumi T., Tate S., Morikawa K. (2004) Domain architectures and characterization of an RNA-binding protein, TLS. J. Biol. Chem. 279, 44834–44840 [DOI] [PubMed] [Google Scholar]
- 24. Lerga A., Hallier M., Delva L., Orvain C., Gallais I., Marie J., Moreau-Gachelin F. (2001) Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 276, 6807–6816 [DOI] [PubMed] [Google Scholar]
- 25. Prasad D. D., Ouchida M., Lee L., Rao V. N., Reddy E. S. (1994) TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene 9, 3717–3729 [PubMed] [Google Scholar]
- 26. Mackenzie I. R., Rademakers R. (2008) The role of transactive response DNA-binding protein-43 in amyotrophic lateral sclerosis and frontotemporal dementia. Curr. Opin. Neurol. 21, 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo W., Chen Y., Zhou X., Kar A., Ray P., Chen X., Rao E. J., Yang M., Ye H., Zhu L., Liu J., Xu M., Yang Y., Wang C., Zhang D., Bigio E. H., Mesulam M., Shen Y., Xu Q., Fushimi K., Wu J. Y. (2011) An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation, and neurotoxicity. Nat. Struct. Mol. Biol. 18, 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kabashi E., Lin L., Tradewell M. L., Dion P. A., Bercier V., Bourgouin P., Rochefort D., Bel Hadj S., Durham H. D., Vande Velde C., Rouleau G. A., Drapeau P. (2010) Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum. Mol. Genet. 19, 671–683 [DOI] [PubMed] [Google Scholar]
- 29. Winton M. J., Van Deerlin V. M., Kwong L. K., Yuan W., Wood E. M., Yu C. E., Schellenberg G. D., Rademakers R., Caselli R., Karydas A., Trojanowski J. Q., Miller B. L., Lee V. M. (2008) A90V TDP-43 variant results in the aberrant localization of TDP-43 in vitro. FEBS Lett. 582, 2252–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barmada S. J., Skibinski G., Korb E., Rao E. J., Wu J. Y., Finkbeiner S. (2010) Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci. 30, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu-Yesucevitz L., Bilgutay A., Zhang Y. J., Vanderweyde T., Vanderwyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., Petrucelli L., Wolozin B. (2010) Tar DNA-binding protein-43 (TDP-43) associates with stress granules. Analysis of cultured cells and pathological brain tissue. PLoS One 5, e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ling S. C., Albuquerque C. P., Han J. S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D. W. (2010) ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl. Acad. Sci. U.S.A. 107, 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M. E., Mackenzie I. R., Capell A., Schmid B., Neumann M., Haass C. (2010) ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 29, 2841–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gal J., Zhang J., Kwinter D. M., Zhai J., Jia H., Jia J., Zhu H. (2011) Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol. Aging 32, 2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kino Y., Washizu C., Aquilanti E., Okuno M., Kurosawa M., Yamada M., Doi H., Nukina N. (2011) Intracellular localization and splicing regulation of FUS/TLS are variably affected by amyotrophic lateral sclerosis-linked mutations. Nucleic Acids Res. 39, 2781–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ito D., Seki M., Tsunoda Y., Uchiyama H., Suzuki N. (2011) Nuclear transport impairment of amyotrophic lateral sclerosis-linked mutations in FUS/TLS. Ann. Neurol. 69, 152–162 [DOI] [PubMed] [Google Scholar]
- 37. Chiò A., Restagno G., Brunetti M., Ossola I., Calvo A., Mora G., Sabatelli M., Monsurrò M. R., Battistini S., Mandrioli J., Salvi F., Spataro R., Schymick J., Traynor B. J., La Bella V., and ITALSGEN Consortium (2009) Two Italian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation. Neurobiol. Aging 30, 1272–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang E. J., Zhang J., Geser F., Trojanowski J. Q., Strober J. B., Dickson D. W., Brown R. H., Jr., Shapiro B. E., Lomen-Hoerth C. (2010) Extensive FUS-immunoreactive pathology in juvenile amyotrophic lateral sclerosis with basophilic inclusions. Brain Pathol. 20, 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bäumer D., Hilton D., Paine S. M., Turner M. R., Lowe J., Talbot K., Ansorge O. (2010) Juvenile ALS with basophilic inclusions is a FUS proteinopathy with FUS mutations. Neurology 75, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dormann D., Haass C. (2011) TDP-43 and FUS. A nuclear affair. Trends Neurosci. 34, 339–348 [DOI] [PubMed] [Google Scholar]
- 41. Bosco D. A., Lemay N., Ko H. K., Zhou H., Burke C., Kwiatkowski T. J., Jr., Sapp P., McKenna-Yasek D., Brown R. H., Jr., Hayward L. J. (2010) Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet. 19, 4160–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fujita K., Ito H., Nakano S., Kinoshita Y., Wate R., Kusaka H. (2008) Immunohistochemical identification of messenger RNA-related proteins in basophilic inclusions of adult-onset atypical motor neuron disease. Acta Neuropathol. 116, 439–445 [DOI] [PubMed] [Google Scholar]
- 43. Anderson P., Kedersha N. (2006) RNA granules. J. Cell Biol. 172, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kedersha N., Anderson P. (2007) Mammalian stress granules and processing bodies. Methods Enzymol. 431, 61–81 [DOI] [PubMed] [Google Scholar]
- 45. Kayali F., Montie H. L., Rafols J. A., DeGracia D. J. (2005) Prolonged translation arrest in reperfused hippocampal cornu Ammonis 1 is mediated by stress granules. Neuroscience 134, 1223–1245 [DOI] [PubMed] [Google Scholar]
- 46. DeGracia D. J., Rudolph J., Roberts G. G., Rafols J. A., Wang J. (2007) Convergence of stress granules and protein aggregates in hippocampal cornu ammonis 1 at later reperfusion following global brain ischemia. Neuroscience 146, 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moeller B. J., Cao Y., Li C. Y., Dewhirst M. W. (2004) Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors. Role of reoxygenation, free radicals, and stress granules. Cancer Cell 5, 429–441 [DOI] [PubMed] [Google Scholar]
- 48. Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., Anderson P. (2004) Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Furukawa Y., Kaneko K., Matsumoto G., Kurosawa M., Nukina N. (2009) Cross-seeding fibrillation of Q/N-rich proteins offers new pathomechanism of polyglutamine diseases. J. Neurosci. 29, 5153–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., Ratti A. (2009) TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111, 1051–1061 [DOI] [PubMed] [Google Scholar]
- 51. Dewey C. M., Cenik B., Sephton C. F., Dries D. R., Mayer P., 3rd, Good S. K., Johnson B. A., Herz J., Yu G. (2011) TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol. Cell Biol. 31, 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moisse K., Volkening K., Leystra-Lantz C., Welch I., Hill T., Strong M. J. (2009) Divergent patterns of cytosolic TDP-43 and neuronal progranulin expression following axotomy. Implications for TDP-43 in the physiological response to neuronal injury. Brain Res. 1249, 202–211 [DOI] [PubMed] [Google Scholar]
- 53. Meyerowitz J., Parker S. J., Vella L. J., Ng D. Ch., Price K. A., Liddell J. R., Caragounis A., Li Q. X., Masters C. L., Nonaka T., Hasegawa M., Bogoyevitch M. A., Kanninen K. M., Crouch P. J., White A. R. (2011) c-Jun N-terminal kinase controls TDP-43 accumulation in stress granules induced by oxidative stress. Mol. Neurodegener. 6, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McDonald K. K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G. A., Vande Velde C. (2011) TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 20, 1400–1410 [DOI] [PubMed] [Google Scholar]
- 55. Dewey C. M., Cenik B., Sephton C. F., Johnson B. A., Herz J., Yu G. (2012) TDP-43 aggregation in neurodegeneration. Are stress granules the key? Brain Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Freibaum B. D., Chitta R. K., High A. A., Taylor J. P. (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 9, 1104–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Volkening K., Leystra-Lantz C., Yang W., Jaffee H., Strong M. J. (2009) Tar DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins, and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS). Brain Res. 1305, 168–182 [DOI] [PubMed] [Google Scholar]
- 58. Kaech S., Banker G. (2006) Culturing hippocampal neurons. Nat. Protoc. 1, 2406–2415 [DOI] [PubMed] [Google Scholar]
- 59. Igaz L. M., Kwong L. K., Xu Y., Truax A. C., Uryu K., Neumann M., Clark C. M., Elman L. B., Miller B. L., Grossman M., McCluskey L. F., Trojanowski J. Q., Lee V. M. (2008) Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am. J. Pathol. 173, 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Neumann M., Kwong L. K., Lee E. B., Kremmer E., Flatley A., Xu Y., Forman M. S., Troost D., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2009) Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 117, 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dormann D., Capell A., Carlson A. M., Shankaran S. S., Rodde R., Neumann M., Kremmer E., Matsuwaki T., Yamanouchi K., Nishihara M., Haass C. (2009) Proteolytic processing of TAR DNA binding protein-43 by caspases produces C-terminal fragments with disease defining properties independent of progranulin. J. Neurochem. 110, 1082–1094 [DOI] [PubMed] [Google Scholar]
- 62. Winton M. J., Igaz L. M., Wong M. M., Kwong L. K., Trojanowski J. Q., Lee V. M. (2008) Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 283, 13302–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mackenzie I. R., Neumann M., Baborie A., Sampathu D. M., Du Plessis D., Jaros E., Perry R. H., Trojanowski J. Q., Mann D. M., Lee V. M. (2011) A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 122, 111–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kosugi S., Hasebe M., Entani T., Takayama S., Tomita M., Yanagawa H. (2008) Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem. Biol. 15, 940–949 [DOI] [PubMed] [Google Scholar]
- 65. Sato T., Takeuchi S., Saito A., Ding W., Bamba H., Matsuura H., Hisa Y., Tooyama I., Urushitani M. (2009) Axonal ligation induces transient redistribution of TDP-43 in brainstem motor neurons. Neuroscience 164, 1565–1578 [DOI] [PubMed] [Google Scholar]
- 66. Nishimura A. L., Zupunski V., Troakes C., Kathe C., Fratta P., Howell M., Gallo J. M., Hortobágyi T., Shaw C. E., Rogelj B. (2010) Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain 133, 1763–1771 [DOI] [PubMed] [Google Scholar]
- 67. Polymenidou M., Lagier-Tourenne C., Hutt K. R., Huelga S. C., Moran J., Liang T. Y., Ling S. C., Sun E., Wancewicz E., Mazur C., Kordasiewicz H., Sedaghat Y., Donohue J. P., Shiue L., Bennett C. F., Yeo G. W., Cleveland D. W. (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Burd C. G., Dreyfuss G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615–621 [DOI] [PubMed] [Google Scholar]
- 69. Hoell J. I., Larsson E., Runge S., Nusbaum J. D., Duggimpudi S., Farazi T. A., Hafner M., Borkhardt A., Sander C., Tuschl T. (2011) RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Biol. 18, 1428–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim S. H., Shanware N. P., Bowler M. J., Tibbetts R. S. (2010) Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J. Biol. Chem. 285, 34097–34105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Barber S. C., Shaw P. J. (2010) Oxidative stress in ALS. Key role in motor neuron injury and therapeutic target. Free Radic. Biol. Med. 48, 629–641 [DOI] [PubMed] [Google Scholar]
- 72. Saxena S., Caroni P. (2011) Selective neuronal vulnerability in neurodegenerative diseases. From stressor thresholds to degeneration. Neuron 71, 35–48 [DOI] [PubMed] [Google Scholar]
- 73. Quaegebeur A., Lange C., Carmeliet P. (2011) The neurovascular link in health and disease. Molecular mechanisms and therapeutic implications. Neuron 71, 406–424 [DOI] [PubMed] [Google Scholar]
- 74. Polymenidou M., Cleveland D. W. (2011) The seeds of neurodegeneration. Prion-like spreading in ALS. Cell 147, 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang Y. J., Xu Y. F., Dickey C. A., Buratti E., Baralle F., Bailey R., Pickering-Brown S., Dickson D., Petrucelli L. (2007) Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J. Neurosci. 27, 10530–10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.