Background: Streptococcus pneumoniae BgaC is a GH-35 β-galactosidase of specific activity toward β(1,3)-linked galactose and N-acetylglucosamine.
Results: Three aromatic residues, Trp-240, Trp-243, and Tyr-455, determine the substrate specificity of BgaC.
Conclusion: BgaC and other GH-35 β-galactosidases adopt a similar domain organization and catalytic mechanism.
Significance: Provided is the first structural insight into the substrate specificity of β-galactosidase toward the β(1,3)-linked galactosyl bond.
Keywords: Crystal Structure, Galactose, Mutagenesis, Streptococcus pneumoniae, Virulence Factors, beta-Galactosidases, Domain Organization, Substrate Specificity
Abstract
The surface-exposed β-galactosidase BgaC from Streptococcus pneumoniae was reported to be a virulence factor because of its specific hydrolysis activity toward the β(1,3)-linked galactose and N-acetylglucosamine (Galβ(1,3)NAG) moiety of oligosaccharides on the host molecules. Here we report the crystal structure of BgaC at 1.8 Å and its complex with galactose at 1.95 Å. At pH 5.5–8.0, BgaC exists as a stable homodimer, each subunit of which consists of three distinct domains: a catalytic domain of a classic (β/α)8 TIM barrel, followed by two all-β domains (ABDs) of unknown function. The side walls of the TIM β-barrel and a loop extended from the first ABD constitute the active site. Superposition of the galactose-complexed structure to the apo-form revealed significant conformational changes of residues Trp-243 and Tyr-455. Simulation of a putative substrate entrance tunnel and modeling of a complex structure with Galβ(1,3)NAG enabled us to assign three key residues to the specific catalysis. Site-directed mutagenesis in combination with activity assays further proved that residues Trp-240 and Tyr-455 contribute to stabilizing the N-acetylglucosamine moiety, whereas Trp-243 is critical for fixing the galactose ring. Moreover, we propose that BgaC and other galactosidases in the GH-35 family share a common domain organization and a conserved substrate-determinant aromatic residue protruding from the second domain.
Introduction
The Gram-positive human pathogen Streptococcus pneumoniae is the major causative agent of acute pneumonia, otitis media, meningitis, and septicemia, which lead annually to millions of deaths worldwide (1). In the human host, S. pneumoniae encounters a variety of glycoconjugates, including mucin, host defense molecules, and glycans exposed on the epithelial surface. Like other pathogenic microbes, S. pneumoniae produces a series of secreted or surface-associated glycosidases to modify the host glycoconjugates (2–4). Genome sequencing, in combination with exploration of new virulence factors, suggests that a large number of glycosidases are necessary for the full virulence of S. pneumoniae (5, 6). For instance, three surface-exposed glycosidases, neuraminidase NanA, β-galactosidase BgaA, and N-acetyl-hexosaminidase StrH, have been identified to sequentially hydrolyze the glycoconjugates necessary for the colonization and pathogenesis of S. pneumoniae (7, 8). The action of deglycosylation is believed not only to expose the binding sites for further invasion but also to supply an abundant carbon source from the hydrolysis products (9).
Recently, a novel surface-exposed glycohydrolase, BgaC, was identified and shown to have an effect on S. pneumoniae adhesion and virulence to the human host (10, 11). BgaC is a classic β-galactosidase (EC 3.2.1.23) and shows specific hydrolysis activity toward the terminal Galβ(1,3)NAG4 moiety of oligosaccharides (10, 11). Sequence comparison suggests that BgaC falls into the glycosidase family 35 (GH-35), which mainly exists in higher eukaryotes (12). BgaC adopts a sequence and structural organization pattern similar to β-galactosidases from higher eukaryotes (such as mammalian lysosomal β-galactosidase) and microbial pathogens rather than typical prokaryotic β-galactosidases (12). The GH-35 β-galactosidases, which exist widely in all living organisms from bacteria to plants and animals (13–16), hydrolyze β(1,3)- or β(1,4)-galactosyl bonds in poly- and oligosaccharides. Some β-galactosidases are grouped in the glycosidase families GH-1, GH-2, and GH-42 in addition to GH-35 (12, 17–20). All of these families belong to a superfamily (or clan) termed GH-A, all members of which possess a catalytic domain of (α/β)8 TIM barrel fold. Two glutamic acid residues emanating from strands β4 and β7 of the barrel act as proton donor and nucleophile, and thus, this clan is sometimes referred to as the 4/7 superfamily (21). To date, several crystal structures of β-galactosidases have been deposited in the Protein Data Bank (PDB) database. These include Escherichia coli β-gal (22), Arthrobacter sp. C2-2 C221 β-gal (23), Kluyveromyces lactis β-gal (24), Thermus thermophilus A4 β-gal (25), Bacillus circulans sp. alkalophilus β-gal (26), Sulfolobus solfataricus β-gal (27), Penicillium sp. β-gal (28), Bacteroides thetaiotaomicron β-gal,5 Trichoderma reesei β-gal (29), and Homo sapiens β-gal (30) (Table 1). Most of these enzymes possess specific hydrolysis activity toward β(1,4)-galactosyl bond, whereas H. sapiens β-gal shows hydrolysis activity toward both β(1,3)- and β(1,4)-galactosyl bonds (31–33), and the substrate specificity of B. thetaiotaomicron β-gal remains unknown. E. coli β-gal, as a member of GH-2, is one of the most widely investigated β-galactosidases. The structure-based catalytic mechanism of E. coli β-gal has been elucidated (22, 34, 35). However, the mechanism of the substrate specificity toward the β(1,3)-galactosyl bond remains unknown.
TABLE 1.
The β-galactosidases of known structure
| Protein | GH family | PDB code | Reference |
|---|---|---|---|
| S. solfataricus β-gal | GH-1 | 1UWQ | Ref. 27 |
| E. coli β-gal | GH-2 | 1BGM | Ref. 22 |
| Arthrobacter sp. C2-2 C221 β-gal | GH-2 | 1YQ2 | Ref. 23 |
| K. lactis β-gal | GH-2 | 3OBA | Ref. 24 |
| Penicillium sp. β-gal | GH-35 | 1TG7 | Ref. 28 |
| B. thetaiotaomicron β-gal | GH-35 | 3D3A | Unpublisheda |
| T. reesei β-gal | GH-35 | 3OG2 | Ref. 29 |
| H. sapiens β-gal | GH-35 | 3THC | Ref. 30 |
| T. thermophilus A4 β-gal | GH-42 | 1KWG | Ref. 25 |
| B. circulans sp. alkalophilus β-gal | GH-42 | 3TTS | Ref. 26 |
a K. Palani, D. Kumaran, S. K. Burley, and S. Swaminathan, unpublished results.
Here we present the crystal structure of BgaC at 1.8 Å and its complex with galactose at 1.95 Å. BgaC is composed of 595 amino acid residues, sharing a significant sequence homology with other GH-35 members. It contains three domains: a typical TIM barrel catalytic domain, similar to other β-galactosidases, followed by two all-β domains of unknown function. This pattern of domain organization is quite similar to that of B. thetaiotaomicron β-gal (PDB code 3D3A) and H. sapiens β-gal (30). The galactose is stabilized at the center of the catalytic domain by residues conserved in all GH-35 members. A couple of residues at the center of the TIM barrel and a loop extended from the second domain make up the active site. In addition, a putative substrate entrance tunnel identified two important loops surrounding the manually constructed Galβ(1,3)NAG at the active site. Subsequent enzymatic activity assays enabled us to characterize three key residues, Trp-240, Trp-243, and Tyr-455, that contribute to the substrate specificity toward Galβ(1,3)NAG.
EXPERIMENTAL PROCEDURES
Overexpression and Purification of BgaC and Mutants
The coding sequence of BgaC/Sp_0060 was amplified from the genomic DNA of S. pneumoniae TIGR4 and cloned into a pET28a-derived expression vector with an N-terminal His6 tag. The construct was transformed into E. coli strain BL21-RIL (DE3) (Novagen), growing at 37 °C in 2× YT culture medium (5 g of NaCl, 16 g of Bacto-tryptone, and 10 g of yeast extract/liter) containing 30 μg/ml kanamycin and 34 μg/ml chloramphenicol. When the A600 nm reached about 1.0, expression of the recombinant proteins was induced with 0.2 mm isopropyl β-d-1-thiogalactopyranoside (IPTG) for another 20 h at 16 °C before harvesting. Cells were collected and resuspended in 40 ml of lysis buffer (20 mm Tris-Cl, pH 7.5, 100 mm NaCl). After 20 min of sonication and centrifugation at 12,000 × g for 30 min, the supernatant containing the soluble target protein was collected and loaded onto a nickel-NTA column (GE Healthcare) equilibrated with the binding buffer (20 mm Tris-Cl, pH 7.5, 100 mm NaCl). The target protein was eluted with 500 mm imidazole, and further loaded onto a Superdex 200 column (GE Healthcare) pre-equilibrated with 20 mm Tris-Cl, pH 7.5, 100 mm NaCl. Fractions containing the target protein were combined and concentrated to 10 mg/ml for crystallization. Samples for enzymatic activity assays were collected at low concentrations (1 mg/ml). The purity of protein was assessed by SDS-PAGE, and the protein sample was stored at −80 °C. The mutants were expressed, purified, and stored in the same manner as the wild-type protein.
Dynamic Light Scattering
Dynamic light scattering was carried out on a DYNAPRO-99 (Wyatt Technology Corp.) with a 532-nm green laser. Each sample was measured in single-use UV-plastic cuvettes (Wyatt Technology Corp.), first equilibrated for 2 min at 25 °C, after which a time scale of the scattered light intensity fluctuations was measured. The molecular weight was analyzed with the use of the software Dynamic V6 (Wyatt Technology Corp.).
Enzymatic Activity Assays
The enzymatic activity assays of recombinant BgaC and its mutants were conducted using 4-nitrophenyl-β-d-galactopyranoside (PNPG) (Sangon) as substrate and following the previous procedures (36) with minor modifications. The reactions were performed at 37 °C in buffer containing 50 mm Na2HPO4/NaH2PO4, pH 6.5, and initiated by the addition of BgaC. Using a DU800 spectrophotometer (Beckman Coulter), the changes in absorption at 420 nm were monitored continuously, and the increase of chromogenic product 4-nitrophenol was calculated subsequently according to a standard curve of 4-nitrophenol. The final Michaelis-Menten parameters (Vmax and Km) were extracted from these data by nonlinear fitting to the Michaelis-Menten equation with the program Origin 7.5.
Preparation of 1-Phenyl-3-methyl-5-pyrazolone Derivatives of Saccharides
1-Phenyl-3-methyl-5-pyrazolone derivation of saccharides complied with the previously reported procedures (37, 38) with minor changes. Briefly, 10 μl of reaction system was mixed with an equal volume of 0.3 m NaOH and 0.5 m methanol solution of 1-phenyl-3-methyl-5-pyrazolone, respectively. A total volume of 30 μl of mixture was placed at 70 °C to react for 35 min and then cooled to room temperature and neutralized with 10 μl of 0.3 m HCl. The obtained solution was dissolved in 100 μl of chloroform. After vigorous shaking and centrifugation, the supernatant containing derivatives was carefully transferred to another 100 μl of chloroform, and this extraction process was repeated three times; then the aqueous layer was diluted to 80 μl with water before HPLC analysis.
HPLC Analysis
The assays toward specific substrate were performed at 37 °C in a 10-μl system containing a buffer of 50 mm Na2HPO4/NaH2PO4, pH 6.5, and the disaccharide Galβ(1,3)NAG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at various concentrations. The reactions were initiated by the addition of enzymes and terminated by mixing with an equal volume of 0.3 m NaOH. After 1-phenyl-3-methyl-5-pyrazolone derivatization as described above, the mixture was centrifuged at 12,000 × g for 10 min, and 10 μl of supernatant was applied to the HPLC system (Agilent 1200 Series). Buffer composed of 20% acetonitrile and 80 mm Na2HPO4/NaH2PO4, pH 7.0, was used for equilibration of the column (Eclipse XDS-C18 column, 4.6 × 150 mm; Agilent) and separation of the components at a flow rate of 1 ml/min. The sugar components of the reaction system were determined by comparison with the retention time of standard monosaccharides. The NAG standard curve was made by quantitative analysis of HPLC with a series of concentrations ranging from 0.1 to 5 mm. The final enzymatic kinetic parameters were calculated based on the yield of NAG.
Crystallization, Data Collection, and Processing
The apo-form BgaC was concentrated to 10 mg/ml by ultrafiltration (Millipore Amicon) for crystallization. Crystals were grown at 289 K using the hanging drop vapor diffusion method, with the initial condition of mixing 1 μl of protein solution with an equal volume of the reservoir solution (30% polyethylene glycol 3350, 0.2 m ammonium acetate, 0.1 m sodium citrate tribasic dehydrate, pH 5.6). The crystals were transferred to cryoprotectant (reservoir solution supplemented with 25% glycerol) and flash-cooled with liquid nitrogen. The crystal of its complex with galactose was obtained by quick cryo-soaking technique. The crystal was immersed in the solution containing 100 mm galactose for about 300 s and mounted in a rayon loop. The diffraction images of the apo-form and galactose-binding form were recorded at 100 K in a liquid nitrogen stream using the beamline at the Shanghai Synchrotron Radiation Facility. The data sets were integrated and scaled with the program HKL2000.
Structure Solution and Refinement
The structure of BgaC was determined by molecular replacement with MOLREP using the coordinates of 38% sequence-identical B. thetaiotaomicron β-galactosidase (PDB code 3D3A) as the search model. The initial model was further refined by using the maximum likelihood method implemented in REFMAC5 as part of the CCP4 program suite and rebuilt interactively by using the σA-weighted electron density maps with coefficients mFo − DFc and mFo − DFc in the program COOT. The galactose complex structure was refined with the Refinement program from PHENIX and rebuilt interactively with COOT. The final model was evaluated with the programs Molprobity and Procheck. The data collection and structure refinement statistics are listed in Table 2. All structure figures were prepared with the program PyMOL (39).
TABLE 2.
Crystal parameters, data collection, and structure refinement statistics
| Data processing | BgaC | BgaC + galactose |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Unit cell (Å) | 79.97, 79.30, 99.39 | 80.35, 82.37, 99.60 |
| Unit cell (degrees) | 90.00, 106.94, 90.00 | 90.00, 106.84, 90.00 |
| Resolution range (Å) | 50.00–1.80 | 50.00–1.95 |
| Unique reflections | 108,735 (10,964)a | 88,435 (8,053) |
| Completeness (%) | 98.8 (100.0) | 96.8 (88.7) |
| 〈I/σ(I)〉 | 14.3 (5.4) | 13.3 (3.7) |
| Rmergeb (%) | 7.0 (24.1) | 6.2 (27.2) |
| Average redundancy | 3.1 (3.0) | 2.4 (2.3) |
| Structure refinement | ||
| Resolution range (Å) | 46.98–1.80 | 41.26–1.95 |
| R-factorc/R-freed (%) | 17.37/20.99 | 20.49/24.30 |
| No. of protein atoms | 9,697 | 9,718 |
| No. of water atoms | 887 | 597 |
| RMSDe bond lengths (Å) | 0.005 | 0.006 |
| RMSD bond angles (degrees) | 0.929 | 0.941 |
| Mean B-factors (Å2) | 19.6 | 40.6 |
| Ramachandran plot (residues)f | ||
| Most favored (%) | 96.8 | 96.9 |
| Additional allowed (%) | 3.2 | 3.1 |
| PDB entry | 4E8D | 4E8C |
a The values in parentheses refer to statistics in the highest bin.
b Rmerge = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity of an observation and 〈I(hkl)〉 is the mean value for its unique reflection. Summations are over all reflections.
c R-factor = Σh|Fo(h) − Fc(h) /ΣhFo(h), where Fo and Fc are the observed and calculated structure-factor amplitudes, respectively.
d R-free was calculated with 5% of the data excluded from the refinement.
e Root mean square deviation from ideal values.
f Categories were defined by Molprobity.
RESULTS AND DISCUSSION
Overall Structure
Each asymmetric unit contains two molecules of BgaC, which are quite similar to each other with an overall root mean square deviation (RMSD) of 0.2 Å over 540 Cα atoms. These two molecules do not form a dimer because of their small, buried interface area of 566 Å2. However, symmetric operation enabled us to define a homodimer of BgaC in the crystal structure with a total buried interface area of 2874 Å2 (Fig. 1A). In fact, BgaC also exists as a dimer in solution, as confirmed by gel filtration chromatography and dynamic light scattering (data not shown). Two subunits in the homodimer are related by a noncrystallographic 2-fold symmetry axis running parallel to the depth direction of each molecule. The dimer interface is composed of two loops and α6 from the catalytic domain in addition to four loops and two β-strands of the first all-β domain (ABD-1). This face-to-face dimer form is quite different from that of H. sapiens β-gal, which forms a back-to-back dimer with an interface made up of the catalytic domain, linker, and ABD-2 (30). Upon increasing the pH to 8.5, the BgaC dimer will dissociate, resulting in the inactivation of the enzyme (data not shown), in agreement with a previous report (11). Further investigation showed that BgaC exists as a stable homodimer at a pH range of 5.5–8.0, indicating the physiological condition to make BgaC active.
FIGURE 1.
Overall structure of BgaC and its comparison with the other two GH-35 β-galactosidases. A, BgaC homodimer. The dimer is viewed along the noncrystallographic 2-fold axis. The three domains are colored in cyan, red, and yellow, respectively. The linker between the first and second domain is colored in blue. B, structural comparison with H. sapiens β-gal and Penicillium sp. β-gal. H. sapiens β-gal and Penicillium sp. β-gal are shown in the same view as BgaC. The corresponding domains are colored as those of BgaC, whereas the second domain and the third domain of Penicillium sp. β-gal are colored in green and orange, respectively.
Each subunit of BgaC is composed of three distinct domains, a catalytic domain patched on one side by two all-β domains (ABDs) (Fig. 1A). The catalytic domain (Thr-2 to Glu-342) adopts a typical (β/α)8 TIM barrel with some distortions. Following the catalytic domain, a stretch of residues, Ser-337 to Ser-365, containing a short β-strand passes through the second ABD (Leu-489 to Lys-591), prior to joining the first ABD (Ser-366 to Pro-488). Both ABDs, which were previously described as a jellyroll fold (40), share a quite similar overall structure, with an RMSD of 6.4 Å over 56 Cα atoms. However, each ABD adopts a β-sandwich composed of a five-stranded β-sheet against a three-strand one, which is somewhat different from the regular β-sandwich with two layers of four-stranded β-sheets. In addition, the stretch linking the catalytic domain and the first ABD contributes a β-strand to the second ABD (Fig. 1A).
Structural Comparison with Other GH-35 β-Galactosidases
To date, four crystal structures of β-galactosidase in the GH-35 family have been deposited in the PDB. Two of them, B. thetaiotaomicron β-gal (PDB code 3D3A) and H. sapiens β-gal (30), possess an overall structure of three domains similar to the 595-residue BgaC, with an RMSD of 1.8 Å and 1.7 Å over Cα atoms, respectively. By contrast, the other two structures, the 1011-residue Penicillium sp. β-gal (28) as well as the 1003-residue T. reesei β-gal (29), are composed of five domains, a catalytic domain wrapped by four ABDs. Structural superposition shows that the first and second ABDs of Penicillium sp. β-gal or T. reesei β-gal are missing in BgaC and replaced by a long stretch connecting the catalytic domain and the last two domains (Fig. 1B). Superposition of the catalytic domain of BgaC against that of Penicillium sp. β-gal and T. reesei β-gal yields the same RMSD of 1.9 Å over 312 Cα atoms and 314 Cα, respectively. The two ABDs of BgaC could be aligned to the last two domains of Penicillium sp. β-gal and T. reesei β-gal, yielding an RMSD of 2.9 and 2.8 Å over 220 and 218 Cα atoms, respectively.
The Active Site
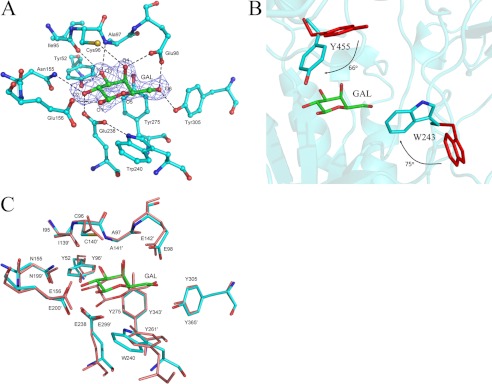
In the 1.95 Å galactose-complexed structure of BgaC, a molecule of galactose fit well into the active site within the TIM barrel (Fig. 2A). The galactose adopts a chair conformation with its O1 in the β-anomer configuration. Four aromatic residues, Tyr-52, Trp-240, Tyr-275, and Tyr-305 form a hydrophobic pocket to accommodate the hexose ring of galactose through stacking interactions. In addition, residues Tyr-52, Ile-95, Cys-96, Ala-97, Glu-98, Asn-155, Glu-156, Glu-238, and Tyr-305 form a hydrogen bond network to fix the hydroxyl groups of galactose.
FIGURE 2.
The active site. A, the galactose binding site of BgaC. The galactose molecule is superimposed with an omit electron density map (mFo − DFc, ϕcalc) contoured at 1 σ. The active site residues are shown as sticks. The polar interactions are indicated by dashed lines. B, induced fit upon galactose binding. The active site residues in the apo-form and galactose-binding form are colored in red and cyan, respectively. The side chains of Trp-243 and Tyr-455 rotate toward galactose, at an angle of 75º and 66º, respectively. C, superposition of the active sites of BgaC (cyan) and Penicillium sp. β-gal (salmon).
Superposition of the complex structure to the apo-form yields an RMSD of 0.20 Å over 551 Cα atoms, indicating no significant conformational changes of the overall structure upon galactose binding. Despite the fact that the galactose-binding residues do not undergo conformational shifts, two residues, Trp-243 and Tyr-455, close to the active site exhibit different conformations (Fig. 2B). As a result of induced fit, the side chains of Trp-243 and Tyr-455 rotate toward galactose, at an angle of 75º and 66º, respectively. Furthermore, assays of enzymatic activity toward the general substrate PNPG showed that the mutants W243A and Y455A have a much higher Km (200- and 20-fold, respectively) but a similar Vmax value compared with the wild type (WT) (Table 3). We also deleted the two ABDs to check whether the catalytic domain alone could execute the hydrolysis activity. Results showed that deletion of the two ABDs completely abolished the activity toward PNPG, which might result from the breaking of the dimer interface and/or the integrity of the active site pocket.
TABLE 3.
Kinetic parameters of BgaC and mutants towards PNPG and Galβ(1–3)NAG
| Substrate enzyme | PNPG |
Galβ(1–3)NAG |
||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| μm | s−1 | ×10−4 s−1·μm−1 | mm | min−1 | min−1·mm−1 | |
| WT | 74.9 ± 2.3 | 3.69 ± 0.0 | 493 ± 18 | 1.66 ± 0.2 | 54.0 ± 1.9 | 32.6 ± 2.0 |
| Y455A | 1370 ± 100 | 2.07 ± 0.0 | 15.2 ± 0.9 | NDa | ND | ND |
| W240A | ND | ND | ND | ND | ND | ND |
| W240F | ND | ND | ND | ND | ND | ND |
| W240Y | 832 ± 68 | 2.23 ± 0.0 | 26.9 ± 1.7 | ND | ND | ND |
| W243A | 15,100 ± 700 | 4.67 ± 0.1 | 3.31 ± 0.0 | ND | ND | ND |
a ND, not determined.
Most active site residues of BgaC could be superimposed to those in the structure of Penicillium sp. β-gal in complex with galactose (28). The proton donor Glu-200′ and the nucleophile Glu-299′ of Penicillium sp. β-gal could be well superimposed to Glu-156 and Glu-238 of BgaC, respectively (Fig. 2C). In addition, Tyr-52, Ile-95, Glu-98, Asn-155, Tyr-275, and Tyr-305 took the same conformations as their counterparts in Penicillium sp. β-gal. Nevertheless, there are some differences at the active site between the two structures. For instance, Asn-140′ of Penicillium sp. β-gal is substituted in BgaC by Cys-96, which forms a hydrogen bond with the O4 of galactose via the thiol group. In addition, Tyr-261′ in Penicillium sp. β-gal is positioned at the loop following the sixth β-strand of the TIM barrel, and its hydroxyl group hydrogen bonds with the Oϵ1 of Glu-299′. However, the counterpart Trp-240 in BgaC comes from the end of the seventh β-strand, and the Nϵ1 of Trp-240 hydrogen bonds with the Oϵ1 of Glu-238 (Fig. 2A). To verify the role of Trp-240 in hydrolysis reaction, we constructed three single mutants, W240A, W240F, and W240Y. The activities toward the general substrate PNPG of both W240A and W240F mutants were not detectable, whereas the W240Y mutant retained 5% hydrolysis activity (Table 3). Multiple-sequence alignment showed that Trp-240 in BgaC is substituted by an aromatic residue, Tyr-270 in H. sapiens β-gal or Tyr-303 in Arabidopsis thaliana β-gal (Fig. 3). Moreover, in the structure of H. sapiens β-gal, the hydroxyl group of Tyr-270 also hydrogen-bonds with the Oϵ1 of the nucleophile Glu-268 (30). We suggest that Trp-240 not only contributes to the hydrophobic pocket but also maintained, via a hydrogen bond, the orientation of the acetyl group of Glu-238, which is crucial for the hydrolysis activity.
FIGURE 3.
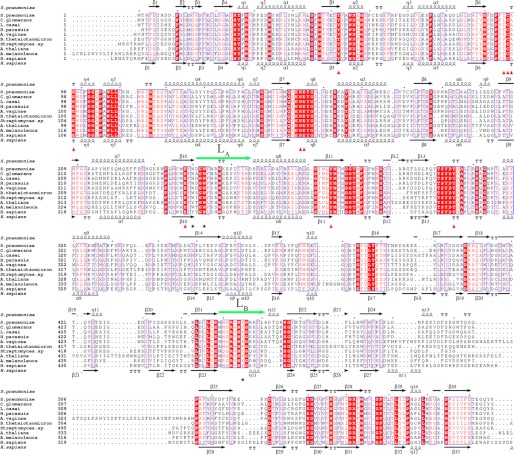
Multiple-sequence alignment of BgaC against other GH-35 β-galactosidases from different organisms. The multisequence alignment was performed with the programs Multalin (45) and Espript (46). The secondary structural elements of BgaC and H. sapiens β-gal are displayed above and below the sequences, respectively. The residues participating in galactose binding are labeled with red triangles. Residues Trp-240, Trp-243, and Tyr-455 in BgaC are marked with black asterisks. The two loops LA and LB are indicated with green arrows. All sequences were downloaded from the NCBI database. The sequences are as follows (NCBI accession numbers in parentheses): S. pneumoniae BgaC (NP_344609.1), Coriobacterium glomerans PW2 β-galactosidase (YP_004373450.1), Lactobacillus paracasei subsp. paracasei 8700:2 glycosyl hydrolase (ZP_04672354.1), Haemophilus parasuis 29755 β-galactosidase (ZP_02478970.1), Atopobium vaginae PB189-T1-4 β-galactosidase family protein (ZP_07319975.1), B. thetaiotaomicron VPI-5482 β-galactosidase (NP_809203.1), Streptomyces sp. Mg1 β-galactosidase (ZP_04999740.1), A. thaliana β-galactosidase 17 (NP_565051.1), A. melanoleuca hypothetical protein PANDA_005062 (EFB12921.1), H. sapiens β-d-galactosidase precursor (AAA51819.1).
Key Residues in the Substrate Specificity toward Galβ(1,3)NAG
To date, all β-galactosidases of known structure are reported to specifically catalyze the β(1,4)-galactosyl bond, with the exceptions of S. solfataricus β-gal from GH-1 and H. sapiens β-gal from GH-35. S. solfataricus β-gal was reported to have a broad spectrum of β-glycosidase substrate specificity toward galactose, glucose, fucose, or xylose (41), whereas H. sapiens β-gal showed hydrolysis activity toward both β(1,3)- and β(1,4)-galactosyl bonds (31).
In the best studied β(1,4)-galactosidase, E. coli β-gal, the substrate-binding site was dissected into two subsites, termed subsite −1 and +1, respectively (42). The aromatic residues at the +1 site were proposed to be critical for substrate specificity. Taking E. coli β-gal as an example, Trp-999 at the +1 site was reported to contribute to stabilizing the direction of +1 glucose by stacking interactions, to facilitate the cleavage of the β(1,4)-galactosyl linkage (43). In the other two GH-2 members, C221 β-gal and K. lactis β-gal, Trp-999 of E. coli β-gal is substituted by Cys-999 and Cys-1001, respectively, which were proposed to have an influence on substrate binding and activity (23, 24). As for A4 β-gal and B. circulans sp. alkalophilus β-gal from GH-42, residues Trp-320 and Trp-315 were proposed to act as the counterpart of Trp-999 in E. coli β-gal (25, 26).
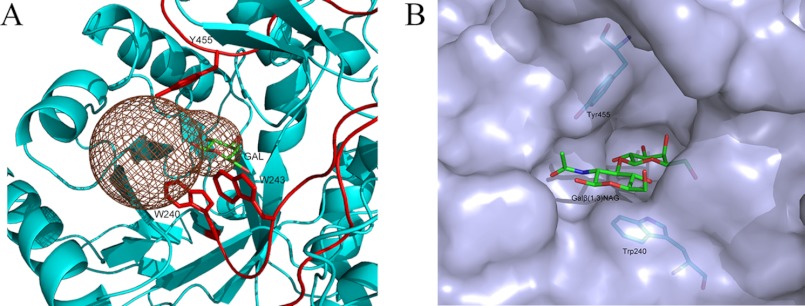
However, the catalytic mechanism of β(1,3)-galactosidases remains unclear, although the structures of H. sapiens β-gal (30) and B. thetaiotaomicron β-gal (PDB code 3D3A) are known. To decipher the structural basis of the substrate specificity toward the β(1,3)-galactosyl bond, we attempted to solve the complex structure of BgaC with the substrate Galβ(1,3)NAG, but we did not succeed. As an alternative, we calculated a putative substrate entrance tunnel with the program CAVER (available on the World Wide Web). As shown in Fig. 4A, a dumbbell-shaped tunnel from the surface to the active site pocket was guarded by two loops, LA (Trp-240 to Asp-255) and LB (Glu-448 to Ala-461). Loop LA between β10 and α8 came from the catalytic domain, whereas LB connecting β21 and η12 protruded from the first ABD (Fig. 3). A close look enabled us to find three aromatic residues, Trp-240, Tyr-455, and Trp-243, at the gorge of the tunnel (Fig. 4A).
FIGURE 4.
A putative model of Galβ(1,3)NAG binding to BgaC. A, a putative substrate entrance tunnel calculated by the program CAVER denoted in brown mesh. Residues Trp-240, Trp-243, and Tyr-455 are shown as sticks. B, a simulated model of Galβ(1,3)NAG in the binding pocket.
Furthermore, by superimposing the Gal moiety of Galβ(1,3)NAG onto the galactose molecule in the BgaC-galactose complex structure and docking the NAG moiety at subsite +1 in an optimal steric geometry, we manually constructed a model of Galβ(1,3)NAG at the active site (Fig. 4B). The model shows that the hexose ring of the NAG moiety was almost in the same plane of the galactose moiety, sandwiched by residues Trp-240 and Tyr-455 on two sides, respectively. The nitrogen atom of the N-acetyl group formed a hydrogen bond with Oϵ2 of Glu-156. The distances from +1 NAG to Trp-240 and Tyr-455 were about 3.0 and 4.5 Å, respectively.
Although both residues Trp-240 and Tyr-455 are generally conserved from bacteria to plants and animals (Fig. 3), Trp-240 of BgaC is substituted by Tyr-270 in H. sapiens β-gal (33) or Tyr-303 in A. thaliana BgaL17 (A. thaliana β-gal) (44), and Tyr-455 of BgaC is replaced by Phe-473 in B. thetaiotaomicron. H. sapiens β-gal was reported to have hydrolysis activity toward various substrates, including the Galβ(1,3)NAG moiety of ganglioside GM1, and the β(1,4)-galactosyl bond in keratin sulfate (31–33). To verify the putative role of Trp-240 and Tyr-455 of BgaC, we determined the enzymatic activities toward Galβ(1,3)NAG of several single mutants. The results indicated that neither mutant W240A nor mutant W240F showed any activity (Table 3). By contrast, the mutant W240Y retained only about 15% activity relative to the WT, with the result that the related kinetic parameters could not be determined (data not shown). The mutant Y455A completely lost its hydrolysis activity (Table 3); however, the activity of the Y455F mutant was comparable with the WT. These results confirmed that both Trp-240 and Tyr-455 play an important role in the substrate binding and activity.
Domain Organization
After a comprehensive comparison of all β(1,3)- and β(1,4)-galactosidases of known structure, we found that all members from GH-35, no matter whether they hydrolyze the β(1,3)- or β(1,4)-galactosyl bond, exhibit a similar domain organization (Fig. 5). Taking BgaC as an example of β(1,3)-galactosidases, the catalytic TIM barrel domain makes up the first domain, where the two key residues, Trp-240 and Trp-243, are located, followed by a long loop that stretches through the second ABD to the first ABD from where the loop LB extends. Loop LB not only contains the substrate specificity determinant residue Tyr-455 but also constitutes a part of the active site pocket (Fig. 1). Similar to BgaC, H. sapiens β-gal and B. thetaiotaomicron β-gal also exhibit an arrangement of three domains. The key residues (Tyr-270 and Trp-273 in H. sapiens β-gal or Trp-261 and Trp-264 in B. thetaiotaomicron β-gal) at the catalytic domain are structurally conserved. Moreover, Tyr-485 of H. sapiens β-gal and Phe-473 of B. thetaiotaomicron β-gal, corresponding to Tyr-455 in BgaC, also reside at a loop that extends from the second ABD. According to the results of multiple-sequence alignment (Fig. 3), the β-galactosidases from Lactobacillus casei, Ailuropoda melanoleuca, and A. thaliana could also be divided into three domains with the same domain organization as BgaC (Fig. 5).
FIGURE 5.
Domain organization of different β-galactosidases from GH-35. The boundaries of each domain were defined according to the previous reports or sequence alignment. The key residues Trp-240, Trp-243, and Tyr-455 of BgaC and their counterparts are labeled. Sp, S. pneumoniae; Hs, H. sapiens; Bt, B. thetaiotaomicron; Lc, L. casei; At, A. thaliana; Am, A. melanoleuca; Psp, Penicillium sp.; Tr, T. reesei; Ac, A. candidus.
With regard to the β(1,4)-galactosidases in GH-35, Penicillium sp. β-gal and T. reesei β-gal also adopt a domain organization similar to that of BgaC, although they both have two extra ABDs. As a counterpart to the first ABD of BgaC, the third ABD of these proteins also extends a loop into the active site. The corresponding residues, Trp-809 in Penicillium sp. β-gal and Trp-811 in T. reesei β-gal, were predicted to play the same roles in the substrate hydrolysis specificity as Tyr-455 in BgaC, according to the structural superposition. The residues Tyr-261 and Phe-265 in Penicillium sp. β-gal or Tyr-260 and Phe-264 in T. reesei β-gal could superimpose well onto Trp-240 and Trp-243 in the catalytic domain of BgaC, respectively. The Aspergillus candidus β-galactosidase was predicted to contain five domains, similar to those of Penicillium sp. β-gal, and three conserved residues, Trp-260, Phe-264, and Trp-806, in the catalytic domain and the third ABD.
Acknowledgments
We thank the staff at the Shanghai Synchrotron Radiation Facility for assistance. We also thank Yong-Xing He and Peng-Chao Guo for help with data collection.
This work was supported by Ministry of Science and Technology of China Grant 2009CB918804 and National Natural Science Foundation of China Grant 30870488.
The atomic coordinates and structure factors (codes 4E8C and 4E8D) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
K. Palani, D. Kumaran, S. K. Burley, and S. Swaminathan, unpublished results.
- Galβ(1,3)NAG
- β(1,3)-linked galactose and N-acetylglucosamine
- NAG
- N-acetylglucosamine
- GH
- glycosidase
- RMSD
- root mean square deviation
- ABD
- all-β domain
- PNPG
- 4-nitrophenyl-β-d-galactopyranoside
- PDB
- Protein Data Bank
- GM1
- monosialotetrahexosyl.
REFERENCES
- 1. Kadioglu A., Weiser J. N., Paton J. C., Andrew P. W. (2008) The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6, 288–301 [DOI] [PubMed] [Google Scholar]
- 2. Andersson B., Gray B., Eden C. S. (1988) Role of attachment for the virulence of Streptococcus pneumoniae. Acta Otolaryngol. Suppl. 454, 163–166 [DOI] [PubMed] [Google Scholar]
- 3. Cámara M., Boulnois G. J., Andrew P. W., Mitchell T. J. (1994) A neuraminidase from Streptococcus pneumoniae has the features of a surface protein. Infect. Immun. 62, 3688–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zähner D., Hakenbeck R. (2000) The Streptococcus pneumoniae β-galactosidase is a surface protein. J. Bacteriol. 182, 5919–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hava D. L., Camilli A. (2002) Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406 [PMC free article] [PubMed] [Google Scholar]
- 6. Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., Heidelberg J., DeBoy R. T., Haft D. H., Dodson R. J., Durkin A. S., Gwinn M., Kolonay J. F., Nelson W. C., Peterson J. D., Umayam L. A., White O., Salzberg S. L., Lewis M. R., Radune D., Holtzapple E., Khouri H., Wolf A. M., Utterback T. R., Hansen C. L., McDonald L. A., Feldblyum T. V., Angiuoli S., Dickinson T., Hickey E. K., Holt I. E., Loftus B. J., Yang F., Smith H. O., Venter J. C., Dougherty B. A., Morrison D. A., Hollingshead S. K., Fraser C. M. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506 [DOI] [PubMed] [Google Scholar]
- 7. King S. J., Hippe K. R., Weiser J. N. (2006) Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59, 961–974 [DOI] [PubMed] [Google Scholar]
- 8. Tong H. H., McIver M. A., Fisher L. M., DeMaria T. F. (1999) Effect of lacto-N-neotetraose, asialoganglioside-GM1, and neuraminidase on adherence of otitis media-associated serotypes of Streptococcus pneumoniae to chinchilla tracheal epithelium. Microb. Pathog. 26, 111–119 [DOI] [PubMed] [Google Scholar]
- 9. Burnaugh A. M., Frantz L. J., King S. J. (2008) Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol 190, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Terra V. S., Homer K. A., Rao S. G., Andrew P. W., Yesilkaya H. (2010) Characterization of novel β-galactosidase activity that contributes to glycoprotein degradation and virulence in Streptococcus pneumoniae. Infect. Immun. 78, 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jeong J. K., Kwon O., Lee Y. M., Oh D. B., Lee J. M., Kim S., Kim E. H., Le T. N., Rhee D. K., Kang H. A. (2009) Characterization of the Streptococcus pneumoniae BgaC protein as a novel surface β-galactosidase with specific hydrolysis activity for the Galβ1–3GlcNAc moiety of oligosaccharides. J. Bacteriol. 191, 3011–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Henrissat B., Davies G. (1997) Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 [DOI] [PubMed] [Google Scholar]
- 13. Dickson R. C., Dickson L. R., Markin J. S. (1979) Purification and properties of an inducible β-galactosidase isolated from the yeast Kluyveromyces lactis. J. Bacteriol 137, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kestwal R. M., Bhide S. V. (2007) Purification of β-galactosidase from Erythrina indica. Involvement of tryptophan in active site. Biochim. Biophys. Acta 1770, 1506–1512 [DOI] [PubMed] [Google Scholar]
- 15. Shaikh S. A., Khire J. M., Khan M. I. (1999) Characterization of a thermostable extracellular beta-galactosidase from a thermophilic fungus Rhizomucor sp. Biochim. Biophys. Acta 1472, 314–322 [DOI] [PubMed] [Google Scholar]
- 16. Wang Z. H., Zeng B., Shibuya H., Johnson G. S., Alroy J., Pastores G. M., Raghavan S., Kolodny E. H. (2000) Isolation and characterization of the normal canine β-galactosidase gene and its mutation in a dog model of GM1-gangliosidosis. J. inherit. Metab. Dis. 23, 593–606 [DOI] [PubMed] [Google Scholar]
- 17. Davies G., Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 18. Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henrissat B., Bairoch A. (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henrissat B., Bairoch A. (1996) Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316, 695–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jenkins J., Lo Leggio L., Harris G., Pickersgill R. (1995) β-glucosidase, β-galactosidase, family A cellulases, family F xylanases, and two barley glycanases form a superfamily of enzymes with 8-fold β/α architecture and with two conserved glutamates near the carboxyl-terminal ends of β-strands four and seven. FEBS Lett. 362, 281–285 [DOI] [PubMed] [Google Scholar]
- 22. Jacobson R. H., Zhang X. J., DuBose R. F., Matthews B. W. (1994) Three-dimensional structure of β-galactosidase from E. coli. Nature 369, 761–766 [DOI] [PubMed] [Google Scholar]
- 23. Skálová T., Dohnálek J., Spiwok V., Lipovová P., Vondrácková E., Petroková H., Dusková J., Strnad H., Králová B., Hasek J. (2005) Cold-active β-galactosidase from Arthrobacter sp. C2-2 forms compact 660-kDa hexamers. Crystal structure at 1.9 Å resolution. J. Mol. Biol. 353, 282–294 [DOI] [PubMed] [Google Scholar]
- 24. Pereira-Rodríguez A., Fernández-Leiro R., González-Siso M. I., Cerdán M. E., Becerra M., Sanz-Aparicio J. (2012) Structural basis of specificity in tetrameric Kluyveromyces lactis β-galactosidase. J. Struct. Biol. 177, 392–401 [DOI] [PubMed] [Google Scholar]
- 25. Hidaka M., Fushinobu S., Ohtsu N., Motoshima H., Matsuzawa H., Shoun H., Wakagi T. (2002) Trimeric crystal structure of the glycoside hydrolase family 42 β-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose. J. Mol. Biol. 322, 79–91 [DOI] [PubMed] [Google Scholar]
- 26. Maksimainen M., Paavilainen S., Hakulinen N., Rouvinen J. (2012) Structural analysis, enzymatic characterization, and catalytic mechanisms of β-galactosidase from Bacillus circulans sp. alkalophilus. FEBS J. 279, 1788–1798 [DOI] [PubMed] [Google Scholar]
- 27. Aguilar C. F., Sanderson I., Moracci M., Ciaramella M., Nucci R., Rossi M., Pearl L. H. (1997) Crystal structure of the β-glycosidase from the hyperthermophilic archeon Sulfolobus solfataricus. Resilience as a key factor in thermostability. J. Mol. Biol. 271, 789–802 [DOI] [PubMed] [Google Scholar]
- 28. Rojas A. L., Nagem R. A., Neustroev K. N., Arand M., Adamska M., Eneyskaya E. V., Kulminskaya A. A., Garratt R. C., Golubev A. M., Polikarpov I. (2004) Crystal structures of β-galactosidase from Penicillium sp. and its complex with galactose. J. Mol. Biol. 343, 1281–1292 [DOI] [PubMed] [Google Scholar]
- 29. Maksimainen M., Hakulinen N., Kallio J. M., Timoharju T., Turunen O., Rouvinen J. (2011) Crystal structures of Trichoderma reesei β-galactosidase reveal conformational changes in the active site. J. Struct. Biol. 174, 156–163 [DOI] [PubMed] [Google Scholar]
- 30. Ohto U., Usui K., Ochi T., Yuki K., Satow Y., Shimizu T. (2012) Crystal structure of human β-galactosidase. The structural basis of GM1 gangliosidosis and Morquio B diseases. J. Biol. Chem. 287, 1801–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Asp N. G., Dahlqvist A. (1972) Human small intestine β-galactosidases. specific assay of three different enzymes. Anal. Biochem. 47, 527–538 [DOI] [PubMed] [Google Scholar]
- 32. Alpers D. H. (1969) Separation and isolation of rat and human intestinal β-galactosidases. J. Biol. Chem. 244, 1238–1246 [PubMed] [Google Scholar]
- 33. Distler J. J., Jourdian G. W. (1973) The purification and properties of β-galactosidase from bovine testes. J. Biol. Chem. 248, 6772–6780 [PubMed] [Google Scholar]
- 34. Juers D. H., Huber R. E., Matthews B. W. (1999) Structural comparisons of TIM barrel proteins suggest functional and evolutionary relationships between β-galactosidase and other glycohydrolases. Protein Sci. 8, 122–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Juers D. H., Jacobson R. H., Wigley D., Zhang X. J., Huber R. E., Tronrud D. E., Matthews B. W. (2000) High resolution refinement of β-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for α-complementation. Protein Sci. 9, 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller J. H. (1972) Experiments in Molecular Genetics, pp. 398–404, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 37. Yang X., Zhao Y., Wang Q., Wang H., Mei Q. (2005) Analysis of the monosaccharide components in Angelica polysaccharides by high performance liquid chromatography. Anal. Sci. 21, 1177–1180 [DOI] [PubMed] [Google Scholar]
- 38. Honda S., Akao E., Suzuki S., Okuda M., Kakehi K., Nakamura J. (1989) High-performance liquid chromatography of reducing carbohydrates as strongly ultraviolet-absorbing and electrochemically sensitive 1-phenyl-3-methyl-5-pyrazolone derivatives. Anal. Biochem. 180, 351–357 [DOI] [PubMed] [Google Scholar]
- 39. DeLano W. L. (2002) The PyMol Molecular Graphics System, Version 0.99rc2, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 40. Stirk H. J., Woolfson D. N., Hutchinson E. G., Thornton J. M. (1992) Depicting topology and handedness in jellyroll structures. FEBS Lett. 308, 1–3 [DOI] [PubMed] [Google Scholar]
- 41. Nucci R., Moracci M., Vaccaro C., Vespa N., Rossi M. (1993) Exo-glucosidase activity and substrate specificity of the β-glycosidase isolated from the extreme thermophile Sulfolobus solfataricus. Biotechnol. Appl. Biochem. 17, 239–250 [PubMed] [Google Scholar]
- 42. Davies G. J., Wilson K. S., Henrissat B. (1997) Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huber R. E., Hakda S., Cheng C., Cupples C. G., Edwards R. A. (2003) Trp-999 of β-galactosidase (Escherichia coli) is a key residue for binding, catalysis, and synthesis of allolactose, the natural lac operon inducer. Biochemistry 42, 1796–1803 [DOI] [PubMed] [Google Scholar]
- 44. Ahn Y. O., Zheng M., Bevan D. R., Esen A., Shiu S. H., Benson J., Peng H. P., Miller J. T., Cheng C. L., Poulton J. E., Shih M. C. (2007) Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 35. Phytochemistry 68, 1510–1520 [DOI] [PubMed] [Google Scholar]
- 45. Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gouet P., Robert X., Courcelle E. (2003) ESPript/ENDscript. Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]