Background: Mutations in the gene of the nuclear polyadenylate-binding protein 1 (PABPN1) lead to oculopharyngeal muscular dystrophy (OPMD) and aggregation of PABPN1.
Results: Fibrillar structures occur independently of the OPMD causing mutation.
Conclusion: OPMD might be caused by processes other than fibrillation.
Significance: Fibrils of full-length PABPN1 have been obtained which might have structures identical to those found in OPMD patients.
Keywords: Amyloid, Biophysics, Nucleotide Repeat Disease, Protein Aggregation, Protein Misfolding, Fibril Formation, OPMD, PABPN1, Polyalanine
Abstract
Oculopharyngeal muscular dystrophy is a late-onset disease caused by an elongation of a natural 10-alanine segment within the N-terminal domain of the nuclear poly(A)-binding protein 1 (PABPN1) to maximally 17 alanines. The disease is characterized by intranuclear deposits consisting primarily of PABPN1. In previous studies, we could show that the N-terminal domain of PABPN1 forms amyloid-like fibrils. Here, we analyze fibril formation of full-length PABPN1. Unexpectedly, fibril formation was independent of the presence of the alanine segment. With regard to fibril formation kinetics and resistance against denaturants, fibrils formed by full-length PABPN1 had completely different properties from those formed by the N-terminal domain. Fourier transformed infrared spectroscopy and limited proteolysis showed that fibrillar PABPN1 has a structure that differs from native PABPN1. Circumstantial evidence is presented that the C-terminal domain is involved in fibril formation.
Introduction
The nuclear poly(A)-binding protein 1 (PABPN1)2 plays an essential role in the processing of pre-mRNA in the nucleus (1, 2). Together with the cleavage and polyadenylation specificity factor and the poly(A) polymerase, PABPN1 stimulates the synthesis of polyadenylate (poly(A)) tails at the 3′-ends of pre-mRNAs (3). In addition, PABPN1 controls the length of the synthesized poly(A) tails with ∼250-adenylate bases (4–7). PABPN1 consists of 306 amino acids and has a predicted molecular mass of 32.8 kDa. The protein is composed of an N-terminal acidic region, a central domain with a conserved RRM domain (RNA recognition motif), and the C-terminal segment (8). The N-terminal domain contains a sequence of 10 consecutive alanine residues. The domain has been shown to be largely unstructured both in its isolated form and in the context of full-length PABPN1 (9). Residues 125–162 form an α-helical segment that precedes the central RNA binding domain (10). The α-helical region is required for the stimulation of the poly(A) polymerase (2). Both the RRM and the C-terminal segment ensure high affinity RNA binding (11). The basic C-terminal domain harbors a nuclear localization signal and 13 asymmetric dimethylated arginine residues (8, 10, 12, 13).
The gene encoding PABPN1 is located on chromosome 14q11.2-q13. This region has been identified to be mutated in patients who suffer from oculopharyngeal muscular dystrophy (OPMD) (14). Thus, PABPN1 has been recognized as a key player in the development of the disease (15). Whereas in the wild-type gene, a (GCN)10 trinucleotide repeat encodes the alanine stretch directly after the start codon, the gene of OPMD patients shows an expansion of the repeat. The expansion leads to a maximum of 7 additional alanines extending the wild-type 10 alanine segment to 17 alanines (15). OPMD is an adult-onset disease that usually presents itself near age 50 and takes a progressive course (16–18). The disease is characterized by a proximal limb weakness, eyelid drooping (ptosis), and severe swallowing difficulties (dysphagia) (18). OPMD biopsy material from muscle fibers shows elongated intranuclear inclusions that consist predominantly of PABPN1 (19, 20).
Currently, the role of the additional alanines on the aggregation behavior of PABPN1 is controversially discussed. We have shown earlier that the purified N-terminal domain (residues 1–125) of PABPN1 forms fibrils with typical amyloid-like characteristics (21, 22). The presence of the polyalanine sequence was absolutely necessary for the conversion of the soluble domain into fibrillar aggregates (21, 22). On the other hand, in vivo studies pointed toward a polyalanine-independent aggregation/assembly of PABPN1 (23). The fact that several oligomerization sites have been identified by independent approaches implicates a high propensity of PABPN1 to form oligomers and/or aggregates: (i) based on the crystal structure of the RNA binding domain, dimerization of the domain has been postulated (24). This assumption has been confirmed by the solution structure of the corresponding domain from Xenopus laevis (25). (ii) Two potential oligomerization sites have been detected via a yeast-two hybrid analysis to search for self-association of PABPN1 fragments. The oligomerization sites have been mapped to amino acids 155–294 and 264–306 (26). To what extent the oligomerization segments contribute to OPMD pathogenesis is still an unresolved question. It is neither clear whether the aggregates found via in vivo approaches represent typical amyloid-like structures or whether the deposits consist of amorphous aggregates containing either native or denatured protein. Despite these uncertainties, evidence for an intrinsic toxicity of overexpressed PABPN1 has been obtained by a Drosophila model for OPMD (27).
To examine the role of PABPN1 conformation and oligomerization in the development of OPMD, a biophysical characterization of full-length and truncated versions of PABPN1 was performed. We show here that in contrast to our findings with the N-terminal domain, fibril formation of full-length PABPN1 takes place independently of the presence of the alanine segment. Rather, evidence has been obtained that the C-terminal 49 amino acids are required for fibril formation. The biophysical characterization presented here shows that fibrils of full-length PABPN1 have completely different properties from those of the N-terminal domain.
EXPERIMENTAL PROCEDURES
Recombinant Expression of PABPN1 Variants
Escherichia coli strain BL21 (DE3) (Novagen) served as host cells. To avoid codon usage problems, either cells contained plasmid pUBS520 (28) in the case of full-length PABPN1, or CodonPlus RP (Stratagene) cells were used for the truncated variants. Recombinant cDNA of full-length PABPN1 variants was in vector pET11a (Novagen). Coding fragments for truncated PABPN1 variants were amplified from this vector and ligated into pET SUMOadapt (29). Recombinant protein production was performed by fermentation in complex medium in a 10-liter bioreactor (BIOSTAT C-DCU; Sartorius Stedim) as described previously (30) with the following modifications: at A600 = 40–50, protein expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The cultivation temperature was reduced to 30 °C to prevent inclusion body formation. Purification of full-length PABPN1 was performed essentially according to a published protocol (31); elution of PABPN1 variants from Blue-Sepharose was achieved by a step gradient with 1 m l-arginine. Truncated PABPN1 variants were expressed as fusions with N-terminal His-SUMO which allowed purification via HisTrap FF crude (GE Healthcare) (5 ml). Fusion proteins were eluted by linear gradients from 20 to 250 mm imidazole in 50 mm Tris-HCl, pH 7.9, 150 mm KCl, 0.5 mm DTT, 10% (v/v) glycerol at a flow rate of 2 ml/min. The His-SUMO fusion was hydrolyzed overnight at 4 °C using SUMO protease (0.05% (v/v) of 0.5 mg/ml protease) followed by dialysis against 50 mm Tris-HCl, pH 7.9, 150 mm KCl, 0.5 mm DTT, 10% (v/v) glycerol to obtain untagged protein (32). To remove the His-tagged SUMO partner, immobilized metal affinity chromatography was repeated. A homogeneous protein species was obtained by anion exchange chromatography using a 5-ml HiTrap Q HP (GE Healthcare). Elution was achieved by a linear gradient from 150 to 500 mm KCl in 50 mm Tris-HCl, pH 7.9, 0.5 mm DTT, 10% (v/v) glycerol, 1 mm EDTA. Conformational homogeneity of purified protein was confirmed by a symmetric elution peak of the last chromatography step (supplemental Fig. S1).
Fluorescence Measurements
Intrinsic tryptophan fluorescence was measured at a protein concentration of 4.5 μm. Fluorescence was recorded in 50 mm Tris-HCl, pH 7.9, 1.5 m KCl, 1 mm EDTA, 1 mm DTT, 10% (v/v) glycerol, at 20 °C with a Jobin Yvon Spex Fluoromax 2. Samples were excited at 295 nm and at a bandwidth of 5 nm. Spectra were accumulated five times by using a 1-nm data pitch and an integration time of 0.2 s. Fibril formation was followed by thioflavin T (ThT) fluorescence. Protein variants were incubated at a concentration of 60 μm in 50 mm Tris-HCl, pH 7.9, 1.5 m KCl, 1 mm EDTA, 1 mm DTT, 10% (v/v) glycerol, at 20 °C. Samples were briefly mixed and then diluted to final concentrations of 2 μm with 50 μm ThT in 50 mm Tris-HCl, pH 7.9, 1.5 m KCl, 1 mm EDTA, 10% (v/v) glycerol. ThT fluorescence was recorded at 482 nm with a bandwidth of 8 nm at 20 °C and upon excitation at 450 nm. Samples were measured in 1-cm cuvettes, and signals were accumulated two times. Prior to fibrillation experiments, samples were centrifuged for 1 h at 105,000 × g (TLX 100.4; Beckman Coulter) and samples were analyzed by UV spectroscopy to exclude the presence of aggregated or fibrillar species.
Solubilization of Fibrils
Fibrillar fractions were isolated from the soluble species via ultracentrifugation (OptimaTM TLX ultracentrifuge) at 50,000 rpm for 1 h. The pellet was incubated with 50 mm Tris-HCl, pH 7.9, 1.5 m KCl, 1 mm EDTA, 10% (v/v) glycerol containing different amounts of guanidinium chloride (GdmCl). For concentrations above 2.5 m GdmCl, 150 mm KCl was used to avoid solubility limits. After incubation for 1 h at 20 °C, solubilized protein was separated from the insoluble fraction by ultracentrifugation (105,000 × g, 1 h). For determination of the protein concentration, the insoluble fraction was solubilized in 6 m GdmCl, and the protein concentration was measured by UV absorption (ϵ280 nm of full-length variants 20,400 m−1cm−1). A typical UV spectrum of PABPN1 variants is shown in supplemental Fig. S2).
Limited Proteolysis
Soluble or fibrillar protein was incubated at room temperature with either trypsin or proteinase K at a mass ratio of 1:600 and 1:1000 (protease:protein) in 50 mm Tris-HCl, pH 7.9, 1.5 m KCl, 1 mm EDTA, 1 mm DTT, 10% (v/v) glycerol. Proteolysis was stopped with either a 5-fold molar excess of aprotinin or PMSF. Analysis of proteolysis products was performed by SDS-PAGE with 14% polyacrylamide gels. For characterization of protease-resistant fragments, limited proteolysis was stopped after 2 h. Subsequently, insoluble fractions were isolated by ultracentrifugation (OptimaTM TLX ultracentrifuge) at 105,000 × g for 1 h. Two washing steps with 50 mm Tris-HCl, pH 7.9, 1.5 m KCl, 1 mm EDTA, 1 mm DTT, 10% (v/v) glycerol were included to remove soluble protein species completely. Insoluble protein was recovered by ultracentrifugation. Finally, the insoluble fractions were dissolved in 6 m GdmCl and loaded on a Jupiter 5 μ C18 300 A (250 × 4.6) HPLC column (Phenomenex). As mobile phase, bidestilled water, 0.1% TFA was used. Protein was eluted from the column by applying a linear gradient up to 100% acetonitrile, 0.1% TFA. Elution profiles were recorded monitoring the absorption of peptide bonds at 215 nm and aromatic amino acids at 280 nm.
RNA Binding Assay
Filter binding assays were carried out as described (4, 11) using oligo(A) (14-mer) as an RNA binding substrate. For the determination of apparent dissociation constants, KD, 8.8 fmol of 32P-labeled oligo(A)-RNA was incubated with increasing concentrations of PABPN1 variants (0–200 nm) in 40 μl of RNA-binding buffer (50 mm Tris-HCl, pH 7.9, 100 mm KCl, 1 mm EDTA, 1 mm DTT, 10% (v/v) glycerol). After 10 min at room temperature, 35 μl of each reaction was applied to nitrocellulose filters (NC 20, 0.2 μm Whatman) which had been preincubated in washing buffer (50 mm Tris-HCl, pH 8, 100 mm NaCl, 1 mm EDTA) containing 5 μg/ml rRNA (Roche Applied Science). After rinsing the filters with 5 ml of ice-cold washing buffer, filter-bound RNA was quantified via scintillation counting.
Size Exclusion Chromatography
For analysis of fibril formation via size exclusion chromatography, 100-μl aliquots of the fibrillation samples were loaded on a SuperdexTM 200 10/300 GL column (GE Healthcare) (25 ml). To ensure an appropriate evaluation of the data, 5 μl of a l-tryptophan standard (0.3 mm) was added to the samples prior the injection. Size exclusion chromatography was performed with an ÄKTApurifier system (GE Healthcare) following elution via absorption at 280 nm. After the runs, chromatograms were normalized to the absorption of the tryptophan standard at an elution volume of 27 ml. As running buffer, 50 mm Tris-HCl, pH 7.9, 1.5 m KCl, 1 mm EDTA, 1 mm DTT, 10% (v/v) glycerol was used.
Miscellaneous
Samples for electron microscopy analysis were prepared as published (22). Infrared spectra were recorded at a Trensor 27 Fourier-transformed infrared (FTIR) spectrometer equipped with a BIO-ATR II cell and a nitrogen-cooled LN-MCT detector. For measurements, 20-μl samples (protein concentration 2–4 mg/ml) were applied on the cells. Spectra were accumulated 64 times at a resolution of 2 cm−1 and 4-fold zero filling.
RESULTS
Fibril Formation of Full-length PABPN1 Does Not Require the Alanine Segment
Previous work from our laboratory had shown that fibril formation of the N-terminal domain of PABPN1 strictly requires the presence of the alanine segment (21). At that time, studies of fibril formation with the full-length protein had not been possible due to the high tendency of PABPN1 to form amorphous aggregates. Here, we resumed the original objective to explore the influence of alanine expansions on the conversion of full-length PABPN1 into fibrillar structures. As in the previous experiments, three variants of PABPN1 which differed in their alanine segments were studied: wild-type protein, WT-PABPN1, with the natural 10 alanine residues; an OPMD-related PABPN1 variant with the most extreme expansion of seven additional alanines, (+7)Ala-PABPN1; and a deletion variant in which the alanine sequence had been deleted, ΔAla-PABPN1 (Fig. 1). Due to the tendency of the protein to aggregate, conditions had to be found which prevented amorphous aggregation and allowed fibril formation studies in vitro. In the presence of 1.5 m KCl, formation of amorphous aggregates could be suppressed. High salt concentrations possibly compensate electrostatic interactions of the oppositely charged N- and C-terminal domains (pI of N-terminal residues 1–114: 4.0; pI of C-terminal residues 257–306: 11.8). The native structure and biological activity of the purified protein were confirmed by fluorescence spectroscopy, circular dichroism, and oligo(A)-RNA binding (see Figs. 3 and 7 and supplemental Figs. S3–S5).
FIGURE 1.
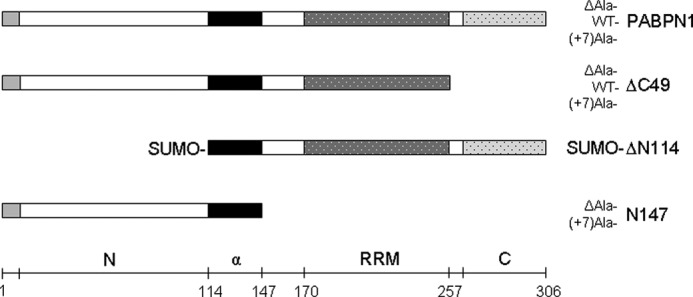
Schematic representation of PABPN1 and deletion variants. Light gray part shows the alanine segment, the following open bar depicts the N-terminal domain. The black bar indicates the α-helical segment that precedes the RRM domain, the C-terminal segment is shown by the dotted bar. N, N-terminal domain; α, α-helical segment; C, C-terminal domain.
FIGURE 3.
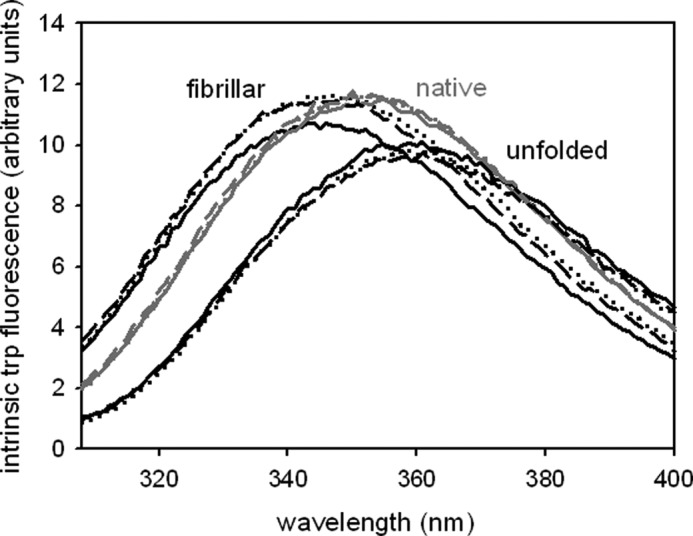
Tryptophan fluorescence analysis. Samples with protein concentrations of 4.5 μm were excited at 295 nm. Emission spectra of native (monomeric), unfolded and fibrillar WT-PABPN1 (solid line), ΔAla-PABPN1 (dotted line), and (+7)Ala-PABPN1 (dashed line) were recorded.
FIGURE 7.
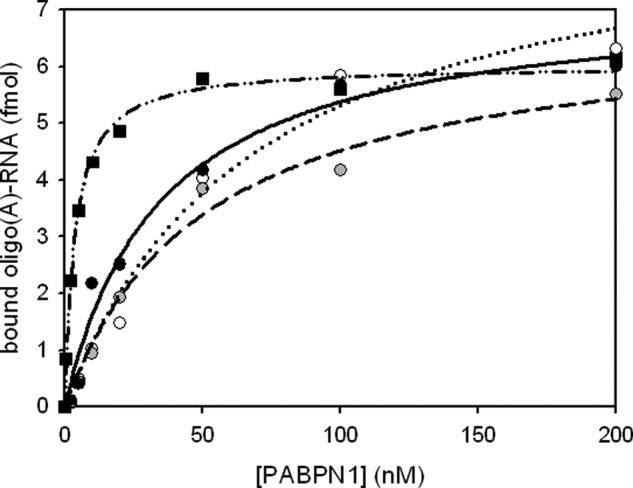
RNA binding assay of fibrillar full-length PABPN1. Filter binding assays were performed with radioactive oligo(A)-RNA to determine the apparent dissociation constants (KD) of fibrillar PABPN1 variants: WT-PABPN1 (black circles), ΔAla-PABPN1 (open circles), and (+7)Ala-PABPN1 (gray circles). For comparison, monomeric WT-PABPN1 (black squares) is shown. Data points were fitted according to Ref. 4. Dashed-dotted line, monomeric WT-PABPN1; solid line, fibrillar WT-PABPN1; dotted line, fibrillar ΔAla-PABPN1; and dashed line, (+7)Ala-PABPN1.
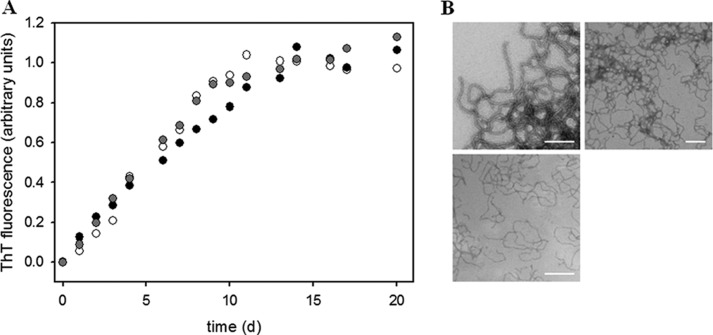
Before fibril formation assays, potentially present nucleation seeds or oligomers were removed by ultracentrifugation. In addition, the absence of aggregated species was controlled by UV spectroscopy (supplemental Fig. S2). Subsequently, fibril formation was tested by ThT fluorescence measurements. In contrast to the N-terminal domains of PABPN1 which in the presence of the N-terminal alanines formed fibrils after a lag phase of approximately 15 days (21), full-length wild-type and (+7)Ala-PABPN1 fibrillized immediately without apparent lag phases (Fig. 2A). Unexpectedly, also the ΔAla-PABPN1 variant gave rise to increased ThT signals which indicated the existence of fibrils. Kinetics of fibril formation of full-length PABPN1 revealed hyperbolic curve shapes which are untypical for the formation of amyloid-like fibrils. However, electron microscopy confirmed the presence of fibrillar structures (Fig. 2B). No difference in the morphologies among the variants could be detected. The kinetics of fibril formation at early time points was analyzed by size exclusion chromatography (supplemental Fig. S6). A time-dependent increase of a peak that corresponded to oligomeric assemblies (>600 kDa) was observed. As with time-dependent ThT fluorescence recordings, also this approach did not show any differences in fibril formation kinetics among the three variants.
FIGURE 2.
Fibril formation of full-length PABPN1. A, time-dependent ThT fluorescence increases monitored at 482 nm. Black circles, WT-PABPN1; open circles, ΔAla-PABPN1; and gray circles, (+7)Ala-PABPN1. Samples were incubated at 20 °C at a protein concentration of 60 μm. B, electron micrographs of fibrillar WT-PABPN1 (upper left), ΔAla-PABPN1 (upper right), and (+7)Ala-PABPN1 (lower). Scale bars, 200 nm.
Fibrils of Full-length PABPN1 Exhibit Structures Different from Soluble PABPN1
The atypical fibril formation kinetics prompted the question as to the nature of the fibrillar species. To clarify whether structural changes had occurred before or during fibril formation, tryptophan fluorescence was monitored. PABPN1 has a single tryptophan in the C-terminal region at position 302 (Trp-302). Changes in the tryptophan emission spectra are an indicator for an altered polypeptide environment and can be used to assess the solvent exposure of tryptophan residues. The soluble forms of the PABPN1 variants exhibited emission spectra with a maximum at 351 nm. In the denatured state, the maxima were shifted toward 360 nm, pointing to a complete solvent exposure of Trp-302. Emission maxima of the fibrillar species were at 344 nm. The blue shift by 7 nm indicates that in the fibrils, Trp-302 is in a more hydrophobic environment than in the soluble state (Fig. 3). Thus, it can be concluded that the region close to Trp-302 has undergone a conformational change upon fibril formation.
Chemical Stability of Fibrillar PABPN1 Differs from Fibrils of N-terminal Domain
A typical feature of amyloid-like fibrils is a considerably higher resistance against chemical or physical influences compared with the monomeric proteins (33–36). Fibrils of the N-terminal domain of PABPN1 had shown an unusual high stability against solubilization with GdmCl and the more chaotropic SCN salt of Gdm (22). To explore the chemical stability of fibrillar PABPN1, solubilization with GdmCl was investigated. After removal of nonfibrillized protein by ultracentrifugation, the insoluble fraction was incubated with GdmCl of various concentrations. Solubilized species were removed from fibrillar species by ultracentrifugation, and the amount of protein present in the pellet fraction was determined (Fig. 4). Quantification of the fibrillar fraction was performed upon addition of 6 m GdmCl that led to complete solubilization of the fibrils. The sigmoidal decrease of fibrillar species at GdmCl concentrations higher than 1.5 m indicated that solubilization might be a cooperative reaction, a finding that currently cannot be explained. The midpoint of solubilization was at approximately 2 m GdmCl. It is noteworthy that fibrils of the N-terminal domain of PABPN1 alone could not be solubilized by these GdmCl concentrations and that even 6 m GdmCl was not sufficient to lead to complete solubilization of these fibrils (22). Thus, fibrils formed by the N-terminal domains and those formed by the full-length PABPN1 must differ in fundamental biophysical and/or structural features. Fibrils of the three PABPN1 variants did not show any differences in their chemical stabilities, indicating that the fibril structures are similar if not identical. Thus, the presence or absence of the alanine segment does not influence the chemical or structural properties of full-length PABPN1 fibrils.
FIGURE 4.
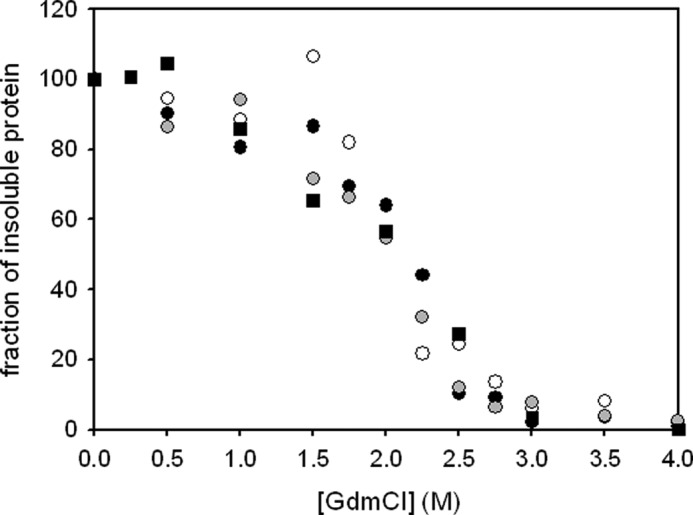
Stability of fibrils against solubilization with GdmCl. The protein fraction that resisted solubilization after 1-h incubation at room temperature has been plotted. Data were normalized to the fraction of insoluble protein after washing with 50 mm Tris-HCl,, pH 7.9, 1.5 m KCl, 1 mm EDTA, 1 mm DTT in the absence of GdmCl. WT-PABPN1 (black circles), ΔAla-PABPN1 (open circles), (+7)Ala-PABPN1 (gray circles) and the truncated variant (SUMO-ΔN114) that lacks the N-terminal domain (black squares) are shown.
FTIR Spectroscopy and Limited Proteolysis Confirm Conformational Differences between Monomeric and Fibrillar Protein
To gain more information about the structural features of fibrils from full-length PABPN1, FTIR spectra were recorded. The usage of attenuated total reflection allowed us to record spectra in aqueous buffer without the replacement of H2O by D2O. Independent of the presence of the alanine sequence, monomeric and fibrillar PABPN1 exhibited similar spectra in the region between wave number 1500 to 1590 cm−1 (Fig. 5). Differences were obvious in the region of wave numbers 1600 cm−1 to 1700 cm−1, an area that provides information about secondary structure contributions through the amide I oscillation. Monomeric protein variants exhibited a high absorption at approximately 1650 cm−1 whereas spectra of fibrillar samples showed a shifted maximum in the range of 1640 cm−1 to 1620 cm−1, indicating an increase of β-sheet structures (37). Thus, the β-sheet content must have increased upon fibrillation. A detailed analysis of FTIR spectra was not performed because quantification of secondary structures via this method is often not reliable. Nevertheless, the FTIR spectra revealed conformational differences between soluble and fibrillar PABPN1.
FIGURE 5.
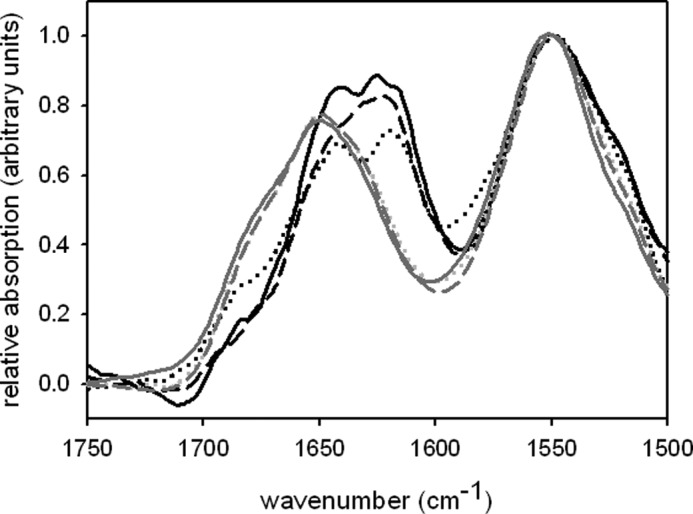
FTIR spectra. IR spectra of the amide I and II regions are shown for monomeric (gray lines) and fibrillar (black lines) protein. Solid line, WT-PABPN1; dotted line, ΔAla-PABPN1; and dashed line, (+7)Ala-PABPN1. Spectra are normalized to the amide II band.
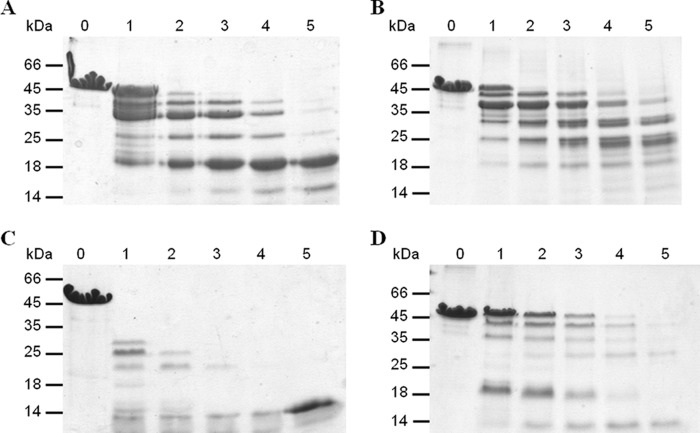
To confirm by an independent method that fibrillar protein had undergone a conformational change, monomeric and fibrillar PABPN1 were subjected to limited proteolysis. Proteolysis was performed with trypsin as a more specific protease, and proteinase K, an unspecific protease (Fig. 6). With both proteases, differences in the proteolysis patterns of soluble and fibrillar protein became evident. After a 3-h incubation of soluble protein with trypsin, a protease-resistant fragment of 18 kDa was the predominating band (Fig. 6A, lane 5). From earlier results3 we know that this band corresponds to the α-helical segment followed by the RRM of PABPN1. The same incubation time of trypsin with fibrils yielded several products migrating at positions corresponding to 25, 30, and 35 kDa (Fig. 6B, lane 5). After a 30-min incubation with proteinase K, monomeric protein had been degraded to products of 14 and 20 kDa (Fig. 6C, lane 2) whereas in the proteolysis pattern of the fibrillar protein, bands of higher molecular mass were visible (Fig. 6D, lane 2). The presence of a weak band of apparent 50 kDa in lane 3 (Fig. 6D) indicates that intact protein is still present after a 1-h proteinase K digestion. The proteolysis of ΔAla-PABPN1 and (+7)Ala-PABPN1 gave rise to comparable digestion patterns (Figs. S7 and S8), indicating that there were no gross differences among the fibrils of the three variants. Taken together, the limited proteolysis experiment clearly shows that structural transitions must have occurred during or upon fibril formation.
FIGURE 6.
Limited proteolysis. Time-dependent degradation of monomeric (A and) and fibrillar WT-PABPN1 (B and D) is shown after 10 min (lanes 1), 30 min (lanes 2), 1 h (lanes 3), 2 h (lanes 4), and 3 h (lanes 5). Lanes 0 indicate the sample without protease. A and B, limited proteolysis with trypsin at a mass ratio of 1:600. C and D, limited proteolysis with proteinase K at a mass ratio of 1:1000.
Despite the fact that the protease resistance was not as pronounced as expected for fibrillar protein, attempts were made to identify those segments of the protein that made up the fibrillar core. To this end, fibrils were first incubated with trypsin for 2 h, isolated by centrifugation, and subsequently solubilized in 6 m GdmCl. Peptidic fragments were separated by HPLC and identified via mass spectrometry (data not shown). The masses obtained by this approach, stemmed predominantly from the C-terminal region of PABPN1, i.e. the last 47 amino acids. We conclude from these data that the C-terminal domain of PABPN1 seems to contribute to the fibril formation of full-length PABPN1 as it was assumed before on the basis of the blue-shifted tryptophan fluorescence in the fibrillar population (Fig. 3). Because, to a certain extent, peptides from all over the PABPN1 were also identified, it is possible that fibrils of PABPN1 do not possess a defined fibrillar core.
Substrate Binding Ability of Fibrillar Protein Is Reduced
Next, the question was addressed as to whether fibrillar PABPN1 is still able to bind RNA. Fibrillar protein was isolated via ultracentrifugation, fibrils were washed intensively to remove soluble PABPN1, and the pellet was resuspended in fibrillation buffer. For determination of the exact protein concentration, an aliquot of the protein solution was solubilized in 6 m GdmCl, and the absorption at 280 nm was measured. To determine the apparent dissociation constant, KD, a titration experiment with oligo(A)-RNA was performed (Fig. 7). The titration experiment revealed KD values that are 1 magnitude higher than those of soluble proteins (Table 1). Thus, it appears that either not all of the protein present in the fibrils is able to bind RNA or that the conformation of PABPN1 in the fibrils impairs high affinity binding of the substrate.
TABLE 1.
Apparent dissociation constants
KD values were obtained from filter binding assays (Fig. 7) as described (4).
| PABPN1 variant | Apparent KD |
|---|---|
| nm | |
| WT-PABPN1 (monomeric) | 4 |
| WT-PABPN1 (fibrillar) | 69 |
| ΔAla-PABPN1 (fibrillar) | 50 |
| (+7)Ala-PABPN1 (fibrillar) | 35 |
| SUMO-ΔN114 (monomeric) | 9 |
In Absence of C-terminal Domain, Fibril Formation Becomes Alanine-dependent
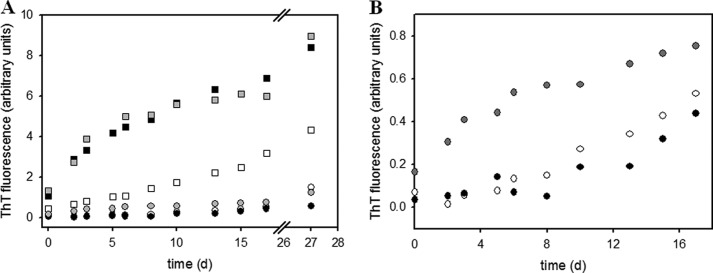
The tryptophan fluorescence spectra of fibrillar protein and the results from the limited proteolysis had already pointed to an involvement of the C-terminal domain of PABPN1 in fibril formation. To investigate in more detail the influence of this domain in the process, a variant in which the last 49 amino acids of PABPN1 were deleted (ΔC49) was studied (11). Again, three different variants were generated that differed in the length of the alanine segments (Fig. 1). When the proteins were incubated in fibrillation buffer at a protein concentration of 60 μm, no increases of the ThT fluorescence could be observed over a time period of 50 days (data not shown). Electron microscopy confirmed the absence of fibrillar structures. At concentrations of 0.5 mm, ThT signals slightly increased and those of (+7)Ala-ΔC variant showing the fastest rise (Fig. 8). This observation prompted the question of whether seeding with the N-terminal domain would accelerate fibril formation of the variants lacking the C-terminal domains. To this end, fibrils of the N-terminal domain with the seven alanine extension (N-(+7)Ala) were converted to seeds and added to the ΔC variants. ThT fluorescence rises were now clearly alanine-dependent (Fig. 8). Yet, no sigmoidal increases were observed, and solubilization with GdmCl (data not shown) revealed a low resistance of the fibrils as in the case with the full-length variants. These observations indicate that the fibrils obtained with the proteins lacking the C-terminal domain had properties and structures similar to those of the full-length protein.
FIGURE 8.
Fibril formation of PABPN1 variants lacking the C-terminal domain. A, samples were incubated at 37 °C at a protein concentration of 0.5 mm. Squares indicate the presence of 5% (w/w) seeds derived from fibrils of the N-terminal domain of PABPN1 (N-(+7)Ala); circles indicate the absence of seeds. Black, WT-ΔC49; white, ΔAla-ΔC49; and gray, (+7)Ala-ΔC49. B, zoom into kinetics until day 17 with unseeded samples.
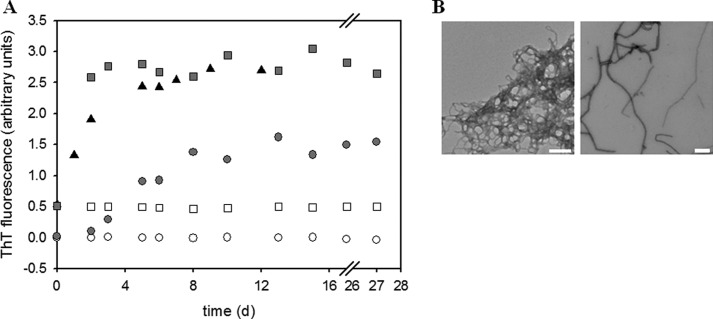
To further analyze which domains within PABPN1 contribute to fibril formation, two more deletion variants were studied: (i) the N-terminal domains possessing the α-helical segment (N147) and only differing in the alanine segment and (ii) a truncated variant in which a large part of the N-terminal domain was deleted (ΔN114) (Fig. 1). Due to the poor solubility of the latter variant and its high tendency to form amorphous aggregates, ΔN114 was analyzed in the context of the N-terminal SUMO fusion partner. First, the correct structure was probed by the ability of the truncated version to bind oligo(A)-RNA (supplemental Fig. S5). The KD of 9 nm showed that the variant had retained its biochemical features with regard to RNA binding. Formation of fibrils was investigated by ThT fluorescence (Fig. 9A). As for full-length PABPN1 variants, hyperbolic kinetics without initial lag phases were recorded for ΔN114. Initial fluorescence signals rose faster than those of fibrillation samples of full-length PABPN1 variants. ThT signals of variant ΔN114 reached a plateau after 10 days, ∼5 days earlier than observed with the full-length PABPN1 variants. In contrast, ThT signals showed sigmoidal rises for (+7)Ala-N147 in the absence of seeds, indicating that fibril formation of this construct followed a similar if not identical mechanism as that of the N-terminal domains (21). In the presence of seeds from the N-(+7)Ala variant, an immediate fluorescence increase was observed. No ThT fluorescence changes were observed for the ΔAla-N147 variant proving for this deletion construct an alanine-dependent fibril formation. The presence of fibrils for (+7)Ala-N147 and ΔN114 was confirmed by electron microscopy (Fig. 9B). Fibrils of ΔN114 exhibited the same low resistance against solubilization with GdmCl as fibrils of full-length PABPN1 whereas those of (+7)Ala-N147 were resistant as fibrils from the N-terminal domain (Table 2). Taken together, these results show that fibril formation of full-length PABPN1 involves the C-terminal domain and that fibrils obtained from the N-terminal domain exhibit properties different from those of protein variants possessing folded structures.
FIGURE 9.
Fibril formation kinetics of truncated PABPN1 variants. A, fibril formation of SUMO-ΔN114 (triangles), ΔAla-N147 (open symbols), and (+7)Ala-N147 (filled symbols). Samples seeded with 5% (w/w) seeds from N-(+7)Ala (21) are indicated by squares, unseeded samples by circles. B, electron micrographs of fibrillar SUMO-ΔN114 (left) and of (+7)Ala-N147 (right).
TABLE 2.
Fibril forming properties of the investigated proteins/protein domains and stabilities of the fibrils against solubilization
| Protein/deletion construct | Fibril formation | Alanine dependence | Resilience against solubilization |
|---|---|---|---|
| PABPN1 | Yes | No | Low |
| ΔC49 | Slowly and only at higher protein concentrations | At elevated protein concentrations | Low |
| ΔN114 | Yes | Low | |
| N147 | Yes | Yes | High |
DISCUSSION
Currently, no data are available which describe the thermodynamics of full-length PABPN1 folding, a fact that hampers studies of conformational transitions of this protein into fibrillar species. Yet with this work, for the first time a conformational transition of full-length PABPN1 into ordered fibrils and an initial characterization of the fibrils could be achieved. The major obstacle to these investigations was the high propensity of PABPN1 to form amorphous aggregates before adopting fibrillar structures. Formation of amorphous aggregates could be suppressed by incubating PABPN1 in buffers with high concentrations of salt, e.g. 1.5 m KCl. We assume that by the salt, interactions of the N- and C-terminal domains, which are oppositely charged, are impaired. When fibril formation of full-length PABPN1 was studied, fibrils were also formed by ΔAla-PABPN1. This observation was in contradiction to our earlier results which had shown that the N-terminal domain converts into fibrils only in the presence of the alanine segment (9, 21, 22). On the other hand, this finding agrees with in vivo studies in which overexpressed PABPN1-GFP fusions accumulated in intranuclear inclusions independently of the presence of the alanine sequence (23). Yet, protein features derived from in vitro and in vivo data can only be compared to a limited extent.
A recent in vitro study with the yeast prion protein Ure2p has shown a similar situation. Whereas the N-terminal domain has been demonstrated to form fibrils with amyloid-like properties, full-length Ure2p assembled into fibrillar structures that did not share the biochemical features with those of the N-terminal domain (38). This result and the data presented in this work illustrate the basic fact that full-length proteins and domains thereof are individual polypeptides with inherently different biochemical properties and tendencies to adopt alternative conformations.
The presence of ordered fibrillar structures of full-length PABPN1 has been confirmed by EM. No differences regarding fibril morphology could be observed between the variants of full-length PABPN1 with varying alanine tracts. Neither could any differences be detected between the fibrils of the truncated versions of PABPN1. Despite the fact that in the EM, fibrils of full-length PABPN1 were indistinguishable from fibrils of the N-terminal domain, several observations suggest that fibrils of full-length PAPBN1 and ΔN114 do not possess the typical amyloid-like structures: (i) fibrils exhibited only a moderate resistance against proteolysis; (ii) solubilization could be achieved with intermediate concentrations of GdmCl; and (iii) ThT fluorescence changes followed nonsigmoidal kinetics, suggesting that nucleation which is generally considered as the rate-limiting step during fibril formation is either not required or plays a minor role before conversion of full-length PABPN1 or ΔN114 into fibrils.
We show here that fibril formation of PABPN1 leads to a 10-fold higher KD toward oligo(A)-RNA that may be caused by a partial or local structural distortion of the RNA binding domain or a reduced accessibility of the domain toward the substrate due to its integration into the fibrils. Another explanation for the reduced RNA binding could consist in a certain proportion of the molecules being in the unfolded state because of steric hindrance in the densely packed fibril. A slightly reduced affinity toward nucleic acid substrates has been observed for typical amyloid-like fibrils from fusions of the N-terminal domain of PABPN1 and the nucleic acid binding model protein CspB (39). Furthermore, a previous NMR analysis of amyloid-like fibrils formed by the N-terminal domain revealed that only a rather short segment comprising ∼35 residues can be sufficient to make up the fibrillar core (40). In such a structure, folded domains could be preserved. Yet, the untypical properties of PAPBN1 fibrils may result from minor conformational changes that could have occurred before fibril formation, such as domain rearrangements. Whether PABPN1 fibrils consist of domain-swapped assemblies as for example shown for ribonuclease A is so far not clear (41). Similarly, it remains to be resolved whether the structures obtained with the in vitro formed fibrils are similar or even identical to those of arising in OPMD muscle nuclei and could be labeled by immunogold-conjugated antibodies (18, 23, 42).
The most unexpected finding of this work was that even in the absence of the alanine segment, fibril formation of the full-length protein can occur and that the complete N-terminal part could be deleted without affecting the propensity of PABPN1 to fibrillize. With these results we confirm in vivo results which had demonstrated that the N-terminal domain of PABPN1 is not the only key player in aggregation formation (23, 43). Conversely, variants in which the C-terminal domain had been deleted formed fibrils very slowly and only when protein concentrations were raised ∼10-fold, i.e. to the three-digit micromolar range. Incubation with seeds from the N-terminal domain possessing the alanine extension accelerated fibril formation. This acceleration by seeds points to a nucleation-based process. However, the seeds from the N-terminal domain did not endow the resulting fibrillar species with the high resistance against solubilization as it has been observed typical for fibrils of the N-terminal domain. Thus, a mechanistic explanation for the seedability is at this stage missing.
Finally, the question remains of why alanine codon expansions in the PABPN1 gene lead to OPMD, although the alanine sequence and even the N-terminal domain seem not to be necessary for fibril formation of PABPN1. An altered protein turnover due to the alanine extension could lead to the observed intranuclear inclusions. As mentioned above, a careful comparison of the in vivo formed fibrils with the fibrils obtained in vitro will add to our understanding in which way deposits of misfolded protein are involved in pathogenesis.
Supplementary Material
Acknowledgments
We thank Senthil Kumar Thangaraj for help with the IR spectrometer, Andrea Sinz and Christian Ihling for performing mass spectrometry analysis, Jay Goodman for polishing the English, and Elmar Wahle for providing plasmids for the truncated PABPN1 variants and facilities for the RNA binding studies.
This work was supported by the SFB610 “Protein Conformational States with Cell Biological and Medical Relevance” Subproject A9.

This article contains supplemental Figs S1–S8.
T. Scheuermann, P. Rücknagel, A. Schierhorn, and E. Schwarz, unpublished observations.
- PABPN1
- poly(A)-binding protein 1
- GdmCl
- guanidinium chloride
- OPMD
- oculopharyngeal muscular dystrophy
- poly(A)
- polyadenylate
- RRM
- RNA recognition motif
- ThT
- thioflavin T.
REFERENCES
- 1. Wahle E. (1991) A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell 66, 759–768 [DOI] [PubMed] [Google Scholar]
- 2. Wahle E. (1995) Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 270, 2800–2808 [DOI] [PubMed] [Google Scholar]
- 3. Bienroth S., Keller W., Wahle E. (1993) Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 12, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wahle E., Lustig A., Jenö P., Maurer P. (1993) Mammalian poly(A)-binding protein II: physical properties and binding to polynucleotides. J. Biol. Chem. 268, 2937–2945 [PubMed] [Google Scholar]
- 5. Colgan D. F., Manley J. L. (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11, 2755–2766 [DOI] [PubMed] [Google Scholar]
- 6. Barabino S. M., Keller W. (1999) Last but not least: regulated poly(A) tail formation. Cell 99, 9–11 [DOI] [PubMed] [Google Scholar]
- 7. Kühn U., Gündel M., Knoth A., Kerwitz Y., Rüdel S., Wahle E. (2009) Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 284, 22803–22814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nemeth A., Krause S., Blank D., Jenny A., Jenö P., Lustig A., Wahle E. (1995) Isolation of genomic and cDNA clones encoding bovine poly(A)-binding protein II. Nucleic Acids Res. 23, 4034–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lodderstedt G., Sachs R., Faust J., Bordusa F., Kühn U., Golbik R., Kerth A., Wahle E., Balbach J., Schwarz E. (2008) Hofmeister salts and potential therapeutic compounds accelerate in vitro fibril formation of the N-terminal domain of PABPN1 containing a disease-causing alanine extension. Biochemistry 47, 2181–2189 [DOI] [PubMed] [Google Scholar]
- 10. Kerwitz Y., Kühn U., Lilie H., Knoth A., Scheuermann T., Friedrich H., Schwarz E., Wahle E. (2003) Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A)-binding protein allosterically regulated by RNA. EMBO J. 22, 3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kühn U., Nemeth A., Meyer S., Wahle E. (2003) The RNA binding domains of the nuclear poly(A)-binding protein. J. Biol. Chem. 278, 16916–16925 [DOI] [PubMed] [Google Scholar]
- 12. Calado A., Kutay U., Kühn U., Wahle E., Carmo-Fonseca M. (2000) Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA 6, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fronz K., Otto S., Kölbel K., Kühn U., Friedrich H., Schierhorn A., Beck-Sickinger A. G., Ostareck-Lederer A., Wahle E. (2008) Promiscuous modification of the nuclear poly(A)-binding protein by multiple protein-arginine methyltransferases does not affect the aggregation behavior. J. Biol. Chem. 283, 20408–20420 [DOI] [PubMed] [Google Scholar]
- 14. Brais B., Xie Y. G., Sanson M., Morgan K., Weissenbach J., Korczyn A. D., Blumen S. C., Fardeau M., Tomé F. M., Bouchard J. P. (1995) The oculopharyngeal muscular dystrophy locus maps to the region of the cardiac α and β myosin heavy chain genes on chromosome 14q11.2-q13. Hum. Mol. Genet. 4, 429–434 [DOI] [PubMed] [Google Scholar]
- 15. Brais B., Bouchard J. P., Xie Y. G., Rochefort D. L., Chrétien N., Tomé F. M., Lafrenière R. G., Rommens J. M., Uyama E., Nohira O., Blumen S., Korczyn A. D., Heutink P., Mathieu J., Duranceau A., Codère F., Fardeau M., Rouleau G. A., Korcyn A. D. (1998) Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 18, 164–167 [DOI] [PubMed] [Google Scholar]
- 16. Fried K., Arlozorov A., Spira R. (1975) Autosomal recessive oculopharyngeal muscular dystrophy. J. Med. Genet. 12, 416–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duranceau A. C., Beauchamp G., Jamieson G. G., Barbeau A. (1983) Oropharyngeal dysphagia and oculopharyngeal muscular dystrophy. Surg. Clin. North Am. 63, 825–832 [DOI] [PubMed] [Google Scholar]
- 18. Brais B., Rouleau G. A., Bouchard J. P., Fardeau M., Tomé F. M. (1999) Oculopharyngeal muscular dystrophy. Semin. Neurol. 19, 59–66 [DOI] [PubMed] [Google Scholar]
- 19. Calado A., Tomé F. M., Brais B., Rouleau G. A., Kühn U., Wahle E., Carmo-Fonseca M. (2000) Nuclear inclusions in oculopharyngeal muscular dystrophy consist of poly(A)-binding protein 2 aggregates which sequester poly(A) RNA. Hum. Mol. Genet. 9, 2321–2328 [DOI] [PubMed] [Google Scholar]
- 20. Tomé F. M., Chateau D., Helbling-Leclerc A., Fardeau M. (1997) Morphological changes in muscle fibers in oculopharyngeal muscular dystrophy. Neuromuscul. Disord. 7, S63–69 [DOI] [PubMed] [Google Scholar]
- 21. Scheuermann T., Schulz B., Blume A., Wahle E., Rudolph R., Schwarz E. (2003) Trinucleotide expansions leading to an extended poly-l-alanine segment in the poly(A)-binding protein PABPN1 cause fibril formation. Protein Sci. 12, 2685–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lodderstedt G., Hess S., Hause G., Scheuermann T., Scheibel T., Schwarz E. (2007) Effect of oculopharyngeal muscular dystrophy-associated extension of seven alanines on the fibrillation properties of the N-terminal domain of PABPN1. FEBS J. 274, 346–355 [DOI] [PubMed] [Google Scholar]
- 23. Tavanez J. P., Calado P., Braga J., Lafarga M., Carmo-Fonseca M. (2005) In vivo aggregation properties of the nuclear poly(A)-binding protein PABPN1. RNA 11, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ge H., Zhou D., Tong S., Gao Y., Teng M., Niu L. (2008) Crystal structure and possible dimerization of the single RRM of human PABPN1. Proteins 71, 1539–1545 [DOI] [PubMed] [Google Scholar]
- 25. Song J., McGivern J. V., Nichols K. W., Markley J. L., Sheets M. D. (2008) Structural basis for RNA recognition by a type II poly(A)-binding protein. Proc. Natl. Acad. Sci. U.S.A. 105, 15317–15322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fan X., Dion P., Laganiere J., Brais B., Rouleau G. A. (2001) Oligomerization of polyalanine-expanded PABPN1 facilitates nuclear protein aggregation that is associated with cell death. Hum. Mol. Genet. 10, 2341–2351 [DOI] [PubMed] [Google Scholar]
- 27. Chartier A., Benoit B., Simonelig M. (2006) A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. EMBO J. 25, 2253–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brinkmann U., Mattes R. E., Buckel P. (1989) High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene 85, 109–114 [DOI] [PubMed] [Google Scholar]
- 29. Bosse-Doenecke E., Weininger U., Gopalswamy M., Balbach J., Knudsen S. M., Rudolph R. (2008) High yield production of recombinant native and modified peptides exemplified by ligands for G-protein coupled receptors. Protein Expr. Purif. 58, 114–121 [DOI] [PubMed] [Google Scholar]
- 30. Schröder-Tittmann K., Bosse-Doenecke E., Reedtz-Runge S., Ihling C., Sinz A., Tittmann K., Rudolph R. (2010) Recombinant expression, in vitro refolding, and biophysical characterization of the human glucagon-like peptide-1 receptor. Biochemistry 49, 7956–7965 [DOI] [PubMed] [Google Scholar]
- 31. Bienroth S., Wahle E., Suter-Crazzolara C., Keller W. (1991) Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J. Biol. Chem. 266, 19768–19776 [PubMed] [Google Scholar]
- 32. Mossessova E., Lima C. D. (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 [DOI] [PubMed] [Google Scholar]
- 33. Scheibel T., Parthasarathy R., Sawicki G., Lin X. M., Jaeger H., Lindquist S. L. (2003) Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc. Natl. Acad. Sci. U.S.A. 100, 4527–4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bocharova O. V., Breydo L., Parfenov A. S., Salnikov V. V., Baskakov I. V. (2005) In vitro conversion of full-length mammalian prion protein produces amyloid form with physical properties of PrP(Sc). J. Mol. Biol. 346, 645–659 [DOI] [PubMed] [Google Scholar]
- 35. Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G. P., Lehrach H., Wanker E. E. (1999) Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology. Proc. Natl. Acad. Sci. U.S.A. 96, 4604–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brown P., Liberski P. P., Wolff A., Gajdusek D. C. (1990) Resistance of scrapie infectivity to steam autoclaving after formaldehyde fixation and limited survival after ashing at 360 degrees C: practical and theoretical implications. J. Infect. Dis. 161, 467–472 [DOI] [PubMed] [Google Scholar]
- 37. Kong J., Yu S. (2007) Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 39, 549–559 [DOI] [PubMed] [Google Scholar]
- 38. Bousset L., Bonnefoy J., Sourigues Y., Wien F., Melki R. (2010) Structure and assembly properties of the N-terminal domain of the prion Ure2p in isolation and in its natural context. PLoS One 5, e9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sackewitz M., von Einem S., Hause G., Wunderlich M., Schmid F. X., Schwarz E. (2008) A folded and functional protein domain in an amyloid-like fibril. Protein Sci. 17, 1044–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sackewitz M., Scheidt H. A., Lodderstedt G., Schierhorn A., Schwarz E., Huster D. (2008) Structural and dynamical characterization of fibrils from a disease-associated alanine expansion domain using proteolysis and solid-state NMR spectroscopy. J. Am. Chem. Soc. 130, 7172–7173 [DOI] [PubMed] [Google Scholar]
- 41. Sambashivan S., Liu Y., Sawaya M. R., Gingery M., Eisenberg D. (2005) Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature 437, 266–269 [DOI] [PubMed] [Google Scholar]
- 42. Berciano M. T., Villagra N. T., Ojeda J. L., Navascues J., Gomes A., Lafarga M., Carmo-Fonseca M. (2004) Oculopharyngeal muscular dystrophy-like nuclear inclusions are present in normal magnocellular neurosecretory neurons of the hypothalamus. Hum. Mol. Genet. 13, 829–838 [DOI] [PubMed] [Google Scholar]
- 43. Verheesen P., de Kluijver A., van Koningsbruggen S., de Brij M., de Haard H. J., van Ommen G. J., van der Maarel S. M., Verrips C. T. (2006) Prevention of oculopharyngeal muscular dystrophy-associated aggregation of nuclear poly(A)-binding protein with a single-domain intracellular antibody. Hum. Mol. Genet. 15, 105–111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.