Abstract
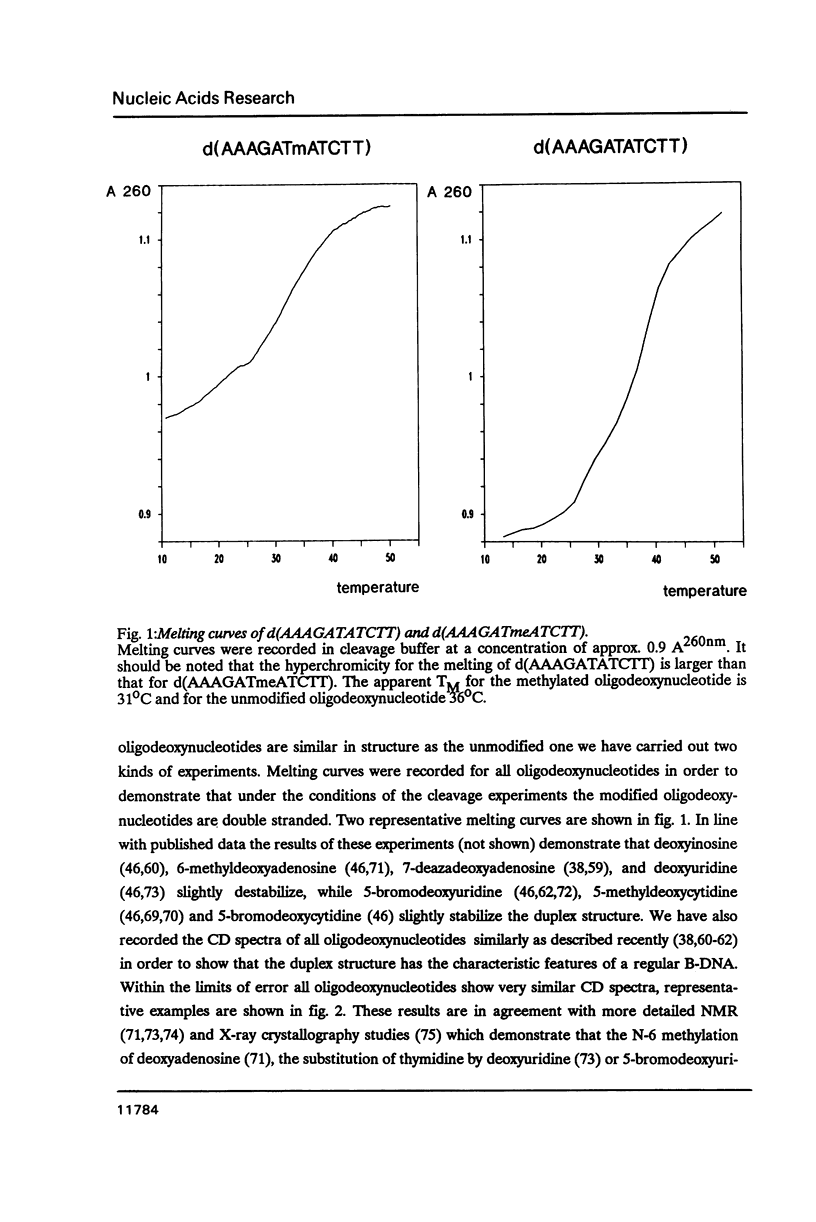
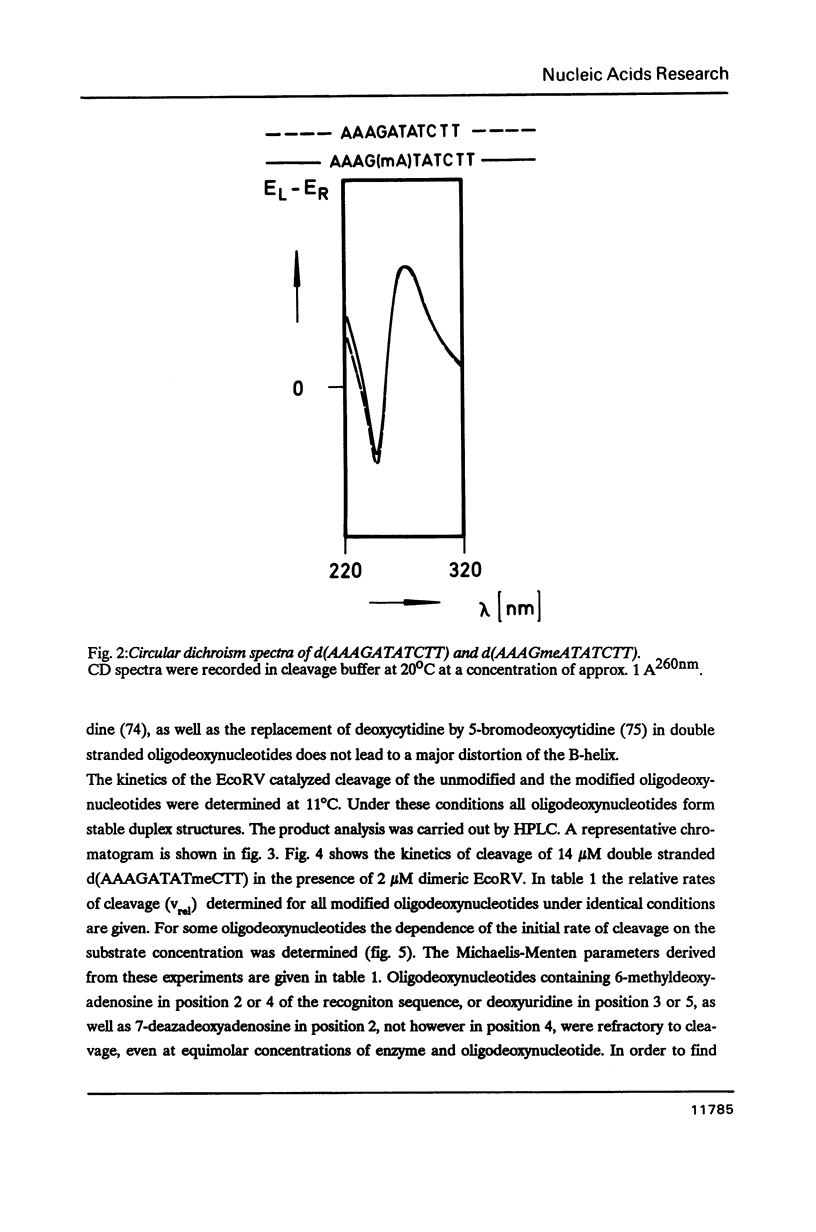
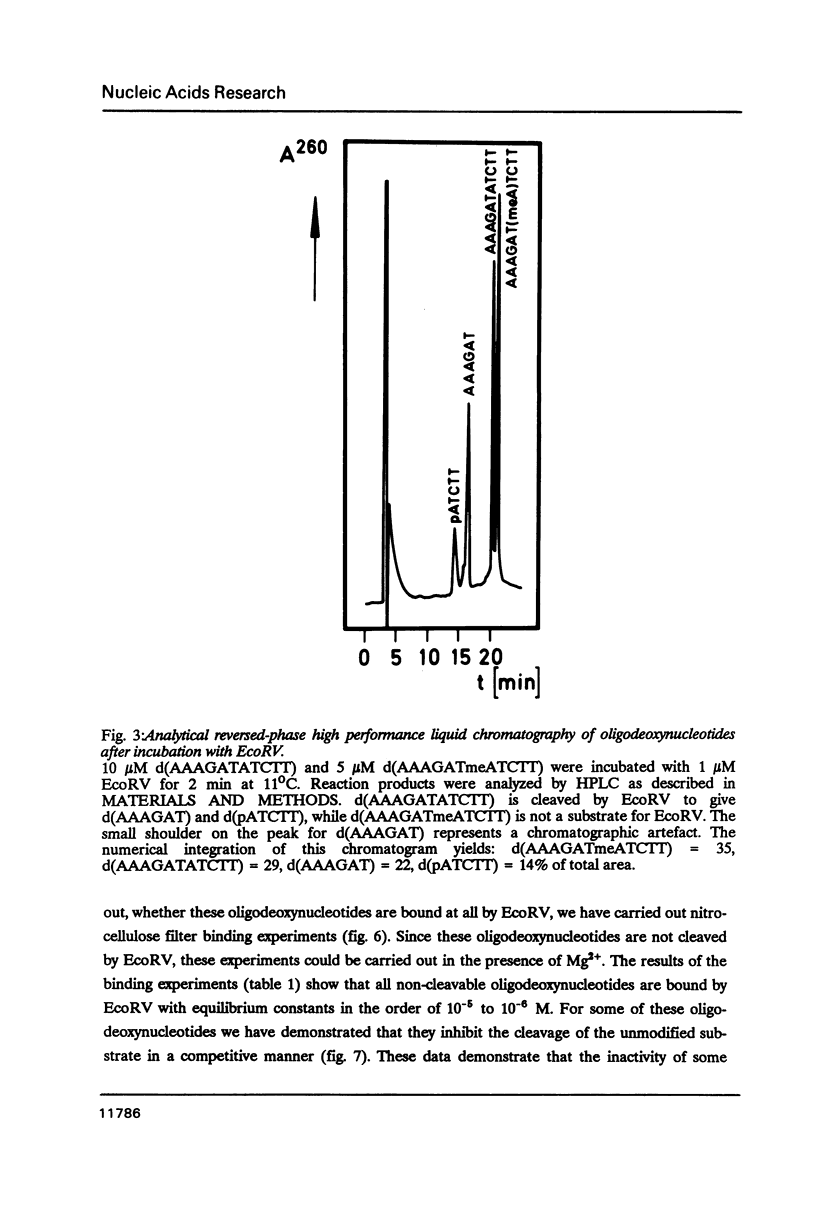
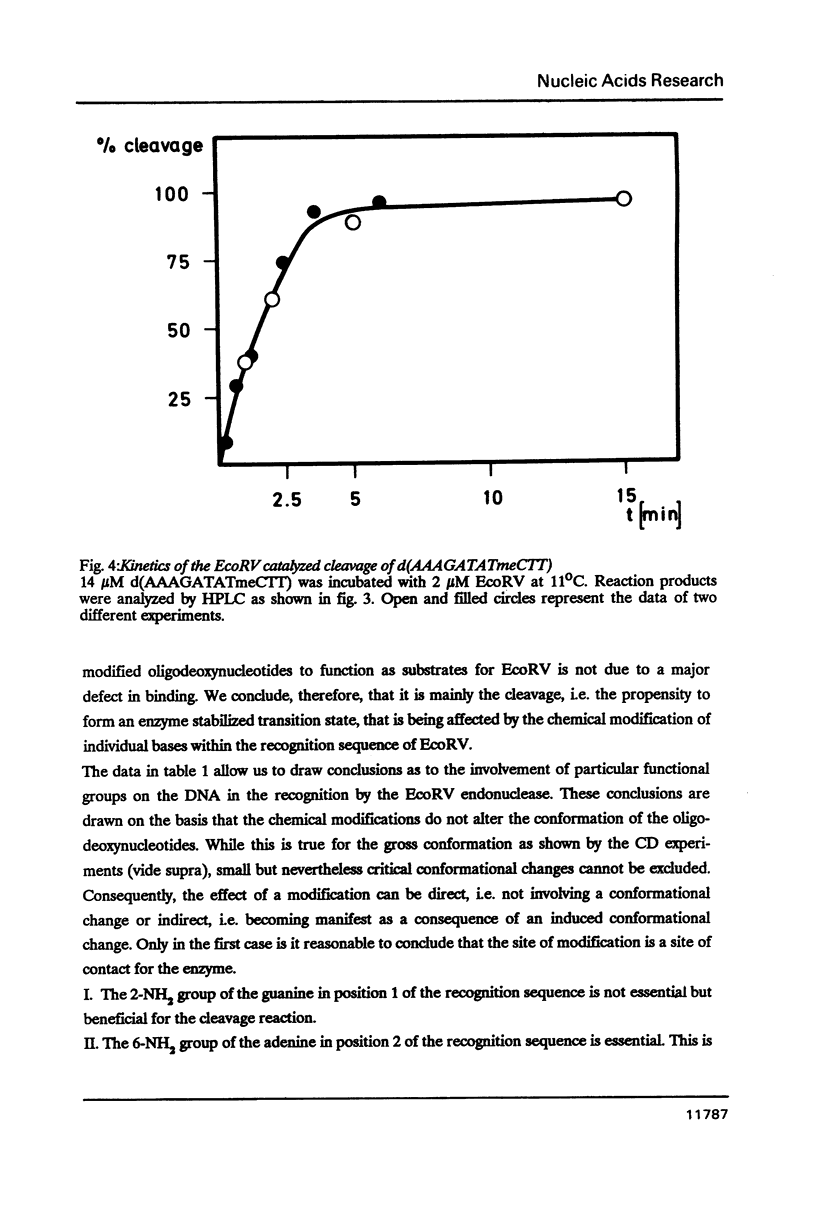
We have prepared a series of undecadeoxynucleotides that contain changes in the functional group pattern present within the EcoRV recognition site - GATATC-. Oligonucleotides were synthesized on solid phase using normal and modified beta-cyanoethylphosphoramidites and analyzed in steady state cleavage experiments with the EcoRV restriction endonuclease. The following groups appear to interact strongly with the enzyme, since their modification or substitution renders the oligonucleotides refractory to cleavage: the exocyclic NH2-groups of both A residues, the N7 of the first A residue, the exocyclic NH2-group of the C residue and the CH3-groups of both T residues. The exocyclic NH-group of the G residue supports effective recognition, since its absence lowers the kcat of the cleavage reaction. The N7 of the second A residue and the C5 position of the C residue apparently are not recognized by EcoRV; their substitution by -CH- or modification with -Br or -CH3, resp., does not considerably change the rate of cleavage. All oligonucleotides investigated compete with the unmodified substrate for binding to the enzyme. We conclude that EcoRV recognizes its substrate presumably through hydrogen bonds to the exocyclic NH2-group and the N7 of the first A residue, the exocyclic NH2-groups of the second A and the C residue, as well as through hydrophobic interactions with both T residues.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alves J., Pingoud A., Haupt W., Langowski J., Peters F., Maass G., Wolff C. The influence of sequences adjacent to the recognition site on the cleavage of oligodeoxynucleotides by the EcoRI endonuclease. Eur J Biochem. 1984 Apr 2;140(1):83–92. doi: 10.1111/j.1432-1033.1984.tb08069.x. [DOI] [PubMed] [Google Scholar]
- Axelson J. T., Bodley J. W., Chen J. Y., Dunlop P. C., Rosenthal L. P., Viskup R. W., Walseth T. F. Anion-exchange chromatography of proteins on AG MP-1 using high-performance liquid chromatography equipment. Anal Biochem. 1984 Nov 1;142(2):373–377. doi: 10.1016/0003-2697(84)90479-2. [DOI] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. An assay for the rates of cleavage of specific sites in DNA by restriction endonucleases: its use to study the cleavage of phage lambda DNA by EcoRI and phage P22 DNA containing thymine or 5-bromouracil by HindIII. Anal Biochem. 1983 Mar;129(2):446–456. doi: 10.1016/0003-2697(83)90575-4. [DOI] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. EcoRI cleavage and methylation of DNAs containing modified pyrimidines in the recogintion sequence. J Biol Chem. 1977 May 25;252(10):3185–3193. [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. Quantitation of the various termini generated by type II restriction endonucleases using the polynucleotide kinase exchange reaction. J Biol Chem. 1979 Apr 10;254(7):2561–2564. [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. The effects of substituted pyrimidines in DNAs on cleavage by sequence-specific endonucleases. J Biol Chem. 1979 Apr 10;254(7):2551–2560. [PubMed] [Google Scholar]
- Bodnar J. W., Zempsky W., Warder D., Bergson C., Ward D. C. Effect of nucleotide analogs on the cleavage of DNA by the restriction enzymes AluI, DdeI, HinfI, RsaI, and TaqI. J Biol Chem. 1983 Dec 25;258(24):15206–15213. [PubMed] [Google Scholar]
- Bougueleret L., Schwarzstein M., Tsugita A., Zabeau M. Characterization of the genes coding for the Eco RV restriction and modification system of Escherichia coli. Nucleic Acids Res. 1984 Apr 25;12(8):3659–3676. doi: 10.1093/nar/12.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Gumport R. I. T4 RNA ligase catalyzed synthesis of base analogue-containing oligodeoxyribonucleotides and a characterization of their thermal stabilities. Nucleic Acids Res. 1985 Dec 20;13(24):8665–8684. doi: 10.1093/nar/13.24.8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Van Cleve M. D., Gumport R. I. The effects of base analogue substitutions on the methylation by the EcoRI modification methylase of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem. 1986 Jun 5;261(16):7279–7286. [PubMed] [Google Scholar]
- Bron S., Luxen E., Venema G. Resistance of bacteriophage H1 to restriction and modification by Bacillus subtilis R. J Virol. 1983 Jun;46(3):703–708. doi: 10.1128/jvi.46.3.703-708.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkus V., Klimasauskas S., Kersulyte D., Vaitkevicius D., Lebionka A., Janulaitis A. Investigation of restriction-modification enzymes from M. varians RFL19 with a new type of specificity toward modification of substrate. Nucleic Acids Res. 1985 Aug 26;13(16):5727–5746. doi: 10.1093/nar/13.16.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkus V., Klimasauskas S., Petrauskiene L., Maneliene Z., Janulaitis A., Minchenkova L. E., Schyolkina A. K. Synthesis and physical characterization of DNA fragments containing N4-methylcytosine and 5-methylcytosine. Nucleic Acids Res. 1987 Oct 26;15(20):8467–8478. doi: 10.1093/nar/15.20.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet A., Kawashima E. DNA containing the base analogue 2-aminoadenine: preparation, use as hybridization probes and cleavage by restriction endonucleases. Nucleic Acids Res. 1988 Jan 11;16(1):305–317. doi: 10.1093/nar/16.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly B. A., Eckstein F., Pingoud A. The stereochemical course of the restriction endonuclease EcoRI-catalyzed reaction. J Biol Chem. 1984 Sep 10;259(17):10760–10763. [PubMed] [Google Scholar]
- Connolly B. A., Potter B. V., Eckstein F., Pingoud A., Grotjahn L. Synthesis and characterization of an octanucleotide containing the EcoRI recognition sequence with a phosphorothioate group at the cleavage site. Biochemistry. 1984 Jul 17;23(15):3443–3453. doi: 10.1021/bi00310a010. [DOI] [PubMed] [Google Scholar]
- D'Arcy A., Brown R. S., Zabeau M., van Resandt R. W., Winkler F. K. Purification and crystallization of the EcoRV restriction endonuclease. J Biol Chem. 1985 Feb 25;260(4):1987–1990. [PubMed] [Google Scholar]
- Delort A. M., Neumann J. M., Molko D., Hervé M., Téoule R., Tran Dinh S. Influence of uracil defect on DNA structure: 1H NMR investigation at 500 MHz. Nucleic Acids Res. 1985 May 10;13(9):3343–3355. doi: 10.1093/nar/13.9.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C. H., Wilson G. A., Young F. E. Biochemical and genetic properties of site-specific restriction endonucleases in Bacillus globigii. J Bacteriol. 1978 Apr;134(1):338–344. doi: 10.1128/jb.134.1.338-344.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer-Hallquist P., Kézdy F. J., Agarwal K. L. Interaction of the HpaI endonuclease with synthetic oligonucleotides. Biochemistry. 1982 Sep 14;21(19):4693–4700. doi: 10.1021/bi00262a027. [DOI] [PubMed] [Google Scholar]
- Fazakerley G. V., Sowers L. C., Eritja R., Kaplan B. E., Goodman M. F. Structural and dynamic properties of a bromouracil-adenine base pair in DNA studied by proton NMR. J Biomol Struct Dyn. 1987 Dec;5(3):639–650. doi: 10.1080/07391102.1987.10506417. [DOI] [PubMed] [Google Scholar]
- Fliess A., Wolfes H., Rosenthal A., Schwellnus K., Blöcker H., Frank R., Pingoud A. Role of thymidine residues in DNA recognition by the EcoRI and EcoRV restriction endonucleases. Nucleic Acids Res. 1986 Apr 25;14(8):3463–3474. doi: 10.1093/nar/14.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res. 1981 Jun 11;9(11):2509–2515. doi: 10.1093/nar/9.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S. E., Lovelady B. M., McCallum S. A. Relaxed specificity of the EcoRV restriction endonuclease. Gene. 1986;41(2-3):173–181. doi: 10.1016/0378-1119(86)90096-x. [DOI] [PubMed] [Google Scholar]
- Hayakawa T., Ono A., Ueda T. Synthesis of decadeoxyribonucleotides containing 5-modified uracils and their interactions with restriction endonucleases Bgl II, Sau 3AI and Mbo I (nucleosides and nucleotides 82). Nucleic Acids Res. 1988 Jun 10;16(11):4761–4776. doi: 10.1093/nar/16.11.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer B., Köster H. On the influence of thymidine analogues on the activity of phage fd promoters in vitro. Nucleic Acids Res. 1980 Dec 20;8(24):6143–6162. doi: 10.1093/nar/8.24.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. H., Farnet C. M., Ehrlich K. C., Ehrlich M. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 1982 Mar 11;10(5):1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Kawamura F., Duffy J. J. Susceptibility of non-thymine containing DNA to four bacterial restriction endonucleases. FEBS Lett. 1975 Jul 15;55(1):278–281. doi: 10.1016/0014-5793(75)81011-8. [DOI] [PubMed] [Google Scholar]
- Ivarie R. Thymine methyls and DNA-protein interactions. Nucleic Acids Res. 1987 Dec 10;15(23):9975–9983. doi: 10.1093/nar/15.23.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarie R. Thymine methyls and DNA-protein interactions. Nucleic Acids Res. 1987 Dec 10;15(23):9975–9983. doi: 10.1093/nar/15.23.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J., Martin D. Restriction endonucleases HindII and TaqI cleave DNA with mismatched nucleotides within their recognition sequences. Nucleic Acids Res. 1986 Mar 11;14(5):1943–1949. doi: 10.1093/nar/14.5.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J., Wood S. G., Martin D., Ubasawa A. Oligonucleotide duplexes containing inosine, 7-deazainosine, tubercidin, nebularine and 7-deazanebularine as substrates for restriction endonucleases HindII, SalI and TaqI. Nucleic Acids Res. 1986 Aug 26;14(16):6579–6590. doi: 10.1093/nar/14.16.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. A., Nierlich D. P. Cleavage of Nonglucosylated Bacteriophage T4 deoxyribonucleic acid by Restriction Endonuclease Eco RI. J Biol Chem. 1975 Mar 25;250(6):2395–2397. [PubMed] [Google Scholar]
- Kessler C., Höltke H. J. Specificity of restriction endonucleases and methylases--a review. Gene. 1986;47(1):1–153. doi: 10.1016/0378-1119(86)90245-3. [DOI] [PubMed] [Google Scholar]
- Kholmina G. V., Rebentish B. A., Skoblov Iu S., Mironov A. A., Iankovskii N. K. Vydelenie i kharakteristika novoi sait-spetsificheskoi éndonukleazy Eco Rv. Dokl Akad Nauk SSSR. 1980;253(2):495–497. [PubMed] [Google Scholar]
- Kita K., Hiraoka N., Kimizuka F., Obayashi A., Kojima H., Takahashi H., Saito H. Interaction of the restriction endonuclease ScaI with its substrates. Nucleic Acids Res. 1985 Oct 11;13(19):7015–7024. doi: 10.1093/nar/13.19.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubareva E. A., Pein C. D., Gromova E. S., Kuznezova S. A., Tashlitzki V. N., Cech D., Shabarova Z. A. The role of modifications in oligonucleotides in sequence recognition by MvaI restriction endonuclease. Eur J Biochem. 1988 Aug 15;175(3):615–618. doi: 10.1111/j.1432-1033.1988.tb14236.x. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Jack W. E., Modrich P. DNA determinants important in sequence recognition by Eco RI endonuclease. J Biol Chem. 1981 Dec 25;256(24):13200–13206. [PubMed] [Google Scholar]
- Lycksell P. O., Gräslund A., Claesens F., McLaughlin L. W., Larsson U., Rigler R. Base pair opening dynamics of a 2-aminopurine substituted Eco RI restriction sequence and its unsubstituted counterpart in oligonucleotides. Nucleic Acids Res. 1987 Nov 11;15(21):9011–9025. doi: 10.1093/nar/15.21.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. B., Rao R. N., Smith H. O. Cloning of restriction and modification genes in E. coli: the HbaII system from Haemophilus haemolyticus. Gene. 1978 Apr;3(2):97–112. doi: 10.1016/0378-1119(78)90054-9. [DOI] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchionni M. A., Roufa R. J. Digestion of 5-bromodeoxyuridine-substituted lambda-DNA by restriction endonucleases. J Biol Chem. 1978 Dec 25;253(24):9075–9081. [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- McLaughlin L. W., Benseler F., Graeser E., Piel N., Scholtissek S. Effects of functional group changes in the EcoRI recognition site on the cleavage reaction catalyzed by the endonuclease. Biochemistry. 1987 Nov 17;26(23):7238–7245. doi: 10.1021/bi00397a007. [DOI] [PubMed] [Google Scholar]
- Miller P. B., Maltman K. L., Warren R. A. Isolation and preliminary characterization of amber mutants of bacteriophage phi W-14 defective in DNA synthesis. J Virol. 1982 Jul;43(1):67–72. doi: 10.1128/jvi.43.1.67-72.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P. B., Wakarchuk W. W., Warren R. A. alpha-Putrescinylthymine and the sensitivity of bacteriophage phi W-14 DNA to restriction endonucleases. Nucleic Acids Res. 1985 Apr 11;13(7):2559–2568. doi: 10.1093/nar/13.7.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., Rubin R. A. Role of the 2-amino group of deoxyguanosine in sequence recognition by EcoRI restriction and modification enzymes. J Biol Chem. 1977 Oct 25;252(20):7273–7278. [PubMed] [Google Scholar]
- Nelson M., McClelland M. The effect of site-specific methylation on restriction-modification enzymes. Nucleic Acids Res. 1987;15 (Suppl):r219–r230. doi: 10.1093/nar/15.suppl.r219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwosu V. U., Connolly B. A., Halford S. E., Garnett J. The cloning, purification and characterization of the Eco RV modification methylase. Nucleic Acids Res. 1988 May 11;16(9):3705–3720. doi: 10.1093/nar/16.9.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka E., Ishino Y., Ibaraki K., Ikehara M. Recognition by restriction endonuclease EcoRI of deoxyoctanucleotides containing modified sugar moieties. Eur J Biochem. 1984 Mar 15;139(3):447–450. doi: 10.1111/j.1432-1033.1984.tb08025.x. [DOI] [PubMed] [Google Scholar]
- Ohtsuka E., Morisawa H., Ikehara M. Studies on deoxynucleic acids and related compounds. IV. Syntheses of an octanucleotide containing a recognition site for restriction enzyme Eco RI and of an arabinosyladenine analog. Chem Pharm Bull (Tokyo) 1982 Mar;30(3):874–880. doi: 10.1248/cpb.30.874. [DOI] [PubMed] [Google Scholar]
- Ono A., Sato M., Ohtani Y., Ueda T. Synthesis of deoxyoligonucleotides containing 7-deazaadenine: recognition and cleavage by restriction endonuclease Bgl II and Sau 3AI (nucleosides and nucleotides Part 55). Nucleic Acids Res. 1984 Dec 11;12(23):8939–8949. doi: 10.1093/nar/12.23.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Ueda T. Minor-groove-modified oligonucleotides: synthesis of decadeoxynucleotides containing hypoxanthine, N2-methylguanine and 3-deazaadenine, and their interactions with restriction endonucleases Bgl II, Sau, 3AI, and Mbo I (Nucleosides and Nucleotides Part 75). Nucleic Acids Res. 1987 Apr 10;15(7):3059–3072. doi: 10.1093/nar/15.7.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Ueda T. Synthesis of decadeoxyribonucleotides containing N6-methyladenine, N4-methylcytosine, and 5-methylcytosine: recognition and cleavage by restriction endonucleases (nucleosides and nucleotides part 74). Nucleic Acids Res. 1987 Jan 12;15(1):219–232. doi: 10.1093/nar/15.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruska J., Horn D. Bromodeoxyuridine substitution in mammalian DNA can both stimulate and inhibit restriction cleavage. Biochem Biophys Res Commun. 1980 Oct 16;96(3):1317–1324. doi: 10.1016/0006-291x(80)90095-9. [DOI] [PubMed] [Google Scholar]
- Petruska J., Horn D. Sequence-specific responses of restriction endonucleases to bromodeoxyuridine substitution in mammalian DNA. Nucleic Acids Res. 1983 Apr 25;11(8):2495–2510. doi: 10.1093/nar/11.8.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Two restriction endonucleases from Bacillus globiggi. Nucleic Acids Res. 1976 Jul;3(7):1747–1760. doi: 10.1093/nar/3.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkel L. J., van der Marel G. A., van Boom J. H., Altona C. Influence of N6-methylation of residue A(5) on the conformational behaviour of d(C-C-G-A-A-T-T-C-G-G) in solution studied by 1H-NMR spectroscopy. 1. The duplex form. Eur J Biochem. 1987 Mar 2;163(2):275–286. doi: 10.1111/j.1432-1033.1987.tb10798.x. [DOI] [PubMed] [Google Scholar]
- Sadowski P. D., Hurwitz J. Enzymatic breakage of deoxyribonucleic acid. II. Purification and properties of endonuclease IV from T4 phage-infected Escherichia coli. J Biol Chem. 1969 Nov 25;244(22):6192–6198. [PubMed] [Google Scholar]
- Schildkraut I., Banner C. D., Rhodes C. S., Parekh S. The cleavage site for the restriction endonuclease EcoRV is 5'-GAT/ATC-3'. Gene. 1984 Mar;27(3):327–329. doi: 10.1016/0378-1119(84)90078-7. [DOI] [PubMed] [Google Scholar]
- Seela F., Kehne A. Palindromic octa- and dodecanucleotides containing 2'-deoxytubercidin: synthesis, hairpin formation, and recognition by the endodeoxyribonuclease EcoRI. Biochemistry. 1987 Apr 21;26(8):2232–2238. doi: 10.1021/bi00382a024. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers L. C., Shaw B. R., Sedwick W. D. Base stacking and molecular polarizability: effect of a methyl group in the 5-position of pyrimidines. Biochem Biophys Res Commun. 1987 Oct 29;148(2):790–794. doi: 10.1016/0006-291x(87)90945-4. [DOI] [PubMed] [Google Scholar]
- Streeck R. E. Single-strand and double-strand cleavage at half-modified and fully modified recognition sites for the restriction nucleases Sau3a and Taqi. Gene. 1980 Dec;12(3-4):267–275. doi: 10.1016/0378-1119(80)90109-2. [DOI] [PubMed] [Google Scholar]
- Szekeres M., Matveyev A. V. Cleavage and sequence recognition of 2,6-diaminopurine-containing DNA by site-specific endonucleases. FEBS Lett. 1987 Sep 28;222(1):89–94. doi: 10.1016/0014-5793(87)80197-7. [DOI] [PubMed] [Google Scholar]
- Vinogradova M. N., Gromova E. S., Griaznova O. I., Isaguliants M. G., Kuznetsova S. A. Vzaimodeistvie fermentov restriktsii i modifikatsii EcoRII s sinteticheskimi fragmentami DNK. IX. Rasshcheplenie substratov s tochechnymi modifikatsiiami v uchastke uznavaniia i prilegaiushchikh posledovatel'nostiakh. Bioorg Khim. 1987 Sep;13(9):1194–1204. [PubMed] [Google Scholar]
- Vinogradova M. N., Gromova E. S., Uporova T. M., Nikol'skaia I. I., Shabarova Z. A. Endonukleaza restriktsii SsoII: vzaimodeistvie s modifitsirovannymi substratami. Dokl Akad Nauk SSSR. 1987;295(3):732–736. [PubMed] [Google Scholar]
- Wiatr C. L., Witmer H. J. Selective protection of 5' ... GGCC ... 3' and 5' ... GCNGC ... 3' sequences by the hypermodified oxopyrimidine in Bacillus subtilis bacteriophage SP10 DNA. J Virol. 1984 Oct;52(1):47–54. doi: 10.1128/jvi.52.1.47-54.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfes H., Fliess A., Pingoud A. A comparison of the structural requirements for DNA cleavage by the isoschizomers HaeIII, BspRI and BsuRI. Eur J Biochem. 1985 Jul 1;150(1):105–110. doi: 10.1111/j.1432-1033.1985.tb08994.x. [DOI] [PubMed] [Google Scholar]
- Woodbury C. P., Jr, Hagenbüchle O., von Hippel P. H. DNA site recognition and reduced specificity of the Eco RI endonuclease. J Biol Chem. 1980 Dec 10;255(23):11534–11548. [PubMed] [Google Scholar]
- Yolov A. A., Gromova E. S., Kubareva E. A., Potapov V. K., Shabarova Z. A. Interaction of EcoRII restriction and modification enzymes with synthetic DNA fragments. V. Study of single-strand cleavages. Nucleic Acids Res. 1985 Dec 20;13(24):8969–8981. doi: 10.1093/nar/13.24.8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolov A. A., Vinogradova M. N., Gromova E. S., Rosenthal A., Cech D., Veiko V. P., Metelev V. G., Kosykh V. G., Buryanov Y. I., Bayev A. A. Interaction of EcoRII restriction and modification enzymes with synthetic DNA fragments. VI. The binding and cleavage of substrates containing nucleotide analogs. Nucleic Acids Res. 1985 Dec 20;13(24):8983–8998. doi: 10.1093/nar/13.24.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]



