Background: TDP-43 is a major pathological hallmark of several neurodegenerative diseases.
Results: TDP-43 interacts with FMRP/STAU1 and binds to the 3′-UTR of SIRT1 mRNA to promote its stability.
Conclusion: TDP-43, FMRP, and STAU1 form a functionally coordinated complex to regulate the expression of SIRT1.
Significance: Adding to our understanding of the mechanistic role of TDP-43 in neurodegenerative diseases.
Keywords: Apoptosis, Molecular Cell Biology, mRNA, Neurodegeneration, Sirt1, TARDBP, TDP-43
Abstract
Despite the identification of the 43 kDa transactive response DNA-binding protein (TDP-43) as a major pathological signatory protein in a wide range of neurodegenerative diseases, the mechanistic role of TDP-43 in neurodegenerative disorders is still poorly understood. Here, we report that TDP-43 is physically associated with fragile X mental retardation protein (FMRP) and Staufen (STAU1) to form a functional complex. Differential microarray analysis revealed that the expression of a collection of functionally important genes including Sirtuin (SIRT1) is regulated by this complex. RNA-immunoprecipitation (RIP) and RNA pull-down assays demonstrated that TDP-43/FMRP/STAU1 specifically binds to the 3′-UTR of SIRT1 mRNA, and that knockdown the expression of any one of these three proteins resulted in the reduction of SIRT1 mRNA and protein. SIRT1 is implicated in double-stranded DNA break repair and is required for cell survival. Indeed, depletion of TDP-43/FMRP/STAU1 sensitizes cells to apoptosis and DNA damages. Collectively, our results revealed a molecular mechanism for the cellular function of TDP-43 and might shed new light on the understanding of the mechanistic role of TDP-43 in neurodegenerative diseases.
Introduction
Transactive response DNA-binding protein (TARDBP/TDP-43)3 is a 43 kDa protein expressed in most tissues (1, 2). It has similar structural properties to the evolutionarily conserved heterogeneous nuclear ribonucleoprotein (hnRNP) (3). In particular, TDP-43 is predominantly a nuclear protein containing two RNA recognition motifs (RRM) and a C-terminal glycine-rich domain (GRD). These domains are known to mediate DNA and RNA recognition as well as protein-protein interactions (4, 5).
TDP-43 has been identified as a major pathological protein aggregate of cytoplasmic ubiquitin-positive inclusions that accumulates in amyotrophic lateral sclerosis (ALS) patients and individuals with frontotemporal lobar degeneration (FTLD) (6, 7). The immunoreactive inclusions of TDP-43 were also observed in a wide range of other neurodegenerative disorders, including Alzheimer disease (AD), Parkinson disease (PD), Huntington disease (HD), and dementia with Lewy bodies (8, 9). Interestingly, in ALS and FTLD, TDP-43 is cleared from the nuclear compartment, suggesting that loss of its normal nuclear function plays a critical role in the pathogenesis of these diseases (6, 7). Additionally, the identification of mutations of TDP-43 that are linked to ALS also supports an argument that TDP-43 directly contributes to the pathogenesis of neurodegenerative disorders (10–12).
Initially found to be capable of binding to the DNA of HIV and of repressing transcription (13, 14), TDP-43 is now highlighted by numerous reports showing that it regulates multiple cellular processes, including cell cycle regulation, apoptosis, as well as microRNA metabolism (15). In particular, as a member of hnRNPs that are capable of binding single-stranded RNAs via their RRMs, TDP-43 has been reported to exert a pleiotropic role in RNA processing events including transcription, splicing, transport, and stability (16, 17). Early in vitro studies showed that TDP-43 preferentially bound RNAs via a GU dinucleotide repeat element (3), while recent analysis of global TDP-43 binding sites showed that the most significant RNA binding occurred at either uninterrupted GU repeats or a GU-rich motif interrupted by a single adenine (18, 19). To date, TDP-43 has been reported to regulate alternative splicing of ciliary neurotrophic factor receptor (CNTFR), MAP-kinase activating death domain (MADD), and myocyte enhancer factor 2D (MEF2D) (20–23). In addition, the mRNA expression/processing of cystic fibrosis transmembrane regulator (CFTR) and apolipoprotein A2 (APOA2) (24–26), survival motor neuron protein 2 (SMN2) (27), and histone deacetylase 6 (HDAC6) (28) was also reported to be regulated by TDP-43.
Despite the advance in the characterization of the biological activity of the TDP-43 protein, there is still much to be learned about the molecular mechanisms underlying the cellular function of TDP-43. Here we report that TDP-43 physically interacts with fragile X mental retardation protein (FMRP) and Staufen (STAU1) to form a functionally coordinated complex. Differential microarray analysis in human neuroblastoma SH-SY5Y cells identified a series of functionally important genes including Sirtuin (SIRT1) that are targeted by this complex. We showed that TDP-43/FMPR/STAU1 complex binds specifically to SIRT1 3′-UTR, and that knockdown the expression of any one of these three proteins resulted in the reduction of SIRT1 mRNA and protein levels. We demonstrated that depletion of TDP-43 sensitizes SH-SY5Y cells to DNA damages and apoptosis.
EXPERIMENTAL PROCEDURES
Cloning and Mutagenesis
Full-length TDP-43 were amplified from cDNA of SH-SY5Y cells and cloned through BamH1/XhoI into pCMV-Tag-2B-Flag vector (Agilent). TDP-43 mutants (D169G, G287S, G290A, G298S, and R361S,) were generated using site-directed mutagenesis according to the manufacturer's instructions (Agilent). TDP-43 deletion mutants including ΔRRM1 (Δ106–176aa), ΔRRM2 (Δ191–262aa), ΔRRM1+ΔRRM2/ΔRRMs (Δ106–262aa), ΔGRN (Δ274–414aa), and ΔNLS (Δ82–98aa) were PCR-amplified and subcloned into pCMV-Tag-2B-Flag vector according to the standard protocols. TDP-43 mutants were generated by quick change site-directed mutagenesis (Agilent).
Antibodies and Reagents
The sources of the antibodies were: anti-FLAG (clone M2), anti-TDP-43 (polyclonal), anti-TDP-43 (monoclonal), anti-α-tubulin, and anti-Flag M2 affinity gel (Sigma); anti-FMRP (Abcam), and anti-SIRT1 (Santa Cruz Biotechnology); anti-STAU1 (MBL); anti-HuR (Santa Cruz Biotechnology). Camptothecin (CPT) was purchased from Sigma. Protein A/G-Sepharose CL-4B beads were from Amersham Biosciences, and protease inhibitor mixture mixture was from Roche Applied Science. All siRNAs were synthesized by Genepharma and the target sequences are listed in supplemental Table S3.
Cell Culture and Transfections
SH-SY5Y human neuroblastoma cells and HEK293T human embryonic kidney cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Transient transfections were performed with Lipofectamine RNAiMAX (for siRNA transfection) and Lipofectamine 2000 (for plasmid transfection) according to the company's specification (Invitrogen).
FPLC Chromatography
SH-SY5Y nuclear extracts were prepared with Nuclear-Cytosol Extraction kit (Applygen Technologies). Approximately 6 mg of nuclear protein was concentrated to 500 μl using a Millipore Ultra free centrifugal filter apparatus (10 kDa nominal molecular mass limit), and then applied to Superose 6 size exclusion column (Amersham Biosciences) that had been equilibrated with PBS and calibrated with protein standards (blue dextran, 2000 kDa; thyroglobulin, 669 kDa; ferritin, 440 kDa; aldolase, 158 kDa; bovine serum albumin, 67 kDa; and Cornonin, 44 kDa; all from Amersham Biosciences). The column was eluted at a flow rate of 0.5 ml/min and fractions were collected every 500 μl.
Immunoblotting and Immunoprecipitation
For immunoblotting, SH-SY5Y were harvested and extracted in RIPA buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.1% SDS, 1% Nonidet P-40, 0.5% sodium deoxycholate) including protease inhibitor mixture (Roche) and centrifuged, and 50 μg of the supernatant was subjected to SDS-PAGE using 8 or 10% polyacrylamide gels and transferred onto nitrocellulose. Membranes were incubated with primary antibodies (anti-SIRT1, FMRP, STAU1, or TDP-43 antibodies) overnight at 4 °C followed by HRP-conjugated secondary antibodies. For immunoprecipitation assays, cellular extracts were incubated with appropriate primary antibodies or normal rabbit/mouse immunoglobin G (IgG) at 4 °C overnight, followed by addition of protein A/G-Sepharose CL-4B beads for 2 h at 4 °C. Beads were then washed and the immune complexes were subjected to SDS-PAGE followed by immunoblotting with secondary antibodies. Immunodetection was performed using enhanced chemiluminescence (ECL System, Amersham Biosciences) according to the manufacturer's instructions.
RNA Isolation and Real-time RT-PCR
RNA was isolated with Trizol (Invitrogen) and treated with DNase I. After reverse transcription with AccessQuick™ RT-PCR System (Promega), real-time PCR was conducted on an ABI 7500 real-time PCR system (Applied Biosystems). cDNA synthesis and amplification were performed according to the manufacturer's instruction. The primers used were listed in supplemental Table S4.
RNA Immunoprecipitation (RIP)
RIP assays were performed using the Magna RIP Kit purchased from Millipore (cat. No. 17-700). SH-SY5Y cells (48 h after transfection) were collected and lysed in 500 μl of RIP lysis buffer supplemented with protease inhibitor and RNase inhibitor. 10% of the total lysate was processed in parallel to obtain the total input sample. After 5 min of incubation on ice, 50 μl of magnetic beads suspension was transferred to each tube. The beads were washed two times using 500 μl of RIP wash buffer, then add ∼5 μg of anti-TDP-43, anti-FMRP, and anti-STAU1 antibodies at room temperature for 30 min. The beads were washed five times with wash buffer, then in 900 μl of immunoprecipitation buffer (0.5 m EDTA, RNase inhibitor, and RIP wash buffer) at 4 °C for overnight. RNA was purified using proteinase K buffer (RIP wash buffer, 10% SDS, and 10 mg/ml proteinase K) and precipitated with Salt Solution I, Salt Solution II, Precipitate Enhancer and then absolute ethanol. The isolated RNA from RIP was analyzed by quantitative RT-PCR. The results were expressed as the relative fold enrichment of the target precipitation as compared with the normal rabbit IgG control. The primers used were listed in supplemental Table S5.
RNA Pull-down Assays
cDNA was used as a template for PCR amplification of different SIRT1 untranslated regions (UTR) and coding regions (CR). All forward primers contained the T7 promoter sequence (T7), CAGAGATGCATAATACGACTCACTATAGGGAGA. To prepare 5′-UTR-A transcript (positions 1 to 53), forward primer, (T7) GTCGAGCGGGAGCAGAG and reverse primer, CTTCCAACTGCCTCTCTGG were used. To prepare CR-B transcript (positions 54 to 754), forward primer, (T7) ATGGCGGACGAGGCG and reverse primer, TTTTTGGTGGTTCTGAAGGATA were used. To prepare CR-C transcript (positions 755 to 1653), forward primer, (T7) GGAAAAAAAGAAAAGATATTAAT and reverse primer, TGAAGAATCTGGTGGTGAA were used. To prepare CR-D transcript (positions 1654 to 2324), forward primer, (T7) GTGATTGTCACACTTTTAGACC and reverse primer, CTATGATTTGTTTGATGGATAGT were used. To prepare 3′-UTR-E transcript (positions 2325 to 2924), forward primer, (T7) TGTAATAATTGTGCAGGTACAGG and reverse primer, TCTCCCCACATATTGTTGACTTCCT were used. To prepare 3′-UTR-F transcript (positions 2925 to 3473), forward primer, (T7) GCACTCGGTTGTCTTTACTT and CCAGTAGAAGTACCATTATTATAGA were used. To prepare 3′-UTR-G transcript (positions 3474 to 4110), forward primer, (T7) GGAGAGTGTAATATTTTGGACTG and TAAGTTAACAGAAAAAAGTCAAATG were used. For biotin pull-down assay, PCR-amplified DNA was used as a template to transcribe biotinylated RNA by using T7 RNA polymerase in the presence of biotin-UTP and was purified as previously described (29).
TUNEL Assays
SH-SY5Y cells were seeded onto 6-well plates for 24 h and transfected with indicated siRNAs using Lipofectamine RNAiMAX (Invitrogen). 12 h later, the cells were transfected with appropriate plasmid constructs using Lipofectamine 2000 (Invitrogen). After 48 h of the plasmid transfection, cells were harvested, and TUNEL assay were performed according to the manufacturer's instructions (TB235, Promega) with a fluorescence method.
Cell Viability Assays
SH-SY5Ycells were seeded onto 24-well plates for 24 h and transfected with indicated siRNAs using Lipofectamine RNAiMAX (Invitrogen). 12 h later, the cells were transfected with appropriate plasmid constructs using Lipofectamine 2000 (Invitrogen). After 24 h of the plasmid transfection, CPT (0.2 μg/ml in DMSO) or DMSO were added for another 24 h and cell viability assays were performed according to the manufacturer's instructions (TB 288, Promega) with a luminescent method.
RESULTS
TDP-43 Is Physically Associated with FMRP and STAU1
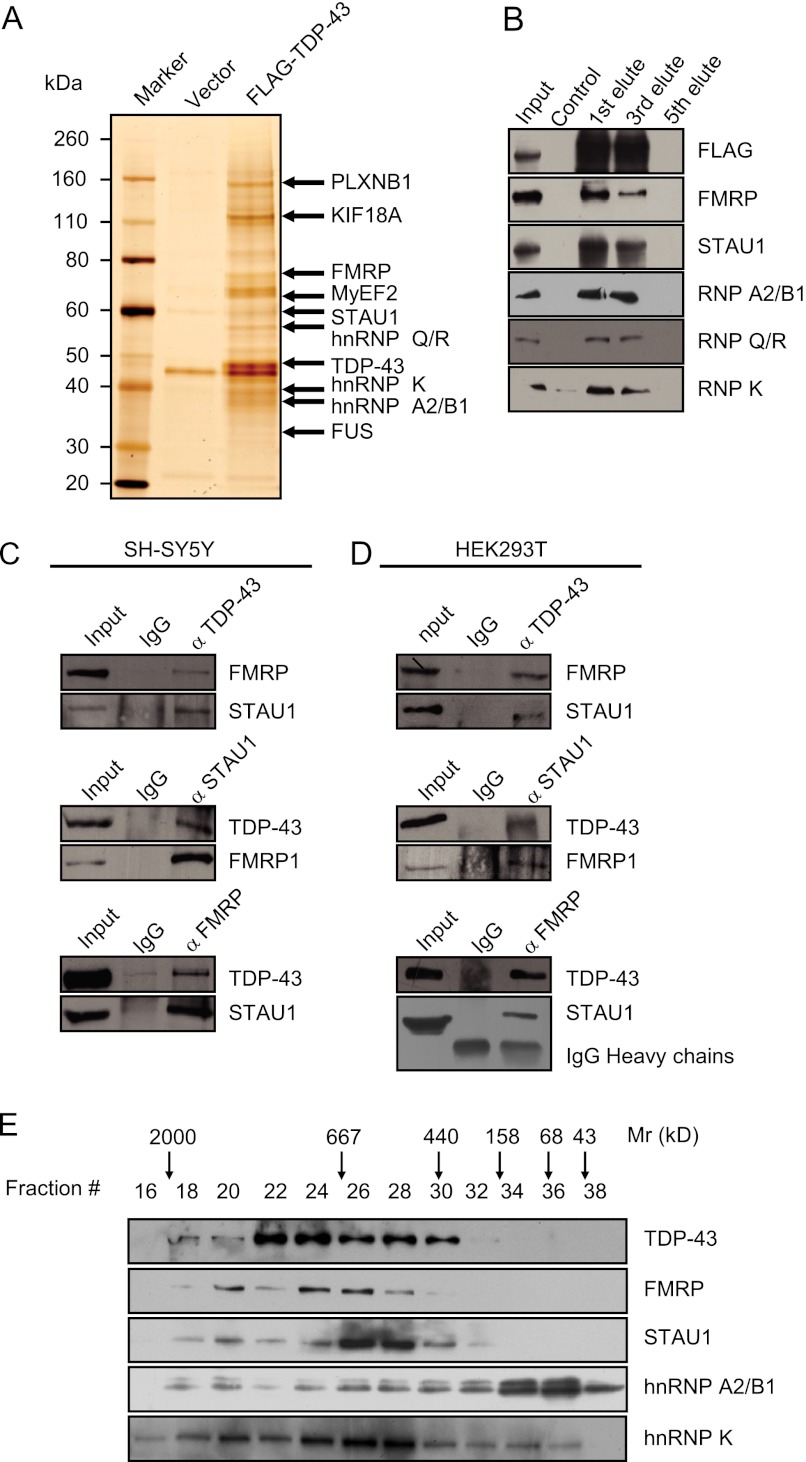
In an effort to further explore the mechanistic role of TDP-43 in neuronal cells, we employed affinity purification and mass spectrometry to identify the proteins that are associated with TDP-43 in vivo. In these experiments, FLAG-tagged TDP-43 (FLAG-TDP-43) was stably expressed in SH-SY5Y cells. Cellular extracts were prepared and subjected to affinity purification using an anti-FLAG affinity gel. Mass spectrometric analysis indicates that TDP-43 co-purified with hnRNP A0, A1, A2/B1, K, L, Q, R, and FUS, as reported previously (30, 31). Interestingly, TDP-43 also co-purified with FMRP and STAU1 (Fig. 1A), two proteins that are reported to be also involved in multiple RNA-processing events (32–35). The presence of FMRP, STAU1, hnRNP A2/B1, hnRNP K, and hnRNP Q/R in the TDP-43-associated protein complex was confirmed by Western blotting analysis (Fig. 1B). The detailed results of the mass spectrometric analysis are provided in supplemental Table S1.
FIGURE 1.
TDP-43 Is associated with FMRP and STAU1. A, mass spectrometry analysis of TDP-43-associated proteins. Nuclear extracts from SH-SY5Y cells stably expressing FLAG-TDP-43 were prepared and subjected to affinity-purification with anti-FLAG M2 affinity gel. The purified protein complex was resolved on SDS-PAGE and silver stained, and the bands were retrieved and analyzed by mass spectrometry. Complete amino acid sequences from mass spectrometry analysis are included in supplemental Table S1. B, Western blotting analysis of the identified proteins in the purified fractions using antibodies against the indicated proteins. C and D, co-immunoprecipitation of TDP-43, FMRP, and STAU1 in neuronal and non-neuronal cell lines. Whole-cell lysates from SH-SY5Y cells (D) or HEK293T cells (E) were prepared and immunoprecipitation was performed with anti-TDP-43, FMRP, or STAU1 followed by immunoblotting with antibodies against indicated proteins. E, co-fractionation TDP-43 associated proteins by FPLC. Nuclear extracts of SH-SY5Y cells were fractionated by a Superose 6 gel filtration column. The fractions were analyzed by Western blotting. Molecular weight standards are shown on top.
To further support the observation that TDP-43 is physically associated with FMRP and STAU1 in vivo, co-immunoprecipitation experiments were performed. In these experiments, total proteins from SH-SY5Y cells were extracted and immunoprecipitated with antibodies against TDP-43 followed by immunoblotting with antibodies against FMRP or STAU1. The results showed that both FMRP and STAU1 could be efficiently co-immunoprecipitated by TDP-43 (Fig. 1C, middle and lower panels). Reciprocal immunoprecipitations with antibodies against FMRP or STAU1 followed by immunoblotting with antibodies against TDP-43 also confirmed that association of TDP-43 with FMRP and STAU1 in vivo (Fig. 1C, upper panel). The association of TDP-43 with FMRP and STAU1 was also detected in human embryonic kidney HEK 293T cells (Fig. 1D). Taken together, these data indicate that a TDP-43/FMRP/STAU1-containing protein complex exists in both neuronal and non-neuronal cell lines. The interaction of TDP-43 with FMRP and STAU1 suggests that these proteins may function in a concerted manner to regulate expression of certain target genes.
To further support the observation that TDP-43, FMRP, and STAU1 exist in the same complex in vivo, protein fractionation experiments were carried out by fast protein liquid chromatography (FPLC). Nuclear extracts derived from SH-SY5Y cells were fractionated by DEAE Sepharose, followed by superpose 6 gel filtration chromatography. Western blotting revealed a major peak at about 669–1000 kDa for TDP-43, and also for the FMRP, STAU1, and hnRNP K, whereas the hnRNP A2/B1 preferred a monomer style (Fig. 1E). Significantly, the elution pattern of TDP-43 was largely overlapped with that of FMRP and STAU1, further supporting the notion that these proteins may form a functionally coordinated complex in vivo.
Identification of TDP-43 Downstream Target Genes
As mentioned above, it is believed that TDP-43 mainly functions as an RNA-binding protein to affect the mRNA processing of target genes (16, 17). To further investigate TDP-43 downstream target genes, RNA microarray analysis was performed in SH-SY5Y cells with the expression of TDP-43 knocked down by RNA interference (RNAi). Total RNA was extracted and hybridized on a Human Genome U133 + 2.0 array (Affymetrix). Compared with control, a total of 258 altered specific mRNA targets by microarray expression profiling in TDP-43-silenced cells were identified (supplemental Table S2). Among the changed genes, FAM73A, HMOX1, NR4A1, VAMP1, ATXN3, GIGYF2, and PFKP were also reported as TDP-43 targets (18, 28, 36, 37).
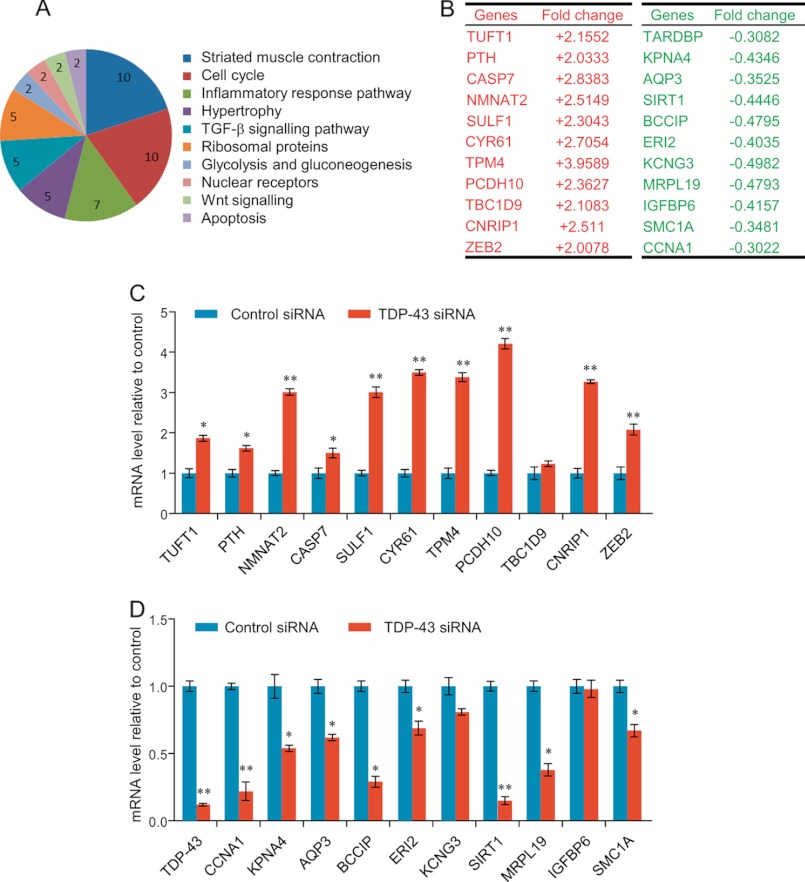
These genes were then classified into cellular signaling pathways using MAS software with a p value cutoff of less than 10−3. These analyses identified several cellular signaling pathways, including cell cycle and apoptosis, which are critically involved in cell proliferation and survival (Fig. 2A).
FIGURE 2.
Identification of TDP-43 target genes. A, classification of the genes identified in mRNA microarray analysis. The statistically significant (p < 0.001) pathways are shown and the numbers indicate the numbers of the pathway-associated genes. B, transcripts are identified by RNA microarray analysis whose levels either increased or decreased in the absence of TDP-43 compared with control-treated cells. C, verification of RNA microarray analysis results by measuring the mRNA expression of the selected genes representing up-regulated mRNAs using quantitative realtime RT-PCR. D, verification of RNA microarray analysis results by measuring the mRNA expression of the selected genes representing down-regulated mRNAs using quantitative realtime RT-PCR. The mRNA levels were normalized to those of GADPH. All the error bars represent S.D. of three independent experiments. *, p < 0.05 and **, <0.01 (one-tailed unpaired t test).
To verify the microarray results, we selected 22 genes among the 258 genes whose expression was altered by TDP-43 knockdown, including 11 up-regulated transcripts and 11 down-regulated transcripts representing each of the classified signaling pathways related to neuronal development, differentiation, proliferation, and survival (Fig. 2B). The mRNA expression of these 22 genes was measured by quantitative real-time reverse transcriptase PCR (qPCR) in SH-SY5Y cells under the knockdown of TDP-43 expression by two different TDP-43-specific siRNAs. The results showed that, upon TDP-43 depletion, 9 of the 11 up-regulated transcripts were increased in an abundance of 1.5–4.5-fold (Fig. 2C) and 9 of the 11 down-regulated transcripts were decreased in an abundance of 1.5–5-fold (Fig. 2D). Therefore, the microarray analysis appears to be a reliable assessment of changes in transcript abundance uponTDP-43 depletion.
Regulation of SIRT1 by TDP-43/FMRP/STAU1 Complex
Neuronal death or elimination of neuronal processes is the characteristic of neurodegenerative diseases (38). After differentiation, the central nervous system has very limited capabilities for both endogenous cell proliferation and regeneration (39). Therefore, it has been speculated that molecular pathways that entail cell proliferation and survival may contribute to anti-neurodegeneration (40, 41).
Notably, among the identified genes whose expression was altered upon TDP-43 knockdown, SIRT1, the NAD+-dependent class III histone deacetylase, was highlighted for its well documented anti-apoptotic and longevity promotion activities, as well as its critical role in neurodegeneration (42, 43). Moreover, we chose SIRT1 for advanced study as our results showed that SIRT1 was the most reduced one upon TDP-43 depletion among the consistently altered target genes.
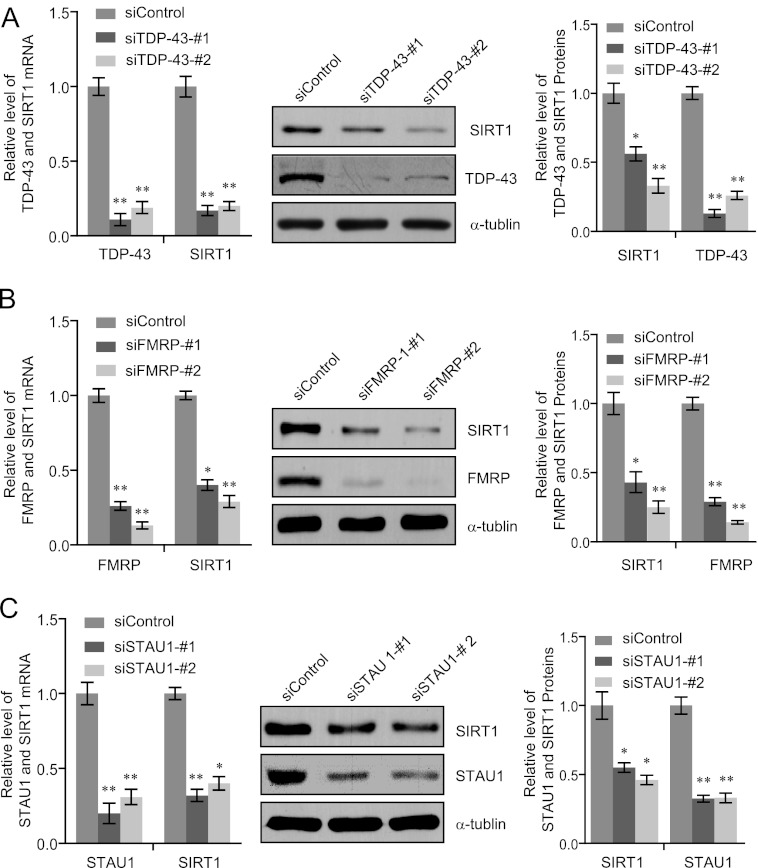
To validate the SIRT1down-regulation by TDP-43 knockdown, we treated SH-SY5Y cells with the same TDP-43 siRNA (siTDP-43-1) and a scrambled control siRNA as for the microarray hybridizations and additionally used one more siRNAs directed against TDP-43 (siTDP-43-2) to avoid possible off-target effects. Both siRNAs specifically reduced TDP-43 expression and led to a significant down-regulation of SIRT1 mRNA, as quantitated by qPCR (Fig. 3A, left panel). This effect was confirmed at the protein level: knockdown of TDP-43 protein was consistently accompanied by a reduced SIRT1 protein level when measured by Western blotting and quantitated using the Quantity One (Bio-Rad) program (Fig. 3A).
FIGURE 3.
Validation of SIRT1 down-regulation in TDP-43, FMRP of STAU1 silenced SH-SY5Y cells. A–C, SH-SY5Y cells were transfected with scrambled control siRNA, TDP-43 siRNA, FMRP siRNA, or STAU siRNA. Total RNAs were prepared and analyzed for SIRT1 expression by RT-PCR (left panels) and Western blotting (middle panels). The intensities of the gel bands were quantitated using the Quantity One (BioRad) program (right panels). The mRNA levels were normalized to those of GADPH and α-tubulin served as a loading control for the Western blotting. Error bars represent S.D. of triplicates. *, p < 0.05 and **, <0.01 (one-tailed unpaired t test).
The finding that TDP-43 is associated with FMRP and STAU1 raises a possibility that TDP-43 acts in concert with these two proteins. In order to test this hypothesis, loss-of-function experiments of FMRP and STAU1 were performed and the effect of FMRP-or STAU1-depletion on the expression of SIRT1 was examined in SH-SY5Y cells. In these experiments, two siRNAs specifically targeted different regions of FMRP mRNA or STAU1 mRNA were used. The results showed that depletion of FMRP in SH-SY5Y cells caused a significant down-regulation of SIRT1 at both mRNA and protein levels (Fig. 3B). Likewise, knockdown of STAU1 resulted in a drastic decrease in both mRNA and protein levels of SIRT1in SH-SY5Y cells (Fig. 3C). These results suggest that TDP-43, FMRP, and STAU1 form a functionally coordinated complex to regulate the expression of SIRT1.
TDP-43-specific Regulation of SIRT1 Expression
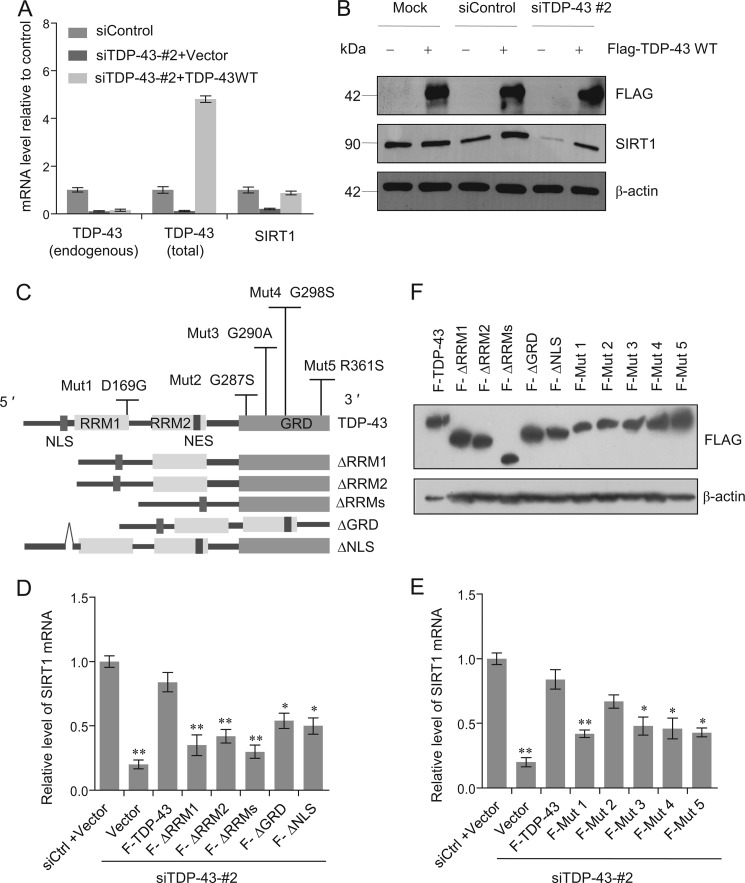
To further consolidate the observation that TDP-43 knockdown was associated with a decrease in the expression of SIRT1 and to test the hypothesis that this effect is TDP-43-dependent, we utilized one of the siRNAs targeting the 3′-UTR of TDP-43 (siTDP-43-2; see Fig. 3A), allowing ectopic re-expression of TDP-43 in SH-SY5Y cells. The experiments revealed that SIRT1 mRNA and protein levels could be restored upon TDP-43 transfection (Fig. 4, A and B), supporting a TDP-43-dependent regulation of SIRT1.
FIGURE 4.
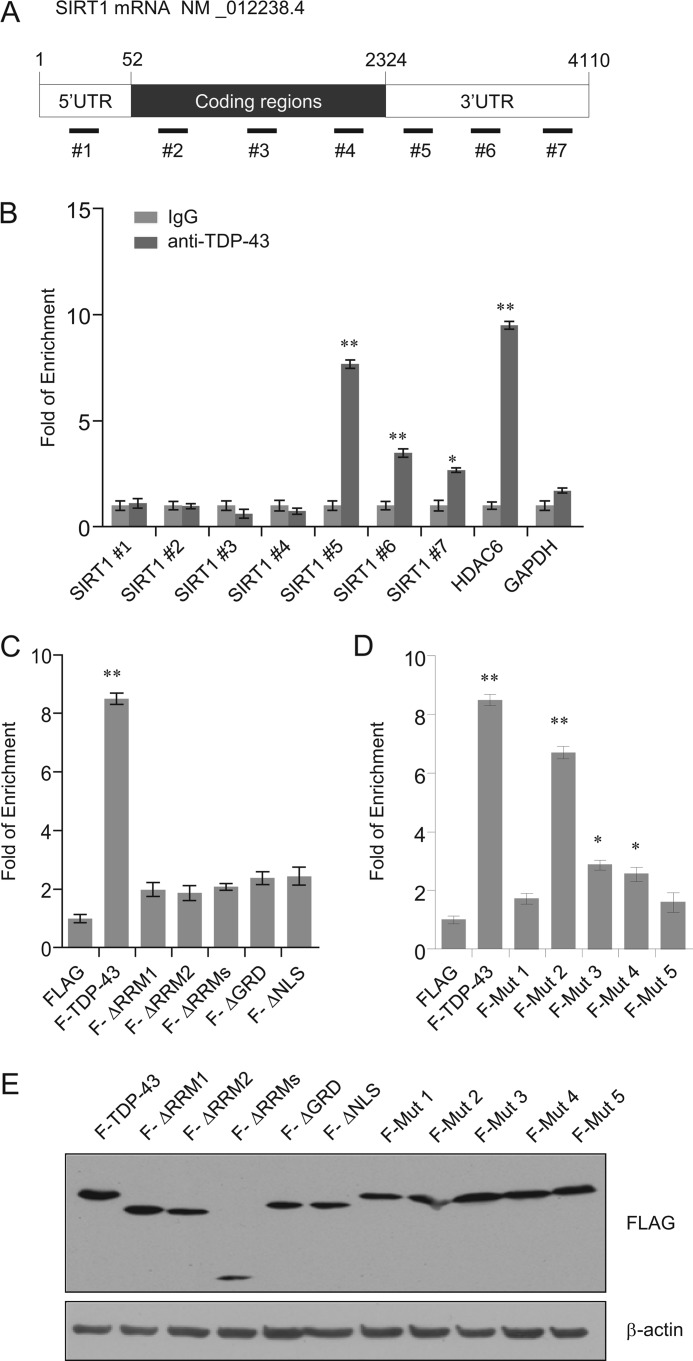
SIRT1 expression is specifically restored by TDP-43 retransfection. A, SH-SY5Y cells were treated with scrambled control siRNA or siRNATDP-43-2. After 24 h cells were transfected with empty vector, or wild type (WT) FLAG -TDP-43 as indicated. Total RNA was extracted and subjected to semi-quantitative RT-PCR amplifying endogenous TDP-43 (primers designed to target the 3′-UTR), total TDP-43 (primers designed to target the coding region), and SIRT1 as indicated. The mRNA levels were normalized to those of GADPH. B, SH-SY5Y cells were mock transfected, treated with scrambled control siRNA or siRNA TDP-43-2. After 24 h cells were transfected with empty vector (−) and wild type (WT) FLAG -TDP-43 (+). FLAG-TDP-43 and SIRT1expression were analyzed by Western blotting. C, schematic of the TDP-43 deletions and point mutations. D and E, SH-SY5Y cells were treated with scrambled siRNA (siCtrl) or siRNA TDP-43-2. After 12 h cells were transfected with empty vector, FLAG tagged TDP-43 and TDP-43 deletions (D) or TDP-43 point mutations (E) as indicated. F stands for FLAG. Total RNA was extracted and subjected to semi-quantitative RT-PCR amplifying SIRT1. The mRNA levels were normalized to those of GADPH. Error bars represent S.D. of triplicates.*, p < 0.05 and **, <0.01 (one-tailed unpaired t test). F, identical expressions of a serious TDP-43 constructs corresponding to Fig. 4, D and E in SH-SY5Y cells were verified by Western blotting.
TDP-43 contains two RRM domains that are implicated in its specific RNA processing functions. To further investigate the molecular mechanism by which TDP-43/FMRP/STAU1 regulates the expression of SIRT1, we generated TDP-43 mutants lacking RRM1 (ΔRRM1) or/and RRM2 (ΔRRM2) (Fig. 4C). Transfection of these mutants failed to restore SIRT1 mRNA levels upon TDP-43 knockdown in SH-SY5Y cells (Fig. 4D). Likewise, TDP-43 mutants either lacking the C-terminal GRD (ΔGRD) or with an impaired nuclear localization (ΔNLS) also failed to restore SIRT1 expression (Fig. 4D). The GRD has been shown to be responsible for the interaction of TDP-43 with hnRNPs and be necessary for the exon skipping function toward CFTR (3, 24). These experiments indicate that nuclear localization of TDP-43 as well as its nucleic acid and protein binding capacities are all important determinants for SIRT1 regulation. These results also support a notion that an intact TDP-43 must be ensured for its normal functions, which is consistent with previous observations (3, 31, 44).
To investigate the effects of clinical mutations on the biological functions of TDP-43 in our system, we performed rescuing experiments by transfecting a series of full-length TDP-43 missense mutants into silenced cells (Fig. 4E). In these experiments, most of the disease-associated TDP-43 mutants including D169G, G290A, G298S, and R361S consistently showed a trend of reduced ability to increase the mRNA level of SIRT1 compared with wild type of TDP-43 (Fig. 4E), except for the G287S mutation in GRD of TDP-43, which showed a similar activity in restoring SIRT1 mRNA expression as the wild type TDP-43 (Fig. 4E). The expressions of TDP-43 mutants corresponding to Fig. 4, D and E in SH-SY5Y cells were examined by Western blotting (Fig. 4F). These results suggest that the functional association between TDP-43 and SIRT1 might be pathophysiologically significant.
TDP-43 Specifically Binds to the 3′-UTR of SIRT1 mRNA
To further understand the molecular basis underlying the regulation of SIRT1 expression by TDP-43, we next tested whether TDP-43 could directly bind to SIRT1 transcripts. For this purpose, RNA-immunoprecipitation assays (RIP) were performed using TDP-43 antibodies under conditions that preserved native protein-RNA complexes, followed by detection of the SIRT1 mRNA from immunoprecipitated complexes by quantitative real-time RT-PCR amplification. In addition, to identify the regions of SIRT1 mRNA that are responsible for the TDP-43-SIRT1 mRNA interaction, primers for SIRT1 mRNA corresponding to either the 5′-UTR, the coding region (CR), or 3′-UTR were prepared as probes (Fig. 5A). The results of these experiments showed that compared with control IgG, quantitative RT-PCR product was significantly enriched in anti-TDP-43-immunoprecipitated materials, and much more abundant PCR products were detected in 3′-UTR than in 5′-UTR and CR of the SIRT1 mRNA, suggesting that TDP-43 binding to SIRT1 mRNA was restricted to 3′-UTR (2325–3826 bp). HDAC6 was used as a positive control (45), and housekeeping gene GAPDH served as a negative control (Fig. 5B). Taken together, these experiments support an argument that TDP-43 specifically binds SIRT1 mRNA and that binding occurs in the 3′-UTR of SIRT1 mRNA.
FIGURE 5.
TDP-43 specifically binds to SIRT1 3′-UTR. A, scheme of SIRT1 constructs used for the primers spanning the intron regions in 5′-UTR, coding region (CR) and 3′-UTR. B, SH-SY5Y cells extracts were immunoprecipitated with anti-TDP-43 antibody or control rabbit IgG, and the co-immunoprecipitated RNA was analyzed with quantitative real-time RT-PCR using indicated primers summarized in supplemental Table S5. HDAC6 and GAPDH served as the positive and negative control, respectively. C, SH-SY5Y cells extracts were transfected with empty vector, FLAG tagged TDP-43, TDP-43 deletions (C) or TDP-43 point mutations (D) as indicated. RNA immunoprecipitated with anti-FLAG antibody was analyzed with quantitative real-time RT-PCR using primer sets for 3′-UTR (5 in Fig. 5A). Error bars represent S.D. of triplicates. *, p < 0.05 and **, <0.01 (one-tailed unpaired t test). E, identical expressions of a serious TDP-43 constructs corresponding to Fig. 5, C & D in SH-SY5Y cells were verified by Western blotting.
We then tried to identify the regions of TDP-43 that are responsible for the TDP-43-SIRT1 mRNA interaction using TDP-43 deletion mutants and the 3′-UTR of SIRT1 mRNA (Fig. 5C). Notably, neither ΔRRM1 nor ΔRRM2 was able to bind to SIRT1 3′-UTR, suggesting that the direct SIRT1 mRNA binding is dependent on the two RRM domains of TDP-43. To further assess the effects of clinical mutations on the cellular function of the TDP-43 gene, we transfected the 5 missense mutants into SH-SY5Y cells (see Fig. 4C). In these experiments, D169G, G290A, G298S, and R361S consistently showed a diminished binding to SIRT1 3′-UTR compared with wild type TDP-43 (Fig. 5D). Interestingly, the G287S mutation located in GRD of TDP-43 appeared not to affect the TDP-43 and SIRT1 mRNA interaction (Fig. 5D). The expressions of TDP-43 mutants corresponding to Fig. 5, C and D in SH-SY5Y cells were validated by Western blotting (Fig. 5E). Taken together, these results indicate that the binding of TDP-43 to SIRT1 mRNA is mediated by its RRM1 and RRM2.
FMRP and STAU1 Bind to SIRT1 mRNA 3′-UTR in a TDP-43-dependent Manner
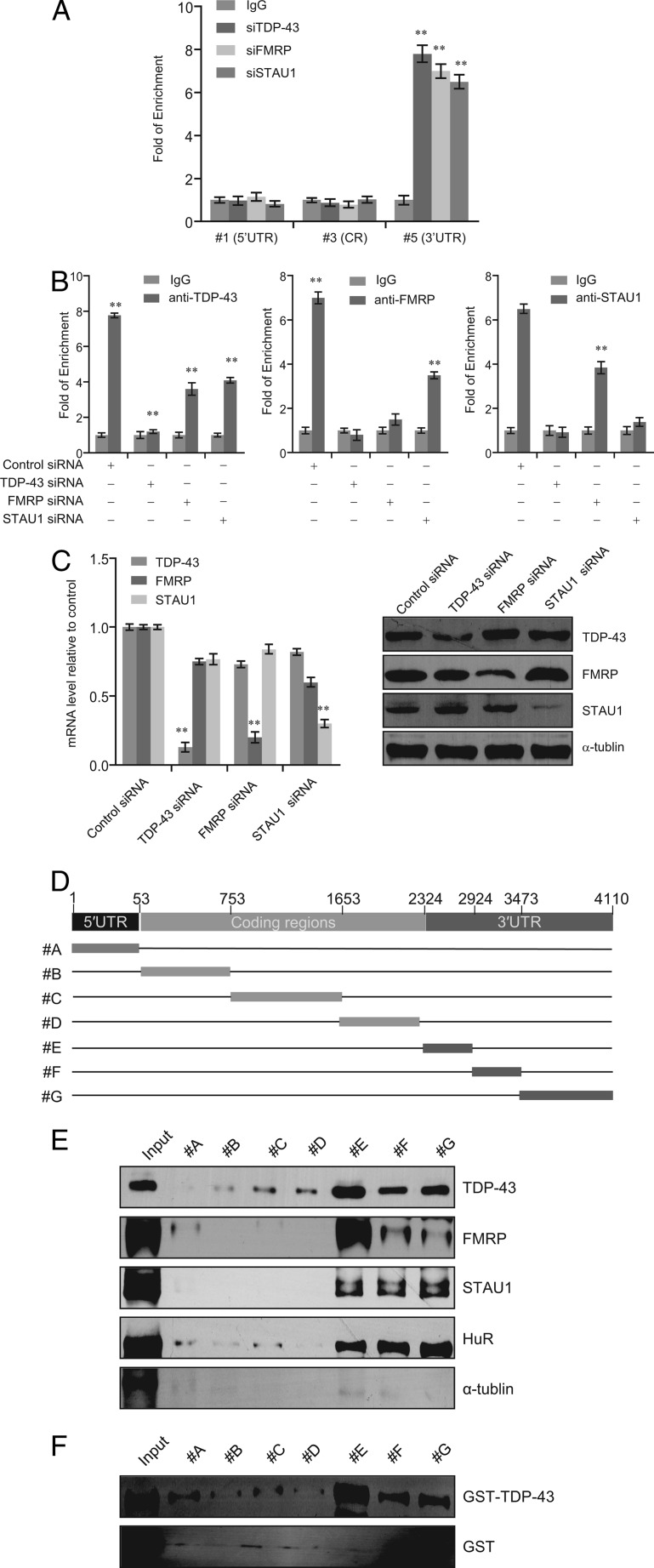
Based on the physical and functional association between TDP-43 and FMRP/STAU1, we sought to determine whether FMRP and STAU1 could also bind to SIRT1 mRNA 3′-UTR. To the end, we performed RIP analysis in SH-SY5Y cells with antibodies against FMRP and STAU1 and with primers targeted the 5′-UTR, CR, or 3′-UTR of SIRT1 mRNA (primer 1, 3, or 5 described in Fig. 5A). The results showed that, similar to TDP-43, both FMRP and STAU1 could specifically bind to SIRT1 mRNA 3′-UTR (Fig. 6A).
FIGURE 6.
FMRP and STAU1 bind to SIRT1 3′-UTR mainly depend on TDP-43. A, SH-SY5Y cells extracts were immunoprecipitated with anti-TDP-43, anti-FMRP, anti-STAU1 antibodies or control rabbit IgG, and the RIP was analyzed with quantitative RT-PCR using primers corresponding to 5′-UTR, coding region (CR) or 3′-UTR of the SIRT1 transcript. B, SH-SY5Y cells were transcfected with scrambled control siRNA, TDP-43 siRNA, FMRP siRNA, or STAU1 siRNA. RIP assays were performed with quantitative RT-PCR amplifying the 3′-UTR of SIRT1 transcript immunoprecipitated by the indicated antibodies. C, knockdown efficiency was validated by real time RT-PCR (left panel) and Western blotting (right panel). Error bars represent S.D. of triplicates. *, p < 0.05 and **, <0.01 (one-tailed unpaired t test). E, schematic of SIRT1 biotinylated probes used in RNA pull-down assays. F, each SIRT1 biotinylated probes was incubated with SH-SY5Y whole cell lysate. The affinity-purified elutes were separated on SDS-PAGE and immunoblotted using antibodies against the indicated proteins. HuR served as a positive control. G, each SIRT1 biotinylated probes was incubated with purified GST-TDP-43 or GST. The affinity purified elutes were separated on SDS-PAGE and immunoblotted using antibodies against TDP-43 and GST.
To further explore the molecular mechanism governing the regulation of SIRT1 mRNA expression by TDP-43/FMRP/STAU1, SH-SY5Y cells were treated with TDP-43-, FMRP-, or STAU1-specific siRNAs or with scrambled control siRNA (Fig. 6B). Using antibodies against TDP-43, FMRP or STAU1, RIP assays showed that the binding of TDP-43, FMRP, and STAU1 to SIRT1 mRNA were all markedly decreased after TDP-43 depletion (Fig. 6B), whereas only marginal decreases in the binding of TDP-43 were detected upon FMRP or STAU1 depletion. These data suggest that the binding of FMRP and STAU1 to the 3′-UTR of SIRT1mRNA is recruited TDP-43 and occurs in a TDP-43-dependent manner. The knockdown efficiency was validated by qPCR and Western blotting (Fig. 6C).
To further investigate the molecular detail involved in the interaction between TDP-43 and the SIRT1 mRNA, biotinylated RNA pull-down assays were performed using 5′UTR (A), coding regions (CR, B to D), and 3′-UTR (E to G) as probes (Fig. 6D). SH-SY5Y cell lysates or purified GST-TDP-43/GST were incubated with SIRT1 RNA in vitro, followed by RNA pull-down assays and Western blotting analysis. As shown in Fig. 6E, TDP-43, FMRP, and STAU1 were pulled down by the biotinylated SIRT1 3′-UTR (especially E). HuR were used as a positive control (46). Moreover, GST-TDP-43, but not the GST tag, could also be pulled down by the SIRT1 3′-UTR (especially E), but not by 5′-UTR and CR (Fig. 6F). These findings indicate that TDP-43 could directly bind SIRT1 mRNA in vivo and in vitro, and that this binding occurs in the SIRT1 3′-UTR.
TDP-43 Knockdown-mediated SIRT1 Down-regulation Sensitizes Cells to Apoptosis and DNA Damages
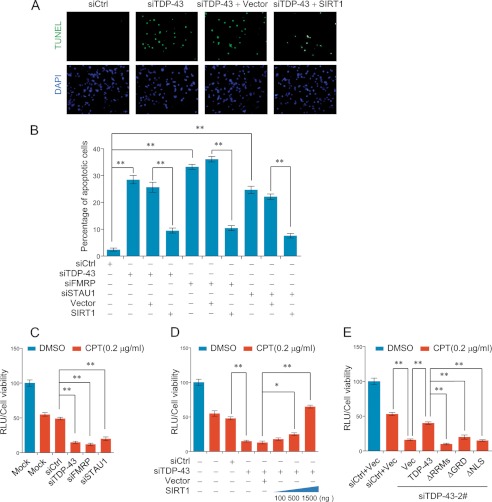
Because SIRT1 inhibits apoptosis and is essential for cell survival (47, 48), we next investigated whether TDP-43/FMRP/STAU1could influence SIRT1-protected apoptosis in SH-SY5Y cells. To this end, SH-SY5Y cells were transiently transfected with either scrambled control siRNA or TDP-43 siRNA for 48 h. TUNEL assays revealed a striking increase in apoptotic cells, 28.76% in TDP-43 RNAi group compared with only 3.65% in control group (p < 0.001) (Fig. 7, A and B). Significantly, the increased cell apoptosis resulted from TDP-43 depletion could be partially rescued through SIRT1 overexpression (from 25.62% to 9.93%, p < 0.001) (Fig. 7A). Similar results were obtained when SH-SY5Y cells were treated with FMRP siRNA or STAU1 siRNA (Fig. 7B). These findings indicated that TDP-43/FMRP/STAU1 complex is required for neuronal cell survival, at least in SH-SY5Y cells, and that this effect might be partially due to its regulation of SIRT1 expression.
FIGURE 7.
TDP-43 knockdown-mediated SIRT1 down-regulation induces apoptosis and sensitizes cells to DNA toxicity. A and B, SH-SY5Ycells were transfected with indicated siRNAs and with appropriate plasmid constructs 12 h later. After 48 h of the plasmid transfection, TUNEL assay were performed with a fluorescence method. For each group, 6 different fields under fluorescence microscopy with 100-fold magnifications were randomly chosen and counted. Representative photos were showed in A and statistical analyzed in B. C–E, SH-SY5Y cells were treated as indicated with presence or absence of camptothecin (CPT, 0.2 μg/ml). 24 h later, cell viability assays were performed with a luminescent method. Error bars represent S.D. of triplicates. *, p < 0.05 and **, <0.01 (one-tailed unpaired t test).
It is reported that SIRT1encourages cell survival by engaged in DNA double-strand breaks (DSB) repair upon DNA damages to promote genomic stability (49, 50). To further validate the neuroprotective role of TDP-43/FMRP/STAU1 complex and its functional association with SIRT1, we examined the cell viability upon exposure to DNA toxicant, camptothecin (CPT). CPT is a cytotoxic quinoline alkaloid which inhibits the DNA enzyme topoisomerase I thus interferes with DNA replication and transcription, leading to DSBs and cell death (51). In these experiments, SH-SY5Y cells in which the expression of TDP-43, FMRP, or STAU1 was knocked down were treated with CPT for 24 h. Cell viability assays by a luminescent method indicated that, compared with control siRNA-treated group, the viability of cells that were treated with siRNA specific for TDP-43, FMRP, or STAU1 was significantly reduced (Fig. 7C). In addition, in TDP-43 depleted SH-SY5Y cells, exogenous expression of SIRT1 could partially rescue the decreased cell viability in a dose-dependent manner (Fig. 7D). Furthermore, in cells that were transfected with TDP-43 siRNA-2 which targets the 3′-UTR of the TDP-43 mRNA (Fig. 3A), re-introduction of the expression of wild type TDP-43 but not the mutants ΔRRMs, ΔGRD, or ΔNLS could partially restore the viability of these cells (Fig. 7E), consistent with our earlier observations that an intact TDP-43 is required in SIRT1 regulation. These results indicated that depletion of TDP-43, FMRP, or STAU1 thus disrupting the TDP-43/FMRP/STAU1 complex impairs SIRT1-dependent DSB repair, leading to neuronal cell death in response to genomic toxicity. Taken together, these data support a notion that an intact TDP-43/FMRP/STAU1 complex is required for the stability of SIRT1 mRNA, and that defects in the integrity of the TDP-43/FMRP/STAU1 complex result in destabilization of SIRT1 mRNA and sensitize cells to apoptosis and DNA toxicity.
DISCUSSION
TDP-43 was identified several years ago as the major constituent of the proteinaceous inclusions that are characteristic of most forms of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) (6). Subsequent studies have confirmed this finding and have shown that dominantly inherited genetic mutations within the gene that encodes TDP-43 are linked with ALS and FTLD phenotypes (10, 11).
Despite the progress toward revealing the full spectrum of TDP-43 pathology in human neurodegenerative diseases, the fundamental question of whether TDP43 dysfunction mediates neurodegeneration through gain of toxic function or a loss of normal function remains unanswered. Nevertheless, there is now an emerging consensus that TDP-43 protein is mechanistically linked to neurodegeneration.
TDP-43 is a 414 aa protein with two RNA recognition motifs, RRM1 and RRM2, and a C-terminal glycine-rich domain. Previous studies have shown that TDP-43 binds several hnRNP proteins such hnRNP A/B and H through its C-terminal tail and participate in mRNA splicing (31, 52). It is believed that RNA binding proteins are the common components of much larger protein complexes required for the stabilization, transport, and metabolism of mRNA. Indeed, we revealed that TDP-43, FMRP, and STAU1, together with several hnRNPs form a functionally coordinated complex in vivo. Structurally, TDP-43, FMRP, STAU1, and hnRNPs contain domains that are implicated in protein-RNA and protein-protein interactions (4, 53), and all of them are associated with neurodegeneration disorders (17, 54, 55), supporting a physical and functional link among them.
Despite accumulating data showing that TDP-43 could bind and stabilize several target genes (18, 28, 36, 37, 56), the significance of these TDP-43 targets with regard to disease pathogenesis remains to be determined. Thus, we performed differential microarray analysis to obtain a cohort of downstream targets. This would provide further insights for the understanding of the pathophysiological functions of TDP-43. Among the target genes identified in our chip assays is SIRT1. SIRT1 plays critical role in neuroprotective actions (57–59). Consistent with previous finding that RRMs is required for TDP-43 binding to 3′-UTR (56), we showed, by RIP assays, that the specific interaction of TDP-43 with SIRT1 mRNA is mediated by RRMs. In addition, we showed that interruption of the integrity of the TDP-43/FMRP/STAU1 complex by knockdown the expression of either TDP-43, FMRP, or STAU1 led to decreased binding of the complex to SIRT1 mRNA, supporting a physical as well as functional connection among these proteins. Significantly, clinical mutations of TDP-43 (D169G, G290A, G298S, and R361S) were associated with a decreased binding of TDP-43 onto SIRT1 mRNA 3′-UTR, further strengthening a functional connection between TDP-43 and SIRT1. Moreover, we showed that TDP-43 could directly bind to the 3′-UTR of SIRT1 mRNA via RNA pull-down assays.
Although our data suggest that FMRP and STAU1 are recruited by TDP-43, the exact role of the each component of the TDP-43/FMRP/STAU1 complex in SIRT1 mRNA processing is currently still unknown. Both TDP-43 and FMRP have been shown to regulate mRNA stability via binding to 3′-UTR of target transcripts (32, 56), but STAU1 is mainly involved in the Staufen-mediated mRNA decay (SMD) when binds to 3′-UTR of target mRNA such as ARF1 (35). Then what might be the purpose of STAU1 being recruited to the TDP-43/FMRP/STAU1 complex? First, different from nonsense-mediated mRNA decay which degrades newly synthesized mRNA for quality control (33), SMD is involved in regulation the stability and quality of natural mRNAs via 3′-UTR binding (35), thus maintaining adequate mRNA level for proper translation; second, STAU1 has been shown to bind tubulin and present in the cytoplasm in association with the rough endoplasmic reticulum, the site of translation (60); the possibility that STAU1 might contribute to SIRT1 mRNA transport and protein synthesis cannot be excluded. Clearly, further investigations are warranted to explore the detailed molecular event involved in the regulation of SIRT1 expression by the TDP-43/FMRP/STAU1 complex.
SIRT1 is a mammalian ortholog of the yeast silent information regulator 2 (Sir2), a NAD+ dependent class III histone deacetylase. This protein has been shown to orchestrate diverse biological processes, including cell differentiation, apoptosis, autophagy, metabolism, and stress response (49, 61). There is also growing evidence that SIRT1 plays critical roles in the pathogenesis of multiple neurodegenerative diseases (42, 49), in which TDP-43 malfunction has been implicated in. In mice models, SIRT1 could suppress the production of toxic product in AD and PD (57, 59, 62), prevent neurodegeneration in AD and ALS, and protect axon against degeneration in Wallerian degeneration (58, 63), consistent with our revelation in this study that TDP-43 and SIRT1 are functionally connected. Moreover, the study that SIRT1 cooperates with E2F1 to regulate apoptotic response to DNA damage (64) and the finding that loss of functional TDP-43 induces apoptosis in a pRb/E2F1-dependent manner (36) further support our argument that the requirement for TDP-43 in neuronal cell survival is partially mediated by SIRT1. Strikingly, recent studies revealed that SIRT1knockdown results in poly Q-expanded aggregation of androgen receptor (AR) and α-synuclein (59, 65). These findings appear to be consistent with the observations that knockdown of TDP-43 leads to the aggregation of neuropathological prion proteins, further supporting our observation that TDP-43 and SIRT1 are functionally connected.
In summary, our experiments revealed that TDP-43 is physically associated with FMRP and STAU1 to form a functionally coordinated complex to co-regulate the expression of SIRT1. Our results indicate that defects in the TDP-43/FMRP/STUA1 complex sensitize neuronal cells to apoptosis and DNA damages in a SIRT1-dependent manner. These findings may shed new light on the understanding of the biological functions and the mechanistic involvement of TDP-43 in neurodegenerative diseases.
Supplementary Material
This work was supported by Grants 90919053 and 31171240 (to Y. W.) and 81030019 (to D. F.) from the National Natural Science Foundation of China.

This article contains supplemental Tables S1–S5.
- TARDBP
- transactive response DNA-binding protein
- ALS
- amyotrophic lateral sclerosis
- CPT
- camptothecin
- DMSO
- dimethyl sulfoxide
- FLTD
- frontotemporal lobar degeneration
- FMRP
- fragile X mental retardation protein
- hnRNP
- heterogeneous nuclear ribonuleoprotein
- GRD
- glycine-rich domain
- RRM
- RNA recognition motif.
REFERENCES
- 1. Wang H. Y., Wang I. F., Bose J., Shen C. K. (2004) Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics 83, 130–139 [DOI] [PubMed] [Google Scholar]
- 2. Ayala Y. M., Pantano S., D'Ambrogio A., Buratti E., Brindisi A., Marchetti C., Romano M., Baralle F. E. (2005) Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J. Mol. Biol. 348, 575–588 [DOI] [PubMed] [Google Scholar]
- 3. Buratti E., Baralle F. E. (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276, 36337–36343 [DOI] [PubMed] [Google Scholar]
- 4. Wang I. F., Wu L. S., Shen C. K. (2008) TDP-43: an emerging new player in neurodegenerative diseases. Trends Mol. Med. 14, 479–485 [DOI] [PubMed] [Google Scholar]
- 5. Kuo P. H., Doudeva L. G., Wang Y. T., Shen C. K., Yuan H. S. (2009) Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 37, 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 7. Davidson Y., Kelley T., Mackenzie I. R., Pickering-Brown S., Du Plessis D., Neary D., Snowden J. S., Mann D. M. (2007) Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol. 113, 521–533 [DOI] [PubMed] [Google Scholar]
- 8. Chen-Plotkin A. S., Lee V. M., Trojanowski J. Q. (2010) TAR DNA-binding protein 43 in neurodegenerative disease. Nat. Reviews. Neurology 6, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cook C., Zhang Y. J., Xu Y. F., Dickson D. W., Petrucelli L. (2008) TDP-43 in neurodegenerative disorders. Exp. Opin. Biol. Ther. 8, 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Deerlin V. M., Leverenz J. B., Bekris L. M., Bird T. D., Yuan W., Elman L. B., Clay D., Wood E. M., Chen-Plotkin A. S., Martinez-Lage M., Steinbart E., McCluskey L., Grossman M., Neumann M., Wu I. L., Yang W. S., Kalb R., Galasko D. R., Montine T. J., Trojanowski J. Q., Lee V. M., Schellenberg G. D., Yu C. E. (2008) TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 7, 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., Miller C. C., Nicholson G., Shaw C. E. (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barmada S. J., Finkbeiner S. (2010) Pathogenic TARDBP mutations in amyotrophic lateral sclerosis and frontotemporal dementia: disease-associated pathways. Rev. Neurosci. 21, 251–272 [DOI] [PubMed] [Google Scholar]
- 13. Acharya K. K., Govind C. K., Shore A. N., Stoler M. H., Reddi P. P. (2006) cis-requirement for the maintenance of round spermatid-specific transcription. Dev. Biol. 295, 781–790 [DOI] [PubMed] [Google Scholar]
- 14. Ou S. H., Wu F., Harrich D., García-Martinez L. F., Gaynor R. B. (1995) Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 69, 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fiesel F. C., Kahle P. J. (2011) TDP-43 and FUS/TLS: cellular functions and implications for neurodegeneration. FEBS J. 278, 3550–3568 [DOI] [PubMed] [Google Scholar]
- 16. Lagier-Tourenne C., Polymenidou M., Cleveland D. W. (2010) TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Human Mol. Gen. 19, R46–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee E. B., Lee V. M., Trojanowski J. Q. (2012) Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nature Rev. Neuroscience 13, 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Polymenidou M., Lagier-Tourenne C., Hutt K. R., Huelga S. C., Moran J., Liang T. Y., Ling S. C., Sun E., Wancewicz E., Mazur C., Kordasiewicz H., Sedaghat Y., Donohue J. P., Shiue L., Bennett C. F., Yeo G. W., Cleveland D. W. (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nature Neurosci. 14, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sephton C. F., Cenik C., Kucukural A., Dammer E. B., Cenik B., Han Y., Dewey C. M., Roth F. P., Herz J., Peng J., Moore M. J., Yu G. (2011) Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 286, 1204–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang Q., She H., Gearing M., Colla E., Lee M., Shacka J. J., Mao Z. (2009) Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science 323, 124–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Del Villar K., Miller C. A. (2004) Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer's disease brain and hippocampal neurons. Proc. Natl. Acad. Sci. U.S.A. 101, 4210–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duberley R. M., Johnson I. P., Anand P., Swash M., Martin J., Leigh P. N., Zeman S. (1995) Ciliary neurotrophic factor receptor expression in spinal cord and motor cortex in amyotrophic lateral sclerosis. J. Neurol. Sci. 129, suppl. 109–113 [DOI] [PubMed] [Google Scholar]
- 23. Tollervey J. R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A. L., Zupunski V., Patani R., Chandran S., Rot G., Zupan B., Shaw C. E., Ule J. (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buratti E., Brindisi A., Pagani F., Baralle F. E. (2004) Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am. J. Human Genet. 74, 1322–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buratti E., Dörk T., Zuccato E., Pagani F., Romano M., Baralle F. E. (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 20, 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mercado P. A., Ayala Y. M., Romano M., Buratti E., Baralle F. E. (2005) Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 33, 6000–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bose J. K., Wang I. F., Hung L., Tarn W. Y., Shen C. K. (2008) TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J. Biol. Chem. 283, 28852–28859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fiesel F. C., Voigt A., Weber S. S., Van den Haute C., Waldenmaier A., Görner K., Walter M., Anderson M. L., Kern J. V., Rasse T. M., Schmidt T., Springer W., Kirchner R., Bonin M., Neumann M., Baekelandt V., Alunni-Fabbroni M., Schulz J. B., Kahle P. J. (2010) Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 29, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang W., Martindale J. L., Yang X., Chrest F. J., Gorospe M. (2005) Increased stability of the p16 mRNA with replicative senescence. EMBO Rep. 6, 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ling S. C., Albuquerque C. P., Han J. S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D. W. (2010) ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl. Acad. Sci. U.S.A. 107, 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y. M., Baralle F. E. (2005) TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 280, 37572–37584 [DOI] [PubMed] [Google Scholar]
- 32. Zalfa F., Eleuteri B., Dickson K. S., Mercaldo V., De Rubeis S., di Penta A., Tabolacci E., Chiurazzi P., Neri G., Grant S. G., Bagni C. (2007) A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nature Neurosci. 10, 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim Y. K., Furic L., Parisien M., Major F., DesGroseillers L., Maquat L. E. (2007) Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 26, 2670–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee E. K., Kim H. H., Kuwano Y., Abdelmohsen K., Srikantan S., Subaran S. S., Gleichmann M., Mughal M. R., Martindale J. L., Yang X., Worley P. F., Mattson M. P., Gorospe M. (2010) hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 17, 732–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim Y. K., Furic L., Desgroseillers L., Maquat L. E. (2005) Mammalian Staufen1 recruits Upf1 to specific mRNA 3′-UTRs so as to elicit mRNA decay. Cell 120, 195–208 [DOI] [PubMed] [Google Scholar]
- 36. Ayala Y. M., Misteli T., Baralle F. E. (2008) TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc. Natl. Acad. Sci. U.S.A. 105, 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chiang P. M., Ling J., Jeong Y. H., Price D. L., Aja S. M., Wong P. C. (2010) Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 16320–16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuan J., Yankner B. A. (2000) Apoptosis in the nervous system. Nature 407, 802–809 [DOI] [PubMed] [Google Scholar]
- 39. Rossi F., Cattaneo E. (2002) Opinion: neural stem cell therapy for neurological diseases: dreams and reality. Nature Reviews. Neuroscience 3, 401–409 [DOI] [PubMed] [Google Scholar]
- 40. Koo E. H., Kopan R. (2004) Potential role of presenilin-regulated signaling pathways in sporadic neurodegeneration. Nature Med. 10, S26–33 [DOI] [PubMed] [Google Scholar]
- 41. Friedlander R. M. (2003) Apoptosis and caspases in neurodegenerative diseases. New England J. Med. 348, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 42. Gan L., Mucke L. (2008) Paths of convergence: sirtuins in aging and neurodegeneration. Neuron 58, 10–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haigis M. C., Guarente L. P. (2006) Mammalian sirtuins–emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20, 2913–2921 [DOI] [PubMed] [Google Scholar]
- 44. Ayala Y. M., Zago P., D'Ambrogio A., Xu Y. F., Petrucelli L., Buratti E., Baralle F. E. (2008) Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Science 121, 3778–3785 [DOI] [PubMed] [Google Scholar]
- 45. Kim S. H., Shanware N. P., Bowler M. J., Tibbetts R. S. (2010) Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J. Biol. Chem. 285, 34097–34105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abdelmohsen K., Pullmann R., Jr., Lal A., Kim H. H., Galban S., Yang X., Blethrow J. D., Walker M., Shubert J., Gillespie D. A., Furneaux H., Gorospe M. (2007) Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25, 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. (2004) Mammalian SIRT1 represses forkhead transcription factors. Cell 116, 551–563 [DOI] [PubMed] [Google Scholar]
- 48. Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 49. Finkel T., Deng C. X., Mostoslavsky R. (2009) Recent progress in the biology and physiology of sirtuins. Nature 460, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oberdoerffer P., Michan S., McVay M., Mostoslavsky R., Vann J., Park S. K., Hartlerode A., Stegmuller J., Hafner A., Loerch P., Wright S. M., Mills K. D., Bonni A., Yankner B. A., Scully R., Prolla T. A., Alt F. W., Sinclair D. A. (2008) SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135, 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hsiang Y. H., Lihou M. G., Liu L. F. (1989) Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Research 49, 5077–5082 [PubMed] [Google Scholar]
- 52. Martinez-Contreras R., Cloutier P., Shkreta L., Fisette J. F., Revil T., Chabot B. (2007) hnRNP proteins and splicing control. Advances Exp. Med. Biol. 623, 123–147 [DOI] [PubMed] [Google Scholar]
- 53. Kiebler M. A., DesGroseillers L. (2000) Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron 25, 19–28 [DOI] [PubMed] [Google Scholar]
- 54. Martinez Tosar L. J., Thomas M. G., Baez M. V., Ibanez I., Chernomoretz A., Boccaccio G. L. (2012) Staufen: from embryo polarity to cellular stress and neurodegeneration. Front. Biosci. 4, 432–452 [DOI] [PubMed] [Google Scholar]
- 55. Sokol D. K., Maloney B., Long J. M., Ray B., Lahiri D. K. (2011) Autism, Alzheimer disease, and fragile X: APP, FMRP, and mGluR5 are molecular links. Neurology 76, 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ayala Y. M., De Conti L., Avendaño-Vazquez S. E., Dhir A., Romano M., D'Ambrogio A., Tollervey J., Ule J., Baralle M., Buratti E., Baralle F. E. (2011) TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 30, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Donmez G., Wang D., Cohen D. E., Guarente L. (2010) SIRT1 suppresses β-amyloid production by activating the α-secretase gene ADAM10. Cell 142, 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58. Kim D., Nguyen M. D., Dobbin M. M., Fischer A., Sananbenesi F., Rodgers J. T., Delalle I., Baur J. A., Sui G., Armour S. M., Puigserver P., Sinclair D. A., Tsai L. H. (2007) SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 26, 3169–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Donmez G., Arun A., Chung C. Y., McLean P. J., Lindquist S., Guarente L. (2012) SIRT1 protects against α-synuclein aggregation by activating molecular chaperones. J. Neurosci. 32, 124–132 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60. Kiebler M. A., Hemraj I., Verkade P., Köhrmann M., Fortes P., Marión R. M., Ortín J., Dotti C. G. (1999) The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J. Neurosci. 19, 288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 62. Outeiro T. F., Kontopoulos E., Altmann S. M., Kufareva I., Strathearn K. E., Amore A. M., Volk C. B., Maxwell M. M., Rochet J. C., McLean P. J., Young A. B., Abagyan R., Feany M. B., Hyman B. T., Kazantsev A. G. (2007) Sirtuin 2 inhibitors rescue α-synuclein-mediated toxicity in models of Parkinson's disease. Science 317, 516–519 [DOI] [PubMed] [Google Scholar]
- 63. Araki T., Sasaki Y., Milbrandt J. (2004) Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 64. Wang C., Chen L., Hou X., Li Z., Kabra N., Ma Y., Nemoto S., Finkel T., Gu W., Cress W. D., Chen J. (2006) Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat. Cell Biol. 8, 1025–1031 [DOI] [PubMed] [Google Scholar]
- 65. Montie H. L., Pestell R. G., Merry D. E. (2011) SIRT1 modulates aggregation and toxicity through deacetylation of the androgen receptor in cell models of SBMA. J. Neurosci. 31, 17425–17436 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.