Background: Bacterial steroid monooxygenase degrades progesterone.
Results: The crystallographic and mutagenesis analysis outline a robust active-site scaffold, capable of performing Baeyer-Villiger oxidations on chemically diverse molecules.
Conclusion: This and related enzymes represent a fascinating case for the comparative evaluation of user tailored protein engineering with enzyme variants arising through evolution.
Significance: These findings highlight the biocatalytic potential of Baeyer-Villiger monooxygenases.
Keywords: Enzyme Structure, Flavin, Oxidation-Reduction, Protein Engineering, Steroid, Baeyer-Villiger, Monooxygenase
Abstract
Steroid monooxygenase (STMO) from Rhodococcus rhodochrous catalyzes the Baeyer-Villiger conversion of progesterone into progesterone acetate using FAD as prosthetic group and NADPH as reducing cofactor. The enzyme shares high sequence similarity with well characterized Baeyer-Villiger monooxygenases, including phenylacetone monooxygenase and cyclohexanone monooxygenase. The comparative biochemical and structural analysis of STMO can be particularly insightful with regard to the understanding of the substrate-specificity properties of Baeyer-Villiger monooxygenases that are emerging as promising tools in biocatalytic applications and as targets for prodrug activation. The crystal structures of STMO in the native, NADP+-bound, and two mutant forms reveal structural details on this microbial steroid-degrading enzyme. The binding of the nicotinamide ring of NADP+ is shifted with respect to the flavin compared with that observed in other monooxygenases of the same class. This finding fully supports the idea that NADP(H) adopts various positions during the catalytic cycle to perform its multiple functions in catalysis. The active site closely resembles that of phenylacetone monooxygenase. This observation led us to discover that STMO is capable of acting also on phenylacetone, which implies an impressive level of substrate promiscuity. The investigation of six mutants that target residues on the surface of the substrate-binding site reveals that enzymatic conversions of both progesterone and phenylacetone are largely insensitive to relatively drastic amino acid changes, with some mutants even displaying enhanced activity on progesterone. These features possibly reflect the fact that these enzymes are continuously evolving to acquire new activities, depending on the emerging availabilities of new compounds in the living environment.
Introduction
Baeyer-Villiger monooxygenases (BVMO)3 use flavin cofactors to catalyze the insertion of an oxygen atom into ketone substrates to generate esters and/or lactones (1). NADPH and molecular oxygen represent the electron and oxygen-atom donors in the reaction, respectively. Since their discovery more than 60 years ago (2, 3), these enzymes have attracted considerable interest for their potential applicability in industrial biocatalysis, their role as (pro)drug targets, and their value as model systems to study how enzymes carry out a classical organic chemistry reaction (4).
In mechanistic terms, BVMOs display two key features: (i) the activation of molecular oxygen through formation of a covalent intermediate, the flavin-peroxide, formed by the reaction of the reduced flavin with oxygen, and (ii) the occurrence of a tetrahedral Criegee intermediate in substrate oxygenation, which is analogous to the intermediate of nonenzymatic Baeyer-Villiger oxidations (5–7). NADP(H) plays a key role in the whole process by functioning not only as a reductant of the flavin but also as an integral part of the active site that promotes flavin-peroxide and Criegee intermediate formation and stabilization. In more detail, the reaction starts with the two-electron reduction of the flavin by NADPH. Next, the reduced prosthetic group reacts with oxygen to generate the flavin-peroxide, which is responsible for the insertion of an oxygen atom into the substrate to generate the oxygenated product. The reaction ends with the regeneration of the oxidized flavin and the release of NADP+. Importantly, the flavin-peroxide forms only if NADP+ is bound to the active site. Indeed, an artificial electron-donor (such as dithionite) does not enable the enzyme to perform the catalytic reaction. These features imply that NADP(H) must have at least two binding modes; one competent in flavin reduction and one capable of promoting formation and stabilization of the crucial flavin-peroxide.
Cyclohexanone monooxygenase (CHMO) was the first BVMO that could be produced in a recombinant manner and has been subject to in-depth enzymological studies (5). In 1999, steroid monooxygenase (STMO) from Rhodococcus rhodochrous was the second BVMO for which the gene was cloned and used for heterologous expression (8). However, so far no subsequent studies on this bacterial steroid degrading monooxygenase have been reported. More recently, phenylacetone monooxygenase (PAMO) became another model system especially after its three-dimensional crystal structure was solved (9). An excellent review about the history and state of the art of the field has been recently published by Lau and co-workers (1).
Genome sequencing has led to the discovery of a huge number of BVMOs in bacteria and fungi (10). The enzymes display a bewildering diversity with regard to substrate preferences that can range from small molecules, such as small aromatic and aliphatic compounds, to large macrocyclic compounds typically used as intermediates in the biosynthesis of complex metabolites such as alkaloids and antibiotics. The crystal structures of several BVMOs have been solved in the past few years including those of CHMO (11), PAMO (9), 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetyl-CoA monooxygenase (OTEMO) (12), MtmOIV (a Baeyer-Villiger monooxygenase involved in mithramycin biosynthesis) (13), and a NAD(P)H-dependent monooxygenase from a marine bacterium that is more closely related to flavin-containing monoxygenases (also known as FMOs) (14). The common features shared by these crystal structures have established a solid structural framework to study enzyme-mediated Baeyer-Villiger oxidations, especially with regard to the existence of conformational changes and the role of NADP(H) and conserved residues for intermediate stabilization (7, 11). Furthermore, the combination of structural and enzyme engineering studies has provided important hints about the protein elements that determine the substrate selectivity of the enzymes. For instance, a loop in direct contact with the pyrimidine ring of the flavin has been shown to be crucial for substrate binding (15). On the other hand, especially with the aim of exploiting these enzymes for biocatalytic purposes, we need to further understand and rationalize their substrate specificity properties.
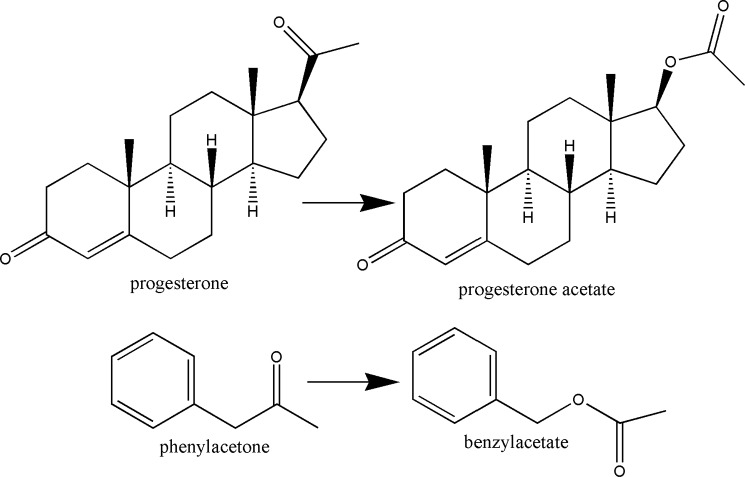
In this context, here we describe the structural elucidation complemented by site-directed mutagenesis studies of STMO from R. rhodochrous, which was originally discovered by Miyamoto et al. (16). This enzyme is capable of converting progesterone into progesterone acetate (Scheme 1). The interest for this enzyme stems from its high sequence homology with the most well characterized BVMOs with known crystal structures (8, 17); STMO has 53% sequence identity with PAMO from Thermobifida fusca, 45% with CHMO from Rhodococcus sp. strain HI-31, and 43% with OTEMO from Pseudomonas putida. The inspiration for our study was to compare these closely related enzymes, which exhibit so diverse substrate preferences ranging from a simple aromatic (phenylacetone) molecule to a steroid. The close evolutionary relatedness adds value to our approach because it allows the identification of structural elements that direct substrate binding and selectivity. Our analysis reveals that especially PAMO and STMO have highly similar active sites to the point that STMO is shown to be active also on phenylacetone. Such promiscuity in substrate preference has interesting implications for BVMO evolution, function, redesign, and biocatalytic applications.
SCHEME 1.
Reactions catalyzed by STMO (insertion of an oxygen between C17 and C20 carbons of 11 α- and 11β-hydroxyprogesterone) and PAMO.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
The gene coding for STMO from R. rhodochrous (8) was cloned into a ChampionTM pET-vector (Invitrogen) for overexpression in Escherichia coli. The resulting protein contained a N-terminal His6-tagged SUMO, which can be removed by SUMO protease. The plasmid was transformed into E. coli strain BL21(DE3) CodonPlus. The resulting colonies were preinoculated into LB broth supplemented with kanamycin (50 μg/ml) and chloramphenicol (50 μg/ml) and grown overnight at 37 °C. Cultures were inoculated and grown in 2-liter Erlenmeyer flasks at 37 °C in 750 ml of LB medium until A600 was 0.3–0.5. Then, isopropyl β-d-1-thiogalactoside was added to a final concentration of 0.3 mm for the induction of expression at 17 °C for 16 h. Cells were harvested by centrifugation. The cell pellet was resuspended in a buffer containing 50 mm HEPES, pH 8.0, and 500 mm NaCl added with 25 μg/ml of DNase and 2 mm PMSF, and lysed by sonication. Insoluble debris was removed by centrifugation at 70,000 × g for 40 min at 4 °C and the soluble lysate clarified by filtration through a 0.45-μm syringe filter. The resulting supernatant was loaded on a Ni2+-affinity column (5 ml HisTrapTM FF, GE Healthcare) by an Åkta purifier (GE Healthcare) and washed with the resuspending buffer added with 5 mm imidazole to remove the main part of contaminants. Elution of the protein occurred at 100 mm imidazole. STMO-containing fractions were dialyzed overnight at 4 °C in 50 mm HEPES/HCl, pH 8.0, 500 mm NaCl in the presence of SUMO protease to remove the N-terminal tag. A further Ni2+-affinity column separated the native STMO from the cleaved tag and the protease. To increase the sample homogeneity for crystallization purposes, a final step of size exclusion chromatography (Superdex 75 16/60, GE Healthcare) was performed in 50 mm HEPES/HCl, pH 7.5, 100 mm NaCl. The protein purity was assessed by SDS-PAGE and by measurement of the ratio in the absorbance at 280 and 457 nm, which was about 9 for pure samples.
Mutagenesis
Site-directed mutagenesis was performed using the PfuUltra Hotstart PCR Master Mix (Agilent) according to the manufacturer's instructions. Primer sequences were generated using the QuikChange tool available from Agilent and are available upon request. As the mutants tended to release FAD, the purification protocol was slightly modified. All the buffers used to resuspend the cell pellet and for affinity chromatography were added with 6% (w/v) glycerol, whereas the amount of NaCl was lowered to 200 mm. Moreover, a small excess of FAD (100–300 μm) was added to the samples before sonication and during dialysis.
Crystallization
For crystallization experiments, the purified proteins were dialyzed against 15 mm HEPES/HCl, pH 7.5, 30 mm NaCl, and concentrated up to 15–20 mg/ml. STMO crystals were grown by the vapor diffusion method at 295 K. Initial crystallization conditions were screened by the sitting-drop technique using sparse matrix kits (Crystal Screen and Crystal Screen 2, Hampton Research, CA) with an automated device (Oryx8, Douglas Instruments). Positive hits were refined using the hanging drop vapor-diffusion technique to identify the optimal crystallization conditions: 2–4 μl of protein solution mixed with equal volumes of reservoir solution consisting of 1.8–2.2 m MgSO4, 0.1 m MES/HCl, pH 6.2–6.5. The protein solution was supplemented with 0.5 mm FAD and, if needed, with 2–5 mm NADP+. Yellow, diffraction quality crystals grew in 3–4 weeks. Crystals were directly flash-cooled in liquid nitrogen, without any cryo-protecting agent in addition to MgSO4.
X-ray Diffraction Data Collection and Structure Determination
Diffraction data of the native crystals were collected at 100 K on the PXII beamline at SLS (Villigen, Switzerland) and on the ID23-2 and ID14-4 beamlines at ESRF (Grenoble, France). All the data were processed with MOSFLM (18) and programs of the CCP4 suite (19). The structure of STMO was solved using the diffraction data of unliganded wild-type enzyme by molecular replacement with PHASER (20) using PAMO as search model (PDB code 1W4X (9)). The model was refined manually through successive rounds using Coot (21), whereas maximum likelihood refinement were carried out by Refmac5 (22). The STMO apoenzyme model was used as a template to solve the other STMO structures. All final models were validated with MolProbity, which did not detect any outliers in the Ramachandran plot (23). Data collection and refinement statistics are listed in Table 1. Pictures were generated with PyMOL.
TABLE 1.
Crystallographic data collection and refinement statistics
| STMOapo | STMOox | STMO-K295A | STMO-T345L | |
|---|---|---|---|---|
| PDB code | 4AOX | 4AOS | 4AP1 | 4AP3 |
| Unit cell (a, b, c) (Å) | 79.76, 79.76, 222.98 | 81.56, 81.56, 224.52 | 82.16, 82.16, 228.81 | 81.75, 81.75, 228.07 |
| Space group | P 41 21 2 | P 41 21 2 | P 41 21 2 | P 41 21 2 |
| Resolution (Å) | 2.4 | 2.5 | 2.9 | 2.4 |
| Rsyma,b (%) | 10.2 (47.2) | 6.7 (15.1) | 10.5 (50.8) | 7.3 (26.5) |
| Completenessb (%) | 99.9 (100.0) | 99.5 (99.6) | 99.9 (100.0) | 99.5 (100.0) |
| Unique reflections | 28,420 | 26,952 | 17,319 | 31,403 |
| Redundancyb | 6.8 (7.4) | 12.6 (13.4) | 7.9 (8.1) | 7.7 (7.7) |
| I/σb | 11.4 (3.5) | 25.1 (14.3) | 14.4 (4.1) | 16.6 (6.1) |
| No. of atoms | 4324 | 4349 | 4022 | 4232 |
| Rcrystc (%) | 22.5 | 19.9 | 20.2 | 20.9 |
| Rfreec (%) | 25.2 | 23.6 | 24.9 | 23.9 |
| Root mean square bond length (Å) | 0.020 | 0.021 | 0.015 | 0.021 |
| Root mean square bond angles (°) | 1.94 | 1.93 | 1.48 | 1.92 |
a Rsym = Σ|Ii − 〈I〉|/ΣIi, where Ii is the intensity of ith observation and 〈I〉 is the mean intensity of the reflection.
b Values in parentheses are for reflections in the highest resolution shell.
c Rcryst = Σ|Fobs − Fcalc|/ΣFobs, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rcryst and Rfree were calculated using the working and test set, respectively.
Steady-state Kinetics
Activities of the purified enzymes were determined spectrophotometrically by monitoring the decrease of the NADPH concentration by measuring the absorbance at 340 nm. The reaction mixture (200 or 1000 μl) contained 50 mm Tris/HCl, pH 7.5, 100 μm NADPH, 5% (v/v) Me2SO in the case of phenylacetone or 5% (v/v) dioxane in the case of progesterone, 0–16 mm phenylacetone or 0–75 μm progesterone, and 0.05–0.2 μm enzyme. Kinetic parameters were obtained by fitting the data to Equation 1 using SigmaPlot 11. Standard errors in curve fitting were generally less than 10% and duplos were within 10%. The kcat/Km values for progesterone were determined from the substrate-concentration-dependent rates observed under apparent nonsaturating conditions.
 |
Phenylacetone and progesterone conversion was also checked by gas-chromatography methods, as described (6).
RESULTS
Crystallographic Analysis and Overall Structure
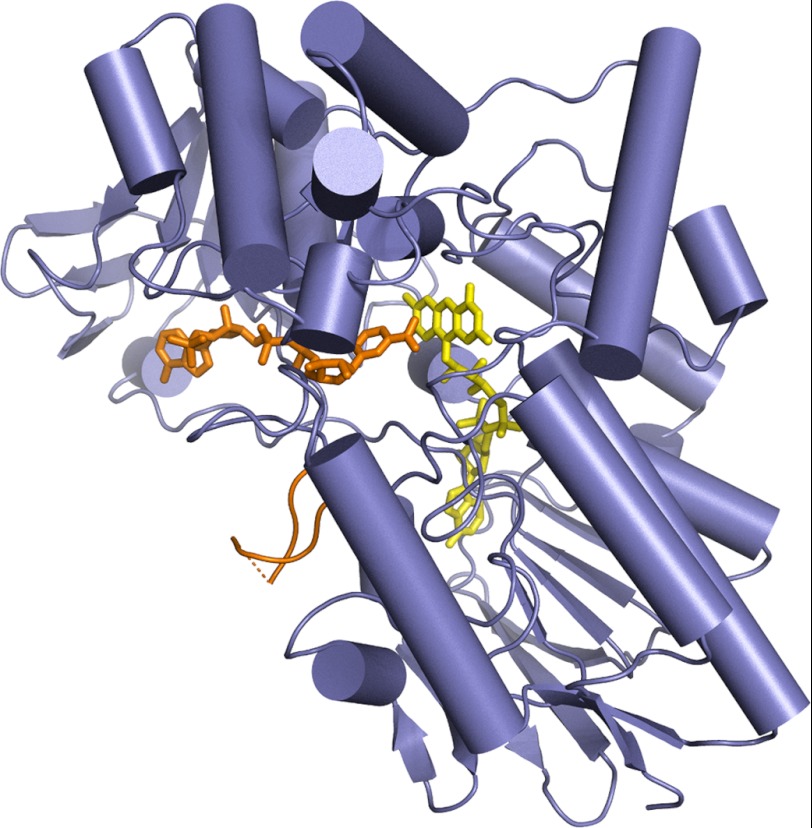
Recombinant STMO from R. rhodochrous was crystallized using magnesium sulfate as precipitant (Table 1). We obtained four crystal structures (2.4–2.9 Å resolution): the wild-type enzyme in the oxidized state and in complex with NADP+, and two active-site mutants K295A and T345L. Except for the amino acid replacements, these four models are essentially indistinguishable (root mean square deviations below 0.2 Å for Cα atoms) and we shall refer mainly to the NADP+ complex for the analysis of the enzyme three-dimensional structure (Figs. 1 and supplemental Fig. S1).
FIGURE 1.
Overall structure of R. rhodochrous STMO. FAD and the NADP+ are depicted in yellow and orange, respectively. The (partly disordered) loop 500–520 is shown in orange. From the comparison with other Baeyer-Villiger enzyme structures, this loop is expected to change conformation to a “closed” state during catalysis (1, 7, 9, 11, 24). The FAD-binding domain (bottom) comprises residues 20–161 and 395–549, whereas the NADP domain is formed by residues 162–394. Residues 1–19 and 510–516 (connected by a dashed line) are disordered.
STMO exhibits the typical two-domain organization of the other known BVMO enzymes, with each domain featuring the dinucleotide-binding topology (Fig. 1). Structural comparisons indicate that FAD- and NADP-binding domains of STMO superimpose onto the corresponding domains of PAMO (7) with root mean square deviations for the Cα atoms of 1.1 and 1.6 Å, respectively (Fig. 2). These close similarities reflect the high 53% sequence identity between the two proteins. A slightly lower but nevertheless strong degree of structural similarity (root mean square deviations below 2.0 Å) is indicated also by comparisons with the known crystal structures of other BVMO enzymes (CHMO and OTEMO (11, 12)), which is in full agreement with their (>40%) sequence identities with STMO. The only variations are small changes in the mutual orientations of the two domains that rotate by 4–6°. Similar domain rotations have been observed in other BVMO structures and probably reflect intrinsic domain flexibility and, possibly, also the different crystal packing environments. As found in the known BVMO structures, the flavin of STMO shows virtually no deviation from planarity (Fig. 4 and supplemental Fig. S1). In the logic of our study, the close similarities highlighted by these comparisons provide an excellent structural framework for exploring the factors that control the diverse substrate selectivities among these enzymes. Along this line, we shall now describe the STMO active site.
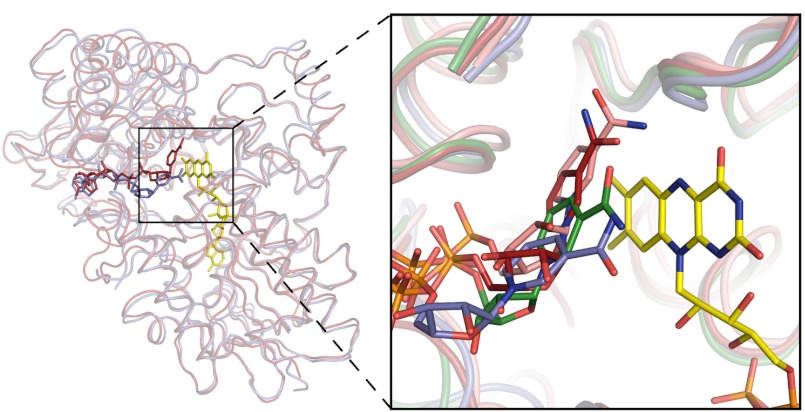
FIGURE 2.
Structural comparison of Baeyer-Villiger monooxygenases. Left, overall superposition between the backbones of STMO (blue) and PAMO (red). The superposition was carried out using the FAD and NADP domains of PAMO as independent units to emphasize the overall similarity of the tertiary structures (root mean square deviations of 1.1 Å for 295 Cα atoms and of 1.6 Å for 232 Cα atoms of the two domains, respectively). Right, close-up view of the active site region. The picture shows the different positions of the nicotinamide ring of NADP+ in STMO (carbons in blue), CHMO (carbons in green; open conformation of PDB entry 3GWF), PAMO (carbons in red; PDB entry 2YLR), and OTEMO (carbons in pink; PDB entry 3UOY, chain A). Cα ribbons of the four proteins are shown together with the FAD of STMO.
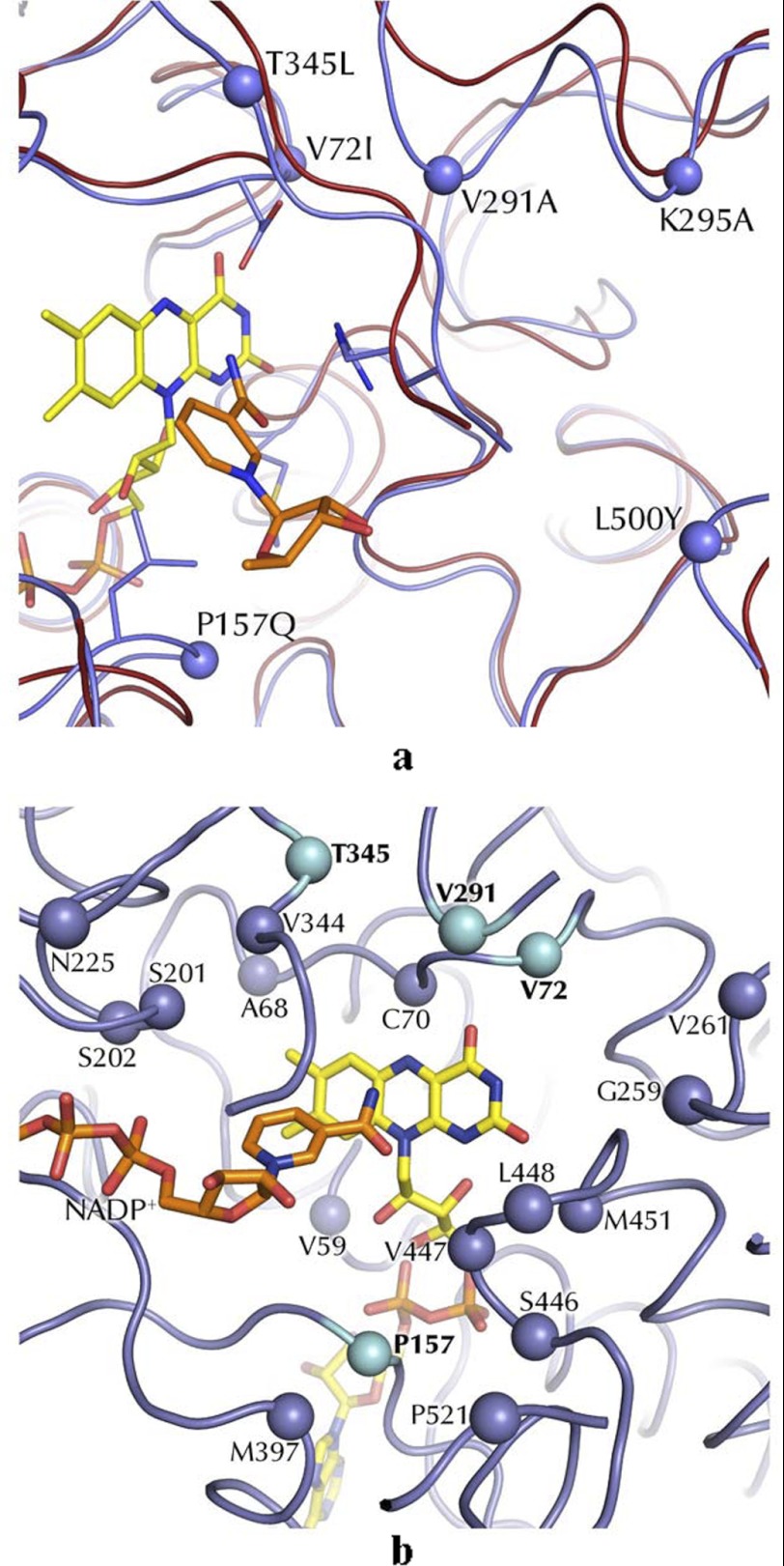
FIGURE 4.
Structural framework to substrate binding in STMO and related enzymes. a, superposition of STMO (blue ribbon) and PAMO (red ribbon) active sites. FAD and NADP+ of STMO are shown. The Cα atoms of the STMO residues targeted by mutagenesis are outlined as blue spheres (Table 3). b, the Cα positions of residues within 8 Å distance from the flavin N5 atoms on the re side of the cofactor (Table 2). The blue spheres (cyan for mutated residues; Table 3) identify STMO residues that have the side chains oriented toward the active site and differ from the corresponding residues of PAMO, CHMO, and OTEMO.
NADP+ Binding and Active Site
The crystal structure of STMO exhibits a globular open pocket at the interface between the two domains to form the binding site for NADP(H) and the steroid substrate (Fig. 1). The two ligand-binding sites are located on opposite walls of the cleft and converge in front of the flavin (Figs. 2 and 3). STMO crystals were also obtained in the presence of an excess of NADP+, which enabled the structure determination of the enzyme bound to the coenzyme ligand (Table 1 and supplemental Fig. S1). NADP+ binds in the typical conformation observed in dinucleotide-binding proteins, with the pyrophosphate group interacting with the N terminus of the α-helix of the βαβ unit of the NADP-binding domain (Figs. 1 and 2). The nicotinamide-ribose group is close to a conserved Arg (Arg-342; Fig. 3), which is essential in catalysis by favoring substrate oxygenation possibly through stabilization of the tetrahedral Criegee intermediate (7).
FIGURE 3.
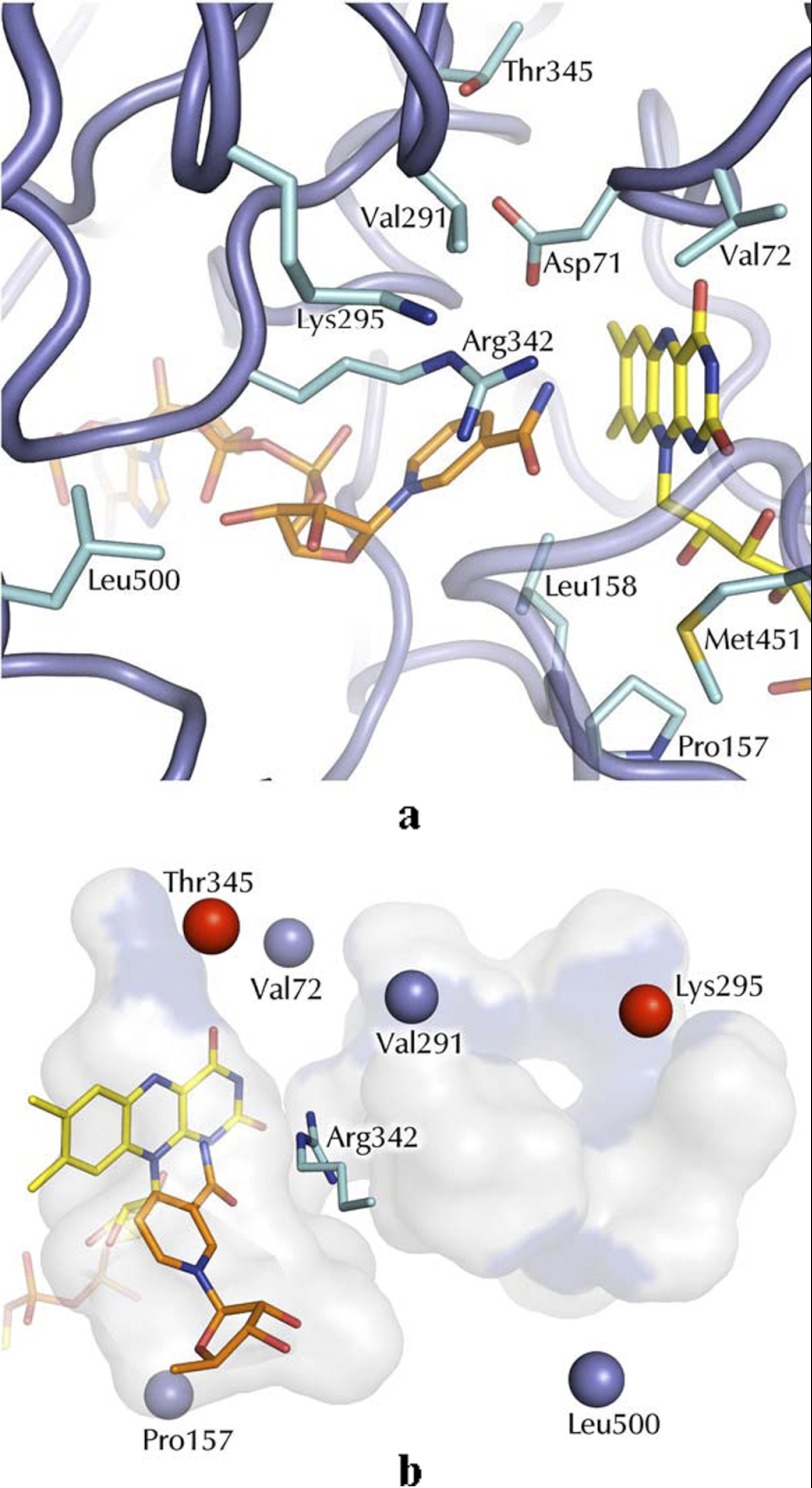
STMO active site. a, the NADP+- and flavin-binding regions. FAD and NADP+ carbons are depicted with yellow and orange carbons, respectively. b, spatial distribution of the sites targeted by our mutagenesis studies (see Table 3) on the active site cavity. The Cα atoms of the mutated residues are shown as blue spheres. Thr-345 and Lys-295 are highlighted in red because mutations targeting these amino acids exhibited strong functional perturbations and were subjected to crystallographic studies (Tables 1 and 3). Bluish areas highlight the mutations exposed on the cavity surface. For the sake of clarity, only the ribose and nicotinamide ring of NADP+ are depicted. Arg-342 is the crucial residue involved in the oxygen-insertion step during catalysis. The cavity was calculated with a probe of 1.3-Å radius using Voidoo (27).
An interesting observation concerns the precise binding mode of the nicotinamide ring with respect to the flavin (Fig. 2). The coenzyme nicotinamide ring is located above the flavin with the carbamide group pointing toward and in direct contact with the central ring of the prosthetic group. The nicotinamide remains partly solvent-accessible and within 4.5 Å distance from Asp-71. The interactions with the solvent and the proximity of the Asp-71 side chain can compensate for the NADP+ positive charge. The functional implications for this conformation are effectively revealed by the comparison of STMO with PAMO, OTEMO, and CHMO structures. The comparative analysis indicates that the nicotinamide of STMO is prominently shifted away from the flavin compared with the other enzymes (Fig. 2). This observation is fully consistent with the “sliding” model proposed by Mirza et al. (11). The nicotinamide, anchored to the protein by the ADP segment of NADPH, is able to slide into the active site to first bind above the flavin with the proper geometry to perform hydride transfer. After this catalytic step, the nicotinamide moves further to eventually adopt the position (red carbons in Fig. 2) that is instrumental to the formation and stabilization of the flavin-peroxide intermediate after reaction of the reduced flavin with dioxygen. NADP+ is locked in this flavin-peroxide promoting conformation by closure of an active site loop, which likely occurs only after flavin reduction (see Refs. 1, 7, 9, 11, and 24 for a discussion on this aspect of catalysis). In STMO, this loop protrudes out of the protein and is partly disordered (residues 510–516; Fig. 1), which is similar to the cases of PAMO in the unligated form (7) and CHMO in the “open” conformation (green in Fig. 2) (11). Thus, the STMO crystals (obtained by co-crystallizing the oxidized enzyme with NADP+) reveal a binding mode that is likely adopted by the reduced nicotinamide in the process of attaining the conformation that affords reduction of the prosthetic FAD cofactor and, possibly, also by the oxidized NADP+ in the process of being released by the enzyme, which is the last step of the reaction.
Substrate-binding Site and Substrate Specificity
The rationale behind this study was to investigate STMO in comparison with PAMO and other well characterized BVMOs to gain insight into the substrate selectivities of these enzymes. In this regard, the crucial starting observation for our analysis was that atomic superpositions clearly indicated that the active site of STMO (Fig. 3a) is closely related to those of other BVMOs, and in particular, to that of PAMO (Fig. 4a and supplemental Fig. S2). As shown in Fig. 4a, the Cα traces of the residues lining the active-site of STMO and PAMO are very similar, with shifts between the Cα positions of homologous residues mostly in the 0.2–0.4 Å range. The only clear variation concerns the loop 286–295 (STMO numbering), whose position is significantly shifted in the two proteins. This loop is located above the active-site pocket and is most likely directly involved in substrate binding (Fig. 4a). Otherwise, the comparative inspection of the 24 residues that are within 8 Å from the flavin N5 atom (Table 2 and Fig. 4b) indicates that 12 side chains are conserved between PAMO and STMO.
TABLE 2.
Analysis of the amino acid variations between the active sites of STMO and related enzymes
All listed residues are within 8 Å distance from the flavin N5 atom on the re side of the cofactor and their side chains are oriented toward the active-site cavity (see Fig. 4b). PAMO, CHMO, or OTEMO residues that are conserved in STMO are highlighted in boldface.
| STMO | PAMO | CHMO | OTEMO |
|---|---|---|---|
| Val-59 | Val-54 | Thr-47 | Thr-47 |
| Ala-68 | Ala-63 | Ala-56 | Cys-56 |
| Cys-70 | Cys-65 | Ser-58 | Leu-58 |
| Asp-71 | Asp-66 | Asp-59 | Asp-59 |
| Val-72a | Ile-67 | Thr-60 | Thr-60 |
| Pro-157a | Gln-152 | Leu-145 | Pro-145 |
| Leu-158 | Leu-153 | Leu-146 | Leu-146 |
| Ser-201 | Ser-196 | Ser-188 | Ala-195 |
| Ser-202 | Ser-197 | Thr-189 | Thr-196 |
| Asn-225 | His-220 | Gln-212 | Asn-219 |
| Val-261 | Thr-256 | Phe-248 | Phe-255 |
| Val-291a | Pro-286 | Phe-279 | Tyr-285 |
| Lys-295a | Ala-290 | Phe-283 | Leu-289 |
| Arg-342 | Arg-337 | Arg-329 | Arg-337 |
| Val-344 | Ile-339 | Leu-331 | Pro-339 |
| Thr-345a | Leu-340 | Cys-332 | Met-340 |
| Met-397 | Leu-392 | Val-385 | Val-392 |
| Ser-446 | Ser-441 | Ser-441 | |
| Val-447 | Ala-442 | Thr-442 | |
| Leu-448 | Leu-443 | Phe-434 | Phe-443 |
| Met-451 | Met-446 | Leu-437 | Val-446 |
| Leu-500a | Tyr-495 | Phe-486 | Leu-495 |
| Trp-506 | Trp-501 | Trp-492 | Trp-501 |
| Pro-521 | Leu-516 | Phe-507 | Val-519 |
a Residues targeted by mutagenesis in this study.
On these bases, we probed STMO and PAMO for their reactivity toward their reciprocal reference substrates, progesterone and phenylacetone. PAMO was found to be unable to convert progesterone but, remarkably, we observed that STMO displays significant Baeyer-Villiger activity on phenylacetone. Although the Km value is >10 times higher compared with that displayed by PAMO, these experiments unambiguously indicate that STMO can now be reported to be a relatively efficient phenylacetone monooxygenase (Table 3). Consequently, the active site of STMO features a rather impressive level of promiscuity in its substrate preference considering the vastly different nature of progesterone and phenylacetone compounds.
TABLE 3.
Kinetic parameters for STMO mutants
| WT | V72I | P157Q | V291A | K295A | T345L | L500Y | PAMOa | |
|---|---|---|---|---|---|---|---|---|
| Phenylacetone | ||||||||
| kcat (s−1) | 1.5 | 2.7 | 0.6 | 1.8 | 3.0 | 0.2 | 1.0 | 3.1 |
| Km (mm) | 1.0 | 0.6 | 0.2 | 0.8 | 2.6 | 0.2 | 0.5 | 0.08 |
| kcat/Km (mm−1 s−1) | 1.5 | 4.7 | 2.6 | 2.2 | 1.2 | 1.2 | 2.1 | 39 |
| Progesteroneb | ||||||||
| kcat/Km (mm−1 s−1) | 1.8 | 3.9 | 1.7 | 1.4 | 5.2 | <0.1 | 4.7 | <0.1 |
| Uncouplingc (s−1) | 0.08 | 0.11 | 0.09 | 0.04 | 0.13 | 0.10 | 0.10 | 0.02 |
a From Ref. 6.
b Due to solubility limitations, the kcat and Km values for progesterone could not be determined.
c Uncoupling refers to NADPH oxidation in the absence of phenylacetone or progesterone substrates.
A Mutagenesis Study of STMO Active Site
The logical question raised by the above described findings concerned the structural basis of these broad substrate activities and the possibility to modulate them by site-directed mutagenesis. We have repeatedly attempted the determination of the STMO structure in complex with progesterone or related molecules. Many experimental strategies were tried but the poor solubility of these compounds prevented success in these experiments.
For this reason, we adopted a different approach based on mutating side chains that are part of the active site. The strategy for choosing the mutations was to probe different types of variations targeting both hydrophobic (Val-72, Pro-157, Val-291, Leu-500) and hydrophilic (Lys-295, Thr-345) residues and with different degrees of conservations in the other structurally characterized BVMO enzymes (Tables 2 and 3). Furthermore, the sites of mutations were located in different areas of the active site, particularly with regard to their distance from the flavin ring (see Figs. 3b and 4, a and b). Thus, V72I mimics PAMO (which has Ile in this position) and affects a residue in direct contact with the pyrimidine ring of the flavin. This residue is in further contact with Val-291, which is positioned on the only small active-site segment whose backbone conformation differs significantly between STMO and PAMO (Fig. 4a). Its conservative replacement to Ala was designed to probe the role of this differing area of the active site in substrate selectivity. Pro-157, Lys-295, Thr-345, and Leu-500 were mutated with the idea of targeting residues that, by being further away from the flavin, could have a role in binding the bulky progesterone thereby having a specific role in substrate specificity of STMO (Fig. 3b). The mutants were all evaluated for their activities on phenylacetone and progesterone, whose poor solubility allowed us only to accurately measure kcat/Km values (Table 3). Furthermore, two mutants (K295A and T345L) were subjected to crystallographic studies (Table 1). In this way, a comprehensive analysis of the substrate binding region could be obtained.
The main and, to a certain extent, most surprising theme emerging from the analysis is that the effect of the mutations on enzymatic activities is relatively modest. In all cases, mutations did not affect the NADPH oxidase (uncoupling) activity, i.e. the consumption of NADPH in the absence of the organic substrate caused by the NADPH-mediated flavin reduction followed by reoxidation to generate hydrogen peroxide. This indicates that flavin reactivity in terms of both reduction by NADPH and oxygen-mediated reoxidation is unaffected by the mutations, which is consistent with the strategy of targeting residues that are part of the substrate-binding site and not expected to have a direct role in redox catalysis. Similar to the wild-type protein, kcat values for progesterone and phenylacetone for most of the mutants were at least 10 times higher than the rates of uncoupling (Table 3). This is in full agreement with the general functional properties of BVMOs. These enzymes stabilize the flavin-peroxide intermediate that, without a substrate, decays slowly to generate hydrogen peroxide, whereas the decay is greatly accelerated by the binding of the oxygen-accepting substrate. Furthermore, the mutant proteins exhibited minor alterations on the activity on phenylacetone in comparison to the wild-type enzyme. Likewise, using progesterone as substrate did not highlight drastic changes, although it is of note that three (V72I, K295A, L500Y) mutants displayed higher activity on this compound, with K295A being three times more active than the wild-type enzyme (Table 3). The only clear exception is T345L, which turned out to make STMO inactive against progesterone without altering the catalytic efficiency for phenylacetone (Table 3). On this basis, the crystal structures of the two more functionally perturbed mutants, T345L and K295A, were solved (Table 1 and Figs. 3b and 4a). They did not show any conformational change except for the replacement of the side chains, suggesting that the analyzed mutations, by affecting residues exposed on the surface of the active-site cleft, do not generally cause significant structural perturbations.
These data are intriguing because at least some of the mutations (P157Q, K295A, L500Y) correspond to rather drastic side chain replacements that can be expected to alter charge distribution and hydrophobicity of the walls lining the binding site. Inspection of the three-dimensional structure shows that bulky progesterone can be accommodated in the active site in various orientations and poses that would be compatible with catalysis. We refrain from presenting an in silico model of a bound progesterone given the many uncertainties, above all concerning the exact conformational state of the substrate-bound enzyme (e.g. the conformation of loop 510–516, which is disordered in our crystals of STMO, and uncertainty of how NADP+ is bound in the substrate-enzyme complex; Fig. 1). Despite this limitation, the lack of activity of T345L on progesterone indicates that this residue is probably involved in forming a productive enzyme-progesterone complex. The bulkier Leu side chain of the mutant may hinder proper binding of the steroid in the active site. This effect is instead seemingly absent for the smaller phenylacetone substrate. This is the only clue from these studies for the presence of a relatively specific enzyme-substrate interaction that can greatly alter substrate binding. Indeed, the limited effects on steady-state parameters by all other mutations indicate that none of the targeted side chains and binding site areas (at least individually) are essential or even truly crucial for the recognition and catalytically productive binding of both progesterone and phenylacetone.
DISCUSSION
BVMOs have a fascinating mechanism that has been clarified by several studies (1, 7). The enzymatic function is based on the dual role of NADP(H), which acts as flavin reductant as well as an integral part of the active site that promotes oxygen activation and oxygenation. This mechanism implies that the enzyme must exist in different functional/conformational states including one that promotes NADPH-mediated reduction of the flavin and one that promotes formation and stabilization of the oxygen-activating flavin-peroxide intermediate, which is essential for substrate oxygenation. These catalytic steps are brought about by conformational changes, at least locally affecting the active site (11, 12). The structure of STMO is fully consistent with this picture by showing a NADP+-binding mode in which the nicotinamide is shifted outward, away from the flavin (Fig. 2). In particular, this observation provides a further indication in support of the idea that NADP(H) must move above the flavin along the catalytic cycle to carry out its dual function in catalysis. A certain degree of flexibility between the two protein domains may be further instrumental to the attainment of the active site conformations that are responsible for performing the various catalytic steps and accommodating the substrates.
A remarkable finding of our study is the fact that STMO, initially discovered as progesterone monooxygenase, is also active on phenylacetone (Scheme 1 and Table 3). This feature is matched by the close structural similarities exhibited by STMO and PAMO active sites (Figs. 2–4). Moreover, of the six substrate-binding site mutants that were generated, only one had a clear effect on activity on at least one of the two substrates. All the other mutants display steady-state parameters that are comparable with those of wild-type STMO, highlighting a remarkable robustness of the system (17) (Table 3). These findings are especially puzzling in light of the self-evident differences between phenylacetone and progesterone in terms of size, bulkiness, hydrophobicity, and hydrogen-bonding groups (Scheme 1). The mutagenesis, kinetic, and structural data do not offer clues for the presence of elements or niches in the binding site that recognize specific chemical and structural features on these substrates. Rather, the emerging idea is that the binding site, which is mainly (but not exclusively) decorated by small aliphatic side chains (Table 2), functions as a sort of general hydrophobic selectivity filter.
This notion is supported by the enhanced activity against progesterone (but not phenylacetone) displayed by K295A, which removes a positive charge from a site of the binding pocket that is at a distance of 8 Å from the flavin (Fig. 3). Both progesterone and phenylacetone can be oxygenated, apparently because their ketone group linked to a hydrophobic/aliphatic moiety that can gain access to the reacting center in front of the flavin. This does not imply that any molecule can be processed by any BVMO. For example, PAMO is not active on progesterone. Likewise, cyclohexanone, the substrate of the closely related CHMO, does not show any detectable Bayer-Villiger conversion by STMO or PAMO (25). The lower degree of hydrophobicity but relatively higher degree of conformational flexibility of this cyclic aliphatic molecule may require more specific interactions to be productively bound by the enzyme. On the other hand, as shown by a recent extensive substrate-profile analysis of BVMOs (24), STMO displays some activity on cyclohexanone derivatives such as 2-phenylcyclohexanone and 2-propylcyclohexanone that carry hydrophobic substituents. In other words, there is substrate selectivity and preference, with, however, a considerable level of promiscuity as effectively indicated by the acceptance of both progesterone and phenylacetone by STMO. A key point that will be subject to further studies is the dynamics and flexibility of the active site, especially with regard to the role of conformational adaptability (26). The substrate preference is apparently the result of the balance of relatively nonspecific, mostly hydrophobic, interactions that the groups on the surface of the binding site are able to establish with the substrate ligands.
These features likely reflect the plasticity of these enzymes that keep evolving in the microbial world in response to the emerging availabilities of new compounds. It remains fascinating that the relatively limited number of mutations can enable an enzyme to acquire a catalytic function on a very different molecule, possibly retaining the ability to act on the original substrate. These findings support even further the idea that the BVMO enzymes of the STMO/CHMO/PAMO group are particularly attractive for user-tailored protein engineering studies. Their robust scaffold and promiscuity in substrate preferences indicate that they have a clear evolutionary potential to be exploited for generation of variants with activities on desired substrates, suited for Baeyer-Villiger oxidations.
Supplementary Material
Acknowledgments
We thank Dale E. Edmondson and Roberto Orru (Emory University) for helpful advice. We acknowledge the European Synchrotron Radiation Facility and the Swiss Light Source for beam time and excellent support during x-ray data collection.
This work was supported by European Union Project Oxygreen number 212281 and Fondazione Cariplo number 2008.3148.

This article contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 4AOX, 4AOS, 4AP1, and 4AP3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- BVMO
- Baeyer-Villiger monooxygenase
- STMO
- steroid monooxygenase
- PAMO
- phenylacetone monooxygenase
- CHMO
- cyclohexanone monooxygenase
- OTEMO
- 2-oxo-Δ3–4,5,5-trimethylcyclopentenylacetyl-CoA monooxygenase.
REFERENCES
- 1. Leisch H., Morley K., Lau P. C. (2011) Baeyer-Villiger monooxygenases. More than just green chemistry. Chem. Rev. 111, 4165–4222 [DOI] [PubMed] [Google Scholar]
- 2. Turfitt G. E. (1948) The microbiological degradation of steroids. 4. Fission of the steroid molecule. Biochem. J. 42, 376–383 [PMC free article] [PubMed] [Google Scholar]
- 3. Peterson G. E., Thoma R. W., Perlman D., Fried J. (1957) Metabolism of progesterone by Cylindrocarpon radicicola and Streptomyces lavendulae. J. Bacteriol. 74, 684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Torres Pazmiño D. E., Dudek H. M., Fraaije M. W. (2010) Baeyer-Villiger monooxygenases. Recent advances and future challenges. Curr. Opin. Chem. Biol. 14, 138–144 [DOI] [PubMed] [Google Scholar]
- 5. Sheng D., Ballou D. P., Massey V. (2001) Mechanistic studies of cyclohexanone monooxygenase. Chemical properties of intermediates involved in catalysis. Biochemistry 40, 11156–11167 [DOI] [PubMed] [Google Scholar]
- 6. Torres Pazmiño D. E., Baas B. J., Janssen D. B., Fraaije M. W. (2008) Kinetic mechanism of phenylacetone monooxygenase from Thermobifida fusca. Biochemistry 47, 4082–4093 [DOI] [PubMed] [Google Scholar]
- 7. Orru R., Dudek H. M., Martinoli C., Torres Pazmiño D. E., Royant A., Weik M., Fraaije M. W., Mattevi A. (2011) Snapshots of enzymatic Baeyer-Villiger catalysis. Oxygen activation and intermediate stabilization. J. Biol. Chem. 286, 29284–29291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morii S., Sawamoto S., Yamauchi Y., Miyamoto M., Iwami M., Itagaki E. (1999) Steroid monooxygenase of Rhodococcus rhodochrous. Sequencing of the genomic DNA, and hyperexpression, purification, and characterization of the recombinant enzyme. J. Biochem. 126, 624–631 [DOI] [PubMed] [Google Scholar]
- 9. Malito E., Alfieri A., Fraaije M. W., Mattevi A. (2004) Crystal structure of a Baeyer-Villiger monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 101, 13157–13162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fraaije M. W., Wu J., Heuts D. P., van Hellemond E. W., Spelberg J. H., Janssen D. B. (2005) Discovery of a thermostable Baeyer-Villiger monooxygenase by genome mining. Appl. Microbiol. Biotechnol. 66, 393–400 [DOI] [PubMed] [Google Scholar]
- 11. Mirza I. A., Yachnin B. J., Wang S., Grosse S., Bergeron H., Imura A., Iwaki H., Hasegawa Y., Lau P. C., Berghuis A. M. (2009) Crystal structures of cyclohexanone monooxygenase reveal complex domain movements and a sliding cofactor. J. Am. Chem. Soc. 131, 8848–8854 [DOI] [PubMed] [Google Scholar]
- 12. Leisch H., Shi R., Grosse S., Morley K., Bergeron H., Cygler M., Iwaki H., Hasegawa Y., Lau P. C. (2012) Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-δ3–4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453. Appl. Environ. Microbiol. 78, 2200–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beam M. P., Bosserman M. A., Noinaj N., Wehenkel M., Rohr J. (2009) Crystal structure of Baeyer-Villiger monooxygenase MtmOIV, the key enzyme of the mithramycin biosynthetic pathway. Biochemistry 48, 4476–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jensen C. N., Cartwright J., Ward J., Hart S., Turkenburg J. P., Ali S. T., Allen M. J., Grogan G. (2012) A flavoprotein monooxygenase that catalyses a Baeyer-Villiger reaction and thioether oxidation using NADH as the nicotinamide cofactor. ChemBioChem 13, 872–878 [DOI] [PubMed] [Google Scholar]
- 15. Reetz M. T., Wu S. (2009) Laboratory evolution of robust and enantioselective Baeyer-Villiger monooxygenases for asymmetric catalysis. J. Am. Chem. Soc. 131, 15424–15432 [DOI] [PubMed] [Google Scholar]
- 16. Miyamoto M., Matsumoto J., Iwaya T., Itagaki E. (1995) Bacterial steroid monooxygenase catalyzing the Baeyer-Villiger oxidation of C21-ketosteroids from Rhodococcus rhodochrous. The isolation and characterization. Biochim. Biophys. Acta. 1251, 115–124 [DOI] [PubMed] [Google Scholar]
- 17. van Beek H. L., de Gonzalo G., Fraaije M. W. (2012) Blending Baeyer-Villiger monooxygenases. Using a robust BVMO as a scaffold for creating chimeric enzymes with novel catalytic properties. Chem. Commun. 48, 3288–3290 [DOI] [PubMed] [Google Scholar]
- 18. Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 19. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polyak I., Reetz M. T., Thiel W. (2012) Quantum mechanical/molecular mechanical study on the mechanism of the enzymatic Baeyer-Villiger reaction. J. Am. Chem. Soc. 134, 2732–2741 [DOI] [PubMed] [Google Scholar]
- 25. Riebel A., Dudek H. M., de Gonzalo G., Stepniak P., Rychlewski L., Fraaije M. W. (2012) Expanding the set of rhodococcal Baeyer-Villiger monooxygenases by high-throughput cloning, expression and substrate screening. Appl. Microbiol. Biotechnol. 10, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu S., Acevedo J. P., Reetz M. T. (2010) Induced allostery in the directed evolution of an enantioselective Baeyer-Villiger monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 107, 2775–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kleywegt G. J., Jones T. A. (1994) Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 50, 178–185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.