Background: LST1/A was a poorly characterized protein encoded in the MHCIII locus.
Results: LST1/A is a myeloid cell-specific transmembrane adaptor associated with the tetraspanin-enriched microdomains that inhibits signaling by recruiting the protein tyrosine phosphatases SHP-1/SHP-2.
Conclusion: LST1/A is a potential negative regulator of myeloid cell signaling.
Significance: Negative regulation of signal transduction is crucial for controlling the cell response.
Keywords: Adaptor Proteins, Major Histocompatibility Complex (MHC), Myeloid Cell, Signal Transduction, Tetraspanins, LST1, SHP-1, SHP-2
Abstract
Transmembrane adaptor proteins are membrane-anchored proteins consisting of a short extracellular part, a transmembrane domain, and a cytoplasmic part with various protein-protein interaction motifs but lacking any enzymatic activity. They participate in the regulation of various signaling pathways by recruiting other proteins to the proximity of cellular membranes where the signaling is often initiated and propagated. In this work, we show that LST1/A, an incompletely characterized protein encoded by MHCIII locus, is a palmitoylated transmembrane adaptor protein. It is expressed specifically in leukocytes of the myeloid lineage, where it localizes to the tetraspanin-enriched microdomains. In addition, it binds SHP-1 and SHP-2 phosphatases in a phosphotyrosine-dependent manner, facilitating their recruitment to the plasma membrane. These data suggest a role for LST1/A in negative regulation of signal propagation.
Introduction
The human gene LST1 and its mouse homologue Lst1 (also known as B144) are located in the MHC class III locus, which is rich in immunologically important genes (1–4). The human LST1 primary transcript has been reported to undergo extensive alternative splicing resulting in a large number of splice variants. However, only limited alternative splicing has been reported for the mouse homologue (5, 6). Translation of LST1 splice isoforms would lead to the synthesis of various soluble and membrane-bound protein products. However, it remains unclear how many LST1 splice variants really exist at the protein level. So far, the only protein product clearly detected in vivo is encoded by the LST1/A isoform, the only isoform conserved between human and mouse (7, 8).
Another unresolved issue is the expression pattern of LST1. Several studies reported enrichment or specific expression of LST1 in leukocytes or leukocyte-rich tissues (1, 3, 7), whereas others indicated that the expression of LST1 is relatively ubiquitous (5, 8). Moreover, it seems that production of particular isoforms could be differentially regulated, which further complicates expression analysis (5, 9).
Although the LST1 gene has been studied extensively at the gene level and/or the mRNA level, the biological functions of the protein product(s) are largely unknown. A single published phenotypic study reported that overexpression of LST1/A in various human cell lines resulted in the formation of filopodia-like structures (7). Nevertheless, no molecular mechanism explaining how LST1 could induce the changes in cell morphology has been proposed, and thus, the physiological function of LST1/A is still unknown.
In this study, we thoroughly characterize the LST1/A isoform with the emphasis on bioinformatic analysis of its amino acid sequence, expression profile, biochemical characterization, and binding partners. Based on these data, we propose a role for LST1/A in negative regulation of cellular signaling.
EXPERIMENTAL PROCEDURES
Bioinformatics
The in silico search in the human genome for proteins possessing the features characteristic of known transmembrane adaptor proteins (TRAPs)4 has been published previously (10). Sequence alignment was performed using ClustalW2 (11).
Antibodies and Reagents
Antibodies to the following antigens were used: Erk2 (rabbit), SHP-1 (rabbit), SHP-2 (rabbit) (all from Santa Cruz Biotechnology, Santa Cruz, CA); phosphotyrosine (mouse, clone 4G10, Upstate Biotechnologies); FLAG tag (mouse, clone M2), GAPDH (rabbit), anti-mouse IgG-HRP (goat), anti-rabbit Ig-HRP (goat) (all from Sigma-Aldrich); CD25 (mouse IgG1, MEM-181, in-house); CD4-FITC (Beckman Coulter, Indianapolis, IN); CD8-FITC (Abd Serotec, Kidlington, UK); CD14-FITC, CD19-FITC, CD3-FITC, and CD56-FITC (all from Exbio, Vestec, Czech Republic); Thy1.1-FITC (His-51, eBioscience, San Diego, CA); mouse affinity-purified IgG (Jackson ImmunoResearch Laboratories, West Grove, PA).
The monoclonal antibody, LST1/02 (IgG1), against human LST1/A was generated by standard techniques. Briefly, F1 (BALB/c × B10.A) hybrid mice were immunized intrasplenically with a human LST1/A peptide (amino acids 75–90) conjugated to activated mcKLH (Pierce, Thermo Fisher Scientific) according to the manufacturer's instructions. Hybridomas were prepared using Sp2/0 myeloma cells as fusion partners and selected by the standard protocol.
Cell Lines and Primary Cells
All cell lines were cultured in the indicated media supplemented with 10% fetal bovine serum, 2 mm glutamine, 20 μg/ml gentamycin, 50 μg/ml streptomycin, and 104 units/ml penicillin at 37 °C in 5% CO2. The Jurkat, Ramos, THP-1, U937 (all from American Type Culture Collection, Manassas, VA), and HeLa (kindly provided by Dr. D. Stanek, Institute of Molecular Genetics, Academy of Sciences of the Czech Republic, Prague, Czech Republic) cell lines were cultivated in RPMI 1640. The Hek293FT (Invitrogen), Phoenix Ampho (Origene, Rockville, MD), and K562 (UHKT cell line collection, Prague, Czech Republic) cell lines were cultivated in DMEM. HL-60 cells (UHKT cell line collection) were cultivated in Iscove's Modified Dulbecco's Medium.
Human peripheral blood mononuclear cells were prepared from buffy coats obtained from healthy donors at Thomayer University Hospital (Prague, Czech Republic) using Ficoll-Paque PLUS (GE Healthcare) gradient centrifugation (900 × g for 30 min). To isolate various subpopulations, T cells, CD4+ cells, CD8+ cells, B cells, NK cells, and monocytes were labeled with anti-CD3, anti-CD4, anti-CD8, anti-CD19, anti-CD56, and anti-CD14 FITC-conjugated antibodies, respectively, followed by anti-FITC magnetic bead labeling (Miltenyi Biotec, Bergisch Gladbach, Germany). The cells were sorted using an AutoMACS magnetic cell sorter (Miltenyi Biotec), and the purity of the samples was determined by flow cytometry to be >90%. For isolation of granulocytes, a gradient composed of Histopaque-1119 (Sigma-Aldrich), overlaid by Ficoll-Paque PLUS, and freshly collected blood from healthy donors (diluted in PBS) was prepared and centrifuged (900 × g for 30 min). Granulocytes were collected from the interface between Ficoll-Paque PLUS and Histopaque-1119 layers, followed by erythrocyte lysis in ACK buffer (150 mm NH4Cl, 0.1 mm EDTA, 1 mm KHCO3). Where applicable, the procedures were performed after obtaining an informed consent from the donors, and in accordance with local ethical guidelines.
DNA Cloning and Transfection
The vector pcDNA3.1 containing the human LST1/A sequence was kindly provided by Dr. S. M. Weissman (Yale School of Medicine, New Haven, CT). For transient transfection experiments, the LST1/A coding sequence was cloned into the pXJ41 vector. For immunoprecipitation experiments, the construct consisting of human LST1/A followed by a FLAG coding sequence at the C terminus was cloned into MSCV-IRES-Thy1.1, a vector based on the MSCV backbone (Clontech) with the Thy1.1 reporter sequence. The tyrosine to phenylalanine mutant of LST1/A-FLAG (2YF) was prepared using PCR mutagenesis. The CD25-LST1/A-FLAG (WT and 2YF) chimeras were prepared by cloning the sequence of the extracellular part of human CD25 (coding amino acids 1–240) followed by the entire sequence of LST1/A-FLAG without the initial methionine into MSCV-IRES-Thy1.1. To prepare LST1/A-enhanced green fluorescent protein (EGFP), LST1/A sequence was cloned into pEGFP-N3 (Clontech Laboratories), and LST1/A-EGFP was subsequently ligated into MSCV-IThy1.1. All constructs were sequenced. Lyn in the pcDNA3.1 vector (kindly provided by Dr. S. Watson University of Birmingham, Birmingham, UK), Syk in pRK5 (kindly provided by Dr W. Kolanus, University of Cologne, Germany), and mouse Btk in pCDEF3 (kindly provided by Dr M. Tomlinson, University of Birmingham, Birmingham, UK), and NTAL in pFLAG-CMV (12) were used for a kinase co-transfection assay.
Lipofectamine 2000 reagent (Invitrogen) was used according to the manufacturer's instructions for transfection of both HEK293FT (for verification of the antibody specificity and the kinase co-transfection assay) and Phoenix Ampho cells (for production of viral particles using the MSCV-IRES-Thy1.1 vector). Retrovirus-containing supernatants were supplemented with polybrene (10 μg/ml, Sigma-Aldrich) and added to the cells. The cells were then centrifuged at 1,250 × g for 90 min. Infected cells were labeled with the anti-Thy1.1-FITC antibody followed by anti-FITC magnetic beads and isolated by the AutoMACS magnetic cell sorter separator.
RNA and Quantitative RT-PCR
RNA was isolated from human cell lines and sorted blood subpopulations using a mini RNA purification kit (Zymo Research). Contaminating DNA was removed using a DNA-free kit (Ambion, Austin, TX). A human normal tissue FirstChoice RNA Survey Panel and a lymph node FirstChoice Total RNA were purchased from Ambion. Human hippocampal total RNA was purchased from BioChain Institute. Reverse transcription, quantitative RT-PCR, and data analysis were performed as described previously (10). Primers specific for the human LST1/A isoform were as follows: forward, 5′-TGG AGA GGA GCT GGA CCC AGG-3′; reverse, 5′-TGG GTT GAG GAA GGT GTC TGG-3′. The unique specificity of these primers for the LST1/A isoform was confirmed by sequencing of the PCR product.
Palmitoylation Assay
Palmitoylation of LST1/A was examined using the acyl-biotinyl exchange chemistry-based method (13). Briefly, plasma membranes from 5 × 107 THP-1 cells were isolated and incubated with N-ethylmaleimide to block free thiols. Subsequently, palmitate residues were removed by hydroxylamine (untreated samples served as a control), and the resulting free thiol groups were labeled with biotin-HPDP (N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide, Pierce, Thermo Fisher Scientific). Biotinylated proteins were then immunoprecipitated on streptavidin-agarose beads, eluted with 2× concentrated SDS-PAGE sample buffer, and analyzed by Western blotting.
Cell Activation and Inhibitor Studies
Pervanadate solution was prepared by addition of hydrogen peroxide (final concentration, 0.27%) to 10 mm sodium orthovanadate, followed by 15 min of incubation at room temperature. For pervanadate-induced activation, cells (5 × 107 cells/ml) were incubated in serum-free RPMI 1640 medium with 10% pervanadate solution for 20 min. For antibody-induced activation, cells (5 × 107 cells/ml) were resuspended in serum-free RPMI 1640 medium, incubated with the indicated primary antibodies (20 μg/ml) for 30 min, washed, and activated for 2 min by goat anti-mouse antibody (10 μg/ml). For inhibitor studies, cells (107 cells/ml) were incubated with 2 μm Syk inhibitor IV (BAY 61-3606) or 10 μm PP2 (both from Calbiochem, Merck, Darmstadt, Germany) for 30 min at 37 °C.
Cell Lysis and Immunoprecipitation
To obtain whole cell SDS lysates, cells in serum-free RPMI1640 (5 × 107 cells/ml) were lysed by adding an equal volume of 2× concentrated SDS-PAGE sample buffer, followed by sonication.
For immunoprecipitation experiments, 5 × 107 cells were lysed in 1 ml of ice-cold lysis buffer (1 mm Pefabloc, 5 mm iodoacetamide, 1 mm sodium orthovanadate, 100 mm NaCl, 50 mm NaF, 2 mm EDTA, 20 mm Tris, pH 7.5) containing 1% dodecylmaltoside and incubated for 10 min on ice. Nuclei and debris were removed by centrifugation, and the resulting lysate was subjected to immunoprecipitation by the anti-FLAG antibody (2 μg/ml), the anti-LST1/A antibody (LST1/02, 10 μg/ml), or the isotype-matched control monoclonal antibody (anti-CD20, MEM-97, 10 μg/ml) followed by incubation with protein A/G PLUS-Sepharose (Santa Cruz Biotechnology). Immunoisolated material was eluted with 2× concentrated SDS-PAGE sample buffer containing 1% DTT and analyzed by Western blotting.
Gel Filtration and Flotation
Cells were lysed in lysis buffer containing the indicated detergent for 30 min. For flotation in a sucrose density gradient, 0.5-ml aliquots of cell lysates were mixed with 0.5 ml of 80% (w/v) sucrose in lysis buffer in an ultracentrifuge tube. Then, they were overlaid with 3.5 ml of 35% sucrose followed by 0.5 ml of lysis buffer. Samples were subjected to ultracentrifugation (200,000 × g for 20 h), and 0.6-ml fractions were collected from the top of the gradient. For gel filtration, lysates were first precleared by low-speed centrifugation (400 × g for 5 min) to remove insoluble material. 0.2-ml aliquots of the resulting supernatants were then loaded on columns containing 2 ml of Sepharose 4B pre-equilibrated in the corresponding lysis buffer. Subsequently, the lysis buffer was loaded continuously on the columns and concomitantly, 0.2-ml fractions were collected. All fractions were mixed with an equal volume of 2× concentrated SDS-PAGE sample buffer and analyzed by Western blotting.
Microscopy
HeLa cells were grown on cover slips overnight, and THP-1 cells were allowed to adhere to polylysine-coated coverslips for 15 min before processing. All samples were fixed with 4% (w/v) formaldehyde for 15 min at room temperature and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) for 5 min. Blocking was performed in 2.5% BSA and 10% goat serum (Sigma-Aldrich) in PBS for 30 min. Cells were incubated with phalloidin-tetramethylrhodamine B isothiocyanate (500 ng/ml, Sigma-Aldrich) for 30 min and washed twice with PBS. The DNA dye, Hoechst 33258 (Invitrogen), was used to visualize nuclei. Images were captured with a Leica SP5 confocal microscope using the 63× objective lens (Leica Microsystems, Mannheim, Germany).
Flow Cytometry
Cells were stained with the indicated antibodies in PBS supplemented with 1% BSA and 10% human serum on ice for 30 min, washed, resuspended in RPMI 1640, and analyzed by a BD LSR II (BD Biosciences). For calcium measurement, cells were loaded with 5 μm Fura-Red dye (Invitrogen) in RPMI 1640 at 37 °C for 30 min. Subsequently, the cells were labeled with the indicated antibodies (50 μg/ml mouse IgG alone or with 10 μg/ml anti-CD25) on ice for 20 min. Cells were prewarmed for 5 min at 37 °C, and the relative intracellular calcium concentration was measured as a ratio of Fura-Red fluorescence intensities elicited by excitation wavelengths 405 and 488 nm. 30 s after the beginning of the measurement, Fc receptors were stimulated with goat anti-mouse antibody (10 μg/ml) and 4 min after the beginning of the measurement, 1 μg/ml ionomycin (Sigma-Aldrich) was added. Data were analyzed using FlowJo software (TreeStar).
RESULTS
LST1/A Is Transmembrane Adaptor Protein Containing ITIM and ITIM-like Motifs
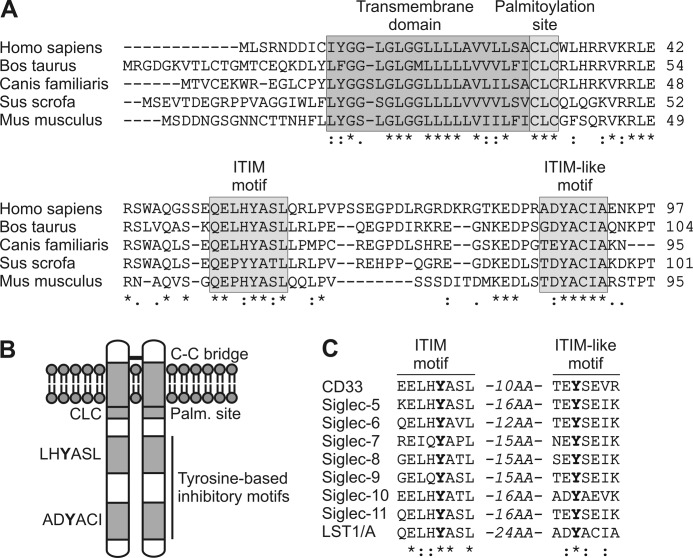
LST1/A was identified as a potential TRAP during our human genome-wide in silico screen described previously (10). Indeed, the LST1/A protein structure exhibits key features of a TRAP (14). First, it possesses only a very short extracellular domain (including a possibly dimerizing cysteine), followed by a transmembrane domain and a comparatively larger cytoplasmic tail (7, 8). Second, the protein lacks any domain with predicted enzymatic activity. Instead, its intracellular part contains a conserved potential palmitoylation site and two putative phospho-tyrosine-based interaction motifs (Fig. 1, A and B). Orthologues of human LST1/A predicted in various mammalian species share a high level of homology in the transmembrane domain, the palmitoylation site, and both of the predicted phosphotyrosine-based interaction motifs (Fig. 1A).
FIGURE 1.
Structure of LST1/A. A, alignment of amino acid sequences of LST1/A orthologues from human (Homo sapiens, GenBankTM accession number: NP_995311), cattle (Bos taurus, XP_002702922), dog (Canis familiaris, XP_848911), pig (Sus scrofa, XP_002702922), and mouse (Mus musculus, O08843). The transmembrane domain, the putative palmitoylation site, ITIM, and ITIM-like motifs are highlighted. B, graphical depiction of human LST1/A dimer structure with a disulfide bridge. C, alignment of amino acid sequences of ITIM and ITIM-like region of human LST1/A with a corresponding region from members of CD33-related Siglec family. Identical (*) and more (:) or less (.) similar amino acids at particular positions in all sequences are indicated. The distance in amino acids (AA) separating these motifs is shown.
Further examination of the human LST1/A sequence revealed that the membrane-proximal phosphotyrosine-based interaction motif constitutes a canonical immunoreceptor tyrosine-based inhibitory motif (ITIM), defined as (L/V/I)xYxx(L/V), where x can be any amino acid (15), and the membrane-distal phosphotyrosine element partially fulfills the criteria for an ITIM-like motif, defined as (D/E)YxE(V/I)(R/K) (Fig. 1, A and B) (16).
The tandem ITIM and ITIM-like elements strongly resemble a signaling inhibitory platform that has been found in the members of the CD33-related Siglec (sialic acid-binding Ig-like lectin receptor) family (16). The amino acid sequence of the ITIM motif is highly conserved among most CD33-related Siglecs and LST1/A. Also, substantial differences between LST1/A and CD33-related Siglecs were identified in the ITIM-like region (particularly at the +2 and +4 positions) (Fig. 1C). This degree of homology between LST1/A and CD33-related Siglecs suggested that LST1/A might function as an inhibitory TRAP, which could recruit some of the negative regulators of signal transduction that were described as phosphotyrosine-dependent binding partners of CD33-related Siglecs, namely SHP-1 (also known as PTPN6), SHP-2 (also known as PTPN11), SOCS3, and Cbl (16, 17).
LST1/A Is Specifically Expressed in Myeloid Cells
Conflicting results on the expression pattern of LST1 have been reported (1, 3, 5, 7, 8). Thus, we revisited this issue using a quantitative comparison of LST1/A expression levels in various cell subsets and tissues at both the mRNA and protein levels. Primers for specific RT-PCR amplification of the LST1/A isoform were designed, and a mouse monoclonal antibody against an epitope in the intracellular part of human LST1/A was developed (Fig. 2A). Quantitative RT-PCR performed on a panel of human tissue mRNA samples identified LST1/A mRNA in a number of lymphoid and non-lymphoid organs and tissues. By far, the highest levels of the transcript were detected in the spleen, suggesting the enrichment of LST1/A in leukocytes (Fig. 2B). A more detailed analysis of blood leukocyte subpopulations revealed specific expression of both LST1/A mRNA and protein in the cells of myeloid origin (monocytes and granulocytes) but not in lymphocytes (B cells, T cells, and NK cells) (Fig. 2C). Analogously, expression of LST1/A was detected in the cell lines derived from myeloid leukocytes (i.e. THP-1, U937, and HL60), but not in the cell lines of epithelial or lymphoid lineages (Fig. 2D). Collectively, these data indicate that LST1/A is expressed predominantly in leukocytes of the myeloid lineage.
FIGURE 2.
Expression analysis of LST1/A. A, Hek293FT were transiently transfected with LST1/A or an empty vector, lysed, and subjected to Western blotting using the newly developed antibody to LST1/A. Staining for GAPDH served as a loading control. B, expression of LST1/A mRNA in various human tissues was analyzed by quantitative RT-PCR. PCR amplifications were carried out in triplicates and mean Ct values were normalized to the GAPDH mRNA level. The dashed horizontal line indicates median from all tested tissue samples. C, expression of LST1/A in the isolated human peripheral leukocyte subsets was analyzed at the transcript level using quantitative RT-PCR (mean Ct values of triplicates were normalized to GAPDH) or at the protein level using Western blotting. Staining for Erk2 served as a loading control. D, expression of LST1/A in the indicated cell lines was analyzed as in C with GAPDH as a loading control for Western blotting. E, lysates of Hek293FT cells transfected with LST1/A and lysates of THP-1 cells were separated under non-reducing or reducing conditions and subjected to Western blotting with the LST1/A antibody.
To fully confirm that our antibody is specific for the LST1/A isoform, we compared the lysates from HEK293FT cells transfected with a vector encoding the LST1/A isoform with lysates from THP-1 cells. Upon treatment of the samples with a reducing agent DTT, the molecular weight of both transfected and endogenous LST1/A proteins were the same (Fig. 2E). In addition, staining with our LST1/A antibody revealed a substantial shift in the apparent molecular weight of LST1/A under the non-reducing conditions, confirming the prediction that LST1/A molecules form disulfide-based covalent dimers (Fig. 2E).
LST1/A Resides in Tetraspanin-enriched Microdomains
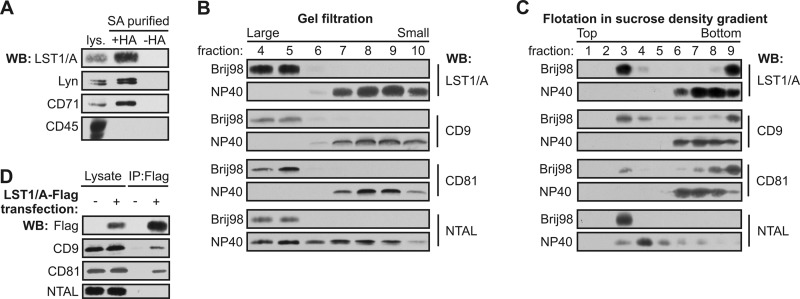
Because LST1/A contains a potential palmitylation site, we examined whether endogenous LST1/A is palmitoylated, using the acyl-biotinyl exchange chemistry-based method (13). LST1/A and positive controls (Lyn and CD71) were palmitoylated, whereas CD45 (a negative control) was not (Fig. 3A). Palmitoylation of various transmembrane proteins could facilitate their localization to lateral membrane compartments, including membrane rafts or tetraspanin-enriched microdomains (TEMs) (18–20).
FIGURE 3.
Biochemical characterization of LST1/A. A, palmitoylation of endogenous LST1/A in membranes isolated from THP-1 cells was examined using the acyl-biotinyl exchange chemistry method. Treatment of a sample with hydroxylamine (+HA) enabled exchange of palmitoyl residues for biotin and subsequent isolation on streptavidin (SA). A sample untreated with hydroxylamine (−HA) served as a negative control. Stainings for palmitoylated Lyn and transferrin receptor (CD71) and non-palmitoylated CD45 were used as positive and negative controls, respectively. B and C, THP-1 cells were lysed in Brij98 or Nonidet P-40 (NP40) and the lysates were fractionated by gel filtration on Sepharose 4B (B) or by flotation in the sucrose density gradient (C). Samples were analyzed by Western blotting using antibodies to LST1/A, tetraspanins CD9 and CD81, and the raft protein NTAL. D, THP-1 cells stably transfected with LST1/A-FLAG or an empty vector were solubilized in the CHAPS detergent and subjected to immunoprecipitation using the anti-FLAG antibody. Samples were stained for FLAG, CD9, CD81, and NTAL. WB, Western blot.
TEMs and lipid rafts can be distinguished based on their differential solubility in various detergents. Although both of these membrane compartments are resistant to mild detergents, such as Brij98 and CHAPS, TEMs, but not lipid rafts, are readily dissolved by more stringent detergents including Nonidet P-40 and Triton X-100 (12, 20–22). Similarly to tetraspanins CD9 and CD81, LST1/A was present in large complexes resistant to Brij98 and CHAPS, as detected by gel filtration on Sepharose 4B (Fig. 3B). LST1/A also was found in the low density fractions after sucrose density gradient centrifugation upon lysis in these mild detergents (Fig. 3C). However, unlike NTAL-containing lipid rafts, LST1/A-containing complexes were disrupted completely by Nonidet P-40 lysis (Fig. 3, B and C).
These biochemical characteristics indicated that LST1/A could be a component of TEMs. To further test this hypothesis, THP-1 cells stably expressing FLAG-tagged LST1/A were lysed in TEM-preserving detergent CHAPS and then subjected to immunoprecipitation by the anti-FLAG antibody. Tetraspanins CD9 and CD81, but not the raft protein NTAL, co-precipitated with LST1/A-FLAG (Fig. 3D), providing evidence for association of LST1/A with tetraspanins. These data demonstrate that LST1/A is a palmitoylated protein localized in TEMs.
LST1/A Localizes to Membrane Filopodia but Does Not Induce Their Formation
Thus far, only one study analyzed the potential function of LST1/A, showing that overexpression of LST1/A leads to filopodia formation in various human cell lines (7). To verify this, C-terminally EGFP-tagged LST1/A was expressed in THP-1 and HeLa cells. Confocal microscopy confirmed that LST1/A-EGFP was localized in the plasma membrane (Fig. 4A). To investigate whether LST1/A induces changes in cell morphology, the actin cytoskeleton was visualized by phalloidin staining. We observed clear localization of LST1/A-EGFP in actin-rich membrane protrusions. In contrast to previously published data (7), the filopodia-like structures were similarly abundant in both LST1/A-EGFP expressing and non-expressing cells as shown by the actin staining (Fig. 4B and supplemental Fig. 1). Therefore, the data did not confirm the role of LST1/A in the formation of filopodia or any other membrane protrusions.
FIGURE 4.
Subcellular localization of LST1/A. A, THP-1 cells were stably transfected with LST1/A-EGFP, fixed, and permeabilized, and nuclei were stained with Hoechst 33258. A representative cell with membrane localized LST1/A is shown. B, HeLa and THP-1 cells were transiently transfected with LST1/A-EGFP, allowed to adhere to a cover glass, fixed, and permeabilized, and the actin cytoskeleton was stained with phalloidin. Note the very similar appearance of the cells expressing and not expressing LST1/A-EGFP present in the same field. All samples were analyzed by a confocal microscope. Scale bar, 10 μm. For additional images, see supplemental Fig. 1.
LST1/A Binds SHP-1 and SHP-2 via Its ITIM and ITIM-like Element
The functions of TRAPs are largely dependent on their capacity to bind various signaling molecules. LST1/A contains two predicted phosphotyrosine-based interaction motifs that could potentially interact with SH2 domains of intracellular proteins. Due to the apparent similarity between the ITIM and ITIM-like pairs present in LST1/A and the CD33-related Siglecs, the ability of LST1/A to bind the previously described ITIM interacting SH2-containing proteins, including SHP-1, SHP-2, SOCS3, Cbl-b, and SHIP1, was tested (16, 23, 24).
For this purpose, THP-1 cells lines stably expressing C-terminally FLAG-tagged wild type (WT) LST1/A or mutant LST1/A with both tyrosines mutated to phenylalanines (2YF) were prepared. Immunoprecipitation using the anti-FLAG antibody followed by Western blotting showed that LST1/A-FLAG WT, but not the 2YF mutant, was constitutively phosphorylated in resting THP-1 cells. Moreover, only the LST1/A WT constitutively associated with protein tyrosine phosphatases, SHP-1 and SHP-2 in quiescent cells (Fig. 5A). Induction of strong global tyrosine phosphorylation by pervanadate treatment substantially enhanced LST1/A-FLAG tyrosine phosphorylation along with increased SHP-2 and especially SHP-1 binding. The observed phosphorylated band corresponding to LST1/A in the pervanadate treated LST1/A 2YF sample could represent either phosphorylated FLAG tag (of the exogenous LST1/A-FLAG) or endogenous LST1/A co-precipitated as a result of its dimerization with the exogenous FLAG-tagged LST1/A 2YF. This possibility would also explain the detectable amount of SHP-1 co-precipitated with LST1/A 2YF after pervanadate treatment (Fig. 5A). Additional explanations include post-lysis phosphorylation of tyrosine 11 in the transmembrane domain. To further analyze this issue, we employed a Jurkat T cell line, which does not express any endogenous LST1/A (Fig. 2D). We stably transduced these cells with LST1/A WT and LST1/A 2YF constructs and followed their phosphorylation and interactions after pervanadate-mediated activation. Under these conditions, LST1/A 2YF mutant was not phosphorylated and did not interact with the SHP-1 and SHP-2 phosphatases, whereas strong tyrosine phosphorylation, as well as SHP-1 and SHP-2 binding, were readily observed for the LST1/A WT protein (Fig. 5B). This experiment suggested that most likely explanation for the presence of tyrosine phosphorylated band and SHP-1 and SHP-2 phosphatases in LST1/A-FLAG 2YF immunoprecipitates was the dimerization with non-mutated endogenous protein. SOCS3, Cbl-b, and SHIP1 did not co-precipitate with LST1/A WT or 2YF in THP-1 cells, suggesting that their SH2 domains are incapable of binding to the phosphorylated tyrosines of LST1/A (data not shown).
FIGURE 5.
Association of LST1/A with SHP-1 and SHP-2. A, THP-1 cells and the THP-1-derived cell lines stably expressing C-terminally FLAG-tagged LST1/A wild type (WT) or LST1/A with both tyrosines mutated to phenylalanines (2YF) were treated with pervanadate or left untreated. Lysates were subjected to immunoprecipitation using the anti-FLAG antibody. The immunoprecipitated material was analyzed by Western blotting (WB) using antibodies to SHP-1, SHP-2, phosphotyrosine (pTyr), and LST1/A. B, Jurkat cells and Jurkat-derived cell lines stably expressing Flag-tagged LST1/A WT and 2YF mutant were treated with pervanadate, lysed, and subjected to immunoprecipitation with anti-FLAG antibody followed by immunoblotting with antibodies to SHP-1, SHP-2, phosphotyrosine, and LST1/A. C, lysates from human peripheral blood monocytes were subjected to immunoprecipitation with the anti-LST1/A antibody or with the isotype-matched control antibody followed by Western blotting using antibodies to SHP-1 (upper panel), SHP-2 (lower panel), phosphotyrosine (pTyr), and LST1/A. D, HEK293FT cells were transfected or not with a plasmid encoding LST1/A or NTAL together with a Lyn, Syk, or Btk encoding vector. After 24 h, cells were lysed, and phosphorylation of LST1/A or NTAL was analyzed by Western blotting. E, THP-1 cells expressing wild-type FLAG-tagged LST1/A were treated with 10 μm PP2 or 2 μm Syk inhibitor IV (S-IV) or left untreated. Lysates from these cells were subjected to immunoprecipitation with anti-FLAG antibody followed by immunoblotting with phosphotyrosine and anti-FLAG (LST1/A) antibodies.
To reproduce these results in a more physiological setting, endogenous LST1/A was immunoprecipitated from primary human monocytes. Similar to the previous experiment, LST1/A was constitutively phosphorylated in the non-stimulated monocytes and co-precipitated with SHP-1 and SHP-2 (Fig. 5C). Taken together, these results imply that a subset of LST1/A molecules are phosphorylated at the ITIM and/or ITIM-like motifs in the resting cells and bind SHP-1 and SHP-2 in a phosphotyrosine-dependent manner. An increase in LST1/A phosphorylation leads to enhanced interaction with the protein tyrosine phosphatases.
To identify the kinase phosphorylating LST1/A, we co-expressed LST1/A in Hek293FT cells with a protein tyrosine kinase from Src (Lyn), Syk (Syk), or Tec (Btk) family. Only Lyn, but not Syk or Btk phosphorylated LST1/A in this assay (Fig. 5D), indicating that Lyn (and possibly other Src family kinases) are responsible for LST1/A phosphorylation. In contrast, another TRAP, NTAL, was phosphorylated mainly by Syk in this assay, which is in agreement with previously published data (12). In addition, a specific inhibitor of Src family kinases, PP2, substantially reduced the constitutive LST1/A phosphorylation observed in THP-1 cells (Fig. 5E). Syk inhibitor IV, a specific inhibitor of Syk did not have any effect on LST1/A phosphorylation under these conditions (Fig. 5E).
LST1/A Is Able to Negatively Regulate Activating Signals Emanating from Surface Receptors
The ability of LST1/A to recruit protein tyrosine phosphatases SHP-1 and SHP-2 to the proximity of the plasma membrane suggested that LST1/A could act as a negative regulator of intracellular signaling pathways. LST1/A contains only a very short extracellular domain, and thus, it is very unlikely that it functions as a receptor. However, it is possible that LST1/A functionally associates with a so far unknown receptor(s) and that it helps to transduce their inhibitory signals.
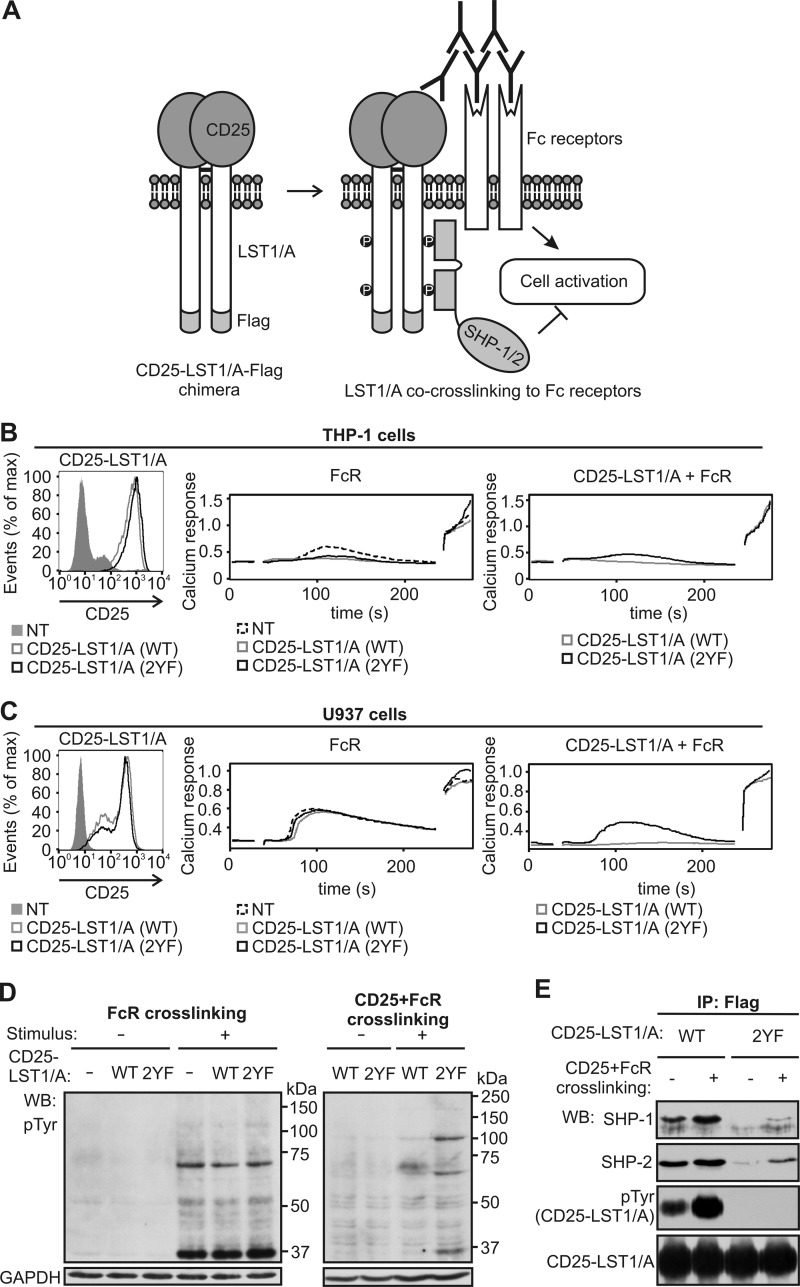
To test the ability of LST1/A to inhibit cellular signaling pathways, we prepared a plasmid coding for a chimeric protein consisting of extracellular part of CD25 fused to LST1/A-FLAG. This chimera could be extracellularly co-cross-linked to any other surface protein such as Fc receptors, and the effect of LST1/A on downstream signaling pathways could be observed (Fig. 6A). First, we prepared THP-1 cells stably expressing CD25-LST1/A-FLAG WT or 2YF chimeras via retroviral transduction (Fig. 6B, left panel). Both cell lines exhibited reduced calcium response after Fc receptor stimulation, compared with non-transfected cells, probably as a consequence of the transduction procedure (Fig. 6B, middle panel). However, co-cross-linking of CD25-LST1 WT, but not the 2YF mutant, with Fc receptors further inhibited Fc receptor-mediated calcium influx, leading to almost complete loss of the response (Fig. 6B, right panel). Due to the problems with the effects of retroviral transduction on calcium flux in transduced THP-1 cells, the U937-derived cell lines stably expressing CD25-LST1/A WT or 2YF were established (Fig. 6C, left panel). In this case, the viral transduction did not affect Fc receptor signaling, but the inhibitory effects of CD25-LST1/A WT on the Fc receptor-mediated calcium influx still were observed clearly (Fig. 6C, middle and right panels).
FIGURE 6.
Crosslinking of the CD25-LST1/A chimera with Fc receptors inhibits signaling. A, schematic representation of the CD25-LST1/A-FLAG chimeric construct. Cross-linking of this chimera with Fc receptors is expected to inhibit Fc receptor-mediated signaling events due to the association of LST1/A with SHP-1 and/or SHP-2 phosphatases. B and C, THP-1 (B) or U937 (C) cells were left non-transfected or stably transfected with CD25-LST1/A(WT)-FLAG or with CD25-LST1/A(2YF)-FLAG, where both tyrosines in the LST1/A sequence were mutated to phenylalanines. The cells were analyzed for expression of CD25 (left panel) and for calcium response after cross-linking of Fc receptors alone (middle panel) or in combination with CD25-LST1/A (right panel); ionomycin was added after 4 min to measure maximum calcium response. Mean relative calcium concentration in time is shown. D, U937 cells expressing CD25-LST1/A(WT)-FLAG or CD25-LST1/A(2YF)-FLAG were stimulated either by cross-linking of Fc receptors (left panel) or by co-cross-linking of Fc receptors and CD25-LST1/A (right panel), solubilized, and analyzed by Western blotting using the phosphotyrosine-specific antibody (pTyr). Staining for GAPDH served as a loading control. E, U937 cells expressing the CD25-LST1/A(WT)-FLAG or CD25-LST1/A(2YF)-FLAG chimera were activated by co-cross-linking of CD25-LST1/A to Fc receptors, solubilized, and immunoprecipitated with the anti-FLAG antibody. Samples were analyzed by Western blotting using antibodies to SHP-1, SHP-2, phosphotyrosine (pTyr), and LST1/A.
Accordingly, expression of CD25-LST1/A WT or 2YF did not influence the overall tyrosine phosphorylation induced by cross-linking of Fc receptors alone (Fig. 6D, left panel). However, Fc receptor co-cross-linking with WT chimeras resulted in a substantial reduction of overall tyrosine phosphorylation in the whole cell lysates when compared with co-engagement with the 2YF chimera. (Fig. 6D, right panel). These results indicate that LST1/A might act as a negative regulator of signaling via ITIM and/or ITIM-like dependent recruitment of SHP-1 and/or SHP-2 to a specific plasma membrane compartment.
Immunoprecipitation of the chimeras showed that co-cross-linking with Fc receptors increased tyrosine phosphorylation of the CD25-LST1/A WT and enhanced SHP-1 and SHP-2 binding (Fig. 6E). CD25-LST1/A 2YF was not tyrosine-phosphorylated, and it co-precipitated with only a very small amount of the protein tyrosine phosphatases, probably via dimerization with endogenous LST1/A as discussed above (Fig. 6E).
DISCUSSION
The LST1/A protein was identified as a potential novel TRAP in our large scale in silico search (10). The intracellular sequence of this protein contains one canonical ITIM and an additional ITIM-like motif, which constitute a structure highly homologous to tandem phospho-tyrosine elements in the majority of CD33-related Siglecs. Interestingly, LST1/A was missed in previous genome-wide screens aimed to identify all ITIM containing transmembrane proteins, most likely because LST1/A lacks a signal peptide (15, 25). In this study, we describe the generation of a monoclonal antibody to human LST1/A and the expression and biochemical and functional analysis of LST1/A.
Our anti-LST1/A antibody was generated after immunization of mice with the peptide corresponding to amino acids 75–90 of human LST1/A. Its specificity has been verified using cells transfected with the LST1/A construct. In addition, the apparent molecular weights of the transfected LST1/A and of the endogenous LST1 protein recognized by our antibody were identical. The peptide used for the immunization was predicted to be a part of several other putative cytoplasmic LST1 isoforms identified at the mRNA level, expected to have lower molecular weight than LST1/A. However, we did not observe any additional LST1 isoforms by Western blotting when lysates and immunoprecipitates from a variety of primary cells and cell lines under both reducing and non-reducing conditions were analyzed (Fig. 2 and data not shown). Moreover, using our antibody, we detected disulfide-based LST1/A dimers. Of the putative isoforms containing the sequence of the immunogen, only LST1/A is predicted to form such dimers. Collectively, these observations provide strong evidence that the endogenous isoform of LST1 detected by our antibody is LST1/A.
Due to the inconsistencies in the previously published data on LST1 expression profile (1, 3, 5–8), we carried out a thorough expression analysis of LST1/A. It revealed that LST1/A is substantially enriched in the spleen, peripheral blood monocytes, and granulocytes, as well as the cell lines of myeloid origin, indicating that LST1/A is expressed predominantly in the leukocytes of myeloid lineage. Although this conclusion is only in partial agreement with previously published studies (1, 3, 5–8), we consider our results reliable for several reasons. 1) We analyzed LST1/A expression profile in primary tissues and blood subpopulations, as well as in cell lines both at the protein and the mRNA levels and obtained a set of mutually supporting data. 2) In contrast to previous reports (1, 3, 5–6), we concentrated on the LST1/A isoform, excluding possible interference of other splice-variants with the analysis. 3) Instead of semi-quantitative mRNA detection (5–6), we employed a quantitative RT-PCR method to monitor the mRNA level. 4) Two gene expression databases, BioGPS (26) and Gene Enrichment Profiler (27) support our data.
Thus far, the only study at least partially addressing the issue of the biological role of LST1/A suggested that LST1/A overexpression leads to the formation of filopodia, although the mechanism of this phenomenon has not been proposed (7). In agreement with this study, we observed LST1/A localization to filopodia-like structures in a number of cell types. However, when we used LST1/A-independent filopodia visualization, we failed to detect any differences in the abundance of these structures between non-transfected and LST1/A transfected cells.
As a starting point for the elucidation of LST1/A function, we used bioinformatic analysis of the LST1/A amino acid sequence, which revealed the presence of two evolutionarily conserved tyrosine-based motifs strongly resembling the ITIM and ITIM-like sequences in the CD33-related Siglec receptor family. Siglecs are receptors binding sialic acid residues present in cell surface glycoproteins both in cis (on the surface of the same cell) and in trans (on another cell) (16). Members of the CD33 family are expressed mainly on various leukocyte subpopulations (17), and their function is to negatively regulate various aspects of leukocyte biology, including induction of apoptosis, inhibition of proliferation, and inhibition of immunoreceptor-induced cell activation (16) by recruiting various inhibitory molecules to their ITIM and/or ITIM-like motifs (28–30). Indeed, we observed that LST1/A bound protein tyrosine phosphatases, SHP-1 and SHP-2, in a phosphotyrosine dependent manner. Importantly, at least a fraction of LST1/A molecules was phosphorylated in resting cells (likely by Src family kinases) and constitutively associated with both protein tyrosine phosphatases. On the other hand, neither of the other binding partners of CD33-related Siglecs (SOC3, Cbl-b) nor a typical ITIM-interacting molecule (SHIP1) co-precipitated with LST1/A.
The important difference between LST1/A and Siglecs or other ITIM containing inhibitory receptors (e.g. FcγRIIb) is that the extracellular part of LST1/A is very short, and it is therefore highly unlikely that LST1/A would function as a cellular receptor per se. However, it could be functionally associated with a so far unidentified receptor(s) and could serve as an intracellular transducer of the inhibitory signal. Despite numerous attempts, we were unable to either identify a receptor, which would be associated with and/or use LST1/A as a signal transducing unit, or discover a pathway inhibited by LST1/A using myeloid cancer cell lines as a model (data not shown). One possible clue might be LST1/A localization in TEMs, where a number of important adhesion molecules and receptors (including integrins, growth factor receptors, and MHCII) are localized (18, 31, 32). It is therefore possible that LST1/A might be functionally coupled to a receptor residing in these types of microdomains.
To provide evidence that LST1/A has the ability to act as a negative regulator of signal transduction cascades downstream of surface receptors, we prepared a chimeric construct composed of the extracellular part of CD25 fused to LST1/A. As expected, we observed that cross-linking of the CD25-LST1/A chimera with Fc receptors led to the inhibition of Fc receptor-mediated calcium influx and protein tyrosine phosphorylation in the THP-1 and U937 monocytic cell lines, due to the association of LST1/A with the SHP-1 and SHP-2 phosphatases.
SHP-1 and SHP-2 are multifunctional protein tyrosine phosphatases involved in the development and differentiation of hematopoietic cell lineages as well as in the onset of inflammation (33). SHP-1 is expressed mainly in hematopoietic cells and neural tissues (33). Motheaten, viable motheaten, and spin mouse strains bearing loss of function mutations in the SHP-1 encoding gene (34, 35) exhibit granulocyte and macrophage hyper-responsiveness to cytokines (36) and development of severe autoinflammatory disease (35, 37). Moreover, partial SHP-1 deficiency leads to severe autoimmune disease, which develops even in the absence of B- and T- lymphocytes but is dependent on signaling via innate immune receptors, such as IL-1 and Toll-like receptors (35, 37). Most likely, cells of the myeloid lineage induce the chronic inflammation observed in SHP-1 insufficient animals. SHP-2 is expressed more ubiquitously than SHP-1 (33). In contrast to SHP-1, SHP-2 is not considered an entirely inhibitory protein tyrosine phosphatase in hematopoietic cells, as both positive and negative roles of SHP-2 in cell signaling have been described (38–41). However, in the context of Siglec mediated, and likely also LST1/A-mediated, signaling, its inhibitory function probably prevails.
We have described LST1/A as a new TRAP, which is expressed predominantly in myeloid cells and can function as a negative regulator facilitating the recruitment of SHP-1 and SHP-2 to the plasma membrane. Therefore, it is likely that some of the SHP-1 and/or SHP-2 functions in myeloid cells could be dependent on their binding to LST1/A. Our studies have clarified several unresolved issues concerning this protein and suggested that further analysis of LST1/A should focus on the negative regulation of signaling in myeloid cells. In the following studies, it will be important to determine the signaling pathways that are regulated by LST1/A in vivo. This should be facilitated by construction of lst1 knock-out mice, which is currently underway.
Supplementary Material
This work was supported by Academy of Sciences of the Czech Republic (AVOZ50520514 and RVO68378050), GACR Projects MEM/09/E011 and P302/12/G101, and by Center of Molecular and Cellular Immunology Project 1M0506, Ministry of Education, Youth and Sports of the Czech Republic.

This article contains supplemental Fig. 1.
- TRAP
- transmembrane adaptor protein
- ITIM
- immunoreceptor tyrosine-based inhibitory motif
- Siglec
- sialic acid-binding Ig-like lectin receptor
- TEMs
- tetraspanin-enriched microdomains
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Holzinger I., de Baey A., Messer G., Kick G., Zwierzina H., Weiss E. H. (1995) Cloning and genomic characterization of LST1: A new gene in the human TNF region. Immunogenetics 42, 315–322 [DOI] [PubMed] [Google Scholar]
- 2. Spies T., Blanck G., Bresnahan M., Sands J., Strominger J. L. (1989) A new cluster of genes within the human major histocompatibility complex. Science 243, 214–217 [DOI] [PubMed] [Google Scholar]
- 3. Tsuge I., Shen F. W., Steinmetz M., Boyse E. A. (1987) A gene in the H-2S:H-2D interval of the major histocompatibility complex which is transcribed in B cells and macrophages. Immunogenetics 26, 378–380 [DOI] [PubMed] [Google Scholar]
- 4. Gruen J. R., Weissman S. M. (2001) Human MHC class III and IV genes and disease associations. Front. Biosci. 6, D960–972 [DOI] [PubMed] [Google Scholar]
- 5. de Baey A., Fellerhoff B., Maier S., Martinozzi S., Weidle U., Weiss E. H. (1997) Complex expression pattern of the TNF region gene LST1 through differential regulation, initiation, and alternative splicing. Genomics 45, 591–600 [DOI] [PubMed] [Google Scholar]
- 6. Rollinger-Holzinger I., Eibl B., Pauly M., Griesser U., Hentges F., Auer B., Pall G., Schratzberger P., Niederwieser D., Weiss E. H., Zwierzina H. (2000) LST1: A gene with extensive alternative splicing and immunomodulatory function. J. Immunol. 164, 3169–3176 [DOI] [PubMed] [Google Scholar]
- 7. Raghunathan A., Sivakamasundari R., Wolenski J., Poddar R., Weissman S. M. (2001) Functional analysis of B144/LST1: A gene in the tumor necrosis factor cluster that induces formation of long filopodia in eukaryotic cells. Exp. Cell Res. 268, 230–244 [DOI] [PubMed] [Google Scholar]
- 8. Schiller C., Nitschké M. J., Seidl A., Kremmer E., Weiss E. H. (2009) Rat monoclonal antibodies specific for LST1 proteins. Hybridoma 28, 281–286 [DOI] [PubMed] [Google Scholar]
- 9. Mulcahy H., O'Rourke K. P., Adams C., Molloy M. G., O'Gara F. (2006) LST1 and NCR3 expression in autoimmune inflammation and in response to IFN-γ, LPS, and microbial infection. Immunogenetics 57, 893–903 [DOI] [PubMed] [Google Scholar]
- 10. Hrdinka M., Dráber P., Stepánek O., Ormsby T., Otáhal P., Angelisová P., Brdicka T., Paces J., Horejsí V., Drbal K. (2011) PRR7 is a transmembrane adaptor protein expressed in activated T cells involved in regulation of T cell receptor signaling and apoptosis. J. Biol. Chem. 286, 19617–19629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 12. Brdicka T., Imrich M., Angelisová P., Brdicková N., Horváth O., Spicka J., Hilgert I., Lusková P., Dráber P., Novák P., Engels N., Wienands J., Simeoni L., Osterreicher J., Aguado E., Malissen M., Schraven B., Horejsí V. (2002) Non-T cell activation linker (NTAL): A transmembrane adaptor protein involved in immunoreceptor signaling. J. Exp. Med. 196, 1617–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wan J., Roth A. F., Bailey A. O., Davis N. G. (2007) Palmitoylated proteins: Purification and identification. Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 14. Horejsí V., Zhang W., Schraven B. (2004) Transmembrane adaptor proteins: Organizers of immunoreceptor signaling. Nat. Rev. Immunol. 4, 603–616 [DOI] [PubMed] [Google Scholar]
- 15. Daëron M., Jaeger S., Du Pasquier L., Vivier E. (2008) Immunoreceptor tyrosine-based inhibition motifs: A quest in the past and future. Immunol. Rev. 224, 11–43 [DOI] [PubMed] [Google Scholar]
- 16. Crocker P. R., Paulson J. C., Varki A. (2007) Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 [DOI] [PubMed] [Google Scholar]
- 17. von Gunten S., Bochner B. S. (2008) Basic and clinical immunology of Siglecs. Ann. N.Y. Acad. Sci. 1143, 61–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Charrin S., le Naour F., Silvie O., Milhiet P. E., Boucheix C., Rubinstein E. (2009) Lateral organization of membrane proteins: Tetraspanins spin their web. Biochem. J. 420, 133–154 [DOI] [PubMed] [Google Scholar]
- 19. Resh M. D. (2006) Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 20. Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., Sánchez-Madrid F. (2009) Tetraspanin-enriched microdomains: A functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 [DOI] [PubMed] [Google Scholar]
- 21. Brown D. A. (2006) Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology 21, 430–439 [DOI] [PubMed] [Google Scholar]
- 22. Draber P., Vonkova I., Stepanek O., Hrdinka M., Kucova M., Skopcova T., Otahal P., Angelisova P., Horejsi V., Yeung M., Weiss A., Brdicka T. (2011) SCIMP, a transmembrane adaptor protein involved in major histocompatibility complex class II signaling. Mol. Cell. Biol. 31, 4550–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Walter R. B., Häusermann P., Raden B. W., Teckchandani A. M., Kamikura D. M., Bernstein I. D., Cooper J. A. (2008) Phosphorylated ITIMs enable ubiquitylation of an inhibitory cell surface receptor. Traffic 9, 267–279 [DOI] [PubMed] [Google Scholar]
- 24. Unkeless J. C., Jin J. (1997) Inhibitory receptors, ITIM sequences, and phosphatases. Curr. Opin. Immunol. 9, 338–343 [DOI] [PubMed] [Google Scholar]
- 25. Staub E., Rosenthal A., Hinzmann B. (2004) Systematic identification of immunoreceptor tyrosine-based inhibitory motifs in the human proteome. Cell Signal. 16, 435–456 [DOI] [PubMed] [Google Scholar]
- 26. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., 3rd, Su A. I. (2009) BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benita Y., Cao Z., Giallourakis C., Li C., Gardet A., Xavier R. J. (2010) Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood 115, 5376–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paul S. P., Taylor L. S., Stansbury E. K., McVicar D. W. (2000) Myeloid-specific human CD33 is an inhibitory receptor with differential ITIM function in recruiting the phosphatases SHP-1 and SHP-2. Blood 96, 483–490 [PubMed] [Google Scholar]
- 29. Avril T., Floyd H., Lopez F., Vivier E., Crocker P. R. (2004) The membrane-proximal immunoreceptor tyrosine-based inhibitory motif is critical for the inhibitory signaling mediated by Siglecs-7 and -9, CD33-related Siglecs expressed on human monocytes and NK cells. J. Immunol. 173, 6841–6849 [DOI] [PubMed] [Google Scholar]
- 30. Ulyanova T., Shah D. D., Thomas M. L. (2001) Molecular cloning of MIS, a myeloid inhibitory siglec, that binds protein-tyrosine phosphatases SHP-1 and SHP-2. J. Biol. Chem. 276, 14451–14458 [DOI] [PubMed] [Google Scholar]
- 31. Angelisová P., Hilgert I., Horejsí V. (1994) Association of four antigens of the tetraspans family (CD37, CD53, TAPA-1, and R2/C33) with MHC class II glycoproteins. Immunogenetics 39, 249–256 [DOI] [PubMed] [Google Scholar]
- 32. Yunta M., Lazo P. A. (2003) Tetraspanin proteins as organizers of membrane microdomains and signaling complexes. Cell Signal. 15, 559–564 [DOI] [PubMed] [Google Scholar]
- 33. Chong Z. Z., Maiese K. (2007) The Src homology 2 domain tyrosine phosphatases SHP-1 and SHP-2: Diversified control of cell growth, inflammation, and injury. Histol. Histopathol. 22, 1251–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsui H. W., Siminovitch K. A., de Souza L., Tsui F. W. (1993) Motheaten and viable motheaten mice have mutations in the haematopoietic cell phosphatase gene. Nat. Genet. 4, 124–129 [DOI] [PubMed] [Google Scholar]
- 35. Croker B. A., Lawson B. R., Rutschmann S., Berger M., Eidenschenk C., Blasius A. L., Moresco E. M., Sovath S., Cengia L., Shultz L. D., Theofilopoulos A. N., Pettersson S., Beutler B. A. (2008) Inflammation and autoimmunity caused by a SHP1 mutation depend on IL-1, MyD88, and a microbial trigger. Proc. Natl. Acad. Sci. U.S.A. 105, 15028–15033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsui F. W., Martin A., Wang J., Tsui H. W. (2006) Investigations into the regulation and function of the SH2 domain-containing protein-tyrosine phosphatase, SHP-1. Immunol. Res. 35, 127–136 [DOI] [PubMed] [Google Scholar]
- 37. Yu C. C., Tsui H. W., Ngan B. Y., Shulman M. J., Wu G. E., Tsui F. W. (1996) B and T cells are not required for the viable motheaten phenotype. J. Exp. Med. 183, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan R. J., Leedy M. B., Munugalavadla V., Voorhorst C. S., Li Y., Yu M., Kapur R. (2005) Human somatic PTPN11 mutations induce hematopoietic-cell hypersensitivity to granulocyte-macrophage colony-stimulating factor. Blood 105, 3737–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bauler T. J., Kamiya N., Lapinski P. E., Langewisch E., Mishina Y., Wilkinson J. E., Feng G. S., King P. D. (2011) Development of severe skeletal defects in induced SHP-2-deficient adult mice: A model of skeletal malformation in humans with SHP-2 mutations. Dis. Model. Mech. 4, 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. An H., Zhao W., Hou J., Zhang Y., Xie Y., Zheng Y., Xu H., Qian C., Zhou J., Yu Y., Liu S., Feng G., Cao X. (2006) SHP-2 phosphatase negatively regulates the TRIF adaptor protein-dependent type I interferon and proinflammatory cytokine production. Immunity 25, 919–928 [DOI] [PubMed] [Google Scholar]
- 41. You M., Yu D. H., Feng G. S. (1999) Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol. Cell. Biol. 19, 2416–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.