Background: Gem, an RGK protein, strongly inhibits P/Q-type Ca2+ channels.
Results: A 12-amino acid C-terminal fragment and a 3-amino acid motif in the core of Gem play an important role in this inhibition.
Conclusion: Gem possesses two candidate inhibitory sites.
Significance: This work identifies key molecular determinants of Ca2+ channel inhibition by RGK proteins, offering opportunities to modulate this inhibition.
Keywords: Calcium Channels, G Proteins, Patch Clamp, Protein Motifs, Signaling, Inside-out, Macropatch, Regulation, Two-electrode Voltage Clamp, Voltage-gated Calcium Channels
Abstract
The RGK family of monomeric GTP-binding proteins potently inhibits high voltage-activated Ca2+ channels. The molecular mechanisms of this inhibition are largely unclear. In Xenopus oocytes, Gem suppresses the activity of P/Q-type Ca2+ channels on the plasma membrane. This is presumed to occur through direct interactions of one or more Gem inhibitory sites and the pore-forming Cav2.1 subunit in a manner dependent on the Ca2+ channel subunit β (Cavβ). In this study we investigated the molecular determinants in Gem that are critical for this inhibition. Like other RGK proteins, Gem contains a conserved Ras-like core and extended N and C termini. A 12-amino acid fragment in the C terminus was found to be crucial for and sufficient to produce Cavβ-dependent inhibition, suggesting that this region forms an inhibitory site. A three-amino acid motif in the core was also found to be critical, possibly forming another inhibitory site. Mutating either site individually did not hamper Gem inhibition, but mutating both sites together completely abolished Gem inhibition without affecting Gem protein expression level or disrupting Gem interaction with Cav2.1 or Cavβ. Mutating Gem residues that are crucial for interactions with previously demonstrated RGK modulators such as calmodulin, 14-3-3, and phosphatidylinositol lipids did not significantly affect Gem inhibition. These results suggest that Gem contains two candidate inhibitory sites, each capable of producing full inhibition of P/Q-type Ca2+ channels.
Introduction
HVA2 Ca2+ channels, including L-, N-, P/Q- and R-type channels, conduct Ca2+ into excitable cells and are important for diverse physiological processes such as neurotransmitter release, hormone secretion, muscle contraction, and gene transcription. They are typically composed of a pore-forming α1 subunit (Cavα1) and auxiliary α2δ and β (Cavβ) subunits. The activity of HVA Ca2+ channels is regulated by protein kinases, trimeric G proteins, membrane lipids, intracellular Ca2+, and various adaptor and signaling proteins (1, 2). Of the latter, the RGK family of Ras-related monomeric small GTPases, including Rad, Rem, Rem2, and Gem/Kir, has emerged as the most potent endogenous protein inhibitors of HVA Ca2+ channels (3–6).
Two modes of action have been proposed for the inhibition of HVA Ca2+ channels by RGK proteins; they are reducing the number of surface channels (7–14) or suppressing the activity of channels on the plasma membrane (13, 15–20). Both forms of inhibition require Cavβ (14, 15, 21), which interacts directly with RGK proteins (7, 10, 16, 17, 19–24). This interaction has been generally assumed to be essential for RGK inhibition (15, 21). We recently showed, however, that Gem inhibition of surface P/Q-type Ca2+ channels expressed in Xenopus oocytes remained intact after the disruption of the Gem-Cavβ interaction and that this inhibition appeared to be caused by direct interactions between Gem and Cavα1 (18). We postulated that Gem and Cavα1 interact via an anchoring site in a Cavβ-independent manner, that Cavβ induces the formation of an inhibitory site on Cavα1, and that an inhibitory site in Gem binds to this Cavα1 site to cause inhibition. The regions involved in these interactions on both Cavα1 and Gem remain to be identified.
The four RGK proteins contain a highly conserved Ras-like core and extended variable N and C termini that are absent in Ras (3, 4). The core contains binding sites for Cavβ (10, 18) and the binding and catalytic sites for guanine nucleotides (25, 26). The N and C termini contain binding sites for calmodulin (CaM), 14-3-3, and phosphatidylinositol lipids, which regulate the subcellular distribution and function of RGK proteins (7–9, 27–31). The role of the interactions with these modulators in RGK inhibition of HVA Ca2+ channels is controversial and is not fully characterized (4, 11–13, 15, 32).
In this study we investigated the molecular determinants in Gem that are important for its inhibition of P/Q-type Ca2+ channels expressed in Xenopus oocytes. We identified two distinct regions in Gem, a 12-amino acid stretch in the C terminus and a 3-amino acid motif in the core, that are critical for Gem inhibitory action and may form two separate inhibitory sites.
EXPERIMENTAL PROCEDURES
Constructs and Cloning
For electrophysiology experiments in Xenopus oocytes, cDNAs encoding various constructs were subcloned into a modified oocyte expression vector pGEMHE. The constructs included rabbit brain Cav2.1 (GenBankTM accession number X57477), rat skeletal muscle α2δ, rat brain β3 (GenBankTM accession number M88751), a mutant β3 named β3_Mut2 bearing the M196A/L200A mutation, and WT or mutated human skeletal muscle Gem (GenBankTM accession number BC022010).
For protein synthesis in Escherichia coli, DE3 bacteria were used for cDNA transformation and protein expression. Gem-(Ser-68–Arg-264) cDNA encoding residues Ser-68 to Arg-264 and Gem-(Ser-68–Lys-276) cDNA encoding residues Ser-68 to Lys-Lys-276 were subcloned into the pAcycDuet vector (Novagen). Peptides GCP1 and GCP2 were purchased from GL Biochem, and peptide GCP1_S was purchased from GenScript. For pulldown assays, GCP1, GCP1_S, and Cav2.1-(Val-1899–Glu-1989) cDNA encoding residues from Val-1899 to Glu-1989 were subcloned into modified pET26b (Novagen) with an MBP tag at the N terminus and His tag at the C terminus. Rattus norvegicus calmodulin (GenBankTM accession number NM_017326) was subcloned into pETDuet-1 (Novagen).
For co-immunoprecipitation experiments in HEK 293T cells (gift of Dr. Hiroaki Matsunami at Duke University), an HA (hemagglutinin) tag was added to the N terminus of full-length Gem cDNA (mutant) with a flexible linker of three glycines in between, and the whole construct was cloned into the pCDNA3.1(−) vector (Invitrogen). An Myc tag was added to the N terminus of full-length rat brain β3 with three glycines in between, and the whole construct was cloned into the pCDNA3.1(−) vector (Invitrogen). Rabbit brain Cav2.1 was subcloned into the p3xFLAG-CMV-7.1 vector (Invitrogen).
Oocyte Preparation and Expression
Ovarian lobes were obtained from adult Xenopus laevis (Xenopus I) under anesthesia. Stages V-VI oocytes were prepared by treatment with 2.5 mg/ml collagenase A for 1.5–2.5 h under 200 rpm shaking in a solution containing 82.4 mm NaCl, 2.5 mm KCl, 1 mm MgCl2, and 5 mm HEPES (pH 7.6) and then rinsed twice (15 min each) with a solution containing 96 mm NaCl, 2.5 mm KCl, 1 mm MgCl2, 5 mm HEPES, 1.8 mm CaCl2, 100 units/ml penicillin, and 100 μg/ml streptomycin (pH 7.6). Single, defolliculated oocytes were individually selected. cRNAs were synthesized in vitro, and varying amounts (0.2–5 ng) were injected into selected oocytes in various combinations. Recordings were performed 3–5 days after injection.
Electrophysiology and Data Analysis
All experiments were performed on Xenopus oocytes prepared 3–5 days after RNA injection. For whole-oocyte recordings by two-electrode voltage-clamp (TEVC), electrodes were filled with 3 mm KCl and had a resistance of 0.5–1 megaohms. The bath solution contained 10 mm BaCl2, 5 mm KCl, 60 mm tetraethylammonium hydroxide, 20 mm NaOH, and 5 mm HEPES (pH 7.4 with HCl). The current was evoked every 6 s by a +10-mV pulse for 40 ms from a holding potential of −80 mV. For inside-out macropatch recordings from oocytes, the electrode had a diameter of 15–30 μm and a resistance of 0.2–0.5 megaohms when filled with a solution containing 45 mm BaCl2, 80 mm KCl, and 10 mm HEPES (pH 7.3 with KOH). The bath (i.e. cytoplasmic) solution contained 125 mm KCl, 4 mm NaCl, 10 mm HEPES, and 10 mm EGTA (pH 7.3 with KOH). 0.3 μm phosphatidylinositol diphosphate and 3 mm Mg-ATP were added freshly to the bath solution to attenuate rundown. Macroscopic currents were evoked from a holding potential at −80 mV by 10-ms depolarizations ranging from −30 to +90 mV in 10-mV increments at a 1-s interval.
To obtain the β-less channels described in Fig. 3F, a fast perfusion speed (∼1.5 ml/min) was used. Wash-off of β3_Mut2 was determined by monitoring the positive shift of the activation curve. To obtain the activation curve, macroscopic currents were evoked by 20-ms depolarizations ranging from −40 to +100 mV in 10-mV increments at a 6-s interval. Tail currents were recorded by repolarization to −40 mV regardless of the preceding test pulse, normalized by that after the depolarization to +100 mV, and plotted against the test potentials.
FIGURE 3.
A 12-amino acid Gem peptide is sufficient to inhibit P/Q-type Ca2+ channels on plasma membrane. A, shown is an amino acid sequence of three Gem-derived peptides. GCP1 and GCP2 correspond to Lys-265–Lys-276 and Asn-277–Leu-296 of Gem, respectively; GCP1_S is a sequence-scrambled version of GCP1. B–D and F, left panels, shown is time course of inhibition of currents (recorded at +20 mV) by 100 μm GCP1 (B), 200 μm GCP2 (C), 5 μm GCP1 (D), or 100 μm GCP1_S (F) in inside-out membrane patches from oocytes expressing Cav2.1, α2δ, and β3. n = 5. E, left panel, shown is time course of inhibition of β-less channels by 100 μm GCP1. Currents (recorded at +20 mV) were obtained from inside-out membrane patches excised from oocytes expressing Cav2.1, α2δ, and β3_Mut2. Before time 0, the patch had been perfused for 5 min such that the channels had lost β3_Mut2 and become β-less. n = 4. B–F, right panels, shown are current traces selected from a representative patch for each condition. Currents were evoked by a depolarization to +20 mV from a holding potential of −80 mV and were obtained immediately before (red), during (blue), or after (green) perfusion of the indicated Gem peptides. The dashed line indicates zero current level. G, left panel, whole oocyte currents were recorded at +10 mV by TEVC from oocytes expressing Cav2.1, α2δ, and β3, without Gem (control), with WT Gem (Gem_WT), or with the indicated mutant Gem. *** p < 0.01 (compared with control). Middle panel, a Western blot shows protein expression of WT Gem and the indicated mutant Gem in the lysates of the oocytes recorded in the left panel. β3 expression was shown as the loading control. Right panel, the bar graph shows the normalized intensity of the corresponding Gem bands in the middle panel.
Currents were sampled at 10 kHz and filtered at 2.5 kHz. All data were analyzed with Clampfit and were represented as the mean ± S.D. (number of observations). Statistical significance was determined using two-tailed Student's t tests.
Protein Synthesis in E. coli
Transformed DE3 bacteria were cultured at 37 °C until A600 reached 0.6 and then induced at room temperature by 0.5∼1 mm isopropyl 1-thio-β-d-galactopyranoside for overnight. Cells were collected at 4000 rpm for 15 min and resuspended in a lysis solution containing 50 mm Tris-HCl, 250 mm NaCl, 2.5% glycerol, and 7 mm β- mercaptoethanol (pH 7.8). Resuspended bacteria were sonicated with a Branson digital sonifier followed by centrifugation at 14,000 rpm for 40 min. For Gem-(Ser-68–Lys-276) and Gem-(Ser-68–Arg-264) protein purification, the supernatant was collected and incubated with nickel-nitrilotriacetic acid His-Bind beads (Novagen) in the presence of 20 mm imidazole at 4 °C for 1 h. Proteins were eluted from the beads with 200 mm imidazole in the lysis solution. For CaM purification, most bacterial proteins were denatured by incubation at 70 °C for 3 min and removed by centrifugation at 13,000 rpm for 30 min. The supernatant was supplemented with 2 mm CaCl2 and 1 mm MgCl2 and then was mixed with phenyl-Superose resin for 1 h. After washing the resin with a buffer containing 2 mm CaCl2 and 1 mm MgCl2, calmodulin protein was eluted with a buffer containing 2 mm EGTA and 1 mm MgCl2. Proteins were concentrated and further purified by a Superdex 75 gel-filtration chromatography (GE Healthcare).
Pulldown Assay
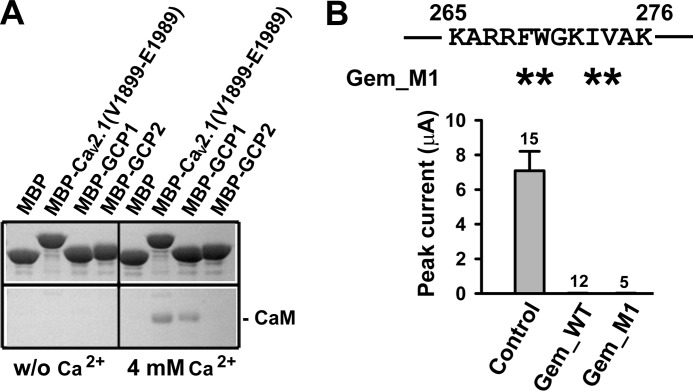
MBP fusion peptides were expressed in DE3 and purified by incubation with amylose resin (New England Biolabs). After elution from the resin with 20 mm maltose, MBP fusion peptides were immobilized on nickel-nitrilotriacetic acid His-Bind beads in PBS buffer with or without Ca2+ (4 mm). The CaM protein bound to peptides was eluted from the beads and denatured at 100 °C for 15 min. The elution was detected with Coomassie Blue staining on SDS-PAGE gel.
Cell Culture and Transfection
HEK 293T cells were maintained at 37 °C in a DMEM medium containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Lipofectamine 2000 (Invitrogen) was used for all transfections.
Oocyte Preparation for Western Blot
Oocytes were collected after TEVC recordings, washed in cold PBS, and lysed with 15–20 μl/oocyte of lysis buffer (PBS, 10% glycerol, 1% Triton X-100, 1–2% protease inhibitor) using trituration through a syringe with a 25-gauge needle (10 times). Lysates were incubated at 4 °C for 1 h while rotating, with brief vortexing every 15–20 min. Aliquots of the lysates were then mixed with 5× SDS and boiled at 55 °C for 15–20 min. 15-μl lysates were loaded onto an acrylamide gel for Western blot.
SDS-PAGE and Western Blot
SDS-PAGE was performed in a Tris-glycine-SDS buffer with 8 or 12% acrylamide gel. Precision Plus Protein All Blue Standards (Bio-Rad) were used as molecular weight markers. For the Western blot, after electrophoresis, the protein gel was transferred to the PVDF membrane and processed with the Odyssey Western-blot kit (Li-Cor). The monoclonal mouse anti-HA antibody HA.11 (Covance), the monoclonal mouse anti-Myc antibody (Sigma), or the monoclonal mouse anti-FLAG antibody M2 (Sigma) was used as the primary antibody (usually overnight incubation at 4 °C). After triple washes with PBS, 1% Tween, membranes were incubated with a secondary antibody (Alexa Fluor 680-conjugated goat anti-mouse IgG; Invitrogen). Images were scanned and analyzed with the Odyssey Infrared Imaging System (Li-COR). The intensity of the protein bands was analyzed with the software ImageJ (NIH).
Co-immunoprecipitation
About 30–48 h later, after transfection, cells were collected in the PBS buffer (Invitrogen (pH 7.2)) at 4 °C. Cell lysates were obtained by adding 1% (V/V) Triton X-100 (Sigma) and 1/50 (V/V) Protease Inhibitor Mixture (Sigma) and rotating at 4 °C for 1 h. Supernatant was obtained by centrifuging at 14,000 rpm for 20 min and was mixed with monoclonal anti-HA antibody (HA.11) coated beads (Covance). After incubation at 4 °C overnight, the beads were spun down and washed 3 times with a solution containing 1 × PBS with a total NaCl concentration of 250 mm and 1/50 (V/V) protease inhibitor mixture (Sigma). The bound proteins were then eluted by adding 0.4 mg/ml HA peptide (Genscript), boiled with an SDS loading buffer for 10 min, and analyzed by SDS-PAGE and Western blot.
RESULTS
12-Amino Acid C-terminal Region in Gem Is Critical for Gem Inhibition
We first carried out systematic deletions to roughly delineate the structural elements of Gem critical for its inhibition of P/Q-type Ca2+ channels expressed in Xenopus oocytes (Fig. 1A). WT or mutant Gem was coexpressed with Cav2.1, α2δ, and β3. Ba2+ was used as the charge carrier, and whole-oocyte currents were recorded by TEVC. WT Gem completely abolished P/Q channel currents (Fig. 1, B–D). The core region of Gem (Gem-(68–247)) was totally ineffective (Fig. 1B, left panel) even though its protein level was much higher than that of WT Gem (Fig. 1B, middle and right panels), indicating that the N and/or C termini were required. Deleting the entire N terminus, however, did not affect Gem inhibition (Gem-(68–296) in Fig. 1C), indicating that the N terminus is not necessary, consistent with previous work showing that the N terminus is not required for Rem2 inhibitory effect (15). In contrast, Gem-(1–264), in which the last 32 amino acids in the C terminus was deleted, was completely inactive (Fig. 1D, left panel) despite its much higher protein level (Fig. 1D, middle and right panels), indicating that this 32-amino acid region is critical for Gem inhibitory action. This result agrees with previous work showing that the corresponding last 32–34 C-terminal residues of Rem and Rad are crucial for their inhibitory effects (20, 21, 23). Another deletion construct, Gem-(68–276), which lacks the last 20 amino acids in the C terminus as well as the entire N terminus, was still fully active (Fig. 1C). The complete lack of effect of Gem-(1–264) and the potent effect of Gem-(68–276) demonstrate that the 12-amino acid region from Lys-265 to Lys-276 in the C terminus is indispensable for Gem inhibition.
FIGURE 1.
Mapping regions of Gem critical for the inhibition of P/Q-type Ca2+ channels. A, shown is a schematic diagram of WT Gem and various truncation mutants. B–D, left panels, shown are whole oocyte currents recorded at +10 mV by TEVC from oocytes expressing Cav2.1, α2δ, and β3 without Gem (control), with WT Gem (Gem_WT), or with the indicated truncated Gem. In these and the following panels showing TEVC currents the number of recordings is indicated above the bar, and all the results shown in each panel were obtained from the same batch of oocytes. B and D, middle panels, a Western blot shows protein expression of WT Gem and the indicated mutant Gem in the lysates of the oocytes recorded in the corresponding left panels. β3 expression is shown as the loading control. B and D, right panels, bar graphs show the normalized intensity of the corresponding Gem bands in the middle panels.
To further test this notion, we applied purified Gem-(Ser-68–Arg-264) and Gem-(Ser-68–Lys-276) proteins to inside-out membrane patches excised from oocytes expressing Cav2.1, α2δ, and β3_GK (the guanylate kinase domain of β3), an approach employed in our previous study (18). Whereas Gem-(Ser-68–Lys-276) acutely and markedly inhibited macroscopic Ba2+ currents conducted through the expressed surface P/Q channels, Gem-(Ser-68–Arg-264) did not produce a significant effect (Fig. 2). This result reinforces the critical importance of the Lys-265–Lys-276 region. The lack of effect of Gem-(Ser-68–Arg-264) is unlikely caused by protein misfolding, as a shorter protein, Gem-(Met-73-Arg-264), was able to be crystallized and exhibited a structure similar to that of canonical Ras (26).
FIGURE 2.
A 12-amino acid region in the C-terminus is crucial for Gem inhibition of P/Q-type Ca2+ channels on plasma membrane. A and B, left panels, shown is time course of inhibition of currents (recorded at +20 mV) by 5 μm purified Gem-(Ser-68–Lys-276) protein (A) or Gem-(Ser-68–Arg-264) protein (B) in inside-out membrane patches from oocytes expressing Cav2.1, α2δ and β3_GK. n = 5. Right panels, current traces selected from a representative patch for each condition are shown. Currents were evoked by a depolarization to +20 mV from a holding potential of −80 mV and were obtained immediately before (red), during (blue), or after (green) perfusion of the indicated purified mutant Gem proteins. The dashed line indicates zero current level.
Lys-265–Lys-276 C-terminal Region in Gem Functions as Candidate Inhibitory Site
How is the Lys-265–Lys-276 region involved in Gem inhibition? One possibility is that it forms an inhibitory site that interacts directly with P/Q channels. To test this hypothesis, a peptide corresponding to this region named GCP1 (Gem C-terminal Peptide 1, Fig. 3A) was synthesized and applied to inside-out membrane patches excised from oocytes expressing Cav2.1, α2δ, and β3. GCP1 at 100 μm greatly reduced the currents (Fig. 3B). In contrast, a peptide corresponding to the last 20 amino acids (Asn-277–Leu-296) of Gem (named GCP2, Fig. 3A) had no effect even at a doubled concentration (Fig. 3C). These results suggest that the inhibitory effect of GCP1 is specific and that the Lys-265–Lys-276 region indeed forms an inhibitory site. The inhibitory effect of GCP1, however, became much weaker when its concentration was decreased to 5 μm (Fig. 3D), a concentration at which purified Gem-(Ser-68–Lys-276) was fully effective (Fig. 2A). An explanation of this discrepancy is that Gem-(Ser-68–Lys-276) can associate with surface P/Q channels through Gem-Cavβ and Gem-Cavα1 interactions, which would greatly increase the effective concentration of the Lys-265–Lys-276 inhibitory site near the channels.
In our previous study we showed that although purified Gem-(Ser-68–Lys-276) protein inhibited surface P/Q channels containing Cavβ, it did not affect channels lacking Cavβ (18). The latter, termed β-less channels, were created by washing off a mutant Cavβ that was able to traffic Cav2.1 to the plasma membrane but dissociated quickly from the surface channel upon vigorous perfusion (18). Using this approach, we found that GCP1, like Gem-(Ser-68–Lys-276), did not inhibit surface β-less P/Q channels (Fig. 3E). This result further demonstrates the specificity of GCP1 and suggests that GCP1 binds to a Cavβ-induced inhibitory site on Cav2.1.
Remarkably, when GCP1 was scrambled into a new peptide named GCP1_S with the same amino acid content but a different sequence, it retained its inhibitory effect (Fig. 3F). This result suggests that the amino acid composition of this region rather than its sequence per se is important for its inhibitory effect.
To examine whether the inhibitory action of GCP1_S was a fortuitous effect of the peptide in isolation, we linked GCP1_S to Gem-(1–264), which as shown in Fig. 1D was inactive. The resulting construct, named Gem-(1–264)+GCP1_S, was able to inhibit P/Q channels, albeit it was slightly less potent than WT Gem was (Fig. 3G, left panel). In contrast, Gem-(1–264)+GCP2, in which the inactive GCP2 peptide was linked to Gem-(1–264), failed to cause inhibition at a similar protein level (Fig. 3G). These results further support GCP1 as a genuine inhibitory site in the full-length Gem.
CaM Binding to Gem Is Not Involved in Gem Inhibition
The identified Lys-265–Lys-276 inhibitory site has been reported to bind CaM (7–9, 14, 27). We confirmed this finding by using purified MBP-tagged GCP1 (MBP_GCP1) to pull down purified CaM in the presence of Ca2+ (Fig. 4A). A fragment of the C terminus of Cav2.1 containing a canonical CaM binding IQ motif, denoted as Cav2.1-(Val-1899–Glu-1989), served as a positive control, whereas MBP-tagged GCP2 (MBP_GCP2) served as a negative control (Fig. 4A). CaM binding to Gem is severely attenuated by mutations of Trp-270 (14). Thus, to examine whether CaM binding to the Lys-265–Lys-276 site plays a role in Gem inhibition, we mutated four large hydrophobic residues (Phe-269, Trp-270, Ile-273, and Val-274) in the Lys-265–Lys-276 region to alanines in the full-length Gem. The resultant Gem mutant (Gem_M1) showed complete inhibition of P/Q channels (Fig. 4B), indicating that CaM binding to the Lys-265–Lys-276 site is not required for Gem inhibition.
FIGURE 4.
CaM binding is not required for Gem inhibition of P/Q-type Ca2+ channels. A, MBP-fused GCP1 peptide pulls down purified CaM in a Ca2+-dependent manner. In the presence of 4 mm Ca2+, CaM interacted with MBP-tagged GCP1 and MBP-tagged Cav2.1-(Val-1899–Glu-1989), which served as a positive control, but not with MBP itself and MBP-tagged GCP2. B, shown are whole-oocyte currents recorded at +10 mV in oocytes expressing Cav2.1, α2δ, and β3 without Gem (control), with WT Gem, or with Gem_M1, in which four hydrophobic residues (asterisks) in the indicated region were mutated to alanine.
Inhibitory Site Exists in Core Region of Gem
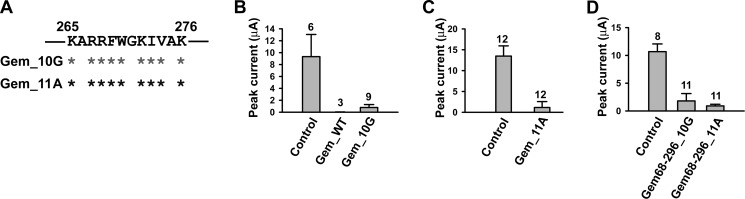
To investigate the role of the Lys-265–Lys-276 site in the inhibition of P/Q channels by full-length Gem, we created two Gem mutants named Gem_10G and Gem_11A, in which 9 of the 12 residues in this region were simultaneously mutated to glycine or alanine, resulting in 10 glycines or 11 alanines in the region (Fig. 5A). Surprisingly, however, both Gem mutants were still capable of inhibiting P/Q channels (Fig. 5, B and C), albeit the magnitude of inhibition by Gem_10G varied in different experiments, ranging from 60 to 100%. The underlying cause for the variation was unclear, but a potential factor was the variation in the effective concentrations of the Gem_10G protein in different batches of oocytes. Regardless of the varying inhibition, the results in Fig. 5, B and C, suggest the existence of an additional inhibitory site in Gem. To map this site, further deletions and point mutations were made in Gem_10G or Gem_11A. Deleting the N terminus of Gem_10G or Gem_11A did not abolish the inhibition (Fig. 5D) nor did simultaneously mutating 4 or 5 amino acids in the C terminus of Gem_10G to alanine either upstream or downstream of Lys-265–Lys-276 (Fig. 6). These results suggest that the additional inhibitory site resides in the core region of Gem.
FIGURE 5.
The Gem Lys-265–Lys-276 region is not the sole inhibitory site in Gem. A, a schematic shows mutations in the Lys-265–Lys-276 region of full-length Gem. Dark and gray asterisks mark residues mutated to alanine and glycine, respectively. B–D, whole-oocyte currents are recorded at +10 mV in oocytes expressing Cav2.1, α2δ, and β3 without Gem (control) or with the indicated Gem mutants.
FIGURE 6.
The C terminus of Gem does not contain a second inhibitory site. A, shown is the amino acid sequence of the last 41 residues (Gln-256–Leu-296) of Gem and Gem_10G. The indicated full-length Gem mutants were all made from Gem_10G, with the asterisks indicating residues that were mutated to alanine. B and C, shown are whole-oocyte currents recorded at +10 mV in oocytes expressing Cav2.1, α2δ, and β3 without Gem (control) or with the indicated Gem mutants. *, p < 0.05; ***, p < 0.01 (compared with Gem_10G).
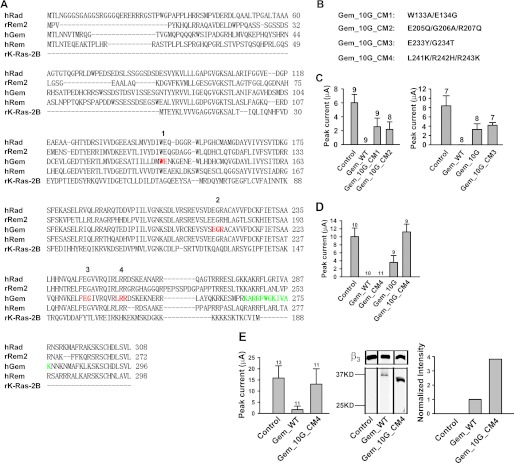
To pinpoint this site, we selectively mutated residues in the core region that are conserved among RGK proteins but are distinct from the corresponding Ras residues (labeled red and 1, 2, 3, and 4 in Fig. 7A). To minimize undesirable structural changes, these residues were mutated to their counterparts in Ras in the Gem_10G background (Fig. 7B). Three of the resulting Gem mutants (Gem_10G_CM1, Gem_10G_CM2, and Gem_10G_CM3) were still able to cause inhibition, albeit less potently than WT Gem but comparable to Gem_10G itself (Fig. 7C), but one mutant, Gem_10G_CM4 (carrying the L241K/R242H/R243K mutation), completely lost its activity (Fig. 7D), indicating that residues Leu-241, Arg-242, and Arg-243 play an important role in Gem inhibition. However, when these residues were mutated in WT Gem (producing a mutant named Gem_CM4), the inhibitory effect was fully preserved (Fig. 7, D and E, left panel). This result suggests that the inhibitory site in the core region and the Lys-265—Lys-276 site in the C terminus can act independently and that each site functioning alone is sufficient to produce full-fledged inhibition.
FIGURE 7.
Identification of a triamino acid motif in the core important for Gem inhibition. A, shown is amino acid sequence alignment of human Rad (BC057815), rat Rem2 (GenBankTM accession number AF084464), human Gem/Kir (BC022010), human Rem (AF084465), and rat K-Ras (BC126086). Red residues are conserved among RGK proteins but not in Ras and were mutated in the Gem_10G background to residues indicated in B. Green residues mark the Lys-265–Lys-276 region. The numbers above the hRad sequence correspond to the names of the mutants in B. B, shown are the name and the mutations of the mutants. CM, core mutation. C and D, and left panel of E, whole-oocyte currents were recorded at +10 mV in oocytes expressing Cav2.1, α2δ, and β3 without Gem (control), with WT Gem, or with the indicated Gem mutants. E, middle panel, a Western blot shows protein expression of WT Gem and Gem_10G_CM4 in the lysates of the oocytes recorded in the corresponding left panel. β3 expression was shown as the loading control. Right panel, a bar graph shows the normalized intensity of the corresponding Gem bands in the middle panel.
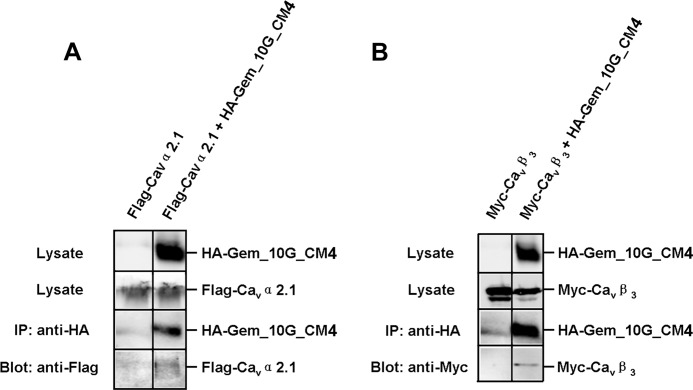
There are at least four possible explanations for the complete lack of inhibition by Gem_10G_CM4. (i) It cannot fold properly. (ii) It expresses poorly in oocytes. (iii) It cannot interact with Cav2.1 or Cavβ. (iv) It no longer possesses a functional inhibitory site. To test the first three possibilities, we examined Gem_10G_CM4 expression in oocytes and its interaction with Cav2.1 or β3 by co-immunoprecipitation in HEK 293T cells. Gem_10G_CM4 protein was expressed abundantly in oocytes (Fig. 7E, middle and right panels), and as shown in Fig. 8, Gem_10G_CM4 was able to associate with Cav2.1 (Fig. 8A) or β3 (Fig. 8B). This result suggests that the first three scenarios are less likely and that Gem_10G_CM4 may not contain a functional inhibitory site.
FIGURE 8.
Gem_10G_CM4 can still associate with Cav2.1 and β3. A and B, a Western blot shows co-immunoprecipitation of HA-Gem_10G_CM4 and FLAG-Cav2.1 (A) or HA-Gem_10G_CM4 and Myc-β3 (B). Immunoprecipitation (IP) of Gem was carried out using an anti-HA antibody from the lysates of HEK 293T cells expressing FLAG-Cav2.1 alone (A, left lane) or Myc-β3 alone (B, left lane), co-expressing HA-Gem_10G_CM4 and FLAG-Cav2.1 (A, right lane), or co-expressing HA-Gem_10G_CM4 and Myc-β3 (B, right lane). Cav2.1 was detected with an anti-FLAG antibody and β3 with an anti-Myc antibody. Each experiment was repeated twice.
DISCUSSION
In our previous study we proposed a “Cavβ-priming” model for Gem inhibition of surface P/Q-type Ca2+ channels (18). This model postulates that Gem associates directly and independently of Cavβ with Cav2.1 through an anchoring site on both Gem and Cav2.1 and that Cavβ induces the formation of an inhibitory site in Cav2.1 where a corresponding inhibitory site in Gem binds to cause inhibition. In this study, through systematic deletions and targeted point mutations, we identify two putative inhibitory sites in Gem (key constructs, their functional effects, and the functional states of the two putative inhibitory sites are summarized in supplemental Fig. 1). One site is formed by a 12-amino acid region (Lys-265–Lys-276) in the C terminus, as evidenced by its ability to directly, acutely, specifically, and in a Cavβ-dependent manner inhibit surface P/Q-type channels on its own as a peptide (Fig. 3). That this 12-amino acid region can inhibit P/Q channels on its own is in line with the report that a Gem fragment containing the last ∼80 residues of the C terminus was sufficient to inhibit P/Q channels (19). Eight of the 12 residues in this 12-amino acid region are fully conserved or highly similar among RGK proteins (Fig. 7A), and this region overlaps with a 13-amino acid region in Rem (corresponding to Phe-271–Arg-283 of human Rem) that was found to be critical for Rem inhibition of L-type channels (23). Thus, it is highly likely that this region also forms an inhibitory site in other RGK proteins.
The Lys-265–Lys-276 region falls within the polybasic motif that has been postulated to be involved in membrane targeting, which has been shown to be critical for Ca2+ channel inhibition by Rem and Rem2 (15, 20, 23). However, the observation that Gem_10G and Gem_11A, in which all the positively charged residues were mutated to either glycine or alanine, were still capable of inhibiting P/Q-type channels (Fig. 5) indicate that either intrinsic membrane targeting is not essential for Gem inhibition or another membrane targeting signal is present elsewhere in Gem.
How the Lys-265–Lys-276 region produces Ca2+ channel inhibition remains to be elucidated. It is notable that the inhibition of surface P/Q channels by the 12-amino acid peptide GCP1 is dependent on Cavβ, just as is the inhibition by the purified Gem-(Ser-68–Lys-276) protein (Fig. 3E). This observation indicates that the Lys-265–Lys-276 region is one of the Gem inhibitory sites that binds to the Cavβ-induced inhibitory pocket in Cav2.1. Because this region contains five positively charged amino acids and because a scrambled peptide (GCP1_S) with the same number of positive charges was still effective in inhibiting P/Q channels either on its own (Fig. 3F) or when linked to Gem-(1–264), an inactive Gem deletion mutant (Fig. 3G), it is likely that the Lys-265–Lys-276 region exerts its inhibitory effect mainly through charge-charge interactions with P/Q channels rather than sequence-specific interactions. In the context of full-length Gem, specificity of the Lys-265–Lys-276 region is probably conferred by specific Gem/Cav2.1 interactions through the anchoring site on both proteins.
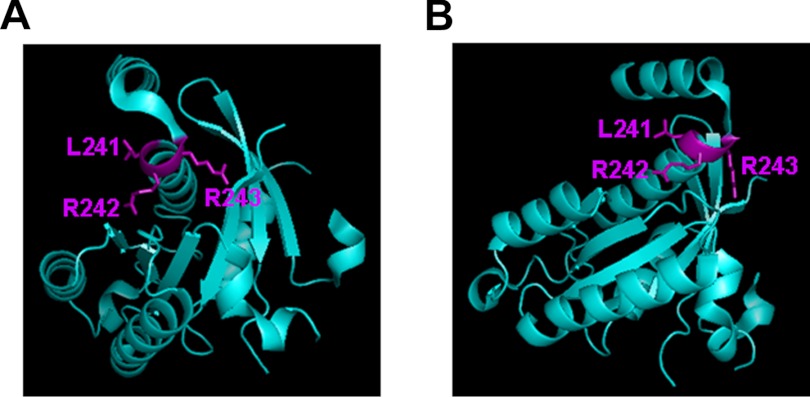
The finding that Gem_10G and Gem_10A, in which the Lys-265–Lys-276 inhibitory site was completely disrupted, was able to fully inhibit P/Q channels (Fig. 5) indicates that the core region of Gem contains another inhibitory site. The existence of an inhibitory site in the core region of other RGK proteins is indicated by previous findings that Rem and Rem2 truncation mutants missing most of the C terminus (including the equivalent Lys-265–Lys-276 inhibitory site) can inhibit Ca2+ channels when they are fused to an exogenous membrane-targeting sequence (15, 20, 23). Our results suggest, but do not prove, that in Gem residues Leu-241/Arg-242/Arg-243 may contribute to form this inhibitory site (Fig. 7). These residues are pivoted at the C-terminal end of an α-helix, with their side chains exposed and ample room to possibly engage in interactions with another protein, as revealed by the crystal structure of a GDP-bound Gem protein fragment (Met-73–Arg-264) containing most of the core (26) (Fig. 9). These residues are completely conserved among RGK proteins but are substituted by other residues in Ras that do not directly regulate HVA Ca2+ channels. Together, these observations suggest that residues Leu-241/Arg-242/Arg-243 form a candidate inhibitory site not only in Gem but also in other RGK proteins. However, more direct evidence is needed to firmly establish this conclusion.
FIGURE 9.
Location and side-chain projection of Leu-241/Arg-242/Arg-243. Two different orientations of a crystal structure (Protein Data Bank ID 2CJW) of a GDP-bound Gem protein fragment (Met-73–Arg-264) are shown. The figures were generated using the program PyMOL.
Our results further show that the core inhibitory site requires the distal C terminus to function properly. Deleting the last 32 C-terminal residues (including Lys-265–Lys-276) rendered this site totally ineffective (Fig. 1D), and mutating various clusters of residues among the last 20 amino acids attenuated its potency (Fig. 6). Furthermore, although the core inhibitory site was capable of producing full inhibition in Gem_10G, this site became completely ineffective in Gem_10GΔC, which lacks the last 20 amino acids but was expressed strongly in oocytes (Fig. 10). These results suggest that the last 20 amino acids are necessary for the proper function of the core inhibitory site. How this distal C-terminal region modulates the function of the core inhibitory site remains to be elucidated. The distance between this region and the core inhibitory site seems to be an important factor, as linking the last 20 amino acids to the inactive Gem-(1–264) did not restore inhibition (Fig. 3G, left panel), although this mutant construct (Gem-(1–264)+GCP2) expressed robustly in oocytes (Fig. 3G, middle and right panels).
FIGURE 10.
The distal C terminus is necessary for the function of the core inhibitory site. Left panel, whole-oocyte currents were recorded at +10 mV in oocytes expressing Cav2.1, α2δ, and β3 without Gem (control), with WT Gem, or with Gem_10GΔC. Middle panel, a Western blot shows protein expression of WT Gem and Gem_10GΔC in the lysates of the oocytes recorded in the corresponding left panel. β3 expression was shown as loading controls. Right panel, a bar graph shows normalized intensity of the corresponding Gem bands in the middle panel.
The notion that RGK proteins contain more than one inhibitory site is in harmony with a report that Rem inhibits L-type Ca2+ channels through three distinct mechanisms, including reducing the number of surface channels, decreasing channel open probability, and immobilizing channel voltage sensors (13). It remains to be investigated how different inhibitory sites are functionally coordinated and regulated.
Various regions of RGK proteins have been found to play a role in the inhibition of HVA Ca2+ channels (15, 19, 21, 23, 24). The N and C termini of RGK proteins contain binding sites for CaM and 14-3-3, and interactions with CaM and 14-3-3 have been shown to regulate the subcellular distribution of RGK proteins (7–9, 28–31). In Gem, these sites include Trp-269 (for CaM) and Ser-22 and Ser-288 (for 14-3-3) (corresponding to Trp-270, Ser-23, and Trp-289 in human Gem) (9, 14, 30, 33). A Gem mutant, W269G (Trp-270 in our studies), which is defective in binding CaM (33), has been reported to weaken Gem regulation of Ca2+ channels (9, 14). Overexpression of CaM in tsA201 cells attenuates Rem inhibition of Cav1.2 currents (34). However, we found that Gem_10G, whose mutations include W270G and are much more severe than W269G, was fully capable of inhibiting P/Q channels (Fig. 5B), indicating that CaM binding to Gem is not required for Gem inhibitory effect. A possible explanation for the discrepancy lies in the observation that the magnitude of RGK regulation of Ca2+ channels depends on the concentration of the RGK protein (35). Our observation that Gem-(Ser-68–Lys-276), which lacks both the N- and C-terminal binding sites for 14-3-3, was fully active in inhibiting P/Q channels (Figs. 1 and 2) indicates that interaction with 14-3-3 is not required for Gem inhibition, in agreement with earlier studies (9, 14).
The Lys-265–Lys-276 region and the Leu-241/Arg-242/Arg-243 tri-amino acid motif do not appear to be involved in the anchoring interaction between Gem and Cav2.1, as Gem_10G_CM4, which contains mutations in both sites, was still able to associate with Cav2.1 in the absence of Cavβ (Fig. 8A). Thus, the anchoring site in Gem remains to be identified. Also remaining to be found are the anchoring site and the Cavβ-induced inhibitory site in Cav2.1.
Supplementary Material
Acknowledgments
We thank Y. Mori for rabbit brain Cav2.1 cDNA, T. Tanabe for α2δ cDNA, E. Perez-Reyes for β3 cDNA, and Dr. H. Matsunami for HEK 293T cells. We also thank our colleagues in the laboratory for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants NS053494 and NS045383 (to J. Y.).

This article contains supplemental Fig. 1.
- HVA
- high voltage-activated
- RGK
- Rem, Rem2, Rad, Gem/Kir
- Cavβ
- calcium channel β subunit
- CaM
- calmodulin
- TEVC
- two-electrode voltage-clamp
- GCP
- Gem C-terminal peptide
- MBP
- maltose-binding protein.
REFERENCES
- 1. Felix R. (2005) Molecular regulation of voltage-gated Ca2+ channels. J. Recept. Signal Transduct. Res. 25, 57–71 [DOI] [PubMed] [Google Scholar]
- 2. Catterall W. A. (2000) Structure and regulation of voltage-gated Ca2+ channels. Annu. Rev. Cell Dev. Biol. 16, 521–555 [DOI] [PubMed] [Google Scholar]
- 3. Kelly K. (2005) The RGK family. A regulatory tail of small GTP-binding proteins. Trends Cell Biol. 15, 640–643 [DOI] [PubMed] [Google Scholar]
- 4. Correll R. N., Pang C., Niedowicz D. M., Finlin B. S., Andres D. A. (2008) The RGK family of GTP-binding proteins. Regulators of voltage-dependent calcium channels and cytoskeleton remodeling. Cell. Signal. 20, 292–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buraei Z., Yang J. (2010) The β subunit of voltage-gated Ca2+ channels. Physiol. Rev. 90, 1461–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Flynn R., Zamponi G. W. (2010) Regulation of calcium channels by RGK proteins. Channels 4, 434–439 [DOI] [PubMed] [Google Scholar]
- 7. Béguin P., Mahalakshmi R. N., Nagashima K., Cher D. H., Ikeda H., Yamada Y., Seino Y., Hunziker W. (2006) Nuclear sequestration of β-subunits by Rad and Rem is controlled by 14-3-3 and calmodulin and reveals a novel mechanism for Ca2+ channel regulation. J. Mol. Biol. 355, 34–46 [DOI] [PubMed] [Google Scholar]
- 8. Béguin P., Mahalakshmi R. N., Nagashima K., Cher D. H., Kuwamura N., Yamada Y., Seino Y., Hunziker W. (2005) Roles of 14-3-3 and calmodulin binding in subcellular localization and function of the small G-protein Rem2. Biochem. J. 390, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Béguin P., Mahalakshmi R. N., Nagashima K., Cher D. H., Takahashi A., Yamada Y., Seino Y., Hunziker W. (2005) 14-3-3 and calmodulin control subcellular distribution of Kir/Gem and its regulation of cell shape and calcium channel activity. J. Cell Sci. 118, 1923–1934 [DOI] [PubMed] [Google Scholar]
- 10. Béguin P., Ng Y. J., Krause C., Mahalakshmi R. N., Ng M. Y., Hunziker W. (2007) RGK small GTP-binding proteins interact with the nucleotide kinase domain of Ca2+ channel β-subunits via an uncommon effector binding domain. J. Biol. Chem. 282, 11509–11520 [DOI] [PubMed] [Google Scholar]
- 11. Sasaki T., Shibasaki T., Béguin P., Nagashima K., Miyazaki M., Seino S. (2005) Direct inhibition of the interaction between α-interaction domain and β-interaction domain of voltage-dependent Ca2+ channels by Gem. J. Biol. Chem. 280, 9308–9312 [DOI] [PubMed] [Google Scholar]
- 12. Yada H., Murata M., Shimoda K., Yuasa S., Kawaguchi H., Ieda M., Adachi T., Murata M., Ogawa S., Fukuda K. (2007) Dominant negative suppression of Rad leads to QT prolongation and causes ventricular arrhythmias via modulation of L-type Ca2+ channels in the heart. Circ. Res. 101, 69–77 [DOI] [PubMed] [Google Scholar]
- 13. Yang T., Xu X., Kernan T., Wu V., Colecraft H. M. (2010) Rem, a member of the RGK GTPases, inhibits recombinant CaV1.2 channels using multiple mechanisms that require distinct conformations of the GTPase. J. Physiol. 588, 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Béguin P., Nagashima K., Gonoi T., Shibasaki T., Takahashi K., Kashima Y., Ozaki N., Geering K., Iwanaga T., Seino S. (2001) Regulation of Ca2+ channel expression at the cell surface by the small G-protein kir/Gem. Nature 411, 701–706 [DOI] [PubMed] [Google Scholar]
- 15. Chen H., Puhl H. L., 3rd, Niu S. L., Mitchell D. C., Ikeda S. R. (2005) Expression of Rem2, an RGK family small GTPase, reduces N-type calcium current without affecting channel surface density. J. Neurosci. 25, 9762–9772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finlin B. S., Correll R. N., Pang C., Crump S. M., Satin J., Andres D. A. (2006) Analysis of the complex between Ca2+ channel β-subunit and the Rem GTPase. J. Biol. Chem. 281, 23557–23566 [DOI] [PubMed] [Google Scholar]
- 17. Finlin B. S., Mosley A. L., Crump S. M., Correll R. N., Ozcan S., Satin J., Andres D. A. (2005) Regulation of L-type Ca2+ channel activity and insulin secretion by the Rem2 GTPase. J. Biol. Chem. 280, 41864–41871 [DOI] [PubMed] [Google Scholar]
- 18. Fan M., Buraei Z., Luo H. R., Levenson-Palmer R., Yang J. (2010) Direct inhibition of P/Q-type voltage-gated Ca2+ channels by Gem does not require a direct Gem/Cavβ interaction. Proc. Natl. Acad. Sci. U.S.A. 107, 14887–14892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leyris J. P., Gondeau C., Charnet A., Delattre C., Rousset M., Cens T., Charnet P. (2009) RGK GTPase-dependent CaV2.1 Ca2+ channel inhibition is independent of CaVβ-subunit-induced current potentiation. FASEB J. 23, 2627–2638 [DOI] [PubMed] [Google Scholar]
- 20. Yang T., Suhail Y., Dalton S., Kernan T., Colecraft H. M. (2007) Genetically encoded molecules for inducibly inactivating CaV channels. Nat. Chem. Biol. 3, 795–804 [DOI] [PubMed] [Google Scholar]
- 21. Finlin B. S., Crump S. M., Satin J., Andres D. A. (2003) Regulation of voltage-gated calcium channel activity by the Rem and Rad GTPases. Proc. Natl. Acad. Sci. U.S.A. 100, 14469–14474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Correll R. N., Botzet G. J., Satin J., Andres D. A., Finlin B. S. (2008) Analysis of the Rem2-voltage-dependent calcium channel β subunit interaction and Rem2 interaction with phosphorylated phosphatidylinositide lipids. Cell. Signal. 20, 400–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Correll R. N., Pang C., Finlin B. S., Dailey A. M., Satin J., Andres D. A. (2007) Plasma membrane targeting is essential for Rem-mediated Ca2+ channel inhibition. J. Biol. Chem. 282, 28431–28440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flynn R., Chen L., Hameed S., Spafford J. D., Zamponi G. W. (2008) Molecular determinants of Rem2 regulation of N-type calcium channels. Biochem. Biophys. Res. Commun. 368, 827–831 [DOI] [PubMed] [Google Scholar]
- 25. Yanuar A., Sakurai S., Kitano K., Hakoshima T. (2006) Crystal structure of human Rad GTPase of the RGK family. Genes Cells 11, 961–968 [DOI] [PubMed] [Google Scholar]
- 26. Splingard A., Ménétrey J., Perderiset M., Cicolari J., Regazzoni K., Hamoudi F., Cabanié L., El Marjou A., Wells A., Houdusse A., de Gunzburg J. (2007) Biochemical and structural characterization of the gem GTPase. J. Biol. Chem. 282, 1905–1915 [DOI] [PubMed] [Google Scholar]
- 27. Moyers J. S., Bilan P. J., Zhu J., Kahn C. R. (1997) Rad and Rad-related GTPases interact with calmodulin and calmodulin-dependent protein kinase II. J. Biol. Chem. 272, 11832–11839 [DOI] [PubMed] [Google Scholar]
- 28. Ward Y., Spinelli B., Quon M. J., Chen H., Ikeda S. R., Kelly K. (2004) Phosphorylation of critical serine residues in Gem separates cytoskeletal reorganization from down-regulation of calcium channel activity. Mol. Cell. Biol. 24, 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Finlin B. S., Andres D. A. (1999) Phosphorylation-dependent association of the Ras-related GTP-binding protein Rem with 14-3-3 proteins. Arch. Biochem. Biophys. 368, 401–412 [DOI] [PubMed] [Google Scholar]
- 30. Mahalakshmi R. N., Nagashima K., Ng M. Y., Inagaki N., Hunziker W., Béguin P. (2007) Nuclear transport of Kir/Gem requires specific signals and importin α5 and is regulated by calmodulin and predicted serine phosphorylations. Traffic 8, 1150–1163 [DOI] [PubMed] [Google Scholar]
- 31. Mahalakshmi R. N., Ng M. Y., Guo K., Qi Z., Hunziker W., Béguin P. (2007) Nuclear localization of endogenous RGK proteins and modulation of cell shape remodeling by regulated nuclear transport. Traffic 8, 1164–1178 [DOI] [PubMed] [Google Scholar]
- 32. Xu X., Marx S. O., Colecraft H. M. (2010) Molecular mechanisms, and selective pharmacological rescue, of Rem-inhibited CaV1.2 channels in heart. Circ. Res. 107, 620–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fischer R., Wei Y., Anagli J., Berchtold M. W. (1996) Calmodulin binds to and inhibits GTP binding of the ras-like GTPase Kir/Gem. J. Biol. Chem. 271, 25067–25070 [DOI] [PubMed] [Google Scholar]
- 34. Pang C., Crump S. M., Jin L., Correll R. N., Finlin B. S., Satin J., Andres D. A. (2010) Rem GTPase interacts with the proximal CaV1.2 C terminus and modulates calcium-dependent channel inactivation. Channels 4, 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seu L., Pitt G. S. (2006) Dose-dependent and isoform-specific modulation of Ca2+ channels by RGK GTPases. J. Gen. Physiol. 128, 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.