Background: The Parkinson-related kinase Pink1 is implicated in mitochondrial quality control.
Results: Integration of multiple targeting signals results in the localization of Pink1 and its processing product to the surface of the outer membrane.
Conclusion: Pink1 follows a unique import pathway that allows probing the functional integrity of mitochondria.
Significance: Determining the Pink1 targeting mode provides the basis for defining its molecular function.
Keywords: Mitochondria, Parkinson Disease, Pink1, Protein Motifs, Protein Translocation, Mitochondrial Targeting Sequence, Stop-transfer
Abstract
The Parkinson disease-associated kinase Pink1 is targeted to mitochondria where it is thought to regulate mitochondrial quality control by promoting the selective autophagic removal of dysfunctional mitochondria. Nevertheless, the targeting mode of Pink1 and its submitochondrial localization are still not conclusively resolved. The aim of this study was to dissect the mitochondrial import pathway of Pink1 by use of a highly sensitive in vitro assay. Mutational analysis of the Pink1 sequence revealed that its N terminus acts as a genuine matrix localization sequence that mediates the initial membrane potential (Δψ)-dependent targeting of the Pink1 precursor to the inner mitochondrial membrane, but it is dispensable for Pink1 import or processing. A hydrophobic segment downstream of the signal sequence impeded complete translocation of Pink1 across the mitochondrial inner membrane. Additionally, the C-terminal end of the protein promoted the retention of Pink1 at the outer membrane. Thus, multiple targeting signals featured by the Pink1 sequence result in the final localization of both the full-length protein and its major Δψ-dependent cleavage product to the cytosolic face of the outer mitochondrial membrane. Full-length Pink1 and deletion constructs resembling the natural Pink1 processing product were found to assemble into membrane potential-sensitive high molecular weight protein complexes at the mitochondrial surface and displayed similar cytoprotective effects when expressed in vivo, indicating that both species are functionally relevant.
Introduction
Mutations in the PTEN-induced putative kinase 1 (PINK1)2 gene give rise to an autosomal recessive form of Parkinson disease (1–3) by a mechanism that until now was still barely understood. The PINK1 gene encodes for a 581-amino acid protein (see Fig. 1A) that comprises a highly conserved kinase domain with homology to the serine/threonine kinases of the Ca2+/calmodulin family (4, 5) and localizes to mitochondria (5–10). Yet, the submitochondrial localization of Pink1 and its targeting mode remain highly controversial, and as previously pointed out (10), clarifying these questions is critical to identify its bona fide substrates and related signaling pathways.
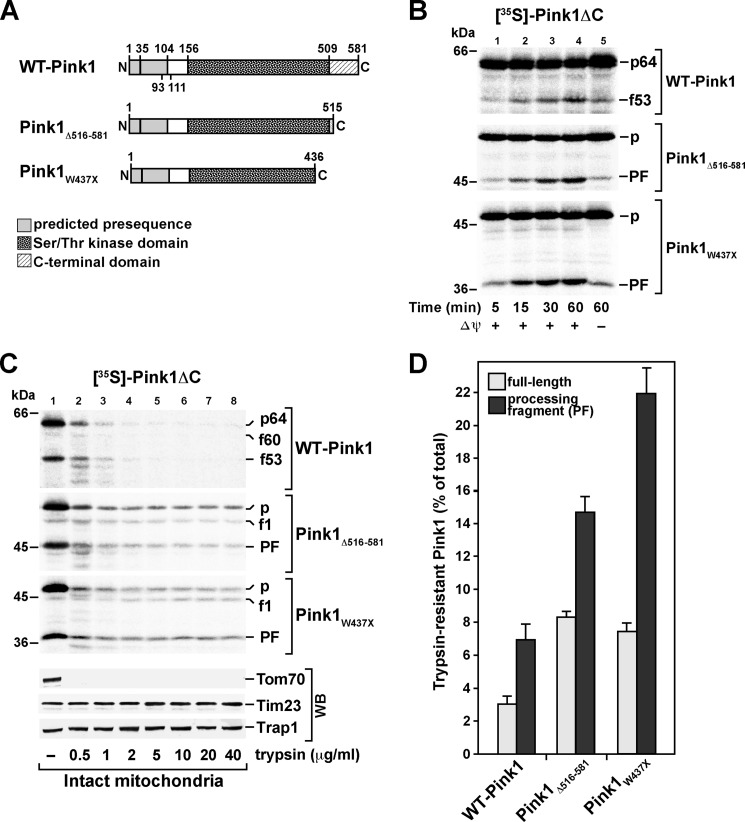
FIGURE 1.
Mitochondrial import of [35S]Pink1 in vitro. A, schematic representation of the wild-type Pink1 domain structure. Previously suggested processing sites consistent with the apparent molecular weight of Pink1f60 and Pink1f53, respectively (positions 35 and 104), and the proposed TMD (residues 93–111) are indicated. B, import of radiolabeled Pink1 into energized mitochondria for the indicated times as described under “Experimental Procedures.” Imported proteins were separated by SDS-PAGE and detected by digital autoradiography. Numbers indicate the approximate molecular weights of the respective Pink1 fragments. The translation product indicated with an asterisk in the input sample (lane 9) indicates a nonspecific labeling reaction of the reticulocyte lysate at ∼48 kDa. The lower panel represents a higher exposure of the autoradiogram for better visualization of Pink1f60. C, mitochondrial import of radiolabeled Pink1 after depletion of Δψ using valinomycin. The lower panel represents a higher exposure of the autoradiogram for better visualization of the processing fragments. D, processing efficiency for Pink1f60 and Pink1f53 as percentage of initial full-length (FL) Pink1p64 amount.
Like most mitochondrial proteins, Pink1 is encoded on the nuclear genome and synthesized as a precursor polypeptide at cytosolic ribosomes. Thus, Pink1 has to be targeted to its final mitochondrial localization in a process that is usually governed by specific signal sequences and accomplished via the protein translocation machinery present in the mitochondrial membranes (11, 12). The most common mitochondrial targeting signals are cleavable N-terminal extensions, which are initially recognized by the cytosol-exposed receptor proteins of the outer membrane. Although the very N terminus of Pink1 has been reported to be sufficient for mitochondrial localization of the protein (7, 8, 13), its actual targeting information and properties remain elusive. In addition, although according to some reports Pink1 primarily localizes to the mitochondrial outer membrane (MOM) (9, 10), it has been found in the intermembrane space (IMS) (7, 14, 15) as well as in the mitochondrial inner membrane (MIM) (7–9, 16). Because of this ambiguity in terms of targeting and localization, it has not been clarified up to now which of the different mitochondrial import pathways is followed by the Pink1 precursor. Besides, the formation of at least one N-terminal Pink1 processing product has been demonstrated in several studies (5, 7, 10, 13, 15, 17, 18).
The situation is confounded by the fact that under basal conditions the endogenous Pink1 protein is only barely detectable (if at all) by most antibodies commercially available (10), indicating that it is expressed at a rather low level in vivo. Thus, the bulk of information on its targeting and localization has been acquired by analysis of different artificial Pink1 reporter constructs that are usually highly overexpressed. To mimic the physiological situation more closely, we decided to study the mitochondrial import pathway of Pink1 by utilizing substoichiometric amounts of the radiolabeled Pink1 precursor protein that was incubated with isolated mitochondria in well established in vitro import assays. The targeting properties of the Pink1 precursor were characterized by analyzing the in vitro import of different deletion mutants as well as fusion constructs containing a dihydrofolate reductase (DHFR) reporter domain. Our findings unequivocally demonstrate that, although following entirely different import pathways, both full-length Pink1 and its major Δψ-dependent cleavage product ultimately localize to the outer leaflet of the MOM. Furthermore, we report the identification of two Δψ-sensitive Pink1-containing high molecular weight protein complexes by blue native gel electrophoresis.
Our results suggest a novel targeting mechanism to the MOM that involves dual Δψ-dependent processing of the Pink1 N terminus. However, whereas the very N terminus of Pink1 is dispensable for mitochondrial import and processing, its C terminus is important for proper localization of Pink1 and its processing product to the outer surface of the MOM. Elucidating its unique import behavior will certainly advance the molecular understanding of mitochondrial Pink1 and aid in reconciling published data on the functional characterization of Pink1.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated FCS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a saturated humidity atmosphere containing 5% CO2. SH-SY5Y cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 15% FCS, 1 mm l-glutamine, 4.5 g/liter glucose, 0.11 g/liter sodium pyruvate, and antibiotics as indicated above. Cells were transiently transfected either by use of the TurboFectTM transfection reagent (Fermentas) or the LTX Lipofectamine reagent (Invitrogen) according to the manufacturers' protocols and harvested 24 h post-transfection. Where indicated, cells were treated with valinomycin for the times indicated. Wild-type (WT) and PARL knock-out (KO) mouse embryonic fibroblasts (19) kindly provided by B. de Strooper (Institute for Neurodegenerative Diseases, KU Leuven, Belgium) were cultured in DMEM supplemented with 10% heat-inactivated FCS, 0.1 mm β-mercaptoethanol (tissue culture grade; Sigma-Aldrich), 2 mm l-glutamine, 50 units/ml penicillin, and 100 μg/ml streptomycin.
Molecular Cloning of Pink1 Constructs
A series of PINK1 deletion constructs (resulting in Pink1Δ1–33, Pink1Δ1–68, Pink1Δ1–83, Pink1Δ1–111, and Pink1Δ516–581) was generated by PCR using the human full-length wild-type cDNA as template and the appropriate oligonucleotide primers. Another set of Pink1 mutants analyzed in this study comprised different N-terminal Pink1 fragments (Pink1(1–33), Pink1(1–68), Pink1(1–83), and Pink1(1–115)) fused to the complete mouse DHFR sequence resulting in the reporter constructs Pink1(1–33)-DHFR, Pink1(1–68)-DHFR, Pink1(1–83)-DHFR, and Pink1(1–115)-DHFR. Amplification of the respective PINK1 fragments was achieved via PCR. Likewise, the DHFR sequence lacking its initial ATG codon was PCR-amplified. For fusion of the PINK1 fragments with the DHFR domain, a BamHI restriction site was introduced at the 3′-end of the PINK1 fragments and at the 5′-end of the DHFR sequence resulting in a GS linker between the Pink1 portions and the DHFR sequence. For in vitro synthesis, the truncated PINK1 versions as well as the DHFR fusion constructs were cloned into the pGEM®-4Z vector (Promega), which contains the bacterial SP6 promoter sequence for transcription. For in vivo expression, the constructs indicated were subcloned into the pcDNA3.1+ vector (Invitrogen) containing the CMV promoter for expression in mammalian cells. Further Pink1 mutants used in this study are the PD-associated Pink1A217D, Pink1G309D, and Pink1W437X as well as the engineered kinase-dead mutant Pink1K219M. The constructs for in vivo expression of these mutants using the pIRES2-EGFP vector (Clontech) have been described previously (20). For in vitro synthesis, the PD mutant constructs were inserted into the pGEM-4Z vector (Promega). All constructs used in this study were sequence-verified by next generation sequencing (GATC Biotech).
Isolation of Mitochondria from Cultured Cells
Isolation of mitochondria from cultured cells was performed essentially as described (21, 22). In brief, after harvesting, cells were twice washed in PBS and resuspended in HMS-A buffer (0.22 m mannitol, 0.07 m sucrose, 0.02 m HEPES, pH 7.6, 1 mm EDTA, 0.1% BSA, 1 mm PMSF). Separation of cell homogenates into the mitochondrial pellet and cytosol was achieved by differential centrifugation. The mitochondrial fraction was washed in HMS-B buffer (0.22 m mannitol, 0.07 m sucrose, 0.02 m HEPES-KOH, pH 7.6, 1 mm EDTA, 1 mm PMSF) and resuspended in HS buffer (20 mm HEPES-KOH, pH 7.6, 250 mm sucrose, 5 mm magnesium acetate, 80 mm potassium acetate, 7.5 mm glutamate, 5 mm malate, 1 mm DTT) for in vitro import experiments.
Import of Radiolabeled Proteins into Isolated Mitochondria
Synthesis and import of radiolabeled proteins into mitochondria in vitro was performed essentially as described (22). Radiolabeled precursor proteins were synthesized by in vitro transcription and translation in the presence of [35S]methionine/cysteine using the TnT-coupled reticulocyte lysate system (Promega). For the import reaction, mitochondria were diluted in HS buffer to a final concentration of 125–250 μg/ml at 30 °C in the presence of 2 mm ATP unless stated otherwise. Where indicated, the membrane potential (Δψ) was dissipated either by addition of 0.5 μm valinomycin alone or by a mixture consisting of 8 μm antimycin A, 0.5 μm valinomycin, and 2 μm oligomycin. The import reaction was terminated by addition of valinomycin and/or placing the samples on ice. Non-imported/protease-accessible mitochondrial proteins were digested by incubation with trypsin for 30 min on ice in the concentrations indicated in the figure legends. Where indicated, proteinase K instead of trypsin was added in the concentrations indicated in the figure legends, and incubation was carried out for 20 min on ice before addition of 1 mm PMSF. After pelleting the mitochondria (12 min, 12,000 × g, 4 °C) and washing in import buffer, samples were analyzed by SDS-PAGE and digital autoradiography. Where indicated, mitochondria were permeabilized using Triton X-100 as described under “Protein Solubility Assay,” and either trypsin or proteinase K was added to the mitochondrial lysates in the concentrations indicated.
Protein Solubility Assay
Isolated mitochondria were resuspended in lysis buffer (30 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5% glycerol, 2 mm EDTA, 0.3% Triton X-100, 1 mm PMSF, 1× protease inhibitor mixture (Roche Applied Science)) and incubated at 4 °C for 30 min with end-over-end rotation. After withdrawal of a total sample, mitochondrial lysates were centrifuged for 30 min at either 100,000, 20,000 or, 12,000 × g. Pellets (P100/20/12) containing Triton-insoluble material were resuspended in SDS sample buffer, and the supernatants (S100/20/12) were trichloroacetic acid (TCA)-precipitated before SDS-PAGE and Western blot.
Hypo-osmotic Swelling of Mitochondria
To disrupt their outer membranes, mitochondria were resuspended in hypo-osmotic buffer (20 mm KPi, pH 7.4, 0.000625% BSA) and incubated on ice for 30 min. After withdrawal of a total sample, mitoplasts (P13) were collected by centrifugation at 13,000 × g for 12 min. The resulting supernatants (S13) were precipitated using TCA, and samples were subjected to SDS-PAGE and Western blotting.
Sodium Carbonate Extraction
To distinguish peripheral from integral membrane proteins, intact mitochondria were resuspended in 0.1 m Na2CO3 and incubated on ice for 30 min. After withdrawal of a total sample, mitochondria were centrifuged at 100,000 × g for 30 min. Pellets (P100) containing integral membrane proteins were resuspended in SDS sample buffer, and the supernatants (S100) were TCA-precipitated before SDS-PAGE and Western blot.
Blue Native PAGE
To analyze mitochondrial proteins/protein complexes via blue native PAGE (23), mitochondria were solubilized in 20 mm Tris-HCl, pH 7.4, 50 mm NaCl, 10% glycerol, 1 mm EDTA, 1% digitonin, 1 mm PMSF. Where indicated, 2 mm ATP and 5 mm MgCl2 instead of EDTA were added in the solubilizing buffer. After a clarifying spin at 20,000 × g to remove non-solubilized material, 10× gel loading buffer (5% (w/v) Coomassie Brilliant Blue G-250, 100 mm Bis-Tris, pH 7.0, 500 mm ϵ-amino-n-caproic acid) was added, and samples were separated by 5–15% blue native PAGE.
Fluorescence-activated Cell Sorting (FACS) Analysis of Cultured Cells via Flow Cytometry
Analysis of Δψ via FACS using the potential-sensitive fluorescent dye tetramethylrhodamine ethyl ester (TMRE) (Molecular Probes, Invitrogen) was performed in HeLa cells overexpressing wild-type Pink1 or the mutants indicated. Cells were transiently transfected with the respective constructs, harvested 24 h post-transfection, and resuspended in PBS containing 0.1% BSA. After 10 min at 37 °C in the presence of 15 μm TMRE, cells were analyzed by flow cytometry (CyFlow® Space CY-S-3001, Partec). Control samples were included by analyzing mock-transfected cells as well as cells treated with the uncoupling K+ ionophore valinomycin.
Immunocytochemistry
HeLa cells were transiently co-transfected with GFP-PARKIN and different PINK1 constructs as indicated following the protocol for LTX Lipofectamine (Invitrogen). 24 h post-transfection, cells were treated with 1 μm valinomycin or vehicle for 1–3 h. Pink1 was stained using an antibody from Novus (BC100-494) at 1:100 dilution. Mitochondria were stained with anti-Tom20 from BD Transduction Laboratories (612278) at 1:400 dilution. Images and quantification were acquired using a Leica PS5 confocal microscope.
Miscellaneous Methods
Standard techniques were applied for SDS-PAGE, Western blot, and immunodecoration. Digital autoradiography was carried out using the FLA 5100 phosphorimaging system (Fujifilm). Quantitative analysis of imported 35S-labeled proteins was performed using MultiGauge software (Fujifilm). In the case of control immunodecorations, samples were transferred to PVDF membrane, and autoradiography of 35S signals was followed by Western blot and immunedecoration. Signal detection was performed by enhanced chemiluminescence (LumiLight, Roche Applied Science) either by developing x-ray films (Fujifilm) or via a charge-coupled device camera (LAS-4000, Fujifilm). Antibodies used were Pink1 (Novus Biologicals, BC100-494), Tom70 (Novus Biologicals), cytochrome c, Smac, Tim23, Tom40, aconitase 2 and α-tubulin (Sigma-Aldrich), F1β and Rieske-Fe/S protein (Molecular Probes), and Tom20 (Santa Cruz Biotechnology for Western blotting and BD Transduction Laboratories for immunocytochemistry). The PARL antibody used was kindly provided by L. Pellegrini (Cambridge Cancer Center, University of Cambridge, Cambridge, UK).
RESULTS
In Vitro Import Assay: Powerful Tool to Study Pink1 Localization, Topology, and Processing
As a prerequisite to our investigation on the mitochondrial import pathway of Pink1 and its submitochondrial localization, we developed an in vitro import assay for this protein. When synthesizing the human Pink1 protein in rabbit reticulocyte lysate in the presence of [35S]methionine/cysteine, three translation products were observed (Fig. 1B, lane 9), the largest of which was ∼64 kDa, representing the full-length precursor protein (Pink1p64). To monitor the import and proteolytic processing of Pink1 in vitro, the radiolabeled Pink1 translation products were incubated with isolated HeLa mitochondria in import buffer supplied with substrates and ATP for various times at 30 °C. Full-length Pink1 efficiently associated with mitochondria (Fig. 1B), and two major protein bands representing putative processing fragments, one of ∼53 kDa and another of ∼44 kDa, accumulated with time. After 30 s of import, a minor fragment of ∼60 kDa was detectable (Pink1f60; Fig. 1B). Quantification of the signals confirmed an increase in the formation of the 53-kDa species (Pink1f53) over time with processing efficiencies of about 51% after 60 min of incubation (Fig. 1D and Table 1). Based on its presence in the reticulocyte lysate and its fast accumulation at mitochondria (Fig. 1, B and C, lane 9 in each), we reasoned that the 44-kDa species (Pink1p44) is generated by an alternative translation start and therefore represents an abnormal precursor protein rather than a genuine processing fragment.
TABLE 1.
Import properties of N-terminal Pink1 deletion constructs
| Assay | WT | ΔN33 | ΔN68 | ΔN83 | ΔN111 |
|---|---|---|---|---|---|
| Import efficiency of full-length Pink1 (% of totala) | |||||
| +Δψ | 87 ± 11 | 73 ± 19 | 48 ± 9 | 35 ± 4 | 35 ± 5 |
| −Δψ | 101 ± 11 | 103 ± 33 | 62 ± 8 | 44 ± 2 | 41 ± 4 |
| Processing efficiency (% of Pink1PF of full-length Pink1a) | |||||
| +Δψ | 55 ± 4 | 69 ± 11 | 56 ± 2 | 55 ± 10 | |
| −Δψ | 18 ± 7 | 54 ± 2 | 48 ± 2 | 14 ± 4 | |
| Carbonate-resistant full-length Pink1 (% of total) | 73 ± 2 | 58 ± 3 | 52 ± 4 | 55 ± 4 | 55 ± 3 |
| Carbonate-resistant processed Pink1PF (% of total) | 51 ± 4 | 55 ± 3 | 49 ± 4 | 50 ± 5 | |
| Solubility in vitro (% of total) | 88 ± 9 | 91 ± 4 | 70 ± 10 | 68 ± 10 | 45 ± 12 |
a After 45 min of incubation as described under “Experimental Procedures.”
To revisit the effect of the Δψ on Pink1 biogenesis in our in vitro assay, we additionally analyzed the import and processing of radiolabeled Pink1 into mitochondria depleted of Δψ. The mitochondrial association of both precursor proteins Pink1p64 and Pink1p44 did not differ between mitochondria with and without Δψ (Fig. 1C). In contrast, formation of the main processing fragment Pink1f53 was dramatically reduced, as compared with energized mitochondria, albeit not entirely abolished (Fig. 1, C and D). In addition, yet another minor Pink1 fragment of ∼55 kDa (Pink1f55) was formed in depolarized mitochondria (Fig. 1C), whereas the minor processing fragment Pink1f60 was no longer detectable (Fig. 1C). Thus, the above results reveal important new aspects regarding Pink1 biogenesis such as the fact that the main 53-kDa cleavage fragment (Pink1f53) is still formed in uncoupled mitochondria. We also analyzed the import behavior of the PD-associated Pink1 mutants Pink1A217D and Pink1G309D. Both of these mutants associated with mitochondria and were processed like the wild-type protein (supplemental Fig. S1). However, in contrast to a former study (7), in the case of Pink1G309D, processing was only observed in energized mitochondria, which might have functional implications for this mutant.
Full-length Pink1 and Its Major Processing Product Localize to Mitochondrial Outer Membrane
To assess the intramitochondrial localization of the different Pink1 species, we incubated isolated HeLa mitochondria with increasing trypsin concentrations after import of radiolabeled Pink1 translation products as described above. Even at the lowest concentration (1 μg/ml), full-length Pink1p64 was degraded by >90% (Fig. 2A). Both the processing fragment Pink1f53 and the Pink1p44 species showed a slightly higher protease resistance but were likewise completely degraded when applying 5–10 μg/ml trypsin to mitochondria (Fig. 2A). Finally, the barely detectable Pink1f60 fragment displayed the highest relative degree of protease resistance. Thus, although displaying slightly differential trypsin susceptibilities, the three major Pink1 species associated with mitochondria; i.e. Pink1p64, Pink1f53, and Pink1p44 all behaved similarly to the endogenous mitochondrial protein Tom70, a MOM receptor whose major portion is exposed to the cytosol. By contrast, Pink1f60 remained largely stable, thus behaving similarly to the IMS-resident Smac or the matrix protein aconitase 2, which were not degraded even in the presence of 20 μg/ml trypsin. To account for neuron-specific factors relevant for the import and processing of Pink1, we imported [35S]Pink1 into mitochondria from the neuron-like SH-SY5Y neuroblastoma cell line. Import and processing as well as protease sensitivity of Pink1 were identical to those in HeLa mitochondria and thus do not seem to occur in a cell-type specific manner (supplemental Fig. S2).
FIGURE 2.
Submitochondrial localization of newly imported 35S-labeled Pink1 and Pink1 transiently overexpressed in vivo. A, protease titration of newly imported Pink1. After import of [35S]Pink1, mitochondria were incubated with trypsin in the concentrations indicated. As a control, immunodecorations against the endogenous proteins Tom70, Smac, and aconitase 2 (Aco2) were carried out. B, protease titration of Pink1 in vivo. Mitochondria isolated from cells transiently overexpressing Pink1 were incubated with trypsin in the concentrations indicated (lanes 1–6) or lysed in 0.3% Triton X-100 (TX-100) before addition of trypsin in the concentrations indicated (lanes 7–12). As a control, immunodecoration against the endogenous proteins Tom70, Tim23, and Rieske-Fe/S protein (RISP) were carried out. WB, Western blot.
The localization results presented above differ from published in vivo data (7, 15, 18). Thus, we reviewed Pink1 localization in vivo by performing a protease titration assay on mitochondria isolated from Pink1-overexpressing HeLa cells. As observed in the in vitro import assay under standard conditions, four different Pink1 species of ∼64, ∼60, ∼53, and ∼44 kDa were detected by a Pink1 antiserum (Fig. 2B, left panel); the specificity of the signals was confirmed by overexpression of Pink1-myc/His6 in HeLa cells and immunodetection using an anti-myc antibody (supplemental Fig. S3). Full-length Pink1 behaved most similarly to the newly imported precursor protein; i.e. it was almost entirely degraded at a concentration of 1 μg/ml trypsin, confirming its exposure to the outer leaflet of the MOM. By contrast, the processing fragment Pink1f53 and the Pink1p44 species remained largely stable in the presence of low trypsin concentrations (1–2 μg/ml) under in vivo conditions. Only at a concentration of 5 μg/ml trypsin or higher were these two fragments degraded. The Pink1f60 fragment displayed significant protease resistance even in the presence of 20 μg/ml trypsin, and as compared with the in vitro import experiments, it was increased. Thus, both our in vitro and in vivo data indicate that full-length Pink1p64 exhibits greater susceptibility to proteolysis as compared with the Pink1f53 fragment (as well as Pink1p44), whereas the highest degree of protease resistance in intact mitochondria was observed for the Pink1f60 fragment.
Differential protease susceptibility of mitochondrial proteins can be due to either differences in submitochondrial localization or conformational stability. To distinguish between these two possibilities, we monitored the protease resistance of the different Pink1 species after solubilization of mitochondria from transiently Pink1-overexpressing HeLa cells using Triton X-100. Although Pink1p64 and Pink1f60 were largely digested in the presence of 1 μg/ml trypsin, both Pink1f53 and Pink1p44 resisted trypsin treatment to a considerable extent after lysis of mitochondria (Fig. 2B, right panel). In contrast, the endogenous control proteins Tim23 and Rieske-Fe/S protein, a matrix-exposed component of respiratory chain complex III, were entirely degraded in the presence of 1–2 μg/ml trypsin, confirming the efficiency of lysis. Use of proteinase K instead of trypsin yielded identical results both in vitro and in vivo (data not shown). Thus, the differential protease sensitivities of Pink1p64 and Pink1f53 (and Pink1p44) as shown above reflect differences in the intrinsic properties of the different Pink1 forms rather than submitochondrial localization differences as was proposed previously (18).
When separating mitochondrial Triton X-100 lysates into soluble and pellet fractions by applying different centrifugation velocities, newly imported radiolabeled Pink1p64 was mainly detected in the soluble fractions (supplemental Fig. S4A). By contrast, even in the in vitro system, both Pink1f53 and Pink1p44 were found in the pellet fraction to a great extent after centrifugation at 100,000 × g (supplemental Fig. S4A, lanes 1–3) and still to some extent at 20,000 × g and even at 12,000 × g (supplemental Fig. S4A, lanes 4–9). Under overexpression conditions, an even larger fraction of both Pink1f53 and Pink1p44 still pelleted at 12,000 × g, whereas Pink1p64 and Pink1f60 were mainly detected in the supernatant fractions as observed in vitro (supplemental Fig. S4B).
Pink1 Processing Is TOM-dependent and It Requires Intramitochondrial ATP and Divalent Cations
Most mitochondrial proteins require the TOM complex for insertion into or crossing of the MOM. Initial recognition of a mitochondrial protein at the MOM is mediated by the two peripheral receptor proteins Tom20 and Tom70 (12). To analyze the involvement of the peripheral outer membrane receptors for targeting of Pink1 to mitochondria, we imported [35S]Pink1 into isolated mitochondria pretreated with trypsin to digest the cytosol-exposed domains of the two receptors. Both Tom20 and Tom70 were degraded at the lowest trypsin concentration (5 μg/ml), whereas the pore-forming β-barrel protein Tom40 was only slightly affected by the addition of protease (Fig. 3A). As another control, we co-imported the matrix-destined model precursor and Tom20 substrate pSu9-DHFR, a fusion protein comprising the F0 subunit 9 (Su9) presequence of the F1F0-ATP synthase from Neurospora crassa and the first 20 residues of the mature protein fused to the complete mouse dihydrofolate reductase (24, 25). Additionally, we co-imported the precursor of the adenine nucleotide carrier 3, a member of the metabolite carrier family (26) and a Tom70 substrate. As judged by the reduction in the mature sized protease-resistant forms, the import of both control proteins was reduced in trypsin-pretreated mitochondria in a concentration-dependent manner (Fig. 3A). In the case of the presequence protein pSu9-DHFR, association of the precursor was likewise reduced as a result of trypsin pretreatment. By contrast, the association of Pink1p64 and of Pink1p44 with mitochondria was not affected. Full-length Pink1 resisted Na2CO3 treatment almost entirely, indicating very stable association or integration into the outer membrane, even in the absence of the MOM surface receptors (Fig. 3B). The generation of Pink1f53, however, displayed a receptor dependence similar to the co-imported control proteins (Fig. 3A).
FIGURE 3.
Dependence of [35S]Pink1 import and processing on mitochondrial outer membrane receptors, ATP, and divalent cations. Radiolabeled Pink1 was incubated with isolated HeLa mitochondria as described for Fig. 1. Imported proteins were separated by SDS-PAGE and detected by autoradiography. As a control, samples were analyzed by Western blot (WB). A, dependence of Pink1 import and processing on the peripheral outer membrane receptors. Isolated mitochondria were incubated with the indicated trypsin concentrations prior to the import reaction (pretreatment). After trypsin inactivation, mitochondria were reisolated. Subsequent import of Pink1 with co-import of pSu9-DHFR (lanes 1–8) or adenine nucleotide carrier 3 precursor (Ant3; lanes 9–16) was performed under standard conditions. Non-imported/protease-accessible proteins were digested by treatment with proteinase K (PK) as described under “Experimental Procedures” (post-treatment). Digital autoradiography was followed by control immunodecoration against the TOM subunits Tom70, Tom40, and Tom20. B, carbonate extraction of newly imported Pink1 in mitochondria pretreated with trypsin as described in A. After co-import of Pink1 and pSu9-DHFR, mitochondria were reisolated and subjected to Na2CO3 extraction as described under “Experimental Procedures.” C, dependence of Pink1 import on ATP. Import of Pink1 and pSu9-DHFR was performed under standard conditions (lanes 1 and 5) or after depleting mitochondria (mitoch.) (lanes 2–4 and 6–8) or both mitochondria and reticulocyte lysate (lanes 3, 4, 7, and 8) of ATP. The Δψ was dissipated by valinomycin (lanes 4 and 8). Samples were treated with trypsin following the import reaction (lanes 5–8) as described under “Experimental Procedures.” D, dependence of Pink1 import on divalent cations. Import of Pink1 with co-import of pSu9-DHFR was performed under standard conditions (lane 1) or in the presence of o-phenanthroline (o-Phen.) in the indicated concentrations (lanes 2–6). E, import of radiolabeled Pink1 into mitochondria of WT and PARL−/− (ko) cells with co-import of pSu9-DHFR as described for Fig. 1. p, precursor; i, intermediate; m, mature; T, total; P, pellet; S, supernatant.
In most cases, once inside the mitochondria, the N-terminal targeting sequence of a mitochondrial protein is cleaved (12). This cleavage requires the translocation of the protein across the mitochondrial inner membrane, which relies on the Δψ and, in most cases, on intramitochondrial ATP. Former studies (7, 15, 17, 18) as well as the results presented above demonstrate the requirement of Δψ for Pink1 import and processing. To assess the requirement of ATP, we incubated [35S]Pink1 with isolated mitochondria that had been depleted of matrix ATP by an excess of apyrase (0.02 unit of apyrase/μg of mitochondrial protein) prior to the import reaction. We also added an excess of the F0-ATPase inhibitor oligomycin (2 μm) to prevent the de novo synthesis of ATP. Import and maturation of the co-imported Su9-DHFR precursor was entirely inhibited under these conditions (Fig. 3C, lower panel, lanes 2, 3, 6, and 7). The Pink1f53 fragment was similarly affected; i.e. its formation was impeded as a result of ATP depletion (Fig. 3C, upper panel, lanes 2 and 3). Instead, an alternative, slightly smaller protease-sensitive cleavage product (Pink1f50) accumulated. When Δψ was then dissipated by valinomycin, we observed a further reduction in Pink1f53 formation, whereas the alternative fragment Pink1f50 occurred as efficiently as in the presence of Δψ (Fig. 3C, upper panel, lane 4). In contrast, Pink1p64 and Pink1p44 remained unaffected by the depletion of ATP or Δψ; their mitochondrial association seemed to be even enhanced in de-energized mitochondria.
The Δψ- and ATP-dependent formation of Pink1f53 and its MOM receptor dependence indicate a cleavage event at the inner leaflet of the MIM or in the matrix. The major processing peptidase in the mitochondrial matrix, matrix processing peptidase (MPP), is a zinc-dependent metallopeptidase and can thus be inactivated by the presence of chelating agents (27, 28). We therefore imported [35S]Pink1 into isolated HeLa mitochondria in the presence of the lipid-soluble divalent metal ion chelator o-phenanthroline, an inhibitor of MPP-mediated processing (29). Maturation of the co-imported Su9-DHFR precursor, which is sequentially processed by the MPP, was inhibited with increasing o-phenanthroline concentrations (Fig. 3D). Despite the fact that we have previously reported that no predicted MPP consensus domain in Pink1 could give rise to a ∼53-kDa fragment (10), o-phenanthroline did abrogate the formation of Pinkf53. Several studies suggest the MIM protease PARL, a member of the rhomboid-like serine proteases (30), to be responsible for the generation of Pink1f53 (15, 18, 31). In keeping with this view, when importing radiolabeled Pink1 into mitochondria isolated from PARL knock-out mouse embryonic fibroblasts, we observed a dramatic reduction in the formation of Pink1f53 (Fig. 3E). However, the generation of Pink1f53 was not completely abolished in mitochondria derived from the PARL−/− mouse embryonic fibroblasts despite the fact that these cells were reported to have no detectable PARL mRNA (19).
Pink1 Sequence Upstream of Kinase Domain Undergoes Dual Processing
To further analyze processing of Pink1, we constructed a series of chimeric proteins containing the complete mouse DHFR as a passenger domain (Fig. 4A). The shortest of these fusion proteins, Pink1(1–33)-DHFR, comprising the N-terminal 33 residues of Pink1 fused to DHFR and thus similar to previously published reporter constructs that revealed residues 1–34 of Pink1 to be sufficient for mitochondrial targeting in vivo (7, 8, 10, 13), was used to investigate the significance of the N terminus for targeting and submitochondrial localization of Pink1 and to elucidate the cleavage site resulting in the formation of Pink1f60. Additionally, Pink1(1–68)-DHFR and Pink1(1–83)-DHFR were constructed to assess the relevance of the N-terminal segment between this first cleavage position and the second cleavage position resulting in Pink1f53 for proper submitochondrial targeting and processing. Finally, we included Pink1(1–115)-DHFR, which reportedly contains the main cleavage site as well as a hydrophobic domain suggested to act as a transmembrane domain (TMD) (7, 10), in our studies. If full-length Pink1 follows the import pathway of a presequence protein, this latter construct should be sufficient to reproduce the mitochondrial import and processing properties of full-length Pink1p64 as it contains the complete sequence preceding the main cleavage site. Consistent with former in vivo results, when incubating the 35S-labeled Pink1-DHFR fusion proteins with isolated HeLa mitochondria, all four constructs were imported and proteolytically processed in a Δψ-dependent manner (Fig. 4B). The Δψ-dependent N-terminal processing products found in all four cases were ∼3 kDa smaller than their full-length counterparts, consistent with cleavage at around residue 30. Of note, the band for the ∼3-kDa shorter products on SDS-PAGE (Fig. 4B) was abundant for Pink1(1–33)-DHFR, Pink1(1–68)-DHFR, and Pink1(1–83)-DHFR but only faint for Pink1(1–115)-DHFR. Previous studies including our own (10) have shown that the cleavage site responsible for generating the ∼53-kDa fragment is around residue 110. Thus, apart from Pink1(1–115)-DHFR, none of the three other fusion proteins contains this particular cleavage site. Consistent with this contention, only the Pink1(1–115)-DHFR construct was associated with a Δψ-dependent processing fragment ∼11 kDa smaller than the full-length construct that most likely corresponds to the main processing product Pink1f53 (Fig. 4B).
FIGURE 4.
Import, processing, and submitochondrial localization of [35S]Pink1-DHFR fusion constructs. A, schematic representation of the wild-type Pink1 domain structure and of Pink1-DHFR fusion constructs consisting of the N-terminal 33, 68, 83, or 115 Pink1 residues fused to the complete mouse DHFR. For wild-type Pink1, previously suggested processing sites consistent with the apparent molecular weight of Pink1f60 and Pink1f53, respectively (positions 35 and 104), and the proposed TMD (residues 93–111) are indicated. B–E, import of radiolabeled Pink1-DHFR fusion constructs into isolated HeLa mitochondria as described for Fig. 1. Samples were separated by SDS-PAGE and analyzed by digital autoradiography. For control immunodecorations, samples were transferred to PVDF membranes that were probed with the indicated antibodies. PF, processing fragment likely corresponding to Pink1f53. C, mitochondria were incubated with 50 μg/ml trypsin after the import reaction. D, submitochondrial localization of newly imported Pink1-DHFR fusion proteins. After import, mitochondria were resuspended in hypo-osmotic buffer in the presence of 25 μg/ml trypsin as described under “Experimental Procedures.” Control immunodecorations against the endogenous proteins Tom40, cytochrome c (Cyt c), Tim23, and aconitase 2 (Aco2) were carried out. Note that cytochrome c is not digestible by trypsin. PF*, processing fragment-derived trypsin fragment. E, carbonate extraction of newly imported radiolabeled Pink1(1–33)-DHFR and Pink1(1–115) in isolated HeLa mitochondria. After import of the fusion constructs, mitochondria were subjected to Na2CO3 extraction as described for Fig. 3B. Digital autoradiography was followed by control immunodecorations against the endogenous proteins Tom40, cytochrome c (Cyt c), Tim23, and aconitase 2 (Aco2). T, total; P, pellet; S, supernatant; OM, outer membrane; WB, Western blot.
The addition of 50 μg/ml trypsin after completion of import in intact mitochondria did not affect the Δψ-dependent cleavage products at all; i.e. as opposed to the wild-type protein, they were all translocated to a protease-protected location (Fig. 4C). To determine their submitochondrial localization, we imported the radiolabeled DHFR fusion proteins into isolated HeLa mitochondria followed by the generation of mitoplasts via hypo-osmotic swelling. At the same time, trypsin was added to digest all proteins outside the matrix compartment. The Δψ-dependent cleavage products were still protease-protected in the case of Pink1(1–33)-DHFR, Pink1(1–68)-DHFR, and Pink1(1–83)-DHFR, indicating that these chimera localize within the matrix (Fig. 4D, lanes 1–9). In contrast, the Pink1(1–115)-DHFR cleavage product became protease-accessible in the swelling assay and was partially digested to a smaller fragment (Fig. 4D, lanes 10–12). In addition, carbonate extraction of Pink1(1–33)-DHFR and Pink1(1–115)-DHFR following the import reaction revealed that both cleavage products observed with the DHFR reporter constructs behaved as soluble proteins (Fig. 4E).
Thus, our data indicate that the N terminus of Pink1, at least up to residue 83, unambiguously represents a bona fide matrix-targeting sequence that suffices to drive the translocation of the C-terminal DHFR domain across both mitochondrial membranes. The Pink1(1–115)-DHFR construct was trypsin-resistant in intact mitochondria, whereas it was protease-sensitive in mitoplasts. Hence, our results also demonstrate that the downstream segment, as present in the case of Pink1(1–115)-DHFR, contains a MIM stop-transfer signal preventing full translocation into the matrix and resulting in the generation of an IMS-located processing product. Finally, our results stress the fact that the sequence of Pink1 between amino acids 1 and 115 is the site of a dual proteolytic processing event (at ∼30 and ∼110).
N-terminally Deleted Pink1 Is Imported and Processed in Mitochondria
The above findings indicate how significant the N terminus of Pink1 is to target the protein to the mitochondria, and yet as previously shown in vivo, the deletion of the N-terminal sequence of Pink1 does not abolish targeting of the protein to the mitochondria (10). To address this apparently divergent set of observations, we constructed a series of deletion mutants lacking the N-terminal 33, 68, 83, and 111 residues, respectively, of the human Pink1 sequence and used them in our in vitro import assay (Fig. 5A). Incubation of isolated HeLa mitochondria with the 35S-labeled Pink1ΔN constructs resulted in the formation of a cleavage product in the case of Pink1Δ1–33, Pink1Δ1–68, and Pink1Δ1–83 very similar in size to Pink1f53 (Fig. 5B, upper panel). Additionally, Pink1Δ1–33 as well as Pink1Δ1–68 formed another, completely Δψ-independent cleavage product (Fig. 5B, arrowheads). Judged by its apparent molecular mass, this minor fragment is the result of cleavage at a position between residue 68 and residue 83 that possibly reflects a non-physiological processing event in the IMS. Processing of the deletion mutants was differentially dependent on the mitochondrial membrane potential. In the case of Pink1Δ1–33 and Pink1Δ1–68, it occurred largely independently of Δψ (Fig. 5B, lanes 6 and 9, and Table 1). Cleavage of Pink1Δ1–83 by contrast required an established Δψ (Fig. 5B, lane 12, and Table 1). Because the Pink1Δ1–83 cleavage product also migrated slightly differently from the cleavage products observed for the two larger mutants and from Pink1f53, it seems to result from a different processing mode. The Pink1Δ1–111 construct was not processed and displayed a migration on SDS-PAGE similar to that of Pink1f53, confirming that cleavage of the full-length protein occurs around residue 110. Incubation of mitochondria with 10 μg/ml trypsin following the import of the different Pink1ΔN constructs resulted in almost complete degradation of both full-length and processed species in all four cases (Fig. 5B, lower panel). The import efficiencies of the different proteins were determined by quantification of the signals obtained for the full-length proteins after 45 min of import in the presence of Δψ (the respective input of precursor proteins was set to 100%). Mean import efficiencies as determined from three independent experiments were about 87% in the case of the full-length wild-type protein (Table 1). N-terminal deletion even up to the major cleavage site (around residue 110) did not abolish the mitochondrial targeting of Pink1 but resulted in a gradual decrease in its import efficiency. Furthermore, we quantified the processing efficiencies of the ΔN mutants in comparison with those of the wild-type protein after 45 min in the presence of Δψ by determining the ratios between the non-processed full-length constructs and the processing products. For the wild-type protein, mean processing efficiencies were at around 55% (Table 1). Processing occurred with comparable efficiencies for all deletion mutants analyzed. Next, we tested the carbonate extractability of the newly imported 35S-labeled Pink1ΔN mutants (Table 1). The Pink1ΔN precursors as well as their cleavage products all behaved similarly with ∼50% found in the P100 fractions. We additionally performed mock extractions in which mitochondria were omitted to exclude aggregation as the reason for pelleting of the deletion constructs. To that end, the 35S-labeled wild-type protein and the Pink1ΔN constructs were resuspended in Na2CO3 with subsequent separation of soluble from insoluble fractions by centrifugation at 100,000 × g. Wild-type Pink1 and Pink1Δ1–33, Pink1Δ1–68, and Pink1Δ1–83 were largely recovered in the soluble fractions (Table 1). By contrast, more than 50% of the Pink1Δ1–111 construct pelleted (Table 1). Thus, the apparent carbonate resistance of the shorter deletion constructs, particularly that of Pink1Δ1–111, which is very close in size to Pink1f53, might reflect their aggregation propensity rather than integration into the MOM.
FIGURE 5.
Import, processing, and submitochondrial localization of 35S-labeled Pink1 N-terminal deletion constructs. A, schematic representation of the wild-type Pink1 domain structure and of Pink1 deletion constructs lacking the N-terminal 33, 68, 83, or 111 residues. For wild-type Pink1, previously suggested processing sites consistent with the apparent molecular weight of Pink1f60 and Pink1f53, respectively (positions 35 and 104), and the proposed TMD (residues 93–111) are indicated. B, import of radiolabeled wild-type Pink1 and Pink1ΔN constructs into isolated HeLa mitochondria with subsequent separation of samples by SDS-PAGE and analysis by digital autoradiography as described for Fig. 1. For the lower panel, mitochondria were incubated with 10 μg/ml trypsin after the import reaction. The additional cleavage products detected in the case of Pink1Δ1–33 and Pink1Δ1–68 are indicated with arrowheads.
Taken together, deletion of the very N-terminal segment neither impeded mitochondrial association of Pink1 nor its proteolytic processing inside mitochondria. However, the efficiencies of mitochondrial targeting and membrane association declined with an increase in the length of the deleted N-terminal portion. Furthermore, the Δψ dependence of the cleavage reaction varied with the different N-terminal truncations, indicating that membrane transport and processing of Pink1 are not as tightly coupled as observed for typical presequence proteins.
Pink1 C Terminus Promotes Outer Membrane Retention
Because N-terminal deletion of Pink1 did not abolish its mitochondrial localization, further targeting information governing its MOM localization must be contained elsewhere in the protein. To analyze potential effects of the C-terminal domain on targeting, submitochondrial localization, and membrane association of Pink1, we made use of a truncation mutant lacking the C-terminal 66 residues of the full-length protein (Pink1Δ516–581). Additionally, we included the PARK6 non-sense mutant Pink1W437X in our analysis (Fig. 6A). Import of [35S]Pink1Δ516–581 into isolated HeLa mitochondria resulted in its Δψ-dependent accumulation and processing to a major fragment of ∼45 kDa, whereas Pink1W437X was Δψ-dependently cleaved to a ∼37-kDa fragment (Fig. 6B, middle and lower panels, lanes 1–4 in each). Hence, both of these cleavage events likely correspond to the processing of wild-type Pink1p64 to Pink1f53 (Fig. 6B, upper panel). Of note, processing efficiencies of the two mutants were increased with the length of the deleted fragment in comparison with the WT protein. To clarify the submitochondrial localization of Pink1Δ516–581 and of Pink1W437X, we performed protease titration assays of the newly imported proteins as described for the wild type. Both C-terminal truncation mutants exhibited enhanced trypsin resistance (Fig. 6, C and D). Quantification of the signals revealed an about 2.5-fold higher stability for both full-length Pink1Δ516–581 and Pink1W437X toward the maximum trypsin concentration (40 μg/ml), whereas the processing fragment likely corresponding to Pink1f53 exhibited 2-fold higher protease resistance in the case of Pink1Δ516–581 and even 3-fold greater resistance in the case of Pink1W437X as compared with the wild-type cleavage product (Fig. 6D). To exclude conformational alterations as the cause for the increase in protease resistance, we monitored the effect of trypsin on the newly imported C-terminal truncation mutants in the presence of Triton X-100. In the mitochondrial detergent lysates, both mutants were entirely degraded at a concentration of 1 μg/ml trypsin as was the wild-type protein (supplemental Fig. S5). The above described differential protease sensitivity of the full-length protein versus the cleavage product in the case of wild-type Pink1 was likewise observed for the two mutants; i.e. the cleavage products displayed higher protease resistance than the non-processed constructs both in intact mitochondria and in detergent lysates.
FIGURE 6.
Import, processing, and submitochondrial localization of 35S-labeled C-terminal Pink1 deletion constructs. A, schematic representation of the wild-type Pink1 domain structure, of a C-terminal Pink1 deletion construct lacking the C-terminal 66 residues (Pink1Δ516–581), and of the PD-related Pink1W437X mutant. For wild-type Pink1, previously suggested processing sites consistent with the apparent molecular weight of Pink1f60 and Pink1f53, respectively (positions 35 and 104), and the proposed TMD (residues 93–111) are indicated. B, import of radiolabeled wild-type Pink1 and C-terminal deletion mutants into isolated HeLa mitochondria as described for Fig. 1 with subsequent separation of samples by SDS-PAGE and analysis by digital autoradiography (p, full-length precursor protein; PF, processing fragment corresponding to Pink1f53). C, protease titration of mitochondria following import of wild-type Pink1 (upper panel), Pink1Δ516–581 (middle panel), and Pink1W437X (lower panel). After the import reaction, mitochondria were incubated with trypsin in the indicated concentrations. As a control, immunodecorations against the endogenous proteins Tom70, Tim23, and Trap1 were carried out (p, full-length precursor protein; f1, intermediate processing fragment corresponding to Pink1f60; PF, processing fragment corresponding to Pink1f53). D, quantification of the trypsin resistance of radiolabeled full-length forms and the main cleavage fragments (processing fragment) of WT-Pink1 and the C-terminal deletion constructs after import in intact mitochondria. Autoradiography signals for the indicated Pink1 fragments in the presence of 40 μg/ml trypsin were quantified and depicted as percentage of the imported amounts without protease treatment. Mean values and S.E. were determined for n = 4 independent experiments. WB, Western blot.
Thus, Pink1Δ516–581 and Pink1W437X are imported into mitochondria and processed in a Δψ-dependent fashion similar to the wild-type protein. The increase in both processing efficiency and protease resistance as compared with wild-type Pink1 indicates a higher proportion of the two C-terminal deletion mutants to fully cross the MOM. Hence, the C terminus of Pink1 influences its mitochondrial targeting and final submitochondrial localization.
Pink1 Assembles into Δψ-sensitive High Molecular Weight Protein Complexes
To complete our study on the biogenesis of Pink1, we assessed whether it may be involved in mitochondrion-associated protein complexes by blue native (BN) PAGE. During the import reaction of 35S-labeled Pink1, either mitochondria were fully energized by the addition of substrates and ATP or the Δψ was disrupted using valinomycin. Subsequently, mitochondria were reisolated and solubilized in digitonin buffer. In energized mitochondria, diffuse Pink1 signals were detected in the size ranges between ∼66 and ∼146 kDa and from ∼480 to ∼700 kDa (Fig. 7A, lanes 1 and 2). The intensities of these signals declined after longer incubation periods with established Δψ. In the samples containing valinomycin during import, distinct signals at ∼700 kDa that increased over time were detected, whereas additional signals between ∼66 and ∼146 kDa were diminished during the incubation period (Fig. 7A, lanes 3 and 4). In control samples denatured in SDS sample buffer, the protein was exclusively found in the monomeric state, confirming the specificity of the signals from the digitonin-solubilized mitochondria (Fig. 7A, lanes 5 and 6).
FIGURE 7.
BN-PAGE of wild-type and mutant Pink1 in vitro and in vivo. A, BN-PAGE of newly imported [35S]Pink1 in isolated HeLa mitochondria. Import of the wild-type Pink1 protein was performed as described for Fig. 1 under standard conditions (lanes 1 and 2) or after dissipation of the Δψ using valinomycin (lanes 3–6). Mitochondria were resuspended in solubilization buffer containing 1% digitonin (lanes 1–4) or in SDS sample buffer (lanes 5 and 6) and subjected to native PAGE. Signals were detected by digital autoradiography. Signals indicated with an asterisk most likely reflect nonspecific signals from the reticulocyte lysate. B, BN-PAGE of mitochondria isolated from SH-SY5Y cells. Wild-type SH-SY5Y cells were cultured in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of 1 μm valinomycin for 12 h. Isolation of mitochondria and preparation of samples for BN-PAGE were as described in A. Proteins were transferred to PVDF membrane and immunodecorated against Pink1, Tom40, and Tim23. C, BN-PAGE of newly imported 35S-labeled wild-type and mutant Pink1. Import of the indicated Pink1 mutant constructs into isolated HeLa mitochondria either in the presence (−Δψ) or in the absence (+Δψ) of valinomycin as well as BN-PAGE were performed as described for A. Signals indicated with an asterisk most likely reflect nonspecific signals from the reticulocyte lysate. D, BN-PAGE of endogenous wild-type Pink1 and of Pink1ΔN overexpressed in SH-SY5Y cells. Wild-type/transfected SH-SY5Y cells were cultured in the absence (+Δψ) or presence (−Δψ) of 1 μm valinomycin for 12 h. Isolation of mitochondria and preparation of samples for BN-PAGE were as described in A. Proteins were analyzed by Western blot using antibodies against Pink1. Signals were adjusted according to the solubilities of the respective constructs.
The mitochondrial association of Pink1 has been shown to increase in response to a drop in the mitochondrial membrane potential (17, 32, 33). Thus, to assess the relevance of Pink1 complex formation in vivo, we cultured SH-SY5Y cells with and without valinomycin for 12 h prior to harvest. Subsequently, mitochondria were isolated and subjected to BN-PAGE as described above. Interestingly, two high molecular mass complexes, one of around 700 kDa as observed after in vitro import and a larger one at around 900 kDa, were detectable by a Pink1-specific antiserum even at endogenous Pink1 expression levels (Fig. 7B, lane 2). In control samples with established Δψ, as expected, no Pink1 signals were detectable by Western blot (Fig. 7B, lane 1). Hence, as observed for the radiolabeled protein, depolarization of mitochondria under physiological conditions resulted in the formation of specific Pink1-containing protein complexes in vivo. Of note, under Pink1-overexpressing conditions in SH-SY5Y cells, the high molecular weight complexes were not only detected in mitochondria of valinomycin-exposed cells but also in those isolated from untreated control cells, albeit in reduced amounts (data not shown). To test whether the Pink1 complexes merely represent translocation intermediates, we immunodetected Tom40 and Tim23, the core components of the TOM complex and the presequence translocase, respectively (Fig. 7B, lanes 3–6). However, no co-migration was detected with Tom40 migrating at around 480 kDa, whereas Tim23 was detected in a complex between 66 and 146 kDa under both conditions. Of note, in polarized mitochondria, Tim23 signals were additionally detected in the range above 1 MDa, potentially representing an interaction of the import machinery with respiratory chain complexes as was recently demonstrated in yeast mitochondria (34).
As demonstrated, Pink1 processing is largely inhibited in the absence of Δψ; i.e. Pink1p64 accumulates at depolarized mitochondria, whereas Pink1f53 is only generated to a minor extent. Although this suggests that the full-length protein is the predominant Pink1 species present in the high molecular weight protein complexes, we wondered whether Pink1f53 is likewise involved in complex formation. We addressed this issue by use of the radiolabeled Pink1Δ1–83 and Pink1Δ1–111 deletion constructs, both of which are close in size to Pink1f53, thus mimicking the behavior of the natural cleavage product. Additionally, the Pink1Δ516–581 mutant was included to check for the requirement of the C-terminal domain for complex formation. The Pink1Δ1–83 construct displayed diffuse and relatively weak initial staining that was even further diminished after longer incubation times in the absence of Δψ (Fig. 7C, lanes 3 and 4). With established Δψ, the deletion mutant was barely detectable (Fig. 7C, lane 9). No higher molecular weight signals were detectable for the newly imported Pink1Δ1–111 either in the presence or in the absence of Δψ (Fig. 7C, lanes 5, 6, and 11). In the case of Pink1Δ516–581, by contrast, complex formation occurred at least as efficiently as for the wild-type protein in the absence of Δψ (Fig. 7C, lanes 7 and 8), whereas in energized mitochondria, it was largely impeded (Fig. 7C, lane 12). Control samples solubilized in SDS sample buffer revealed that the overall low signal strength in the case of the two N-terminal deletion mutants was due to their poor digitonin solubility (data not shown). Hence, possible BN-PAGE signals might have been below the detection limit in the in vitro system. Therefore, we additionally monitored the native state of the three deletion mutants in vivo. To that end, SH-SY5Y cells were transfected with the different deletion constructs, and mitochondria isolated from the transfectants were analyzed by BN-PAGE. Interestingly, both the C-terminal truncation mutant and the two N-terminal deletion constructs were found in the 700-kDa complex in mitochondria prepared from valinomycin-treated cells, whereas no signals were detectable in the size range of the larger complex (Fig. 7D, lanes 3–8).
Hence, neither deletion of the N terminus nor deletion of the extreme C terminus impeded complex formation per se but specifically inhibited the formation of the 900-kDa complex. Consequently, the natural cleavage product Pink1f53 might well contribute to the 700-kDa complex found in isolated mitochondria, whereas the 900-kDa complex that most likely involves additional cytosolic components might exclusively contain the full-length protein.
N-terminally Truncated Pink1 Exhibits Protective Functions in Vivo
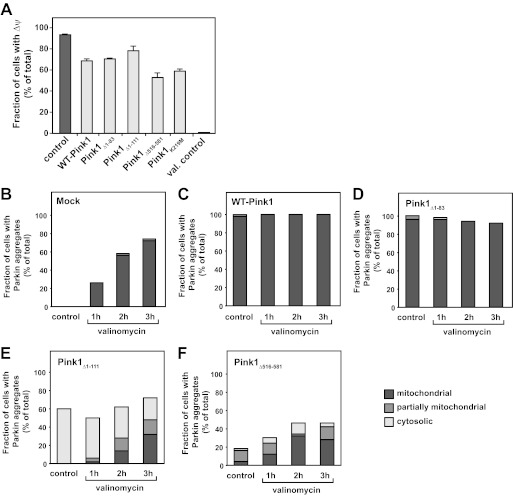
The Δψ-sensitive assembly of Pink1 into multimeric complexes that predominantly accumulate in depolarized mitochondria, i.e. when Pink1 processing is largely impeded, points to potential functional differences between the full-length protein and its main cleavage product Pink1f53. Accordingly, we made use of the Pink1Δ1–83 and Pink1Δ1–111 deletion constructs to study possible functional implications of Pink1 cleavage in vivo. We also included Pink1Δ516–581 in the analysis to examine the functional relevance of the C-terminal domain. Pink1 has repeatedly been implicated in the maintenance of mitochondrial integrity and functionality, which in turn depend on the membrane potential. Thus, we addressed the effect of either WT-Pink1 or the deletion mutants close in size to the main cleavage product Pinkf53 as well as the mutant lacking the C-terminal domain on Δψ in living cells. Transiently transfected cells either overexpressing wild-type Pink1 or the Pink1Δ1–83, Pink1Δ1–111, or Pink1Δ516–581 mutant constructs as well as mock-transfected cells were harvested 24 h post-transfection, and the membrane potential was assessed by FACS using the potential-dependent fluorescent dye TMRE. As shown in Fig. 8A, overexpression of Pink1 under basal conditions (under which endogenous Pink1 is apparently expressed at a very low level) had no protective effect as compared with control cells but rather resulted in a reduction of the number of cells with Δψ (93.3 ± 0.8% in control cells versus 68.5 ± 2.1% in WT Pink1-overexpressing cells), indicating that Pink1 expression levels are tightly regulated and critical for mitochondrial integrity. Overexpression of Pink1Δ1–83 likewise resulted in a ∼20% reduction in Δψ (70.6 ± 0.6%). By contrast, when overexpressing Pink1Δ1–111, Δψ displayed significantly higher values more similar to those of control cells (78.2 ± 3.3%). Deletion of the C-terminal domain resulted in a decrease of TMRE fluorescence by almost 50% (52.9 ± 4.4%), indicating the functional significance of the C terminus. Finally, we included the kinase-deficient engineered Pink1K219M mutant (5, 35), which displayed a reduced fraction of cells with Δψ as compared with both control cells and WT-Pink1-overexpressing cells (58.9 ± 1.9%), a behavior similar to that of the C-terminal deletion mutant.
FIGURE 8.
Functional analysis of Pink1 deletion constructs in vivo. A, analysis of the membrane potential by FACS analysis. HeLa cells transiently transfected with wild-type Pink1 or the deletion constructs indicated were harvested 24 h post-transfection, and Δψ was monitored using the potential-sensitive fluorescent dye TMRE via FACS. As controls, mock-transfected cells and cells transfected with the engineered kinase-deficient Pink1 mutant Pink1K219M were included in the analysis. Mean values and S.E. were determined from four independent experiments performed in duplicates (n = 8). B–F, Parkin translocation to mitochondria in HeLa cells transiently transfected with GFP-Parkin alone (B; Mock) and together with WT-Pink1 (C) or the deletion mutant Pink1Δ1–83 (D), Pink1Δ1–111 (E), or Pink1Δ516–581 (F). Transfected cells were either treated with vehicle (DMSO) or with 1 μm valinomycin for 1–3 h. Mitochondrial co-localization of GFP-Parkin was assayed by staining of transfected cells with anti-Tom20. Images and quantification were acquired on a Leica PS5 confocal microscope. Representative images are shown in supplemental Fig. S6. Quantifications were done from at least three independent experiments with a total of 50 cells per construct and per condition tested. val., valinomycin.
The E3 ubiquitin ligase Parkin has been shown to associate with mitochondria in a Pink1-dependent manner (20, 32, 36–38). Thus, we additionally investigated the effects of our N- and C-terminal deletion mutants on Parkin translocation to mitochondria in response to Δψ dissipation. To that end, HeLa cells were transiently co-transfected with GFP-Parkin and either wild-type Pink1 or the Pink1Δ1–83, Pink1Δ1–111, or Pink1Δ516–581 mutant constructs. Valinomycin was added 24 h post-transfection for another 1–3 h, and Parkin translocation to mitochondria was assessed by immunofluorescence (representative microscopic images are shown in supplemental Fig. S6). Of control cells only transfected with GFP-Parkin, ∼72% displayed GFP-Parkin co-localization with mitochondria after 3 h of valinomycin treatment (Fig. 8B). As shown before (20), overexpression of wild-type Pink1 resulted in Parkin recruitment to mitochondria in ∼100% of co-transfected cells with and without uncoupler (Fig. 8C). Overexpression of Pink1Δ1–83 had an effect similar to that of the wild-type protein with ∼96% of co-transfected cells showing GFP-Parkin in mitochondrion-associated aggregates even in the absence of valinomycin (Fig. 8D). In contrast, deletion of the N-terminal 111 amino acids of Pink1 interfered with Parkin recruitment. Instead, a high proportion of cells with cytoplasmic Parkin aggregates were found both in the absence (∼60%) and in the presence of valinomycin (∼24%) (Fig. 8E). After 3 h of valinomycin treatment, still only ∼32% of GFP-Parkin was found to co-localize with mitochondria. Similarly, C-terminal deletion impaired mitochondrial Parkin recruitment (Fig. 8F); i.e. only ∼30% of co-transfected cells exhibited Parkin translocation to mitochondria in response to valinomycin when overexpressing Pink1Δ516–581.
DISCUSSION
Pink1 has been designated as a mitochondrial kinase (39, 40), but its mode of targeting and import and its submitochondrial localization are still controversial. As published elsewhere (7, 18), Pink1 import and processing display some similarities to those of a presequence protein. However, although involving N-terminal cleavage, our study demonstrates that the mitochondrial import of Pink1 deviates from the typical presequence pathway (11, 12) in several crucial aspects. First, the processing of full-length Pink1 into its main cleavage product Pink1f53 occurs to an atypically low degree (only ∼50%). Second, the formation of Pink1f53 is strongly promoted by the Δψ but not abolished in its absence. And third, both Pink1p64 and Pink1f53 remain protease-accessible from the outside of mitochondria after in vitro import.
Both our in vitro and our in vivo results are consistent with Pink1p64 as well as Pink1f53 being localized to the outer leaflet of the MOM. This is in apparent contradiction to previously published in vitro import assays in which these two Pink1 species were shown to be partially protease-protected (7, 18). Our analysis of the protease susceptibility of Pink1 after detergent permeabilization of mitochondria revealed that the increased protease resistance of Pink1f53 (and Pinkp44) as compared with Pink1p64, is not due to a protection by the mitochondrial membranes. Instead, our results support the notion that certain intrinsic properties of the Pink1f53 fragment (e.g. an increased aggregation propensity) account for its increased protease resistance. Thus, could the low solubility of Pink1f53 result in its apparent protease resistance when expressed in vivo (7, 18)?
The MIM protease PARL, a member of the rhomboid-like serine proteases (30), has been suggested to mediate the formation of Pink1f53 (15, 18, 31). Given that rhomboid proteases are not known to involve metal ions and that we found a profound inhibition of Pink1 cleavage by the metal ion chelator o-phenanthroline, Pink1 likely undergoes a more complex proteolytic processing pattern than hitherto believed. For instance, Pink1f53 might result from a two-step processing; i.e. the first cleavage would be catalyzed by a metal ion-dependent peptidase (e.g. the matrix-localized MPP), and the second step would be catalyzed by PARL. The minor N-terminal fragment Pink1f60 possibly represents a processing intermediate. This molecular weight indication is an approximate value based on the migration behavior on SDS-PAGE and Western blot, respectively. A ∼60-kDa Pink1 fragment has been suggested to represent an alternative MPP-dependent processing product present in PARL-deficient cells (18). In our assay, Pink1f60 was also detectable in wild-type cells, and it behaved as a cleavage product generated in the matrix; i.e. its formation was Δψ-dependent, and it resisted protease treatment. It is also worth noting that Pink1f53 formation was still observed in mitochondria from PARL−/− cells. The Afg3l2 subunit of the MIM protease mAAA has been suggested to catalyze Pink1f53 formation in the absence of PARL (41). Cleavage by the matrix-exposed mAAA would require the Δψ-dependent translocation of Pink1 across the MIM (42). However, the residual Pink1f53 formation in PARL-deficient mitochondria occurred with even increased efficiency in uncoupled mitochondria as compared with mitochondria from wild-type cells. Consequently, mAAA might indirectly contribute to the cleavage of Pink1 as discussed for the mitochondrial fusion factor Opa1 (43). Because Pink1f53 was still detected in the absence of Δψ, Pink1f60 does not seem to represent an essential processing intermediate in the formation of Pink1f53; i.e. the two fragments could alternatively originate from independent cleavage events. Different pools of Pink1f53 depending on the energy status of mitochondria might also exist. The Δψ-dependently generated Pink1f53 would be derived from Pink1f60, whereas in the absence of Δψ, Pink1f53 might be directly generated from Pinkp64. Differential processing modes in dependence of the mitochondrial energy status have also been described for Opa1 (43, 44). It is also conceivable that initial cleavage of the Pink1 N terminus by a metallopeptidase of the MIM or the matrix allows the subsequent PARL-mediated cleavage by means of conformational alterations in Pink1 that result in the exposure of the PARL recognition motif (45, 46). Conversely, in de-energized mitochondria, i.e. in the absence of cleavage of the extreme Pink1 N terminus, the PARL recognition motif might be only poorly accessible. In line with this hypothesis and as demonstrated by analysis of N-terminal Pink1 deletion mutants, the very N terminus of Pink1 promotes the Δψ dependence of the cleavage reaction, most likely due to its high proportion of positive charges, but it is dispensable for the generation of Pink1f53. Furthermore, deletion of the extreme Pink1 N terminus resulted in enhanced Pink1f53 formation when Δψ was dissipated as compared with the wild-type protein (see Table 1).
As demonstrated by our N-terminal Pink1-DHFR reporter constructs, the Pink1 N terminus at least up to residue 83 promotes import into the mitochondrial matrix. The Pink1(1–115)-DHFR construct in which the previously reported main cleavage site (39) is available was mainly processed to an ∼11-kDa smaller fragment as observed for the wild-type protein. This processing product was primarily localized in the IMS, indicating that the N-terminal segment between residues 83 and 115 interferes with complete inner membrane passage, thus supporting the recently suggested role of the hydrophobic stretch between residues 93 and 111 as a “stop-transfer” sequence (10, 47) as found in a subset of presequence proteins destined for the IMS or the MIM (12, 48, 49). Furthermore, the targeting of Pink1(1–115)-DHFR demonstrates that even the complete putative stop-transfer sequence, which was alternatively proposed to act as a TMD (7), was not sufficient to retain the fusion protein in the MOM. The localization of both Pink1p64 and Pink1f53 at the outer leaflet of the MOM together with the inability of the Pink1-DHFR fusion proteins to reproduce the targeting properties of wild-type Pink1 as well as mitochondrial association and cleavage or our N-terminal Pink1 deletion constructs in mitochondria point to the existence of additional targeting information even downstream of the main processing site. In support of this notion, a hybrid construct consisting of the N-terminal presequence of a mitochondrial inner membrane protein fused to the Pink1 C terminus (residues 151–581) is retained at the outer face of the MOM (47). Furthermore, an enhanced protease resistance of C-terminally truncated Pink1 in our import assay points to the significance of the C-terminal domain for Pink1 targeting and submitochondrial localization.
A growing number of studies describe individual mitochondrial import pathways that involve N-terminal cleavage but do not follow the classical presequence pathway. The two NADH-cytochrome b5 reductase forms encoded by the yeast MCR1 gene display a phenomenon that is referred to as alternative topogenesis (50); i.e. one Mcr1 form resides in the MOM, whereas the other form is located in the IMS (51–53). Like full-length Pink1, the Mcr1 N terminus contains a presequence-like segment followed by a rather hydrophobic stretch, thus resembling bipartite signal-containing precursors (12). TOM-mediated MOM translocation and Δψ-dependent cleavage of the N-terminal targeting signal result in the smaller IMS-located Mcr1 isoform. On the other hand, the Mcr1 N terminus behaves similarly to the signal-anchor domains of MOM proteins (54) resulting in its TOM-independent MOM integration. Another well studied example of a dually localized protein encoded by a single gene is represented by the fumarase enzyme (55–58). In this case, all translation products are first imported into mitochondria and processed in the matrix followed by retrotranslocation of a subset of mature fumarase to the cytosol; the precise mechanism allowing for backward movement of a protein through the mitochondrial translocation channels is still elusive.
In light of these examples of atypical mitochondrial import pathways and of our results, we propose the following model to explain the import and processing of Pink1 (Fig. 9). After its cytosolic translation, a fraction of Pink1p64 undergoes rapid folding, hence preventing its efficient translocation across the MOM and giving rise to the full-length protein as observed in vivo under steady-state conditions (59). Because of its bona fide N-terminal matrix-targeting sequence (supplemental Fig. S7), in the presence of Δψ, a subset of Pink1p64 crosses the MOM via the TOM complex and is inserted into the MIM via the TIM23/PAM machinery. N-terminal processing by MPP or another divalent cation-dependent peptidase exposed to the matrix side of the MIM results in the formation of Pink1f60. The MIM protease PARL then catalyzes the generation of Pink1f53, which possibly requires the lateral release of Pink1f60 from the MIM translocon. Thereafter, we suggest that Pink1f53 is released from the mitochondrial import machinery via retrograde translocation potentially driven by the folding of its C-terminal portion in the cytosol as described for fumarase.
FIGURE 9.
Model of Pink1 import and processing in mitochondria. A, in the absence of an inner Δψ, association of full-length Pink1p64 with the MOM occurs independently of protease-accessible surface receptors and possibly even entirely independently of the TOM complex (1a). Rapid folding of Pink1p64 may prevent its translocation into or across the MOM. A small subset of Pink1 precursor proteins is translocated across the outer membrane, most likely via the TOM complex (1b), and reaches a location where processing to Pink1f53 becomes possible (2) even in the absence of Δψ. The extreme C-terminal portion of Pink1 prevents full translocation across the MOM. Pink1f53 is released from the import machinery via retrograde translocation and eventually ends up as a MOM protein associated with the outer face of the membrane or as a soluble cytosolic protein (3). B, in the presence of Δψ, the presequence-like N-terminal segment of Pink1p64 drives the translocation of a subset of Pink1p64 across the MOM via the TOM complex and its insertion into the MIM via the TIM23 machinery (1). Direct release of the inserted Pink1p64 from the translocase complexes results in a MOM-associated form of the full-length protein (2a). Alternatively, the protein is driven into the MIM by the inner Δψ and ATP hydrolysis by the import motor complex. However, based on the stop-transfer properties of the N-terminal hydrophobic segment (amino acids 93–111), the complete translocation into the matrix is prevented (2b). At least for a subset of inserted molecules, N-terminal processing by MPP or another divalent cation-dependent peptidase exposed to the matrix side of the MIM results in the formation of Pink1f60 (3). The MIM protease PARL then catalyzes the generation of Pink1f53, which most likely requires the lateral release of Pink1f60 from the TIM23 complex. Similar to the situation described above (A, 3), Pink1f53 then associates with the MOM as a peripheral membrane protein with the help of the C-terminal segment or is released as a soluble cytosolic protein (4). OM, outer membrane; IM, inner membrane.
Association of Pink1p64 with the MOM occurs independently of protease-accessible surface receptors either by means of the proposed stop-transfer sequence, which possibly serves as a signal-anchor domain, or via its hydrophobic C-terminal segment. Likewise, the combined action of both parts of the protein is conceivable. In de-energized mitochondria, a fraction of Pink1p64 is still translocated across the MOM and processed by PARL albeit with significantly reduced efficiency as compared with that in the presence of Δψ.
When analyzing Pink1 in its native state, we found it present in two high molecular mass protein complexes in the size range of 700 and 900 kDa both in vitro and in vivo. The formation of these complexes was enhanced in the absence of Δψ and might thus be relevant for the recently proposed role of Pink1 in the clearance of depolarized mitochondria via selective autophagy (40, 60). Because the larger complex was only observed under in vivo conditions, the presence of other, so far unidentified cytosolic components is to be expected. Previous reports using overexpressed Pink1 in transfected cells already indicated the presence of Pink1 in higher molecular weight complexes; however, the sizes and compositions of these complexes were not defined (61, 62). Our N-terminal deletion constructs similar in size to Pink1f53 were exclusively found in the smaller of the two complexes, indicating functional differences between Pink1p64 and Pink1f53. Some studies report the full-length protein to be the functional active protein with cleavage representing degradation rather than a processing event (18, 32, 33). However, would it not be odd that a cell undergoes the expense to synthesize a protein species, which, under physiological conditions, is simply destined for degradation after its import into mitochondria and processing by at least two specific proteases? According to other studies, the processed species is considered as the mature functional kinase (31, 35, 37). Provided that the Pink1 complexes described in this study are functionally relevant and given the ability of our N-terminal deletion constructs to assemble into the 700-kDa complex, the mitochondrial processing of Pink1 does not seem to result in a general loss of function. Instead, our analysis indicates that both Pink1p64 and Pink1f53 are functionally relevant. Accordingly, although not excluding some functional redundancy, we suggest that Pink1p64, which particularly accumulates at depolarized mitochondria, might be predominantly involved in the recently proposed mitophagy pathway, whereas the cleavage product Pink1f53 could serve other and so far unknown functions either at the level of mitochondria or in the cytosol. A reported example of a dually targeted enzyme that displays distinct functions in the different compartments has been described for the yeast aconitase 1, which localizes to both mitochondria and the cytosol (50, 63). Interestingly, as demonstrated in this study for Pink1, the C terminus of aconitase 1 serves as a targeting element, and it contributes to the dual localization of the enzyme (64).
The targeting mode of Pink1, and thus the ratio between full-length Pink1 and its main processing product, is mainly dependent on the Δψ, which in turn represents a sensor for the energy status of mitochondria. Interestingly, the Pink1f53:Pink1p64 ratio seems to be physiologically relevant as PD-linked loss-of-function mutations of Pink1 or knockdown of its supposed interaction partner Parkin resulted in its increase in cell culture studies (8, 59). Conversely, a decrease in the Pink1f53:Pink1p64 ratio has been shown to result in the mitochondrial phenotypes described for PD-linked Pink1 mutations (31). Consistent with these data, we observed a stabilization of Δψ when overexpressing Pink1Δ1–111, which is very close in size to Pink1f53. Conversely, Pink1p64 levels at mitochondria are influenced by Δψ under endogenous conditions, and they appear to be a pivotal criterion in the decision of whether the mitophagy pathway involving Parkin translocation to mitochondria is induced or not. Thus, the ratio between full-length Pink1 and its processing product might be of crucial importance in the pathogenesis of PARK6-associated PD. If that is true, elucidating the functional differences between the two Pink1 forms will certainly advance the molecular understanding of Pink1-associated PD and represents a challenging issue for future Pink1 functional studies.
Supplementary Material
Acknowledgments
We thank U. Gerken for expert technical assistance; Drs. N. Pfanner, T. Bender, and E. Schon for critical comments on the manuscript; and Y. Liu for helpful discussion and assistance with the experiments. S. Topka from the Institute for Anatomy, University of Bonn was very helpful in introducing the FACS procedure. The pReceiverM10xPINK1 clone used for in vivo expression of Pink1-myc/His was a kind gift from G. Auburger (Department of Neurology, University Medical School, Frankfurt am Main, Germany).
This work was supported, in whole or in part, by National Institutes of Health Grants NS042269, NS064191, NS38370, NS070276, and NS072182. This work was also supported by a grant from the BONFOR program of the Medical Faculty of the University of Bonn (to W. V.); United States Department of Defense Grants W81XWH-08-1-0522, W81XWH-08-1-0465, and W81XWH-09-1-0245; the Parkinson's Disease Foundation; the Thomas Hartman Foundation For Parkinson's Research; Project A.L.S.; the Muscular Dystrophy Association and P2ALS (to S. P.); and Deutsche Forschungsgemeinschaft Grant 805231 (to M. A. T.).

This article contains supplemental Figs. S1–S7.
- Pink1
- PTEN-induced putative kinase 1
- BN
- blue native
- DHFR
- dihydrofolate reductase
- F1β
- F1 subunit β of the F1F0-ATP synthase
- MIM
- mitochondrial inner membrane
- MOM
- mitochondrial outer membrane
- IMS
- intermembrane space
- Mcr1
- mitochondrial NADH-cytochrome b5 reductase 1
- MPP
- matrix processing peptidase
- PARL
- presenilin-associated rhomboid-like
- PD
- Parkinson disease
- TIM
- translocase of the inner (mitochondrial) membrane
- TMD
- transmembrane domain
- TOM
- translocase of the outer (mitochondrial) membrane
- TMRE
- tetramethylrhodamine ethyl ester
- Trap1
- tumor necrosis factor receptor-associated protein 1
- Δψ
- membrane potential
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Valente E. M., Bentivoglio A. R., Dixon P. H., Ferraris A., Ialongo T., Frontali M., Albanese A., Wood N. W. (2001) Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am. J. Hum. Genet. 68, 895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bentivoglio A. R., Cortelli P., Valente E. M., Ialongo T., Ferraris A., Elia A., Montagna P., Albanese A. (2001) Phenotypic characterisation of autosomal recessive PARK6-linked parkinsonism in three unrelated Italian families. Mov. Disord. 16, 999–1006 [DOI] [PubMed] [Google Scholar]
- 3. Valente E. M., Salvi S., Ialongo T., Marongiu R., Elia A. E., Caputo V., Romito L., Albanese A., Dallapiccola B., Bentivoglio A. R. (2004) PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann. Neurol. 56, 336–341 [DOI] [PubMed] [Google Scholar]
- 4. Unoki M., Nakamura Y. (2001) Growth-suppressive effects of BPOZ and EGR2, two genes involved in the PTEN signaling pathway. Oncogene 20, 4457–4465 [DOI] [PubMed] [Google Scholar]
- 5. Beilina A., Van Der Brug M., Ahmad R., Kesavapany S., Miller D. W., Petsko G. A., Cookson M. R. (2005) Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc. Natl. Acad. Sci. U.S.A. 102, 5703–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., González-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W. P., Latchman D. S., Harvey R. J., Dallapiccola B., Auburger G., Wood N. W. (2004) Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160 [DOI] [PubMed] [Google Scholar]
- 7. Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E. M., Casari G. (2005) Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 14, 3477–3492 [DOI] [PubMed] [Google Scholar]
- 8. Muqit M. M., Abou-Sleiman P. M., Saurin A. T., Harvey K., Gandhi S., Deas E., Eaton S., Payne Smith M. D., Venner K., Matilla A., Healy D. G., Gilks W. P., Lees A. J., Holton J., Revesz T., Parker P. J., Harvey R. J., Wood N. W., Latchman D. S. (2006) Altered cleavage and localization of PINK1 to aggresomes in the presence of proteasomal stress. J. Neurochem. 98, 156–169 [DOI] [PubMed] [Google Scholar]
- 9. Gandhi S., Muqit M. M., Stanyer L., Healy D. G., Abou-Sleiman P. M., Hargreaves I., Heales S., Ganguly M., Parsons L., Lees A. J., Latchman D. S., Holton J. L., Wood N. W., Revesz T. (2006) PINK1 protein in normal human brain and Parkinson's disease. Brain 129, 1720–1731 [DOI] [PubMed] [Google Scholar]
- 10. Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E. A., Przedborski S. (2008) The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 105, 12022–12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pfanner N., Geissler A. (2001) Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2, 339–349 [DOI] [PubMed] [Google Scholar]
- 12. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takatori S., Ito G., Iwatsubo T. (2008) Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci. Lett. 430, 13–17 [DOI] [PubMed] [Google Scholar]
- 14. Plun-Favreau H., Klupsch K., Moisoi N., Gandhi S., Kjaer S., Frith D., Harvey K., Deas E., Harvey R. J., McDonald N., Wood N. W., Martins L. M., Downward J. (2007) The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat. Cell Biol. 9, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 15. Meissner C., Lorenz H., Weihofen A., Selkoe D. J., Lemberg M. K. (2011) The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J. Neurochem. 117, 856–867 [DOI] [PubMed] [Google Scholar]
- 16. Marongiu R., Spencer B., Crews L., Adame A., Patrick C., Trejo M., Dallapiccola B., Valente E. M., Masliah E. (2009) Mutant Pink1 induces mitochondrial dysfunction in a neuronal cell model of Parkinson's disease by disturbing calcium flux. J. Neurochem. 108, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin W., Kang U. J. (2008) Characterization of PINK1 processing, stability, and subcellular localization. J. Neurochem. 106, 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin S. M., Lazarou M., Wang C., Kane L. A., Narendra D. P., Youle R. J. (2010) Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D'Adamio L., Derks C., Dejaegere T., Pellegrini L., D'Hooge R., Scorrano L., De Strooper B. (2006) Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126, 163–175 [DOI] [PubMed] [Google Scholar]
- 20. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnston A. J., Hoogenraad J., Dougan D. A., Truscott K. N., Yano M., Mori M., Hoogenraad N. J., Ryan M. T. (2002) Insertion and assembly of human Tom7 into the preprotein translocase complex of the outer mitochondrial membrane. J. Biol. Chem. 277, 42197–42204 [DOI] [PubMed] [Google Scholar]
- 22. Gispert S., Ricciardi F., Kurz A., Azizov M., Hoepken H. H., Becker D., Voos W., Leuner K., Müller W. E., Kudin A. P., Kunz W. S., Zimmermann A., Roeper J., Wenzel D., Jendrach M., García-Arencíbia M., Fernández-Ruiz J., Huber L., Rohrer H., Barrera M., Reichert A. S., Rüb U., Chen A., Nussbaum R. L., Auburger G. (2009) Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PLoS One 4, e5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schägger H. (2001) Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65, 231–244 [DOI] [PubMed] [Google Scholar]
- 24. Bömer U., Maarse A. C., Martin F., Geissler A., Merlin A., Schönfisch B., Meijer M., Pfanner N., Rassow J. (1998) Separation of structural and dynamic functions of the mitochondrial translocase: Tim44 is crucial for the inner membrane import sites in translocation of tightly folded domains, but not of loosely folded preproteins. EMBO J. 17, 4226–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Voisine C., Craig E. A., Zufall N., von Ahsen O., Pfanner N., Voos W. (1999) The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell 97, 565–574 [DOI] [PubMed] [Google Scholar]
- 26. Palmieri F. (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 447, 689–709 [DOI] [PubMed] [Google Scholar]
- 27. Luciano P., Tokatlidis K., Chambre I., Germanique J. C., Géli V. (1998) The mitochondrial processing peptidase behaves as a zinc-metallopeptidase. J. Mol. Biol. 280, 193–199 [DOI] [PubMed] [Google Scholar]
- 28. Gakh O., Cavadini P., Isaya G. (2002) Mitochondrial processing peptidases. Biochim. Biophys. Acta 1592, 63–77 [DOI] [PubMed] [Google Scholar]
- 29. Nett J. H., Trumpower B. L. (1996) Dissociation of import of the Rieske iron-sulfur protein into Saccharomyces cerevisiae mitochondria from proteolytic processing of the presequence. J. Biol. Chem. 271, 26713–26716 [DOI] [PubMed] [Google Scholar]
- 30. Freeman M. (2008) Rhomboid proteases and their biological functions. Annu. Rev. Genet. 42, 191–210 [DOI] [PubMed] [Google Scholar]
- 31. Deas E., Plun-Favreau H., Gandhi S., Desmond H., Kjaer S., Loh S. H., Renton A. E., Harvey R. J., Whitworth A. J., Martins L. M., Abramov A. Y., Wood N. W. (2011) PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 20, 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wiedemann N., van der Laan M., Hutu D. P., Rehling P., Pfanner N. (2007) Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J. Cell Biol. 179, 1115–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haque M. E., Thomas K. J., D'Souza C., Callaghan S., Kitada T., Slack R. S., Fraser P., Cookson M. R., Tandon A., Park D. S. (2008) Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc. Natl. Acad. Sci. U.S.A. 105, 1716–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dagda R. K., Cherra S. J., 3rd, Kulich S. M., Tandon A., Park D., Chu C. T. (2009) Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 284, 13843–13855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 39. Deas E., Plun-Favreau H., Wood N. W. (2009) PINK1 function in health and disease. EMBO Mol. Med. 1, 152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chu C. T. (2010) Tickled PINK1: mitochondrial homeostasis and autophagy in recessive parkinsonism. Biochim. Biophys. Acta 1802, 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Greene A. W., Grenier K., Aguileta M. A., Muise S., Farazifard R., Haque M. E., McBride H. M., Park D. S., Fon E. A. (2012) Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 13, 378–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nolden M., Ehses S., Koppen M., Bernacchia A., Rugarli E. I., Langer T. (2005) The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123, 277–289 [DOI] [PubMed] [Google Scholar]
- 43. Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J. C., Westermann B., Rugarli E. I., Langer T. (2009) Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187, 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Head B., Griparic L., Amiri M., Gandre-Babbe S., van der Bliek A. M. (2009) Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187, 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lemberg M. K., Martoglio B. (2002) Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10, 735–744 [DOI] [PubMed] [Google Scholar]
- 46. Strisovsky K., Sharpe H. J., Freeman M. (2009) Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol. Cell 36, 1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lin W., Kang U. J. (2010) Structural determinants of PINK1 topology and dual subcellular distribution. BMC Cell Biol. 11, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meier S., Neupert W., Herrmann J. M. (2005) Proline residues of transmembrane domains determine the sorting of inner membrane proteins in mitochondria. J. Cell Biol. 170, 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bohnert M., Rehling P., Guiard B., Herrmann J. M., Pfanner N., van der Laan M. (2010) Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr. Biol. 20, 1227–1232 [DOI] [PubMed] [Google Scholar]
- 50. Yogev O., Pines O. (2011) Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim. Biophys. Acta 1808, 1012–1020 [DOI] [PubMed] [Google Scholar]
- 51. Hahne K., Haucke V., Ramage L., Schatz G. (1994) Incomplete arrest in the outer membrane sorts NADH-cytochrome b5 reductase to two different submitochondrial compartments. Cell 79, 829–839 [DOI] [PubMed] [Google Scholar]
- 52. Haucke V., Ocana C. S., Hönlinger A., Tokatlidis K., Pfanner N., Schatz G. (1997) Analysis of the sorting signals directing NADH-cytochrome b5 reductase to two locations within yeast mitochondria. Mol. Cell. Biol. 17, 4024–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meineke B., Engl G., Kemper C., Vasiljev-Neumeyer A., Paulitschke H., Rapaport D. (2008) The outer membrane form of the mitochondrial protein Mcr1 follows a TOM-independent membrane insertion pathway. FEBS Lett. 582, 855–860 [DOI] [PubMed] [Google Scholar]
- 54. Becker T., Vögtle F. N., Stojanovski D., Meisinger C. (2008) Sorting and assembly of mitochondrial outer membrane proteins. Biochim. Biophys. Acta 1777, 557–563 [DOI] [PubMed] [Google Scholar]
- 55. Yogev O., Naamati A., Pines O. (2011) Fumarase: a paradigm of dual targeting and dual localized functions. FEBS J. 278, 4230–4242 [DOI] [PubMed] [Google Scholar]
- 56. Stein I., Peleg Y., Even-Ram S., Pines O. (1994) The single translation product of the FUM1 gene (fumarase) is processed in mitochondria before being distributed between the cytosol and mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 4770–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Knox C., Sass E., Neupert W., Pines O. (1998) Import into mitochondria, folding and retrograde movement of fumarase in yeast. J. Biol. Chem. 273, 25587–25593 [DOI] [PubMed] [Google Scholar]
- 58. Sass E., Karniely S., Pines O. (2003) Folding of fumarase during mitochondrial import determines its dual targeting in yeast. J. Biol. Chem. 278, 45109–45116 [DOI] [PubMed] [Google Scholar]
- 59. Weihofen A., Ostaszewski B., Minami Y., Selkoe D. J. (2008) Pink1 Parkinson mutations, the Cdc37/Hsp90 chaperones and Parkin all influence the maturation or subcellular distribution of Pink1. Hum. Mol. Genet. 17, 602–616 [DOI] [PubMed] [Google Scholar]
- 60. Deas E., Wood N. W., Plun-Favreau H. (2011) Mitophagy and Parkinson's disease: the PINK1-parkin link. Biochim. Biophys. Acta 1813, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu W., Vives-Bauza C., Acín-Peréz R., Yamamoto A., Tan Y., Li Y., Magrané J., Stavarache M. A., Shaffer S., Chang S., Kaplitt M. G., Huang X. Y., Beal M. F., Manfredi G., Li C. (2009) PINK1 defect causes mitochondrial dysfunction, proteasomal deficit and α-synuclein aggregation in cell culture models of Parkinson's disease. PLoS One 4, e4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thomas K. J., McCoy M. K., Blackinton J., Beilina A., van der Brug M., Sandebring A., Miller D., Maric D., Cedazo-Minguez A., Cookson M. R. (2011) DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 20, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Regev-Rudzki N., Karniely S., Ben-Haim N. N., Pines O. (2005) Yeast aconitase in two locations and two metabolic pathways: seeing small amounts is believing. Mol. Biol. Cell 16, 4163–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ben-Menachem R., Regev-Rudzki N., Pines O. (2011) The aconitase C-terminal domain is an independent dual targeting element. J. Mol. Biol. 409, 113–123 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.