Background: β-Catenin plays diverse temporal roles in liver development.
Results: We report calpain-induced truncated β-catenin during mid- to late- hepatic development that resides in maturing hepatocytes nuclei and at membrane. RNA-seq studies identified novel targets.
Conclusion: This may be a mechanism of the pleiotropic functions of β-catenin in hepatic development.
Significance: Identification of different β-catenin species may have diagnostic implications in differentiating fetal and embryonal hepatoblastomas.
Keywords: β-Catenin, Calpain, Development, Liver, Wnt Pathway
Abstract
Hepatic competence, specification, and liver bud expansion during development depend on precise temporal modulation of the Wnt/β-catenin signaling. Also, loss- and gain-of-function studies have revealed pleiotropic roles of β-catenin in proliferation and hepatocyte and biliary epithelial cell differentiation, but precise mechanisms remain unknown. Here we utilize livers from different stages of murine development to determine β-catenin signaling and downstream targets. Although during early liver development full-length β-catenin is the predominant form, at late stages, where full-length β-catenin localizes to developing biliary epithelial cells only, a 75-kDa truncated β-catenin species is the principal form localizing at the membrane and in the nucleus of differentiating hepatocytes. The truncated species lacks 95 N-terminal amino acids and is transcriptionally active. Our evidence points to proteolytic cleavage of β-catenin by calpain as the mechanism of truncation in cell-free and cell-based assays. Intraperitoneal injection of a short term calpain inhibitor to timed pregnant female mice abrogated β-catenin truncation in the embryonic livers. RNA-seq revealed a unique set of targets transcribed in cells expressing truncated versus full-length β-catenin, consistent with different functionalities. A further investigation using N- and C-terminal-specific β-catenin antibodies on human hepatoblastomas revealed a correlation between full-length versus truncated β-catenin and differentiation status, with embryonal hepatoblastomas expressing full-length β-catenin and fetal hepatoblastomas expressing β-catenin lacking its N terminus. Thus we conclude that calpain-mediated cleavage of β-catenin plays a role in regulating hepatoblast differentiation in mouse and human liver, and the presence of the β-catenin N terminus correlates with differentiation status in hepatoblastomas.
Introduction
Applications of hepatocytes derived from stem cells can range from modeling human disease and toxicity screening tools to cell therapy and regenerative medicine and thus are of high significance (1). Although many attempts have been made to differentiate stem cells from various sources into fully functional hepatocytes suitable for patient transplantation, even the most effective protocols produce cells that retain expression of α-fetoprotein (AFP), a marker of undifferentiated cells, and lack expression of many key cytochrome p450 enzymes critical to mature liver function, revealing that our understanding of this differentiation program is incomplete (2).
A key pathway known to guide liver development is the Wnt/β-catenin pathway. Wnt/β-catenin has a role in patterning the foregut endoderm from which the liver arises, in producing a gradient involved in hepatic induction that patterns the gut along the anterior-posterior axis, and in promoting expansion of bipotential hepatoblasts that form the nascent liver bud (3–12). The importance of the Wnt/β-catenin pathway in liver persists throughout the life of an adult organism, as it plays roles in zonal metabolism (13–16), hepatic progenitor maintenance (17–21), and liver regeneration (22–27), and its dysregulation is observed in many liver cancers (28).
Although compelling evidence supports the role of β-catenin in proliferation of hepatoblasts during liver bud expansion, seemingly conflicting reports exist on the role of β-catenin in hepatoblast differentiation into biliary epithelial cells and hepatocytes. Premature activation of β-catenin via targeted deletion of Adenomatous Polyposis Coli gene product (APC) in the livers of developing mice results in pronounced biliary differentiation of hepatoblasts at the expense of hepatocytes (29) and is consistent with a role for β-catenin activity in promoting hepatoblast differentiation into biliary epithelial cells (29). Hepatoblast-specific, FoxA3-Cre driven β-catenin deletion, however, leads to not only defects in biliary specification of hepatoblasts but also maturation of hepatocytes (12). Embryos possessing the β-catenin deletion die late in gestation, with livers exhibiting abnormalities beginning at approximately embryonic day 13, when hepatoblast differentiation starts to occur. Knock-out livers appear to arrest at this stage, composed of cells exhibiting the high nuclear-to-cytoplasmic ratio and unpolarized morphology reminiscent of uncommitted E13/14 stage hepatoblasts. Knock-out livers show an absence of bile ducts and also expression of the hepatocyte-specific transcription factors C/ebpα and HNF4α as well as reduced expression of several other hepatocyte markers (12). This suggests that β-catenin activity is necessary for both biliary epithelial cell and hepatocyte differentiation, but it is unclear what mechanism could account for both of these observations.
In addition, the role of β-catenin in hepatoblastomas (HB)2 has been reported, and monoallelic mutations and deletions in exon 3 of CTNNB1, the region where β-catenin is targeted for degradation by the proteasome, are present in 50–90% of HB (30–35). Based on the predominant stage of hepatoblasts constituting a HB, they may be classified as either fetal (resembling livers from around days E14.5-E18.5 in developing mouse) or embryonal (resembling the hepatoblasts seen during approximately E11.5-E12.5 of mouse liver development) (36). This classification is significantly correlated with prognosis (37), but histology may not always be sufficient to make that distinction.
In this study we describe a novel truncated 75-kDa β-catenin species that appears in developing liver whose localization and appearance in hepatocytes coincides with hepatocyte maturation. Moreover, we demonstrate that this form of β-catenin is produced post-translationally via proteolytic cleavage of its N-terminal 95 amino acids by calpain and that it localizes to the membranes and nuclei of hepatocytes in late fetal liver. RNA-seq studies on transfected primary hepatocytes reveal known as well as unique target genes of the truncated β-catenin. We further reveal that β-catenin status correlates with hepatoblastoma differentiation, with embryonal morphology correlating with full-length β-catenin, and truncated β-catenin associated with fetal forms. Thus, in summary, we report that calpain-mediated cleavage of β-catenin occurs during normal liver development, producing a truncated species with a distinct function that produces a gene expression pattern unique from that of full-length β-catenin.
EXPERIMENTAL PROCEDURES
Immunoblotting
Whole livers isolated from wild-type C57BL/6 mice at various developmental time points (E12.5, E14.5, E16.5, E17.5, E18.5, and adult) were lysed in Nonidet P-40 buffer (1% Nonidet P-40, 50 mm Tris-HCl, pH 8.0, 150 mm NaCl) subjected to SDS-PAGE and blotted onto PVDF membranes then blocked in 5% nonfat milk in TBST, 5% BSA in TBST, or 5% fish gelatin in TBST. Thereafter, membranes were incubated overnight in the appropriate primary antibody diluted in blocking solution, washed in TBST, incubated with secondary antibody (Millipore) diluted 1:30,000–1:80,000 in blocking buffer, washed, and signal-visualized using ECL. Antibodies used were: C-terminal β-catenin (amino acids 670–781; BD Biosciences #610154), N-terminal β-catenin (amino acids 1–18; Abcam #ab32572), β-catenin (amino acids 29–49) Millipore #06-734), phospho-Tyr-142 β-catenin (Abcam #ab27798), phospho-Tyr-654 β-catenin antibody (custom, Aves Labs), TCF4 (Millipore #05-511, 1:250), focal adhesion kinase (Cell Signaling, #3285, 1:1000), μ-calpain (Cell Signaling #2556, 1:500), and actin (Millipore MAB1501, 1:500).
Northern Blotting
mRNA was isolated from E12.5, E14.5 E16.5, E17.5, and E18.5 livers using Poly(A)PuristTM kit (Ambion). Samples were then pooled, and Northern blot analysis was performed according to standard methods using a full-length, radiolabeled β-catenin DNA probe.
RT-PCR
Whole livers isolated from wild-type C57BL/6 mice at various developmental time points (E12.5, E14.5, E16.5, E17.5, E18.5, and adult) were pooled (n > 3), and total RNA was extracted with TRIzol (Invitrogen) according to the manufacturer's instructions. SuperScript III (Invitrogen) was used to synthesize first strand cDNA from 1 μg of total DNase-treated RNA with oligo(dT)20 primers according to manufacturer's instructions. The cDNA was used as the template for RT-PCR performed with primers complementary to the 5′-UTR (5′-AAG CCC TCG CTC GGT GG-3′) and 3′-UTR (5′-CTGAACCATTTCTATAACCGCATCTGTTG-3′) and SYBR Green PCR Master Mix reagent (SuperArray Bioscience).
Cell Fractionation Studies
Nuclear/cytoplasmic fractions and membrane fractions were extracted using the NE-PER kit and MEM-PER kit (Pierce), respectively, according to the manufacturer's instructions. Protein were boiled in SDS gel loading buffer, loaded onto polyacrylamide gels, and subjected to SDS-PAGE. Although 30 μg of protein was loaded for the nuclear and cytoplasmic fractions, 1 μg of protein was loaded for the membrane fraction.
Immunoprecipitation Studies
500 μg of liver lysates in Nonidet P-40 buffer were diluted to 700 μl in Nonidet P-40 buffer containing protease/phosphatase inhibitors. For β-catenin immunoprecipitations, 20 μl of agarose beads preconjugated to rabbit anti-β-catenin antibody (Santa Cruz, sc-1496-R AC) were added and incubated on an inverter for 1 h at 4 °C. For E-cadherin and TCF4 immunoprecipitations, 2 μg of antibody (TCF4: Millipore, E-cadherin: BD Biosciences #610182) was added to tube and incubated on an inverter 1 h at 4 °C, and then 20 μl of Protein A/G Plus-agarose beads (Santa Cruz, sc-2003) were added and incubated at 4 °C for 1 h on an inverter. All reactions were then spun to collect beads, supernatant was removed, and beads were washed 4 times in 800 μl of Nonidet P-40 buffer; beads were then boiled in 1× SDS loading buffer for loading on gels.
Immunostaining
To prepare tissue for immunohistochemistry, whole livers (or whole embryos in the case of E12.5 embryos) were fixed in 10% buffered formalin followed by 70% ethanol before paraffin embedding. Four- to five-μm-thick paraffin sections were deparaffinized, antigen retrieval was performed by microwaving in citrate buffer (10 mm citric acid, pH 6.0) for 12 min and cooled to room temperature, and endogenous peroxidase activity was quenched by treatment for 7 min with 3% H2O2. Tissue was then blocked by Large Ultra V Block (Labvision) for 5 min followed by incubation in primary antibody diluted in TBST containing 5% serum from the species in which the secondary antibody was raised (normal donkey serum or normal goat serum; Jackson ImmunoResearch) overnight at 4 °C. Sections were washed in PBS then incubated in 1:500 dilution of biotinylated secondary antibody (Millipore) at room temperature for 30 min, washed, and then developed using diaminobenzidine and counterstained in Shandon Instant Hematoxylin. Paraffin sections from 16 HB patients were also used for immunohistochemistry for β-catenin. The HBs were labeled as being positive or negative for nuclear and cytoplasmic β-catenin by a single pathologist. Correlations between staining and tumor subtype were assessed using Fisher's Exact Test, and p < 0.001 was considered significant. Antibodies used for staining were: C-terminal β-catenin (amino acids 670–781; BD Biosciences #610154), N-terminal β-catenin (amino acids 1–18) Abcam #ab32572), and β-catenin (amino acids 29–49) Millipore #06-734).
TOPFLASH Assay
HEK293 cells (∼50% confluent) were fed with fresh medium (antibiotic free, freshly made minimum Eagle's medium + 10% FBS) just before transfection. Cells were transfected with a 3 μl of FuGENE:2 μg of DNA ratio (800 ng of β-catenin (WT, Δ95 β-catenin, or empty vector), 800 ng of TOPFLASH DNA, 400 ng of Renilla) according to Dual Luciferase Reporter (Promega) protocol and read on a luminometer (EG&G Berthold Lumat LB 9507). Luciferase activity was then quenched, and Renilla expression was detected by the addition of 100 μl of STOP-GLO reagent. TOPFLASH values were calculated as ratios of Luciferase signal to Renilla signal.
In Vitro Calpain Assay
Recombinant GST-β-catenin (Millipore) was diluted to 22.5 nm in calpain reaction buffer (10 mm HEPES, pH 8, 2 mm DTT, 1 mm EDTA) in 1.5-ml Eppendorf tubes coated with Silicote (Sigma) and added to a 2.5 nm dilution of recombinant calpain (Calbiochem #208718) in the same buffer, incubated at room temperature for 15 min, and boiled in SDS buffer for 5 min to stop the reaction. Identical reactions were performed in the presence of 10 mm EGTA to inhibit calpain and in the absence of calpain were used as negative controls for β-catenin cleavage by calpain. These samples were loaded onto gels, and immunoblots were performed using anti-β-catenin antibody (1:500 BD Biosciences #610154).
Calpain Activity Assay
Freshly isolated E14.5, E18.5, and adult livers were pooled and lysed in Nonidet P-40 buffer in the absence of protease inhibitors. Protein concentrations were measured by BCA assay and diluted in Hepes/DTT/EDTA buffer (10 mm HEPES, pH 7.2, 10 mm DTT, and 1 mm EDTA). Diluted lysate was added to an equal amount of prepared Calpain-GLO reagent (Promega) and incubated for 15 min, and luminescence was measured on a plate reader (Biotek Synergy HT). Hepes/DTT/EDTA buffer containing 45 nm calpain (Calbiochem #208718) was used as a positive control, and lysates mixed with Calpain-GLO reagent in the presence of EGTA were used as a negative control to ensure specificity of assay for calpain activity.
Animal Studies
Timed pregnant animals obtained from Charles River were injected intraperitoneally at E13.5 with either 12 mg/kg MDL28170 (Sigma) or vehicle (DMSO, Sigma). Embryonic livers were removed 2–3 h later and lysed in Nonidet P-40 buffer containing 2× Halt Protease and Phosphatase Inhibitor Mixture (Thermo Scientific), protein concentration was measured by BCA assay, and equal amounts of liver from DMSO-treated and MDL28170-treated animals were loaded onto gels for SDS-PAGE/immunoblot analysis.
cDNA Library Generation
Primary mouse hepatocytes were plated in serum-containing medium, allowed to adhere for 40 h, then changed to antibiotic-free, serum-free medium and transfected with a 2:1 ratio of Lipofectamine 2000:pAAV (Δ95 or full-length) β-catenin internal ribosomal entry site GFP DNA in 10-cm dishes (60 μl of Lipofectamine, 30 μg of DNA). The medium was changed 5 h after transfection, and cells were harvested 30 h later. Total RNA was extracted from 50 mg of tissues with the TRIzol method (Invitrogen). The extraction procedure was performed according to the manufacturer's recommendation. This was followed by removal of ribosomal RNA using RIBOZERO kit (Nextera, Inc). After phenol/chloroform and ethanol purification of the digested products, random hexamers were used in the first strand cDNA synthesis with 20 ng of RNA and Superscript II TM (Invitrogen). The second strand cDNA synthesis was carried out at 16 °C by adding Escherichia coli DNA ligase, E. coli DNA polymerase I, and RNase H in the reaction. This was followed by the addition of adenosine nucleotide in the 3′ ends of newly synthesized cDNA and ligated with adaptors provided by Illumina RNASeq kit. The ligation products were amplified for 15 cycles of PCR using adaptor primers.
Cluster Generation and Sequencing through Illumina HiSeq2000
The library cDNAs were denatured in 0.1 n NaOH for 5 min and neutralized in hybridization buffer. The samples were loaded onto C-bot amplifier to generate sequencing clusters in a Illumina TruSeq v3 flowcell. This was followed by chemical sequencing in HiSeq2000 sequencer. The detailed procedure follows the recommendations of the manufacturer. The bcl files generated from HiSeq2000 were converted to fastq files using CASSAVA 1.8 software. The fastq files were mapped to mouse genome downloaded from NCBI using Genomebench from CLC BIO, Inc. The coverage analysis was subsequently exported to Excel to generate a spreadsheet.
RESULTS
Expression of a Truncated β-Catenin Species Correlates with Hepatoblast Differentiation
Time of hepatoblast differentiation to hepatocytes and bile ducts during murine liver development is well understood (38, 39). To begin investigating the role of β-catenin in hepatoblast differentiation, we immunoblotted liver lysates from mice at embryonic days 12.5 (E12.5), E14.5, E16.5, E17.5, and E18.5 and also samples taken from adult (>28 days) mouse livers for β-catenin. Although this analysis reveals the presence of full-length (∼97 kDa) β-catenin at E12.5 and E14.5, we also observe a truncated, 75-kDa β-catenin species beginning at E12.5. The truncated species increases during development concurrently with the decrease in full-length protein and exists as the predominant form of β-catenin from E16.5 until birth (Fig. 1A).
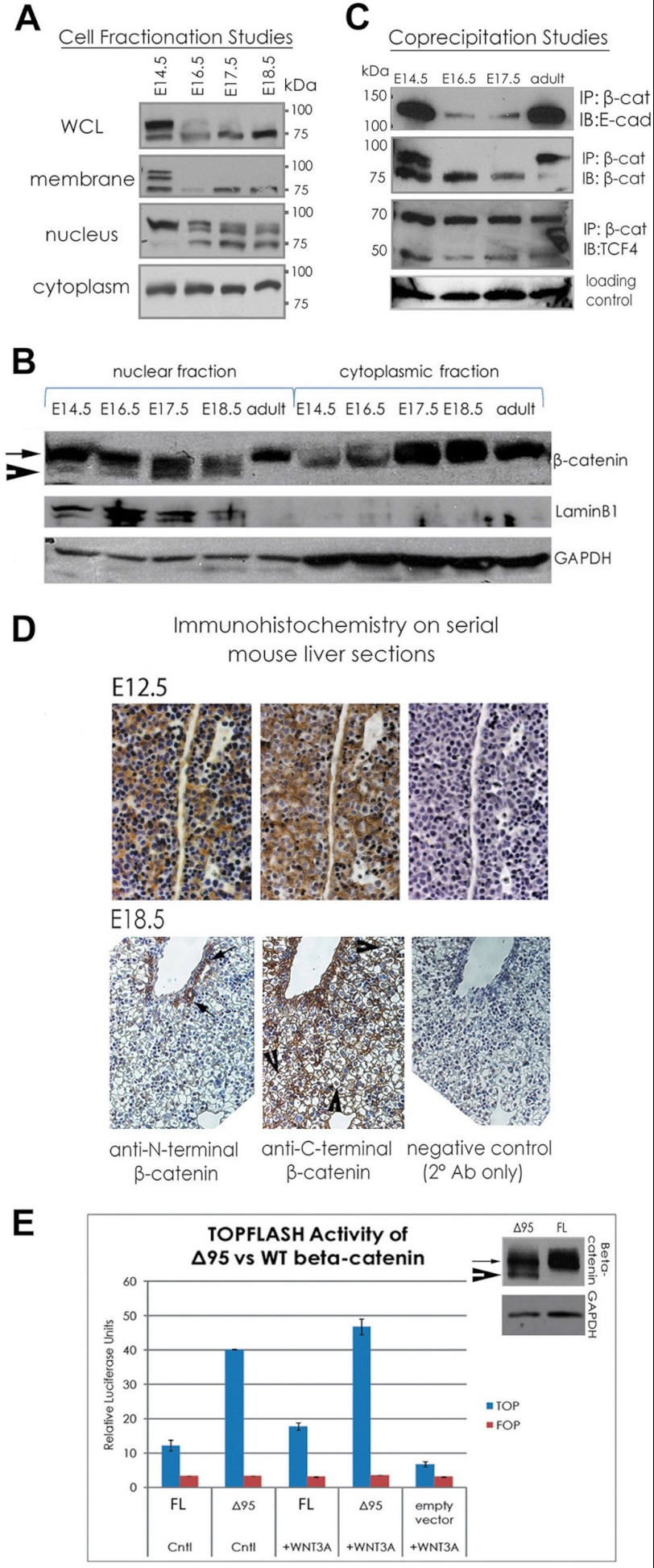
FIGURE 1.
A truncated β-catenin species produced by post-translational cleavage of the N-terminal 95 amino acids is produced in developing liver during hepatoblast differentiation. A, an immunoblot analysis reveals a truncation of β-catenin from 97 to ∼75 kDa occurring as liver develops. B, a Northern blot on pooled mRNA from E12.5, E14.5, E16.5, E17.5, and E18.5 livers probed with a radiolabeled, full-length Ctnnb1 probe reveals only one β-catenin transcript at the expected size of ∼3565 bp. C, RT-PCR on cDNA from E14.5, E18.5, and adult using primers against the 5′-UTR and 3′-UTR of the Ctnnb1 transcript livers also indicates the presence of one mRNA species coding for β-catenin. D, immunoblots on liver lysates from E12.5, E14.5, E16.5, E17.5, and E18.5 using antibodies against different β-catenin epitopes shows that that antibodies targeting amino acids 571–781, phospho-Tyr-654, and phospho-Tyr-142 bind to the truncated β-catenin species, but those targeting amino acids 1–18 or 29–49 do not. E, tandem mass spectrometry on truncated β-catenin protein immunoprecipitated from pooled E18.5 liver lysate detected peptides comprising amino acids 96–125, 134–151, 159–170, 190–199, 486–495, 535–541, 550–564, 647–660, and 673–683.
Truncated β-Catenin Is Not the Result of Alternative Splicing
To determine whether the shorter β-catenin species is the result of alternative splicing, we performed Northern blot and RT-PCR analyses. A Northern blot on pooled mRNA from embryonic days E12.5, E14.5, E16.5, E17.5, and E18.5 using a full-length, radiolabeled β-catenin probe reveals only a single mRNA transcript at the expected size for Ctnnb1 (Fig. 1B). Likewise, RT-PCR on cDNA from E14.5 (both full-length and truncated β-catenin-expressed), E18.5 (predominantly truncated β-catenin-expressed), and adult liver (predominantly full-length β-catenin-expressed) using primers against the 5′-UTR and 3′-UTR of Ctnnb1 produces a single PCR product consistent with the expected full-length transcript length of 3565 bp (Fig. 1C).
Truncated β-Catenin Lacks 95 N-terminal Amino Acids
Next we sought to establish which region of β-catenin was absent in the truncated species. Immunoblotting of embryonic liver samples with antibodies against various β-catenin epitopes (amino acids 571–781, Tyr(P)-654; amino acids 1–18, Tyr(P)-142, amino acids 29–49) reveals that the truncated species retains amino acids 571–781, Tyr(P)-654, and Tyr(P)-142, but not amino acids 1–18 and amino acids 29–49 (Fig. 1D). Tandem mass spectrometry peptide sequencing of immunoprecipitated 75-kDa β-catenin from E18.5 mouse livers detects amino acid 96 as the N-terminal-most amino acid as well as inclusion of amino acids 96–125, 134–151, 159–170, 486–495, 535–541, 550–564, 647–660, and 673–683 (Fig. 1E). Thus, we have identified a 75-kDa truncated β-catenin that is expressed during normal liver development and lacks the N-terminal 95 amino acids.
Truncated β-Catenin Is Present at Membranes and in Nuclei of Hepatocytes in Late Fetal Liver
Cell fractionation experiments were done next to address relative abundance of full-length and the truncated form of β-catenin especially if abundance of one may overshadow the other in whole cell lysates (Fig. 2A). Indeed, only 1 μg of protein loaded for membrane resembles whole cell lysates, suggesting this compartment to contain most of the β-catenin. Blots using 30 μg for other fractions reveal the presence of truncated β-catenin in both the membrane and nuclear fractions (Fig. 2, A and B). In addition, co-immunoprecipitation experiments indicate an association of truncated β-catenin with both its adherens junction binding partner E-cadherin and its nuclear binding partner TCF4 (Fig. 2C). Immunohistochemistry on serial E12.5 and E18.5 liver sections was performed using antibodies against the N terminus (to detect full-length β-catenin) and C terminus of β-catenin (to detect full-length plus truncated β-catenin). Both forms are present at membrane and in the cytoplasm and nuclei of cells at E12.5, whereas at E18.5 full-length β-catenin expression was limited to the nucleus and cytoplasm of bile duct cells, whereas the anti-C-terminal antibody detects those regions but also exhibits abundant membranous and nuclear staining in hepatocytes (Fig. 2D).
FIGURE 2.
Truncated β-catenin is expressed in differentiating hepatocytes and is transcriptionally active (A and C). Cell fractionation studies reveal truncated β-catenin present in membrane and nuclear fractions of liver during E16.5, E17.5, and E18.5, whereas cytoplasmic and nuclear fractions contained full-length β-catenin (B). Co-immunoprecipitation indicates that truncated β-catenin binds both E-cadherin and TCF4. IgG band served as loading control (D). Immunohistochemistry with antibodies against N terminus of β-catenin to detect full-length β-catenin and C terminus to detect both forms of β-catenin reveal that truncated β-catenin (regions positive with anti-C-terminal but not anti-N-terminal antibody) is located in the nuclei and at membranes of hepatocytes. Full-length β-catenin expression is limited to biliary epithelial cells only. E, Δ95 β-catenin transfection of HEK293 cells activates TOPFLASH, supporting a role as a nuclear transactivator (full-length (FL) β-catenin versus Δ95 β-catenin; control (Cntl) medium p < 0.0003; WNT3A medium p < 0.003, Student's t test). The immunoblot (right) on transfected cells shows expression levels of Δ95 and full-length β-catenin in each. IP, immunoprecipitate; WB, Western blot; WCL, whole cell lysates.
Presence of Δ95 β-Catenin Correlates with Δ95 β-Catenin Is Transcriptionally Active
Next, to further explore whether Δ95 β-catenin acts as a dominant-negative or is able to promote expression of β-catenin targets, we performed transient transfections of vectors expressing Δ95-β-catenin, full-length β-catenin, or empty vector into HEK293 cells, which were then treated with conditioned media either from control L cells or those expressing WNT3A. As measured by the TOPFLASH assay, Δ95 β-catenin exhibits the capacity to activate gene expression in both media, further supporting a role for the truncated species in the nucleus (Fig. 2E).
Calpain Is the Protease Responsible for Truncation of β-Catenin in Developing Liver
Because it has been reported that calpain cleavage of the N-terminal 95 amino acids leads to generation of a 75-kDa truncated β-catenin product (40, 41), we explored whether calpain might be responsible for producing this cleavage during development. Western blot revealed the appearance of μ-calpain at E12.5 that increased dramatically between E14.5 and E18.5 and coincided with the presence of 75-kDa species of β-catenin (Fig. 3A). However, because calpain activity is regulated post-translationally, we performed a calpain activity assay on lysates from freshly isolated embryonic livers at E14.5 and E18.5 as well as adult liver. High activity of calpain was indeed observed during development (Fig. 3B). Activity was highest at E14.5, approximately when truncated β-catenin begins to be prominently produced, and is appreciable at E18.5 when truncated form is still significant. Calpain activity is lowest in the adult liver where truncated β-catenin is scarce. As another way to assess calpain activity, we probed embryonic liver lysates for focal adhesion kinase, a known calpain target, and found cleavage of this protein paralleling that of β-catenin (Fig. 3C). To demonstrate the feasibility of cleavage of β-catenin by calpain, we treated GST-tagged recombinant β-catenin protein with recombinant calpain in vitro in a cell-free assay. Such treatment produced a cleavage product of similar molecular weight to the truncated species observed in development (Fig. 3D).
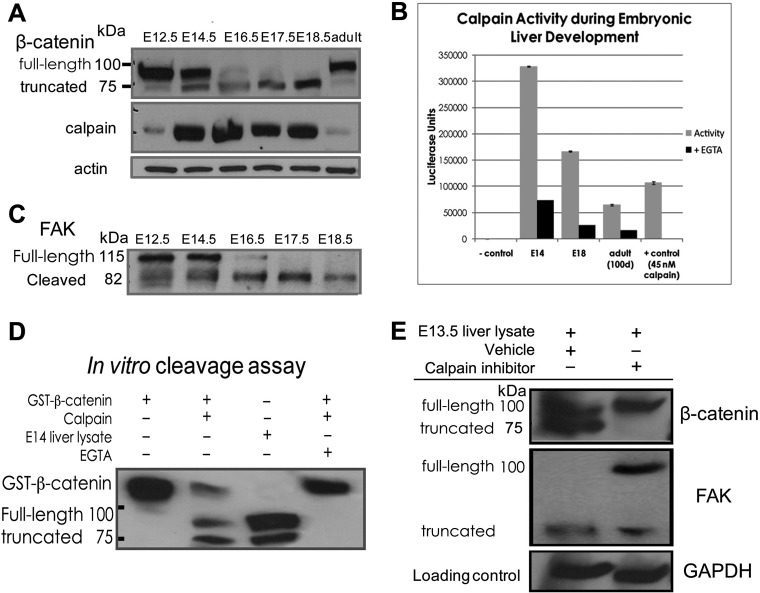
FIGURE 3.
Calpain cleavage of β-catenin is responsible for production of the truncated β-catenin species. A, immunoblot analysis indicates that calpain is expressed in developing liver and that its expression level is highest at times when β-catenin cleavage is observed. B, calpain-GLO activity assay detects active calpain during liver development, with activity highest at E14.5, when significant β-catenin cleavage appears, and persisting at E18.5 when the truncated β-catenin is the predominant form present. Calpain activity is lowest but still present in adult samples when minimal β-catenin cleavage is observed. No calpain activity is seen in samples lacking the addition of liver lysates or purified calpain (far left), and positive control for calpain (45 nm purified enzyme added) is positive for calpain activity (far right). In each case the addition of EGTA, a known calpain inhibitor, results in significant quenching of calpain activity. C, an immunoblot for the known calpain substrate focal adhesion kinase (FAK) shows a cleavage pattern paralleling that of β-catenin and provides further evidence that calpain is active in developing liver. (D). In vitro protease assay combining purified calpain and recombinant GST-tagged β-catenin produces a β-catenin cleavage pattern identical to that seen in developing liver. This cleavage is inhibited by EGTA, a known inhibitor of calpain activity. E, livers from embryos of timed pregnant mice at 13.5 days post-coitus injected intraperitoneally with the calpain inhibitor MDL28170 (12.5 mg/kg) show a lack of truncated β-catenin production relative to vehicle-treated animals. Immunoblot performed in parallel for focal adhesion kinase shows that the calpain-mediated cleavage of focal adhesion kinase is inhibited as well.
Inhibition of Calpain Activity in Developing Embryos Prevents the Production of Truncated β-Catenin
Finally, to directly test the role of calpain in producing truncated β-catenin during in vivo development, we injected timed-pregnant female mice at E13.5 stage of gestational development with the specific calpain inhibitor MDL28170 or vehicle control (DMSO). Three hours after injection, the livers from embryos were assayed for β-catenin by Western blot. A marked absence of β-catenin cleavage was observed in the calpain inhibitor-treated animals but not the DMSO-injected group, consistent with calpain being responsible for the physiological post-translational modification of β-catenin during hepatic development. To verify efficacy of calpain inhibitor, we examined focal adhesion kinase cleavage after injection of inhibitor and DMSO. We observed a dramatic decrease in the cleavage of focal adhesion kinase as indicated by a gain of its uncleaved higher molecular weight form after the inhibitor and not DMSO treatment (Fig. 3E).
Expression of Δ95 and Full-length β-Catenins Led to Transcription of Unique Sets of Targets
To determine whether truncated β-catenin was capable of transcribing different targets than full-length, we transfected primary hepatocytes (Fig. 4A) with both constructs and compared gene expression by RNA-seq. Because Δ95-β-catenin is more stable, as it lacks GSK3β phosphorylation sites, we expected and found higher expression of known TCF targets such as Lect2, cyclinD1 (Ccnd1), regucalcin (Rgn), and glutamine synthetase (Glul) (Fig. 4B). However, we also found 75 genes up-regulated at least 2-fold in Δ95-β-catenin expressing cells relative to full-length (supplemental Table 1). A list of the top 19 genes shows several targets to be histone- and mitosis-regulating genes (Fig. 4C). Interestingly, only one gene, Egr1, was expressed 2-fold more in full-length β-catenin-expressing cells than Δ95-expressing cells.
FIGURE 4.
Identification of unique genes overexpressed in primary murine hepatocytes transfected with truncated β-catenin. A, an immunoblot performed on primary mouse hepatocytes transfected with Δ95 β-catenin (b-cat) and full-length (FL) β-catenin indicates expression of the appropriate proteins. B, RNA-seq analysis reveals up-regulation of several known β-catenin targets after overexpression of Δ95 β-catenin. C, RNA-seq also reveals genes uniquely expressed by Δ95 β-catenin, and the top 20 targets are shown. D, Genomatix search for common transcription factor (TF) binding sites in promoters of potential Δ95 targets reveals binding sites for many factors present at a frequency significantly above what is seen in the genome at large, suggesting one or more of these factors may be co-regulating Δ95 β-catenin targets.
To determine whether Δ95-β-catenin could plausibly be interacting with one (or more) specific transcription factor(s) to promote target gene expression, we used Genomatix to search for common transcription factor binding sites in the top 50 targets. Our analysis revealed 12 candidate transcription factor families for which the binding sites are disproportionately represented in our targets relative to their frequency in the genome (Fig. 4D) (p < 0.01). Two of these were core transcription factors, which is interesting in light of the recent finding that core promoter binding elements play roles in differentiation (42, 43). Also intriguing, 5 of the 12 are transcription factors families with members known to interact with β-catenin: SP1/Klf4 (44), HIF/Hif1α (45), PERO/PPARγ (46), NFκB/NFκB (47), and EGR/Egr1 (48). This suggests that Δ95-β-catenin may be differentially interacting with these factors to eventually determine target genes and biological outcome such as hepatocyte maturation.
β-Catenin Species Distinguish Histological Subtypes of Hepatoblastoma
Because hepatoblastomas arise in prenatal liver, we speculated that if our model were correct and applicable to humans, we might see a relationship between β-catenin species and differentiation status in hepatoblastomas. Hepatoblastomas are classified based on whether they morphologically resemble hepatoblasts (embryonal type) or late prenatal hepatocytes (fetal type), and these classifications are associated with different patient outcomes (36). Using antibodies against the N terminus and C terminus of β-catenin, we investigated whether N-terminally deleted β-catenin expression was associated with the more differentiation fetal morphology in hepatoblastoma patients. We stained 16 HBs, scoring them as positive or negative for each, as well as scoring them histologically as fetal or embryonal (Table 1). Astoundingly, we found that tumors positive for N-terminal β-catenin and C-terminal β-catenin (corresponding to full-length β-catenin expression) were invariably embryonal (8/8), whereas those positive for C-terminal β-catenin but negative for N-terminal β-catenin (corresponding to an N-terminally truncated β-catenin) were always of the fetal subtype (7/7). By Fisher's Exact test (Table 2), this represents a highly significant correlation (p = 0.00016), clearly indicating the utility of N- and C-terminal-specific β-catenin antibodies to distinguish embryonal from fetal HB and supporting a model in which truncated β-catenin promotes hepatoblast differentiation.
TABLE 1.
Summary of immunohistochemistry for N-terminal and C-terminal β-catenin-directed antibodies in hepatoblastoma patients
| Case no. | Histologic subtype | C-terminal nuclear and/or cytoplasmic (+/−) | N-terminal nuclear and/or cytoplasmic (+/−) |
|---|---|---|---|
| HB-83B | E | + | + |
| HB-85 | E | + | + |
| HB-70A | E | + | + |
| HB-50A | E | + | + |
| HB-24 | E | + | + |
| HB-2B | E - 87% | + | + |
| HB-3B | E | + | + |
| HB-39 | E treated | + | + |
| HB-84B | F | + | − |
| HB-83B | F | + | − |
| HB-85 | F | − | − |
| HB-50A | F | + | − |
| HB-25 | F | + | − |
| HB-23 | F | + | − |
| HB-14 | F | + | − |
| HB-41A | F | + | − |
TABLE 2.
Fisher Exact Test for correlation studies of β-catenin immunohistochemistry in hepatoblastomas
| Staining | Histological subtype |
Total casesa | ||
|---|---|---|---|---|
| Embryonal | Fetal | |||
| N-term + | C-term + | 8 | 0 | 8 |
| N-term − | C-term + | 0 | 7 | 7 |
| 8 | 7 | 15 | ||
a p = 0.0002.
DISCUSSION
The results presented here highlight the existence of a novel 75-kDa species of β-catenin that is produced via proteolytic cleavage of the N-terminal 95 amino acids by calpain and whose expression and localization point to a role hepatocyte differentiation. In particular, we observed the truncated β-catenin species localized to the membranes and nuclei of hepatocytes, whereas full-length β-catenin became limited to the nuclei and cytoplasm of biliary epithelial cells. Although further work to characterize the specific function of truncated β-catenin in differentiating hepatocytes is ongoing in our laboratory, we believe at this time that full-length β-catenin may promote biliary epithelial cell differentiation and that calpain is activated in cells fated to become hepatocytes, leading to production of the truncated β-catenin species that promotes the process of hepatocyte differentiation and maturation through expression of a unique set of gene targets (Fig. 5). Reports that both stabilization of (full-length) β-catenin and its deletion lead to a defect in hepatocyte differentiation (12, 29) we believe can be explained by the necessity of the Δ95-β-catenin in this process.
FIGURE 5.

Model. Hepatoblasts in which β-catenin is truncated by calpain become hepatocytes, whereas those in which full-length β-catenin is retained become biliary epithelial cells.
Several lines of evidence support a role for β-catenin in hepatocyte differentiation. Livers of mice with a hepatoblast-specific β-catenin deletion exhibit failures in both hepatocyte as well as biliary epithelial cell differentiation (12), suggesting β-catenin is critical for differentiation of both cell types. In developing human livers, Wnt3a expression is seen in the parenchyma of second trimester livers, correlating with early hepatocyte differentiation (49). Furthermore, although a hepatic progenitor cell line established from day E14.5 mouse liver cells can differentiate into mature hepatocytes via treatment with dexamethasone, this differentiation is inhibited by the Wnt/β-catenin pathway antagonist SFRP3, lending further support to the role of β-catenin in hepatocyte differentiation (50).
Furthermore, the idea that a truncated β-catenin species may play a role in differentiation of a particular cell type is supported in the literature. Expression of a β-catenin species lacking the N-terminal tail in developing mammary gland results in either precocious lobuloalveolar development and differentiation (51) or precocious lateral bud formation, hyperproliferation, and premature differentiation of luminal epithelium during pregnancy (52), depending on the cell type targeted. Intriguingly, this phenotype is different from that induced by Wnt overexpression in the mammary gland, which would instead result in stabilization of full-length β-catenin, and induces ductal hyperbranching (53, 54). Expression of Δ89 β-catenin in the small intestine, however, resulted in increased cell division in undifferentiated cells in the proliferative compartment as well as increased apoptosis and an increase in E-cadherin at adherens junctions but no observable changes in cell fate outcomes (55).
We also uncovered a role for calpain in truncating β-catenin and modifying its activity in developing liver. Although abundant support exists for the role of calpain in differentiation of various cell types, including muscle cells, osteoblasts, and adipocytes (56–63), and there is evidence that β-catenin is a substrate for calpain in differentiating muscle cells (64), no role had been identified for calpain in hepatocyte differentiation. Unfortunately, the calpain inhibitor we employed that crosses the placenta has a very short half-life (2 h) and does not provide inhibition long enough for us to investigate effects on phenotype. Additional studies to characterize the functions of calpain-mediated β-catenin are ongoing.
β-Catenin lacks DNA binding property and relies on other transcription factors to dictate target gene expression for a specific biological response. We found that expression of a Δ95 β-catenin led to significant up-regulation expression of a set of expected and known gene targets based on the stability of the protein. However, there were several uniquely overexpressed genes in Δ95 β-catenin transfected cells, and many of these are histone H3 and H2 proteins. Since these proteins are involved in nucleosome structure and function, which is pertinent for histone-DNA interactions, and chromatin compaction, Δ89 β-catenin may eventually influence the epigenetic regulation of gene expression (65). Other genes like Cenph are known to have a role in spindle formation and chromosomal segregation during cell division (66). In addition, several of these novel targets possessed binding sites for transcription factors known to interact with β-catenin, including Hif1α, Klf4, Egr1, and NFκB. Whether alteration in the structure of β-catenin due to deletion of 95 amino acids at the N terminus allows for differential interaction with these factors to activate the hepatocyte differentiation program will be explored in the future.
Notably, we found a strong correlation between expression of N-terminal-truncated β-catenin and a more differentiated, fetal morphology in hepatoblastomas. These tumors often harbor monoallelic mutations or deletions affecting the N termini of β-catenin gene. In many cases they are missing exon 3 and/or part of exon 4, producing a truncated and stable β-catenin resembling the species described herein (31, 32, 34, 35). Presently, it remains undetermined whether the product of the two mechanisms differs or relates in their functions. However, the relationship between the presence or absence of the β-catenin N terminus and differentiation is still noteworthy, as it supports our model and extends it to a disease and may be a reliable way to distinguish embryonal from fetal HB. In addition, a separate analysis of HBs has also indicated a relationship between N-terminal deletions of β-catenin and “pure fetal” morphology (as opposed to embryonal or mixed fetal/embryonal cases), corroborating our findings. Intriguingly, a recent report indicated that expression of Lect2, one of the putative targets of truncated β-catenin, is increased in a large fraction of hepatoblastomas (67). Based on our findings, we surmise that it may correlate with those expressing N-terminal-truncated β-catenin.
In summation, we believe that our work has important implications for those attempting to produce differentiated hepatocytes ex vivo as well as for the classification of embryonic versus fetal hepatoblastoma. Furthermore, in light of the focus on β-catenin mutations in hepatic cancers, we feel it is important to challenge the idea that point mutations and deletions of exon 3 produce “activated” β-catenins equivalent in function. Our observation that expression of β-catenin lacking its N terminus appears to promote a more differentiated state in developing hepatocytes as well as hepatoblastomas lends evidence in support of the movement away from viewing β-catenin activity as typically “oncogenic” (68) and toward viewing it as highly pleiotropic demanding additional studies.
Supplementary Material
Acknowledgments
We thank John Stoops, Dr. Aaron Bell, and Sucha Singh for technical assistance. We thank Brian Hood and Dr. Tom Conrads for peptide sequencing. We also thank Dr. Roger Clem for assistance with manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R01DK62277 and 1R01CA124414 (to S. P. S. M.). This work was also supported by Rango's Fund for the Enhancement of Pathology Research.

This article contains supplemental Table 1.
- HB
- hepatoblastoma
- TCF
- T-cell factor-4
- TBST
- TBS-Tween.
REFERENCES
- 1. Dalgetty D. M., Medine C. N., Iredale J. P., Hay D. C. (2009) Progress and future challenges in stem cell-derived liver technologies. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G241–G248 [DOI] [PubMed] [Google Scholar]
- 2. Navarro-Alvarez N., Soto-Gutierrez A., Kobayashi N. (2009) Stem cell research and therapy for liver disease. Curr. Stem Cell Res. Ther. 4, 141–146 [DOI] [PubMed] [Google Scholar]
- 3. Finley K. R., Tennessen J., Shawlot W. (2003) The mouse secreted frizzled-related protein 5 gene is expressed in the anterior visceral endoderm and foregut endoderm during early post-implantation development. Gene Expr. Patterns 3, 681–684 [DOI] [PubMed] [Google Scholar]
- 4. Li Y., Rankin S. A., Sinner D., Kenny A. P., Krieg P. A., Zorn A. M. (2008) Sfrp5 coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling. Genes Dev. 22, 3050–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McLin V. A., Rankin S. A., Zorn A. M. (2007) Repression of Wnt/β-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 134, 2207–2217 [DOI] [PubMed] [Google Scholar]
- 6. Pilcher K. E., Krieg P. A. (2002) Expression of the Wnt inhibitor, sFRP5, in the gut endoderm of Xenopus. Gene Expr. Patterns 2, 369–372 [DOI] [PubMed] [Google Scholar]
- 7. Goessling W., North T. E., Lord A. M., Ceol C., Lee S., Weidinger G., Bourque C., Strijbosch R., Haramis A. P., Puder M., Clevers H., Moon R. T., Zon L. I. (2008) APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev. Biol. 320, 161–174 [DOI] [PubMed] [Google Scholar]
- 8. Hussain S. Z., Sneddon T., Tan X., Micsenyi A., Michalopoulos G. K., Monga S. P. (2004) Wnt impacts growth and differentiation in ex vivo liver development. Exp. Cell Res. 292, 157–169 [DOI] [PubMed] [Google Scholar]
- 9. Micsenyi A., Tan X., Sneddon T., Luo J. H., Michalopoulos G. K., Monga S. P. (2004) β-Catenin is temporally regulated during normal liver development. Gastroenterology 126, 1134–1146 [DOI] [PubMed] [Google Scholar]
- 10. Monga S. P., Monga H. K., Tan X., Mulé K., Pediaditakis P., Michalopoulos G. K. (2003) β-Catenin antisense studies in embryonic liver cultures. Role in proliferation, apoptosis, and lineage specification. Gastroenterology 124, 202–216 [DOI] [PubMed] [Google Scholar]
- 11. Suksaweang S., Lin C. M., Jiang T. X., Hughes M. W., Widelitz R. B., Chuong C. M. (2004) Morphogenesis of chicken liver. Identification of localized growth zones and the role of β-catenin/Wnt in size regulation. Dev. Biol. 266, 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tan X., Yuan Y., Zeng G., Apte U., Thompson M. D., Cieply B., Stolz D. B., Michalopoulos G. K., Kaestner K. H., Monga S. P. (2008) β-Catenin deletion in hepatoblasts disrupts hepatic morphogenesis and survival during mouse development. Hepatology 47, 1667–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benhamouche S., Decaens T., Godard C., Chambrey R., Rickman D. S., Moinard C., Vasseur-Cognet M., Kuo C. J., Kahn A., Perret C., Colnot S. (2006) Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev. Cell 10, 759–770 [DOI] [PubMed] [Google Scholar]
- 14. Burke Z. D., Tosh D. (2006) The Wnt/β-catenin pathway. Master regulator of liver zonation? Bioessays 28, 1072–1077 [DOI] [PubMed] [Google Scholar]
- 15. Colletti M., Cicchini C., Conigliaro A., Santangelo L., Alonzi T., Pasquini E., Tripodi M., Amicone L. (2009) Convergence of Wnt signaling on the HNF4α-driven transcription in controlling liver zonation. Gastroenterology 137, 660–672 [DOI] [PubMed] [Google Scholar]
- 16. Giera S., Braeuning A., Köhle C., Bursch W., Metzger U., Buchmann A., Schwarz M. (2010) Wnt/β-catenin signaling activates and determines hepatic zonal expression of glutathione S-transferases in mouse liver. Toxicol. Sci. 115, 22–33 [DOI] [PubMed] [Google Scholar]
- 17. Apte U., Thompson M. D., Cui S., Liu B., Cieply B., Monga S. P. (2008) Wnt/β-catenin signaling mediates oval cell response in rodents. Hepatology 47, 288–295 [DOI] [PubMed] [Google Scholar]
- 18. Hu M., Kurobe M., Jeong Y. J., Fuerer C., Ghole S., Nusse R., Sylvester K. G. (2007) Wnt/β-catenin signaling in murine hepatic transit amplifying progenitor cells. Gastroenterology 133, 1579–1591 [DOI] [PubMed] [Google Scholar]
- 19. Spee B., Carpino G., Schotanus B. A., Katoonizadeh A., Vander Borght S., Gaudio E., Roskams T. (2010) Characterization of the liver progenitor cell niche in liver diseases. Potential involvement of Wnt and Notch signaling. Gut 59, 247–257 [DOI] [PubMed] [Google Scholar]
- 20. Williams J. M., Oh S. H., Jorgensen M., Steiger N., Darwiche H., Shupe T., Petersen B. E. (2010) The role of the Wnt family of secreted proteins in rat oval “stem” cell-based liver regeneration. Wnt1 drives differentiation. Am. J. Pathol. 176, 2732–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang W., Yan H. X., Chen L., Liu Q., He Y. Q., Yu L. X., Zhang S. H., Huang D. D., Tang L., Kong X. N., Chen C., Liu S. Q., Wu M. C., Wang H. Y. (2008) Wnt/β-catenin signaling contributes to activation of normal and tumorigenic liver progenitor cells. Cancer Res. 68, 4287–4295 [DOI] [PubMed] [Google Scholar]
- 22. Apte U., Singh S., Zeng G., Cieply B., Virji M. A., Wu T., Monga S. P. (2009) β-Catenin activation promotes liver regeneration after acetaminophen-induced injury. Am. J. Pathol. 175, 1056–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Monga S. P., Pediaditakis P., Mule K., Stolz D. B., Michalopoulos G. K. (2001) Changes in WNT/β-catenin pathway during regulated growth in rat liver regeneration. Hepatology 33, 1098–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nejak-Bowen K. N., Thompson M. D., Singh S., Bowen W. C., Jr., Dar M. J., Khillan J., Dai C., Monga S. P. (2010) Accelerated liver regeneration and hepatocarcinogenesis in mice overexpressing serine 45 mutant β-catenin. Hepatology 51, 1603–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sekine S., Gutiérrez P. J., Lan B. Y., Feng S., Hebrok M. (2007) Liver-specific loss of β-catenin results in delayed hepatocyte proliferation after partial hepatectomy. Hepatology 45, 361–368 [DOI] [PubMed] [Google Scholar]
- 26. Sodhi D., Micsenyi A., Bowen W. C., Monga D. K., Talavera J. C., Monga S. P. (2005) Morpholino oligonucleotide-triggered β-catenin knockdown compromises normal liver regeneration. J. Hepatol. 43, 132–141 [DOI] [PubMed] [Google Scholar]
- 27. Tan X., Behari J., Cieply B., Michalopoulos G. K., Monga S. P. (2006) Conditional deletion of β-catenin reveals its role in liver growth and regeneration. Gastroenterology 131, 1561–1572 [DOI] [PubMed] [Google Scholar]
- 28. Monga S. P. (2011) Role of Wnt/β-catenin signaling in liver metabolism and cancer. Int. J. Biochem. Cell Biol. 43, 1021–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Decaens T., Godard C., de Reyniès A., Rickman D. S., Tronche F., Couty J. P., Perret C., Colnot S. (2008) Stabilization of β-catenin affects mouse embryonic liver growth and hepatoblast fate. Hepatology 47, 247–258 [DOI] [PubMed] [Google Scholar]
- 30. Curia M. C., Zuckermann M., De Lellis L., Catalano T., Lattanzio R., Aceto G., Veschi S., Cama A., Otte J. B., Piantelli M., Mariani-Costantini R., Cetta F., Battista P. (2008) Sporadic childhood hepatoblastomas show activation of β-catenin, mismatch repair defects and p53 mutations. Mod. Pathol. 21, 7–14 [DOI] [PubMed] [Google Scholar]
- 31. Jeng Y. M., Wu M. Z., Mao T. L., Chang M. H., Hsu H. C. (2000) Somatic mutations of β-catenin play a crucial role in the tumorigenesis of sporadic hepatoblastoma. Cancer Lett. 152, 45–51 [DOI] [PubMed] [Google Scholar]
- 32. Koch A., Denkhaus D., Albrecht S., Leuschner I., von Schweinitz D., Pietsch T. (1999) Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the β-catenin gene. Cancer Res. 59, 269–273 [PubMed] [Google Scholar]
- 33. López-Terrada D., Gunaratne P. H., Adesina A. M., Pulliam J., Hoang D. M., Nguyen Y., Mistretta T. A., Margolin J., Finegold M. J. (2009) Histologic subtypes of hepatoblastoma are characterized by differential canonical Wnt and Notch pathway activation in DLK+ precursors. Hum. Pathol. 40, 783–794 [DOI] [PubMed] [Google Scholar]
- 34. Taniguchi K., Roberts L. R., Aderca I. N., Dong X., Qian C., Murphy L. M., Nagorney D. M., Burgart L. J., Roche P. C., Smith D. I., Ross J. A., Liu W. (2002) Mutational spectrum of β-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene 21, 4863–4871 [DOI] [PubMed] [Google Scholar]
- 35. Wei Y., Fabre M., Branchereau S., Gauthier F., Perilongo G., Buendia M. A. (2000) Activation of β-catenin in epithelial and mesenchymal hepatoblastomas. Oncogene 19, 498–504 [DOI] [PubMed] [Google Scholar]
- 36. Weinberg A. G., Finegold M. J. (1983) Primary hepatic tumors of childhood. Hum. Pathol. 14, 512–537 [DOI] [PubMed] [Google Scholar]
- 37. von Schweinitz D., Hecker H., Schmidt-von-Arndt G., Harms D. (1997) Prognostic factors and staging systems in childhood hepatoblastoma. Int. J. Cancer 74, 593–599 [DOI] [PubMed] [Google Scholar]
- 38. Si-Tayeb K., Lemaigre F. P., Duncan S. A. (2010) Organogenesis and development of the liver. Dev. Cell 18, 175–189 [DOI] [PubMed] [Google Scholar]
- 39. Zorn A. M., Wells J. M. (2009) Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 25, 221–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abe K., Takeichi M. (2007) NMDA receptor activation induces calpain-mediated β-catenin cleavages for triggering gene expression. Neuron 53, 387–397 [DOI] [PubMed] [Google Scholar]
- 41. Rios-Doria J., Kuefer R., Ethier S. P., Day M. L. (2004) Cleavage of β-catenin by calpain in prostate and mammary tumor cells. Cancer Res. 64, 7237–7240 [DOI] [PubMed] [Google Scholar]
- 42. Deato M. D., Tjian R. (2008) An unexpected role of TAFs and TRFs in skeletal muscle differentiation. Switching core promoter complexes. Cold Spring Harb. Symp. Quant. Biol. 73, 217–225 [DOI] [PubMed] [Google Scholar]
- 43. Goodrich J. A., Tjian R. (2010) Unexpected roles for core promoter recognition factors in cell type-specific transcription and gene regulation. Nat. Rev. Genet. 11, 549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McConnell B. B., Bialkowska A. B., Nandan M. O., Ghaleb A. M., Gordon F. J., Yang V. W. (2009) Haploinsufficiency of Krüppel-like factor 5 rescues the tumor-initiating effect of the Apc(Min) mutation in the intestine. Cancer Res. 69, 4125–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kaidi A., Williams A. C., Paraskeva C. (2007) Interaction between β-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat. Cell Biol. 9, 210–217 [DOI] [PubMed] [Google Scholar]
- 46. Liu J., Wang H., Zuo Y., Farmer S. R. (2006) Functional interaction between peroxisome proliferator-activated receptor γ- and β-catenin. Mol. Cell. Biol. 26, 5827–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deng J., Miller S. A., Wang H. Y., Xia W., Wen Y., Zhou B. P., Li Y., Lin S. Y., Hung M. C. (2002) β-Catenin interacts with and inhibits NF-κB in human colon and breast cancer. Cancer Cell 2, 323–334 [DOI] [PubMed] [Google Scholar]
- 48. Lu D., Han C., Wu T. (2012) Microsomal prostaglandin E synthase-1 promotes hepatocarcinogenesis through activation of a novel EGR1/β-catenin signaling axis. Oncogene 31, 842–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hay D. C., Fletcher J., Payne C., Terrace J. D., Gallagher R. C., Snoeys J., Black J. R., Wojtacha D., Samuel K., Hannoun Z., Pryde A., Filippi C., Currie I. S., Forbes S. J., Ross J. A., Newsome P. N., Iredale J. P. (2008) Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 12301–12306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bi Y., Huang J., He Y., Zhu G. H., Su Y., He B. C., Luo J., Wang Y., Kang Q., Luo Q., Chen L., Zuo G. W., Jiang W., Liu B., Shi Q., Tang M., Zhang B. Q., Weng Y., Huang A., Zhou L., Feng T., Luu H. H., Haydon R. C., He T. C., Tang N. (2009) Wnt antagonist SFRP3 inhibits the differentiation of mouse hepatic progenitor cells. J. Cell. Biochem. 108, 295–303 [DOI] [PubMed] [Google Scholar]
- 51. Imbert A., Eelkema R., Jordan S., Feiner H., Cowin P. (2001) Delta N89 β-catenin induces precocious development, differentiation, and neoplasia in mammary gland. J. Cell Biol. 153, 555–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Teulière J., Faraldo M. M., Deugnier M. A., Shtutman M., Ben-Ze'ev A., Thiery J. P., Glukhova M. A. (2005) Targeted activation of β-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development 132, 267–277 [DOI] [PubMed] [Google Scholar]
- 53. Tsukamoto A. S., Grosschedl R., Guzman R. C., Parslow T., Varmus H. E. (1988) Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell 55, 619–625 [DOI] [PubMed] [Google Scholar]
- 54. Lane T. F., Leder P. (1997) Wnt-10b directs hypermorphic development and transformation in mammary glands of male and female mice. Oncogene 15, 2133–2144 [DOI] [PubMed] [Google Scholar]
- 55. Wong M. H., Rubinfeld B., Gordon J. I. (1998) Effects of forced expression of an NH2-terminal truncated β-catenin on mouse intestinal epithelial homeostasis. J. Cell Biol. 141, 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barnoy S., Glaser T., Kosower N. S. (1997) Calpain and calpastatin in myoblast differentiation and fusion. Effects of inhibitors. Biochim. Biophys. Acta 1358, 181–188 [DOI] [PubMed] [Google Scholar]
- 57. Garach-Jehoshua O., Ravid A., Liberman U. A., Reichrath J., Glaser T., Koren R. (1998) Up-regulation of the calcium-dependent protease, calpain, during keratinocyte differentiation. Br. J. Dermatol. 139, 950–957 [DOI] [PubMed] [Google Scholar]
- 58. Li J. J., Xie D. (2007) Cleavage of focal adhesion kinase (FAK) is essential in adipocyte differentiation. Biochem. Biophys. Res. Commun. 357, 648–654 [DOI] [PubMed] [Google Scholar]
- 59. Moyen C., Goudenege S., Poussard S., Sassi A. H., Brustis J. J., Cottin P. (2004) Involvement of micro-calpain (CAPN 1) in muscle cell differentiation. Int. J. Biochem. Cell Biol. 36, 728–743 [DOI] [PubMed] [Google Scholar]
- 60. Murray S. S., Grisanti M. S., Bentley G. V., Kahn A. J., Urist M. R., Murray E. J. (1997) The calpain-calpastatin system and cellular proliferation and differentiation in rodent osteoblastic cells. Exp. Cell Res. 233, 297–309 [DOI] [PubMed] [Google Scholar]
- 61. Patel Y. M., Lane M. D. (1999) Role of calpain in adipocyte differentiation. Proc. Natl. Acad. Sci. U.S.A. 96, 1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ueda Y., Wang M. C., Ou B. R., Huang J., Elce J., Tanaka K., Ichihara A., Forsberg N. E. (1998) Evidence for the participation of the proteasome and calpain in early phases of muscle cell differentiation. Int. J. Biochem. Cell Biol. 30, 679–694 [DOI] [PubMed] [Google Scholar]
- 63. Yajima Y., Kawashima S. (2002) Calpain function in the differentiation of mesenchymal stem cells. Biol. Chem. 383, 757–764 [DOI] [PubMed] [Google Scholar]
- 64. Kramerova I., Kudryashova E., Wu B., Spencer M. J. (2006) Regulation of the M-cadherin-β-catenin complex by calpain 3 during terminal stages of myogenic differentiation. Mol. Cell. Biol. 26, 8437–8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Campos E. I., Reinberg D. (2009) Histones. Annotating chromatin. Annu. Rev. Genet. 43, 559–599 [DOI] [PubMed] [Google Scholar]
- 66. Zhao X., Zhao L., Tian T., Zhang Y., Tong J., Zheng X., Meng A. (2010) Interruption of cenph causes mitotic failure and embryonic death, and its haploinsufficiency suppresses cancer in zebrafish. J. Biol. Chem. 285, 27924–27934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ovejero C., Cavard C., Périanin A., Hakvoort T., Vermeulen J., Godard C., Fabre M., Chafey P., Suzuki K., Romagnolo B., Yamagoe S., Perret C. (2004) Identification of the leukocyte cell-derived chemotaxin 2 as a direct target gene of β-catenin in the liver. Hepatology 40, 167–176 [DOI] [PubMed] [Google Scholar]
- 68. Lucero O. M., Dawson D. W., Moon R. T., Chien A. J. (2010) A re-evaluation of the “oncogenic” nature of Wnt/β-catenin signaling in melanoma and other cancers. Curr. Oncol. Rep. 12, 314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.