Background: Cascade is a prokaryotic immune complex that targets foreign DNA.
Results: We present the crystal structure of CasA, the largest subunit of Cascade, and analyze its contribution to DNA target binding.
Conclusion: CasA is necessary for the DNA target binding by Cascade.
Significance: Learning how each subunit of Cascade functions is crucial for understanding how this immune complex works.
Keywords: Crystal Structure, DNA, DNA-binding Protein, RNA, RNA-binding Proteins, CRISPR, CasA
Abstract
Prokaryotes make use of small RNAs encoded by CRISPR (clustered regularly interspaced short palindromic repeat) loci to provide immunity against bacteriophage or plasmid invasion. In Escherichia coli, the CRISPR-associated complex for antiviral defense (Cascade) utilizes these RNAs to target foreign DNA for destruction. CasA, the largest subunit of Cascade, is essential for its function. Here we report the crystal structure of Thermus thermophilus CasA. The structure is composed of two domains that are arranged in a chair-like conformation with a novel fold forming the larger N-terminal domain. Docking of the crystal structure into cryo-electron microscopy maps reveals two loops in CasA that likely have important functions in DNA target binding. Finally, DNA binding experiments show that CasA is essential for binding of Cascade to DNA target.
Introduction
In response to attack by invasive genetic elements, such as bacteriophages or plasmids, prokaryotes integrate short fragments of foreign DNA into their chromosome at sites of clustered regularly interspaced short palindromic repeats (CRISPRs)2 (1–3). RNA transcribed from these CRISPR arrays is cleaved within each repeat sequence to yield a library of small CRISPR-derived RNAs (crRNAs), each of which contains complementary sequence to the invading element (4–10). After packaging into CRISPR-associated (Cas) protein complexes, mature crRNAs act as guides that detect and mediate destruction of foreign nucleic acid (2, 10–16). For a recent review of CRISPR/Cas biology, see Ref. 17.
The CRISPR-associated complex for antiviral defense (Cascade) mediates DNA target recognition by the CRISPR/Cas system in Escherichia coli (10). Cascade is a 405-kDa ribonucleoprotein complex composed of an unequal stoichiometry of five, functionally essential protein subunits (CasA1, CasB2, CasC6, CasD1, and CasE1) and a 61-nucleotide crRNA (13). Cascade binding to DNA target not only requires base-pairing between the crRNA and its complementary sequence in the DNA target (the protospacer) but also depends on a protospacer adjacent motif (PAM) (18). In E. coli targets, the 3-nucleotide PAM sequence (5′-CAT-3′) is found upstream of the protospacer (18). CRISPR arrays lack a PAM sequence, ensuring that the host genome is not targeted and thus serving as a mechanism for self versus nonself recognition (18–22). Once bound to target, Cascade is believed to recruit the Cas3 nuclease that then destroys the DNA target (10, 23–25).
Two cryo-electron microscopy (cryo-EM) reconstructions of E. coli Cascade, one bound to a single-stranded target that spans the protospacer but not the PAM sequence (the protospacer target) and the other unbound, have provided detailed insight into the organization of its subunits (15). Overall, Cascade has a sea horse-shaped architecture (see Fig. 1A) with a backbone formed by a helical-filament of six copies of CasC wrapped around the extended crRNA. The head is capped by CasE at the 3′-end of the crRNA, and CasA and CasD cap the tail at the 5′-end. Two copies of CasB sit on the inner surface of the CasC-crRNA spine, directly connecting the head and the tail of the complex. The reconstruction of Cascade bound to protospacer target reveals a concerted conformational change involving CasA, CasB, and CasE that could be a signal to recruit Cas3 (15).
FIGURE 1.
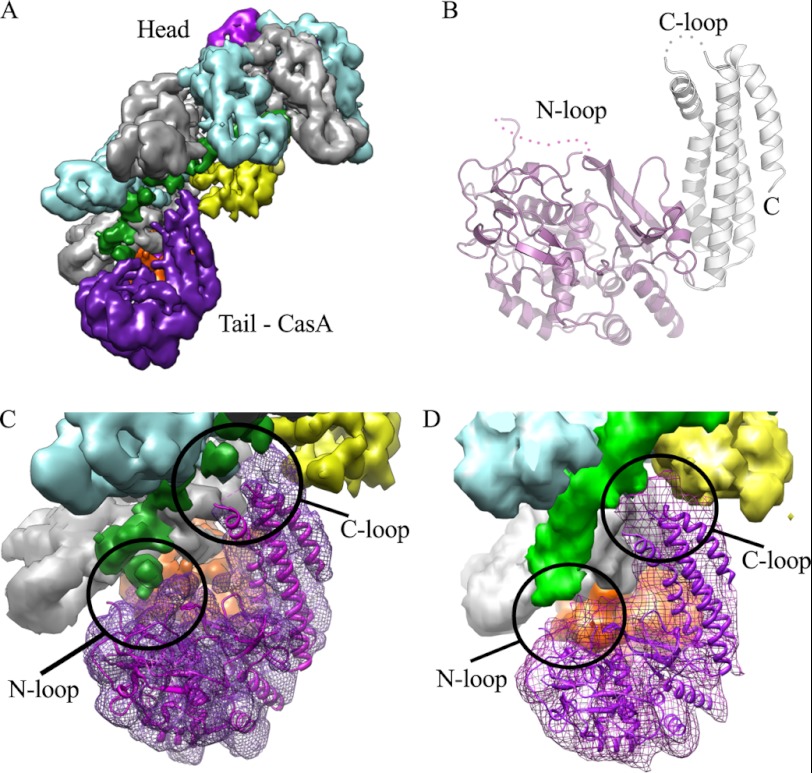
Structure of TthCasA. A, cryo-EM reconstruction of Cascade. Coloring is as in Ref. 15 as follows: magenta, CasA; yellow, CasB; cyan and gray, CasC; orange, CasD; and pink, CasE. CasA is located at the tail of the structure. B, ribbon representation of the crystal structure of CasA. The N-domain is colored magenta, and the C-domain is colored white. The N- and C-loops are labeled. C, fit of the crystal structure of CasA into the cryo-EM map of Cascade. D, fit of the crystal structure of CasA into the cryo-EM map of Cascade bound to protospacer target. C and D are colored as in A.
Despite recent advances in the understanding of our organization and biochemistry of Cascade (6, 8, 10, 13, 15, 18), the functions of many of its subunits are still poorly understood. To better understand the role of CasA, we have determined the crystal structure of CasA, docked this structure into cryo-EM reconstructions (15), and compared the DNA binding properties Cascade with those of a subcomplex of Cascade lacking the CasA subunit (CasBCDE).
EXPERIMENTAL PROCEDURES
Cloning and Protein Expression
The cloning and expression strategy was similar to that described previously (10, 13). Thus, all genes were amplified from genomic DNA (American Type Culture Collection) and directionally cloned into a series of expression vectors (Table 1). An E. coli CRISPR array consisting of seven identical spacers (sequence: 5′-CCAGTGATAAGTGGAATGCCATGTGGGCTGTC-3′) was synthesized by GeneArt. All proteins were overexpressed in the T7Express strain of E. coli (New England Biolabs). Cells were grown in LB medium, supplemented with the appropriate antibiotic(s) (Table 1), at 37 °C to an A600 of 0.3–0.5, and subsequently protein expression was induced with 0.2 mm isopropyl-β-d-thiogalactopyranoside overnight at 20 °C.
TABLE 1.
Plasmids used in these studies
| Vector | Relevant Features |
|---|---|
| pRSFDuet-1b (KanR)a | Two multiple cloning sites (mcs1 and mcs2)b |
| pBAT4 (AmpR)c | No tags |
| pHAT4 (AmpR)c | Encodes for an N-terminal His-tag, followed by a TEV protease site |
| pMAT11 (AmpR)c | Encodes for an N-terminal His-MBP (maltose-binding protein) tag, followed by a TEV protease site |
| Clone | Vector | Genes |
|---|---|---|
| pCRISPRd | pRSFDuet-1b | mcs1: CRISPR (7x spacer) mcs2: empty |
| pCRISPR-Ad | pRSFDuet-1b | mcs1: CRISPR (7x spacer) mcs2: CasA |
| pBCDEd | pHAT4 | E. coli casB-casC-casD-CasE |
| pTthCasA | pBAT4 | T. thermophilus CasA |
| pEcoCasA | pMAT11 | E. coli CasA |
a From Novagen.
b mcs1 can encode for an N-terminal His tag, however all cloning here removes this tag.
c From Ref. 36.
d Cells used to express Cascade were made by transformation with pCRISPR-A and pBCDE. Cells used to express CasBCDE+crRNA were made by transformation with pCRISPR and pBCDE.
Purification of E. coli Proteins
E. coli CasA, Cascade, and the CasBCDE-crRNA subcomplex were all purified using the same protocol. Harvested cells were lysed in buffer L (20 mm Tris-HCl, pH 8.0, 100 mm NaCl and 10% glycerol), clarified, and then loaded onto a 5-ml immobilized metal affinity chromatography column (Bio-Rad). The column was then washed with 10 mm imidazole before the protein of interest was eluted with 250 mm imidazole. N-terminal tags were removed by treatment with tobacco etch virus (TEV) protease overnight at 4 °C. Samples were then desalted to remove imidazole and then reapplied to immobilized metal affinity chromatography resin to remove the His-tagged TEV protease, any cleaved tag, or any remaining tagged protein. Samples were then concentrated and loaded on a HiLoad 26/60 S200 size-exclusion column (GE Healthcare) pre-equilibrated with Buffer A (20 mm Tris-HCl, pH 8.0, 200 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine). As seen previously, all proteins eluted as symmetrical peaks at their expected molecular weights (13).
Purification of Thermus thermophilus CasA
Harvested cells were lysed in buffer L, and the clarified lysate was heat-treated at 70 °C for 10 min. Following centrifugation, the sample was adjusted to 1.5 m ammonium sulfate and loaded onto a 5-ml Fast Flow Phe column (GE Healthcare) pre-equilibrated with 40 mm Tris-HCl, pH 7.5, 1.5 m ammonium sulfate, 10% glycerol. Protein was eluted with a linear gradient of 1.5–0 m ammonium sulfate. The relevant fractions were pooled, and the protein was further purified over a 5-ml Fast Flow Q column (GE Healthcare) before finally being loaded on a HiLoad 26/60 S200 column (GE Healthcare) pre-equilibrated with Buffer A. The final purified protein was concentrated to ∼30 mg/ml using Ultracel 10K centrifugal filter unit (Millipore).
Crystallization of T. thermophilus CasA
Crystals of T. thermophilus CasA were obtained by the sitting-drop vapor diffusion method, mixing 1 μl of CasA at ∼30 mg/ml with 1 μl of precipitant solution of 0.1 m MOPS, pH 7.4, and 2.3 m sodium acetate. For stabilization and cryoprotection, crystals were transferred to a solution of 0.1 m MOPS, pH 7.4, and 3.5 m sodium acetate. Crystals were flash-frozen in liquid nitrogen. Platinum derivatives were obtained by soaking crystals in a solution containing 0.1 m MOPS, pH 7.4, 3.5 m sodium acetate, and 20 mm K2PtCl4 for 3 h.
Structure Determination
X-ray diffraction data were collected at either beamline 9.2 at the Stanford Synchrotron Radiation Light Source (SSRL) or beamline X25 at the National Synchrotron Light Source (NSLS). Data were processed with HKL2000 (26). SHELX (27) was used to find the positions of the platinum sites. Phases were calculated using SOLVE (28) and improved by solvent flattening and noncrystallographic symmetry averaging in RESOLVE (28). Iterative model building and refinement were carried out in COOT (29) and PHENIX (30).
Cryo-electron Microscopy Map Fitting and Preparation of Figures
Rigid-body docking of the T. thermophilus CasA crystal structure into the cryo-electron microscopy density of E. coli Cascade was performed with Chimera (31). All structure panels were generated using PyMOL (32) or Chimera (31).
DNA Binding Experiments
Binding assays contained 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 10% glycerol. All oligonucleotides were gel-purified. dsDNA was made by annealing oligonucleotide A (5′-TCAATCTACAAAATTGAGCAAATCAGACAGCCCACATGGCATTCCACTTATCACTGGCATTGCTTTCGAGCTTGCCGATCAGCTT-3′) with oligonucleotide B (5′-AAGCTGATCGGCAAGCTCGAAAGCAATGCCAGTGATAAGTGGAATGCCATGTGGGCTGTCTGATTTGCTCAATTTTGTAGATTGA-3′). Trace amounts (5–200 pm) of 5′-end 32P-labled dsDNA were incubated with an increasing concentration of Cascade or CasBCDE for 1 h at 37 °C, prior to electrophoresis through a 5% polyacrylamide gel. In experiments with saturating CasA, 250 nm was confirmed to be saturating as repeating these experiments with 1 μm CasA (data not shown) gave the same results. DNA was visualized by phosphorimaging and quantified using Image Gauge (Fuji). As described before (18), fraction of DNA bound was plotted versus protein concentration and fit to a one-site binding isotherm, using the GraphPad Prism software. Reported Kd values are the average of three replicates.
RESULTS
Crystal Structure of CasA
To gain a more detailed understanding of CasA (also known as CRISPR-subtype E. coli 1 (Cse1) or YgcL), we determined its crystal structure. Initial attempts to crystallize the E. coli protein were unsuccessful. We therefore expressed and purified the homolog from T. thermophilus HB8 (TthCasA). We chose this organism because the sequence of TthCasA has ∼50% similarity with E. coli CasA, and crystal structures of both TthCasB (33) and TthCasE (34) have been determined. Crystals of TthCasA were obtained by vapor diffusion using a precipitant solution containing sodium acetate. The crystals belonged to the space group P21 (a = 93.9 Å, b = 47.9 Å, c = 129.2 Å, and α = γ = 90°, β = 97.52°) and contain two monomers in the asymmetric unit. The structure was determined by single isomorphous replacement, utilizing platinum-soaked crystals, and the structure was refined to 2.4 Å resolution with an Rwork of 19.4% and an Rfree of 25.3%. Additional data collection, phasing, and refinement statistics are given in Table 2. The final model displayed good geometry and contained all of the TthCasA sequence with the exception of the first 4 N-terminal and last 6 C-terminal residues, as well as two internal loops formed by residues 129–142 (N-loop) and residues 405–409 (C-loop). The structures of the two monomers of TthCasA in the asymmetric unit are virtually identical with a root mean square deviation of 0.1 Å over 473 Cα atoms.
TABLE 2.
Data collection, processing, and phasing statistics
| Native | Pt Soak | |
|---|---|---|
| Data collection | ||
| Resolution (Å)a | 2.37 (2.48–2.37) | 2.79 (2.89–2.79) |
| Rsymb | 10.1 (63.4) | 6.6 (31.7) |
| I/σa | 11.2 (2.2) | 11.1 (2.4) |
| Redundancya | 3.7 (3.6) | 2.0 (1.9) |
| Completeness (%)a | 98.5 (97.6) | 98.7 (96.7) |
| Wilson B factor | 50.70 | |
| Mean figure of merit | 0.32 | |
| Heavy atom sites | 6 | |
| Refinement | ||
| Resolution (Å) | 46–2.37 | |
| Rwork (%)c | 19.4 | |
| Rfree (%)c | 25.3 | |
| r.m.s.d.d bond (Å) | 0.01 | |
| r.m.s.d.d angle (°) | 1.2 | |
| No. of atoms | 7828 | |
| B-factors | 63.5 | |
| Ramachandran plot | ||
| Most favored (%) | 90.3 | |
| Additional allowed (%) | 9.7 | |
a The values in parentheses are for the highest resolution shell.
b Rsym is Σ|Io − I|/ΣIo, where Io is the intensity of an individual reflection, and I is the mean intensity for multiply recorded reflections.
c Rwork is ‖Fo − Fx‖/Fo, where Fo is an observed amplitude and Fx a calculated amplitude; Rfree is the same statistic calculated over a subset of the data that has not been used for refinement.
d r.m.s.d., root mean square deviation.
Overall the structure of TthCasA can be divided into two domains corresponding to the N- and C-terminal parts of the polypeptide chain (Fig. 1B). The two domains are arranged in a chair-like conformation, with the N-domain forming the seat and the C-domain forming the backrest (Fig. 1B). The larger N-domain includes residues 1–364 and is composed of 11 β-strands and 9 α-helices. A search of the structural database using the DALI server found no significant matches, suggesting that the N-domain has a novel fold. The smaller C-domain includes residues 365–502 and is formed by five α-helices. Four of these α-helices form an up-down-up-down four-helix bundle. The fifth, smaller α-helix is located in a flexible loop (the C-loop) separating the first and second helix of the bundle.
Docking the Crystal Structure of TthCasA into the Cryo-EM maps of Cascade
To gain further insight into the role of CasA, we rigid-body fit the crystal structure of TthCasA into the cryo-EM maps of Cascade with and without bound protospacer target (15). The crystal structure aligned well into both maps, and α-helices in the crystal structure aligned with the corresponding rods of density in the cryo-EM maps (Fig. 1, C and D). The quality of the fit into both cryo-EM maps suggests that there is no significant change in the relative orientation of the two domains of CasA, observed in the crystal structure, upon binding to Cascade.
In the cryo-EM map of Cascade with no bound target, the CasA N-domain sits adjacent to CasD, and contiguous cryo-EM density suggests that the N-loop of CasA contacts with the 5′-end of crRNA (Fig. 1C). The C-domain of CasA contacts the fifth and sixth CasC subunits as well as the neighboring CasB subunit (Fig. 1C). Upon binding to protospacer target, the CasA, CasB, and CasE subunits undergo a concerted conformational change (15). In the cryo-EM map of Cascade bound to protospacer target, the movement of CasA is such that the N-loop appears to no longer interact with the crRNA, whereas the C-loop now makes new contacts with the crRNA-protospacer duplex (Fig. 1D).
DNA Binding by Cascade
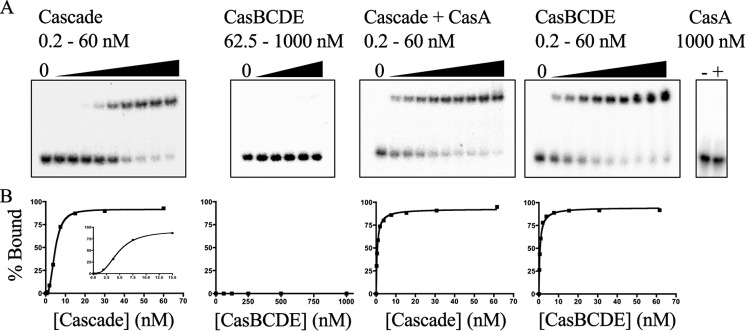
Binding of Cascade to nonself target relies on the recognition of a PAM (18). Previous studies on the role of CasA in this binding were performed before the E. coli PAM was identified (13). We therefore examined the role of CasA in Cascade binding to an 85-bp dsDNA target containing protospacer and functional PAM sequences. A fixed concentration of dsDNA target was incubated with increasing concentrations of either Cascade or CasBCDE, and complex formation with dsDNA was analyzed by native gel electrophoresis (Fig. 2A). CasBCDE did not bind dsDNA target, whereas Cascade did. The amount of complex formed between dsDNA target and Cascade exhibited a sigmoidal dependence on Cascade concentration (Fig. 2B). There are at least two possible explanations for the sigmoidal binding curve, either (i) cooperative binding between multiple sites on Cascade or the dsDNA or (ii) the existence of two equilibriums, one between Cascade and dsDNA and the other between a single subunit of Cascade and the rest of the complex. Because the stoichiometry between Cascade and dsDNA target is thought to be 1:1 (13, 15) and CasA was seen to dissociate from Cascade during competitive ssDNA binding experiments (13), the sigmoidal dependence is more likely the result of dissociation of CasA from Cascade at low concentrations. To confirm this hypothesis, we repeated the above binding experiments in the presence of saturating concentrations of CasA (250 nm). Under these conditions, the amount of complex formed between Cascade and dsDNA target exhibited a hyperbolic dependence on Cascade concentration (Fig. 2B) with an apparent dissociation equilibrium constant (Kd) of 0.54 ± 0.1 nm. The addition of a saturating concentration of CasA to CasBCDE rescued binding of this complex to dsDNA target (Fig. 2A) and also displayed a hyperbolic dependence on CasBCDE concentration with a Kd indistinguishable from Cascade in the presence of saturating concentration of CasA (Fig. 2B). In control experiments, CasA alone was not able to bind dsDNA target (13) (Fig. 2A).
FIGURE 2.
dsDNA target binding by Cascade. A, representative gel shift assays for Cascade and CasBCDE in the absence and presence of saturating concentrations (250 nm) of CasA. Also shown is a gel shift assay of CasA alone (1000 nm). The concentrations of the titrated species are given above each gel. B, binding curves measured from each of the assays in A. For the Cascade titration in the absence of CasA, the inset is a zoomed-in view of the binding curve, highlighting it sigmoidal character.
DISCUSSION
The crystal structure of TthCasA reveals a two-domain protein with a novel N-terminal fold and a C-terminal four-helix bundle. A prominent feature of this structure is two disordered loops, one in the N-domain and another in the C-domain, termed the N-loop and C-loop, respectively. Docking the crystal structure of TthCasA into the cryo-EM maps of Cascade suggests that these loops become ordered when CasA binds Cascade and that they make significant contacts with the crRNA and the protospacer target. In the absence of target, the N-loop makes contact with the 5′-end of the crRNA, but upon Cascade binding to protospacer target, the N-loop disengages from the crRNA (Fig. 1D) (15). The C-loop makes little or no contacts with the crRNA in the absence of target but does make extensive contacts with the crRNA-protospacer duplex when Cascade is bound to protospacer target (Fig. 1D). Thus, both of these loops appear to make key contributions to the specific structural states that correlate with Cascade target binding.
The PAM plays a critical role in self versus nonself recognition (18–22). The PAM is found in nonself DNA targets but not in the host sequence, CRISPR loci. Recent DNA binding experiments have demonstrated that mutations in the PAM sequence decrease the affinity of Cascade for DNA target (18), suggesting a direct interaction between Cascade and the PAM. The N-loop of CasA may mediate this critical interaction. If a longer nonself target, including the PAM sequence, were modeled onto Cascade, the projected path of the target would position the PAM adjacent to the site where the N-loop of the crystal structure of CasA docks into the EM map (Fig. 1D).
Our DNA binding experiments show that CasA is essential for specific binding of Cascade to nonself target (Fig. 2). Taken together with the observation that CasA dissociates from the complex at low concentrations, this suggests that CasA expression levels may provide an opportunity for regulation of the activity of Cascade within the cell. Cascade would not be able to bind dsDNA target at low expression levels of CasA, but at high expression levels, Cascade could bind DNA target and signal its destruction by Cas3. Confirmation of this model will require measurement of the cellular concentrations of the individual Cascade subunits.
In summary, we have shown here that the CasA subunit of Cascade is essential for nonself target binding. We present the crystal structure of CasA and its fit into cryo-EM maps of Cascade bound and unbound to protospacer target. This structural analysis reveals two loops in CasA that are likely key sensors for dsDNA target binding.
While this manuscript was in preparation, a similar analysis of CasA was published by Doudna and colleagues (35). This manuscript independently presents similar results but also experimentally confirms the role of the N-loop in both PAM binding and additionally in the control of nonspecific DNA binding by Cascade.
Acknowledgments
We thank Annie Héroux for help with data collection at the NSLS and Jennifer M. Kavran for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM097330 (to S. B.).
The atomic coordinates and structure factors (code 4F3E) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CRISPR
- clustered regularly interspaced short palindromic repeats
- Cas
- CRISPR-associated
- crRNA
- CRISPR-RNA
- Cascade
- CRISPR-associated complex for antiviral defense
- PAM
- protospacer adjacent motif
- TEV
- tobacco etch virus.
REFERENCES
- 1. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., Horvath P. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 2. Garneau J. E., Dupuis M. È., Villion M., Romero D. A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A. H., Moineau S. (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 [DOI] [PubMed] [Google Scholar]
- 3. Andersson A. F., Banfield J. F. (2008) Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320, 1047–1050 [DOI] [PubMed] [Google Scholar]
- 4. Carte J., Wang R., Li H., Terns R. M., Terns M. P. (2008) Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22, 3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., Pirzada Z. A., Eckert M. R., Vogel J., Charpentier E. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gesner E. M., Schellenberg M. J., Garside E. L., George M. M., Macmillan A. M. (2011) Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Mol. Biol. 18, 688–692 [DOI] [PubMed] [Google Scholar]
- 7. Haurwitz R. E., Jinek M., Wiedenheft B., Zhou K., Doudna J. A. (2010) Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329, 1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sashital D. G., Jinek M., Doudna J. A. (2011) An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat. Struct. Mol. Biol. 18, 680–687 [DOI] [PubMed] [Google Scholar]
- 9. Wang R., Preamplume G., Terns M. P., Terns R. M., Li H. (2011) Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure 19, 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brouns S. J., Jore M. M., Lundgren M., Westra E. R., Slijkhuis R. J., Snijders A. P., Dickman M. J., Makarova K. S., Koonin E. V., van der Oost J. (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horvath P., Barrangou R. (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170 [DOI] [PubMed] [Google Scholar]
- 12. Hale C. R., Zhao P., Olson S., Duff M. O., Graveley B. R., Wells L., Terns R. M., Terns M. P. (2009) RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jore M. M., Lundgren M., van Duijn E., Bultema J. B., Westra E. R., Waghmare S. P., Wiedenheft B., Pul U., Wurm R., Wagner R., Beijer M. R., Barendregt A., Zhou K., Snijders A. P., Dickman M. J., Doudna J. A., Boekema E. J., Heck A. J., van der Oost J., Brouns S. J. (2011) Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 18, 529–536 [DOI] [PubMed] [Google Scholar]
- 14. Lintner N. G., Kerou M., Brumfield S. K., Graham S., Liu H., Naismith J. H., Sdano M., Peng N., She Q., Copié V., Young M. J., White M. F., Lawrence C. M. (2011) Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J. Biol. Chem. 286, 21643–21656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wiedenheft B., Lander G. C., Zhou K., Jore M. M., Brouns S. J., van der Oost J., Doudna J. A., Nogales E. (2011) Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477, 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wiedenheft B., van Duijn E., Bultema J. B., Bultema J., Waghmare S. P., Waghmare S., Zhou K., Barendregt A., Westphal W., Heck A. J., Heck A., Boekema E. J., Boekema E., Dickman M. J., Dickman M., Doudna J. A. (2011) RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 10092–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wiedenheft B., Sternberg S. H., Doudna J. A. (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338 [DOI] [PubMed] [Google Scholar]
- 18. Semenova E., Jore M. M., Datsenko K. A., Semenova A., Westra E. R., Wanner B., van der Oost J., Brouns S. J., Severinov K. (2011) Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 108, 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deveau H., Barrangou R., Garneau J. E., Labonté J., Fremaux C., Boyaval P., Romero D. A., Horvath P., Moineau S. (2008) Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gudbergsdottir S., Deng L., Chen Z., Jensen J. V., Jensen L. R., She Q., Garrett R. A. (2011) Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 79, 35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marraffini L. A., Sontheimer E. J. (2010) Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mojica F. J., Díez-Villaseñor C., García-Martínez J., Almendros C. (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740 [DOI] [PubMed] [Google Scholar]
- 23. Mulepati S., Bailey S. (2011) Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3). J. Biol. Chem. 286, 31896–31903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beloglazova N., Petit P., Flick R., Brown G., Savchenko A., Yakunin A. F. (2011) Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 30, 4616–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sinkunas T., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. (2011) Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30, 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 28. Terwilliger T. (2004) SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 11, 49–52 [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goddard T. D., Huang C. C., Ferrin T. E. (2007) Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 [DOI] [PubMed] [Google Scholar]
- 32. Delano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC, New York [Google Scholar]
- 33. Agari Y., Yokoyama S., Kuramitsu S., Shinkai A. (2008) X-ray crystal structure of a CRISPR-associated protein, Cse2, from Thermus thermophilus HB8. Proteins 73, 1063–1067 [DOI] [PubMed] [Google Scholar]
- 34. Ebihara A., Yao M., Masui R., Tanaka I., Yokoyama S., Kuramitsu S. (2006) Crystal structure of hypothetical protein TTHB192 from Thermus thermophilus HB8 reveals a new protein family with an RNA recognition motif-like domain. Protein Sci. 15, 1494–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sashital D. G., Wiedenheft B., Doudna J. A. (2012) Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peränen J., Rikkonen M., Hyvönen M., Kääriäinen L. (1996) T7 vectors with a modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373 [DOI] [PubMed] [Google Scholar]