Abstract
P2X receptors are trimeric ATP-activated ion channels permeable to Na+, K+ and Ca+2. The seven P2X receptor subtypes are implicated in physiological processes that include modulation of synaptic transmission, contraction of smooth muscle, secretion of chemical transmitters and regulation of immune responses. Despite the importance of P2X receptors in cellular physiology, the three-dimensional composition of the ATP binding site, the structural mechanism of ATP-dependent ion channel gating and the architecture of the open ion channel pore are unknown. Here we report the crystal structure of the zebrafish P2X4 receptor in complex with ATP and a new structure of the apo receptor. The agonist-bound structure reveals a previously unseen ATP binding motif and an open ion channel pore. ATP binding induces cleft closure of the nucleotide binding pocket, flexing of the lower body β-sheet and a radial expansion of the extracellular vestibule. The structural widening of the extracellular vestibule is directly coupled to the opening of the ion channel pore by way of an iris-like expansion of the transmembrane helices. The structural delineation of the ATP binding site and the ion channel pore, together with the conformational changes associated with ion channel gating, will stimulate development of new pharmacological agents.
Adenosine-5′-triphosphate (ATP), best known for its roles in energy metabolism, intracellular signaling, biosynthetic reactions and active transport, is a crucial extracellular signaling molecule 1 that binds to two different classes of ATP receptors: ionotropic P2X receptors2 and G-protein-coupled P2Y receptors3. P2X receptors are found exclusively in eukaryotes, expressed throughout the human body including the nervous, cardiovascular and immune systems, and are implicated in a wide range of physiological processes that include synaptic transmission, smooth muscle contraction, taste, nociception and inflammation4,5. Accordingly, P2X receptors hold great interest as new therapeutic targets for inflammatory, cardiovascular and neuronal disease6.
P2X receptors are trimeric assemblies composed of seven distinct subunit subtypes (P2X1-7)7 that associate to form homomeric and heteromeric complexes8,9. All subunits share a common topology, with intracellular termini, two transmembrane domains and a large, glycosylated and disulfide-rich extracellular domain2,10. The extracellular domain harbors binding sites for ATP, competitive antagonists and modulatory metal ions, whereas the transmembrane domains form a non-selective cation channel11. Gating properties of the ion channel by agonist varies dramatically with receptor subtype, with P2X2, P2X4 and P2X7 homomeric channels showing slow desensitization and P2X1 and P2X3 channels exhibiting rapid desensitization7. While the P2X receptors are ostensibly non-selective cation permeable ion channels, multiple studies suggest that P2X receptors, especially upon prolonged ATP application, are permeable to large organic cations such as N-methyl-D-glucamine (NMDG) 12,13. The pharmacology of P2X receptor subtypes is also divergent, with the sole common feature being activation by ATP and variable yet striking allosteric modulation by metal ions, protons and lipophilic small molecules14. At present, we know little about the location and composition of the ATP binding site, the conformational changes that ensue upon ATP binding, and the nature of the open, ion conducting pore of P2X receptors.
The zebrafish P2X4 receptor15 was recently crystallized in an apo, closed state, and the resulting structure revealed the chalice-shaped trimeric architecture of P2X receptors 16, defined the protein fold of the extracellular domains and illuminated the structure of the closed ion channel pore. The closed state structure, in combination with previous mutational studies 17–22, suggested a location for three non-canonical and intersubunit ATP binding sites. However, the absence of an experimental crystal structure in complex with ATP meant that the binding site for ATP, the mechanism of ATP-dependent gating and the nature of the open ion channel pore remained speculative. Here we report the crystal structures of the slowly desensitizing P2X4 receptor in the presence and absence of ATP at 2.8 and 2.9 Å resolution, respectively. The ATP-bound structure reveals a previously unseen ATP binding motif and an open pore conformation. Most importantly, a comparison of the two structures suggests how ATP binding is coupled to ion channel gating, thus providing the first structural insights into the agonist-induced activation mechanism of P2X receptors based on atomic resolution crystal structures.
Crystallization and structure determination
Initial crystals of the ΔP2X4-B - ATP complex, utilizing the construct employed in the x-ray structure determination of the apo state14, diffracted to only 7 Å resolution. We therefore screened new constructs of the P2X4 receptor, in combination with additional P2X receptor orthologs, by fluorescence-detection size-exclusion chromatography (FSEC)23. Deletion of several additional residues from the C-terminus and return of residue 51 to the native Phe yielded ΔP2X4-C, a construct that starts at Ser28, ends at Lys365 (ΔN27/ΔC24/N78K/N187R) and possesses a sharp and symmetric FSEC elution profile (Supplementary Fig. 1a). The ΔP2X4-C construct exhibits ATP binding and gating activities similar to the wild-type receptor 16, as judged by filter binding and two-electrode voltage clamp experiments, respectively (Supplementary Figs. 1b, c, d, e, f). Importantly, ΔP2X4-C yields crystals in the presence of ATP that diffract x-rays to 2.8 Å resolution. The resulting structure was solved by molecular replacement by utilizing the previously solved ΔP2X4-B structure as a search probe and refined to good crystallographic statistics and stereochemistry (Supplementary Table 1 and Supplementary Figs. 2, 3 and 4).
We also measured x-ray diffraction data from apo state crystals of the ΔP2X4-B construct at a higher resolution (2.9 Å) than that obtained from crystals used in the initial structure determination at 3.1 Å resolution16. Analysis of electron density maps derived from the higher resolution apo and ATP-bound data sets allowed us to correct an incorrect registration of residues 88–97, by one residue, present in the initial apo state structure (Supplementary Fig. 5; Supplementary Table 1). Because the new ΔP2X4-B structure (termed ΔP2X4-B2) is more accurate than the original structure (termed ΔP2X4-B1), we use the former in comparisons with the ATP-bound state.
Architecture
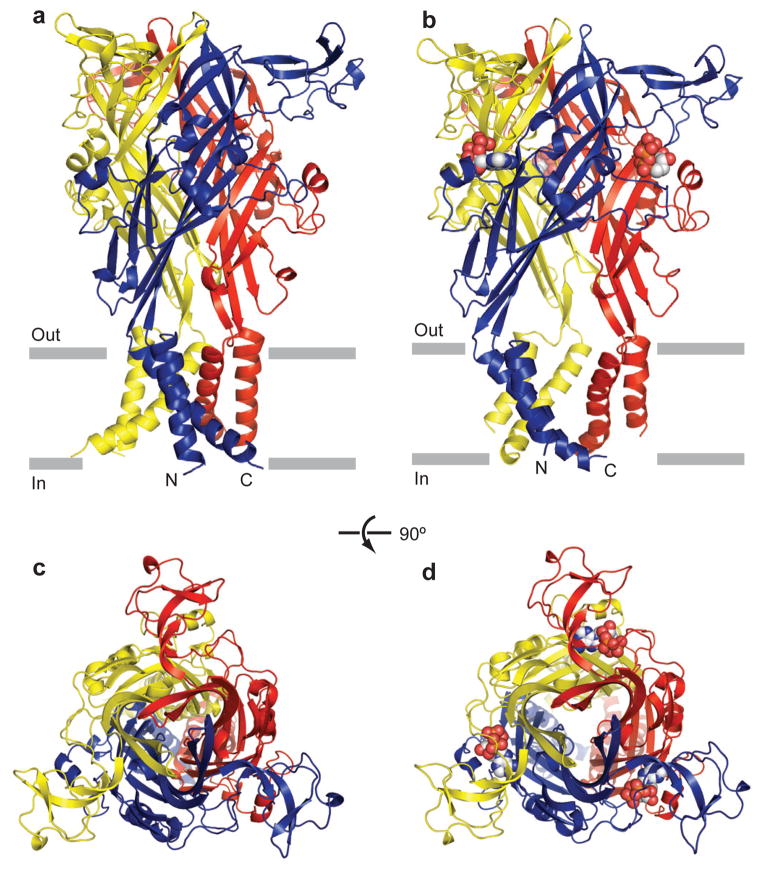
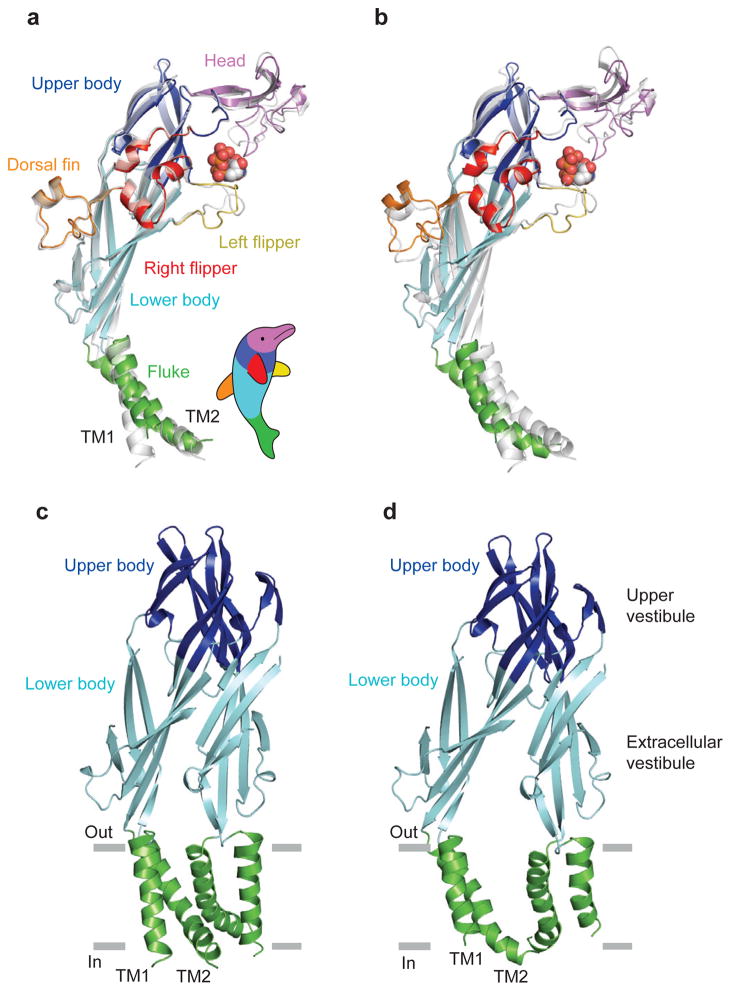
The ATP-bound ΔP2X4-C receptor adopts a chalice-shaped, homotrimetric architecture consisting of a large hydrophilic and glycosylated extracellular domain, a transmembrane (TM) domain composed of 6 α-helices, and short intracellular amino and carboxy termini (Fig. 1). Each subunit resembles the shape of a dolphin, with the TM helices and the extracellular region akin to the flukes and the body, respectively (Fig. 2a). The protein fold and overall structure of an ATP-complexed ΔP2X4-C subunit is similar to that of an apo ΔP2X4-B2 subunit (Figs. 1 and 2a), as illustrated by a root mean squared deviation (r.m.s.d.) of 1.8 Å for the Cα atom position following superposition16. Superposition of the ATP-bound and apo trimers, however, yields r.m.s.d. values of 3.2 Å for Cα atoms, thus demonstrating that substantial conformational changes are associated with the binding of ATP, with some of the largest differences found at and adjacent to the ATP binding sites in the extracellular domain and within the ion conducting TM domain (Fig. 2b). In the context of the trimeric receptor, superposition of the apo, closed state with the ATP-bound state demonstrates that the upper body domain does not undergo substantial conformational changes upon ATP binding, thus suggesting that the upper body domain is a relatively rigid ‘scaffold’ or ‘brace’ (Figs. 2c, d). The lower body domain, by contrast, does not superimpose well upon comparison of the apo and ATP bound states because of an outward ‘flexing’ of the body domain (Figs. 2c, d). This conformational difference between the two states expands the lower region of the extracellular vestibule, increasing the separation between Cα atoms of Asp59 residues on adjacent subunits from 15.0 Å in the apo state to 25.5 Å in the ATP-bound form. The dramatic movement of the lower body domains, in turn, directly expands the TM helices (Figs. 2c, d). Overall, analysis of the ATP-bound structure, in combination with comparison to the apo structure, allows us to define the agonist binding site and to propose a mechanism by which agonist binding leads to ion channel gating in P2X receptors.
Figure 1. The architectures of zebrafish P2X4.
a, b, zebrafish ΔP2X4-B2 (a) and ΔP2X4-C (b) trimer structures viewed parallel to the membrane. Each subunit is shown in a different color. ATP is shown in sphere representation. c, d, zebrafish ΔP2X4-B2(c) and ΔP2X4-C (d) trimer structures viewed from the extracellular side.
Figure 2. Analysis of conformational difference between the ΔP2X4-C and ΔP2X4-B2.
Each structural feature of the dolphin-shaped P2X4 subunit is coloured differently. a, b, Superpositions between the ΔP2X4-C and ΔP2X4-B2 (grey) using Cα positions of the protomers (a) and trimers (b). c, d The TM and the body domains of ΔP2X4-B2 (c) and ΔP2X4-C (d). Only two subunits in the foreground are shown.
ATP binding site
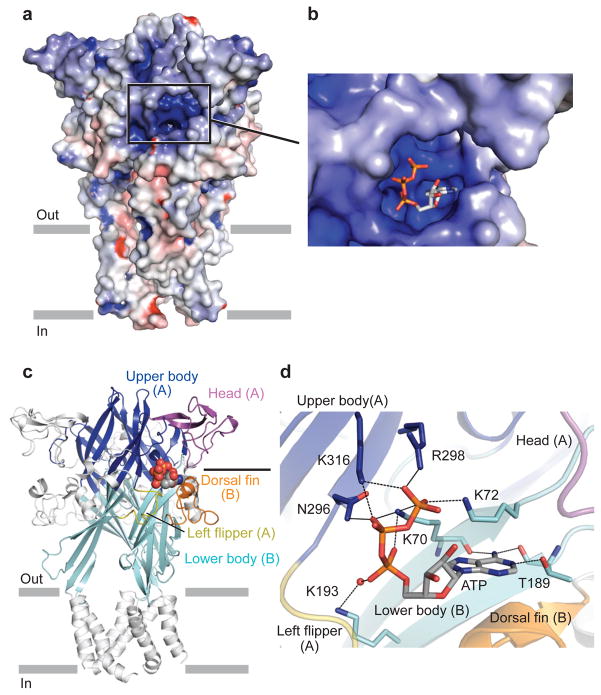
Inspection of electron density maps derived from ΔP2X4-C - ATP cocrystals immediately revealed a prominent feature consistent with the molecular composition of an ATP molecule (Fig. 3 and Supplementary Figs. 6 and 7). Application of the crystallographic symmetry inherent in the R32 space group of this crystal form yields three equivalent ATP binding sites at each of the three pairs of subunit interfaces in the trimeric receptor, sites located ~40 Å from the extracellular boundary of the TM domain. The intersubunit ATP-binding pocket19,24, lined with multiple positively charged residues (Figs. 1b, and 3a, b), is cradled by the head domain (chain A), upper body (chain A), lower body (chain B), left flipper (chain A) and the dorsal fin (chain B) (Fig. 3c). ATP is recognized by the upper (chain A) and lower (chain B) body domains through extensive hydrophilic interactions (Fig. 3d) derived from an underlying protein fold that is, to the best of our knowledge, different from any other known ATP binding motif. The head domain, left flipper and dorsal fin participate in several additional direct contacts with ATP (Fig. 3d and Supplementary Fig 7), consistent with their essential participation in the ATP binding site (Fig. 2).
Figure 3. ATP binding site.
a, b, An electrostatic potential surface of the ΔP2X4-C (a), contoured from −10 kT (red) to +10 kT (blue) (dielectric constant:80), and its close-up view (b). c, The regions forming the ATP-binding pocket are coloured as in Fig. 2a. The ATP molecule is shown in sphere representation. d, Close-up view of the ATP-binding site. The oxygen atom from the glycerol molecule is shown in sphere representation. Black dashed lines indicate hydrogen bonding (<3.3Å).
The bound ATP molecule adopts a U-shaped structure with the β- and γ-phosphates folded toward the adenine ring and the base-sugar constellation in an ‘anti’ conformation (Fig. 3d) -- a conformation previously observed in class II aminoacyl tRNA synthetases25. In the ΔP2X4-C complex with ATP the U-shaped conformation of negatively charged phosphate groups participate in salt bridge and hydrogen bonding interactions, with a cluster of highly conserved basic and polar residues emanating from the two subunits that together form the agonist binding pocket (Fig. 3d). Using the ATP molecule bound at the agonist site between the A and B subunit as a paradigm, Lys70 (chain B) occupies a crucial site because its ammonium group resides at the center of the triphosphate ‘U’, forming interactions with oxygen atoms on the α, β and γ phosphate groups. Asn296 and Lys316 from chain A mediate additional contacts with β-phosphate groups, while Lys 72 (chain B), Arg298 (chain A) and Lys316 (chain A) participate in interactions with the γ-phosphate (Fig. 3d).
The extensive intersubunit interactions with the β- or γ-phosphate groups provide a plausible explanation for why adenosine diphosphate (ADP) and adenosine monophosphate (AMP) have very weak or no effect on the activation of P2X receptors26. Yet the triphosphate moiety is not entirely buried in the agonist binding pocket and the β and γ groups are partially exposed to solvent, consistent with the observation that diadenosine polyphosphates are full or partial agonists at rat P2X receptors27,28. Interactions between the receptor and ATP are also mediated by solvent molecules, and in the crystal structure a glycerol molecule bridges interactions between Lys193 and the α phosphate (GLC1) (Fig. 3d and Supplementary Fig. 8a) and a second glycerol molecule participates in interactions with the β and γ phosphates (Supplementary Fig. 8b). Under physiological conditions we suggest that multiple water molecules, instead of the glycerol molecules, occupy these sites.
The adenine base of ATP is deeply buried in the ATP binding pocket (Fig. 3b) and is recognized by three hydrogen bonds with the side chain of Thr189 and the main chain carbonyl oxygen atoms of Lys70 and Thr189 in the lower body (Fig. 3c, d and Supplementary Fig. 7a). All these residues are strictly conserved16 and have been implicated in the ATP-dependent gating of P2X receptors 17,18–22 and in the adenine base recognition 29. There are also hydrophobic interactions between the adenine base and Leu191 in the lower body and Ile232 in the dorsal fin (Supplementary Fig. 7b). The hydrophobicity of these residues is conserved16, and Leu186 in rat P2X2, corresponding to Leu191 in zebrafish P2X4, is suggested to be involved in the recognition of the adenine base30.
The ribose ring of ATP is recognized only by Leu217 in the dorsal fin through hydrophobic interactions (Supplementary Fig. 7b) and the O2 and O3 atoms of the ribose ring are solvent-accessible (Figs. 3b). Consistent with the solvent exposure of the ribose O2 and O3 atoms, ribose-modified ATP analogues can activate or inhibit P2X receptors31,32.
Base specificity
P2X receptors preferentially recognize ATP whereas cytidine-5′-triphosphate (CTP) has at most a weak effect on receptor activity33,34 and guanosine-5′-triphosphate (GTP) and uridine-5′-triphosphate (UTP) do not activate P2X receptors 26. To understand the mechanism by which P2X receptors achieve this specificity, we superimposed GTP, CTP and UTP onto the ATP molecule bound to the ΔP2X4-C structure and we measured the binding of the nucleotide triphosphate molecules by way of a competition assay using 3H-ATP (Supplementary Fig. 9).
CTP and ATP harbor similar ‘amidine’ functional groups within their base ring structures and on the basis of our superposition, CTP can form hydrogen bonds between the base N4 atom and the carbonyl oxygen atoms of Lys70 and possibly Thr189 (Supplementary Fig. 9b). Because the base of CTP is smaller than that of ATP, however, the N3 atom is too far from the side chain of Thr189 to form a hydrogen bond. In addition, the smaller cytidine base does not fill the agonist binding pocket and the resulting cavity likely further diminishes the extent by which CTP can bind to and activate P2X receptors. GTP and UTP, by contrast with ATP and CTP, possess nearly reciprocal hydrogen bonding groups on their base rings (Supplementary Figs. 9a, b, c, d) and are therefore unable to form favorable hydrogen bonding interactions with carbonyl oxygen atoms of Lys70 and Thr189, thus providing a chemical explanation for why GTP and UTP bind with low affinity to P2X receptors.
Open pore conformation
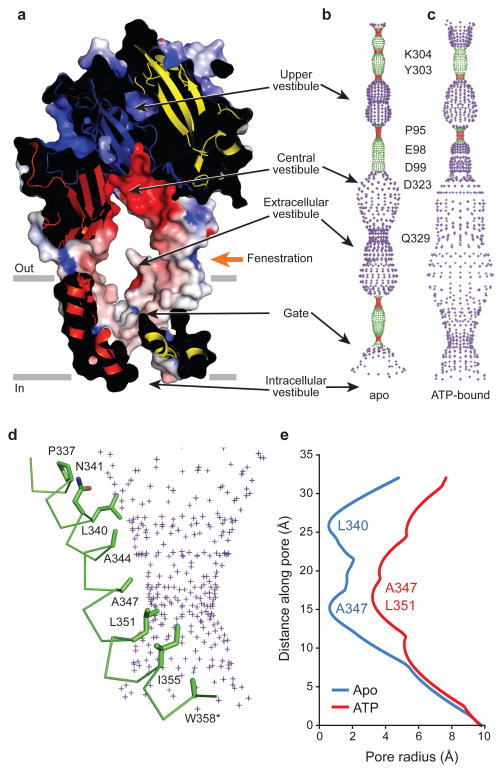
The ΔP2X4-C structure reveals an uninterrupted, continuous transmembrane pore (Fig. 4). The pore is lined primarily by TM2 with residues Leu340, Ala344, Ala347, Leu351 and Ile355 exposed to the putative ion permeation pathway (Fig. 4d), an observation consistent with cysteine-accessibility studies 35–39. In comparison to the ΔP2X4-B2 apo state structure, where Leu340 and Ala347 define the extracellular and intracellular boundaries of the closed ion channel gate, respectively, the most narrow diameters of the ion conductive pathway in the ΔP2X4-C structure are at Ala347 and Leu351 (~7 Å), observations in accord with experiments on the permeability of P2X receptors to organic cations 40,41 (Figs. 4d, e and Supplementary Fig. 10). Thus, we propose that this ATP bound structure represents an activated, open channel conformation.
Figure 4. The transmembrane pore.
a, A section of an electrostatic potential surface of the ΔP2X4-C, contoured as in Fig 3a. Pore-lining surfaces of ΔP2X4-B2 (b) and ΔP2X4-C (c). Each color indicates a different radius range from the pore center (red:<1.15Å, green: 1.15–2.3 Å, and purple:>2.3Å). d, Pore-lining residues of ΔP2X4-C shown in stick representation with the pore-lining surface. e, Pore radius for ΔP2X4-C and ΔP2X4-B2 along the pore center axis.
P2X4 receptors are well known for undergoing so-called ‘pore dilation’ upon prolonged application of ATP, a process defined by increased permeability to large organic cations such as NMDG, with a mean diameter of 7.3 Å12,13,42. To probe the functional properties of the ΔP2X4-C ion channel pore, we carried out two-electrode voltage clamp studies, examining whether prolonged application of ATP leads to pore dilation. Upon a 5-minute application of saturating ATP, we find that the evoked current remains constant, suggesting that the ΔP2X4-C construct represents a non pore-dilated open state (Supplementary Figs. 1g, h).
Interestingly, there are no hydrophilic residues lining the middle of the pore (Fig. 4d). Therefore, water molecules coordinated to permeating cations likely interact with main chain carbonyl oxygen and nitrogen atoms. For the rat P2X2 receptor, however, a threonine implicated in ion selectivity is at the position equivalent to Ala347 in zebrafish P2X4 (Figs. 4d, e)16, near the constriction region in the pore, thus suggesting that this protein side chain interacts directly with permeant hydrated cations 43,44.
Ion access to the pore
The previous ΔP2X4-B1 structure suggested two possible pathways by which cations might access the ion channel: a central pathway, along the three-fold axis of symmetry, and a lateral pathway through the fenestrations ‘above’ the ion channel pore (Figs. 4a–c)16. In the open-state, ATP-bound ΔP2X4-C structure, the pathway along the three-fold axis of symmetry is too small to allow for ion permeation, whereas the lateral fenestrations in the extracellular vestibule of ΔP2X4-C are wide open (Figs. 4a–c)16. Therefore, the lateral fenestrations are the pathways by which hydrated ions enter and exit the receptor, in agreement with recent cysteine-accessibility, cysteine-based cross-linking and computational studies45,46. Once ions pass through the fenestrations, the highly acidic central vestibule attracts cations and repels anions, thus concentrating cations close to the entrance of the ion channel pore46.
Structural transition in the TM domain
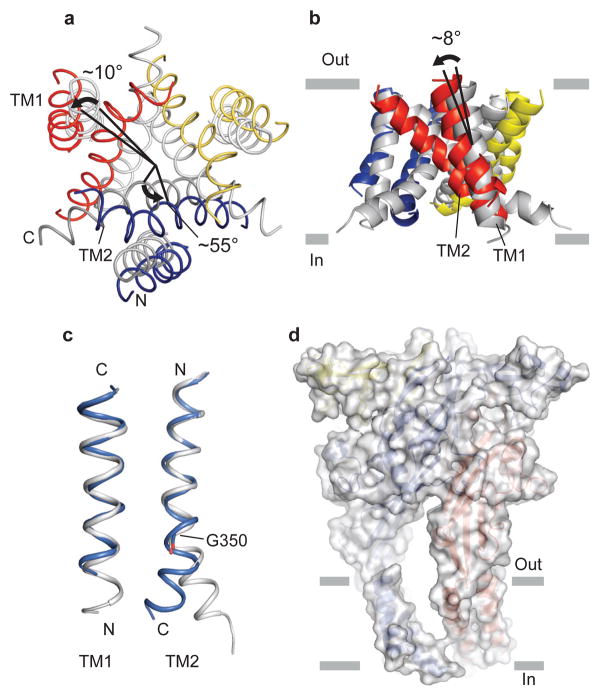
A structural comparison of the TM regions of the closed and open states shows that the TM helices rearrange in an iris-like movement in going from the closed to the open state (Fig. 5). Relative to the closed state structure, TM1 and TM2 rotate by ~10° and ~55° counterclockwise about a pore-center axis perpendicular to the membrane plane, and increase their tilt angle by ~8° and ~2° about an axis parallel to the membrane plane, respectively (Figs. 5a, b and Supplementary Movie S1). The consequence of the iris-like movement of TM1 and TM2 helices is that the helices move away from the central axis by ~3 Å to expand an ion conductive pore (Supplementary Fig. 10 and Supplementary Movie S1).
Figure 5. Structural transition in the TM domain.
a, b, The TM region of ΔP2X4-C and ΔP2X4-B2 (grey), viewed from the intracellular side (a) and from parallel to the membrane (b). ΔP2X4-B2 is superimposed on ΔP2X4-C using Cα positions of the trimer. The black arrows and bars denote the rotation of the TM helices (a) and the orientation of the TM helices (b), respectively. c, Close-up view of the TM helices. TM helices of ΔP2X4-B2 (grey) are superimposed on those of ΔP2X4-C using Cα positions of residue 36–55 for TM1 and residue 334–349 for TM2, respectively. Gly350 is shown in stick representation. d, A surface model of the ΔP2X4-C trimer with the cartoon representation inside.
The transition from closed to open pore greatly alters intra- and intersubunit interactions between the TM helices (Supplementary Fig. 11). In the apo state of the receptor, multiple interactions between TM2 helices that include contacts between Leu340, Leu346 and Ala347 stabilize the closed conformation of the pore (Supplementary Fig. 11c). In the ATP-bound open state, these interactions are ruptured as the TM helices move away from the three-fold axis. Accompanying the radial movement of the TM helices from the three-fold axis is a kinking of the TM2 helix that allows for new contacts to form between subunits involving Leu346 and Ile355, stabilizing the wide-opening pore conformation (Supplementary Fig. 11d). The kink in TM2 is localized to Gly350 (Fig. 5c, Supplementary Fig. 11 and Supplementary Movie S1), a conserved residue previously suggested to function as a gating hinge12,47. Gly350 is associated with weak electron density (Supplementary Fig. 4b), an observation consistent with its role as a flexible hinge.
The movement of the TM helices away from the three-fold axis creates striking ‘gaps’ between the TM helices of adjacent subunits, voids that must be occupied by lipid molecules when the channel resides in its native membrane environment (Fig. 5d and Supplementary Figs. 11c, d). Interestingly, residues lining the ‘gaps’ between subunits and near the kink in TM2 include amino acids implicated in interaction with ivermectin48,49, a positive allosteric modulator of P2X4 receptors (Supplementary Fig. 12) 50. We therefore suggest that ivermectin, and perhaps endogenous lipids, may occupy the ‘gap’ between TM helices in the open state and by so doing allosterically modulate the activity of the channel.
Mechanism of activation
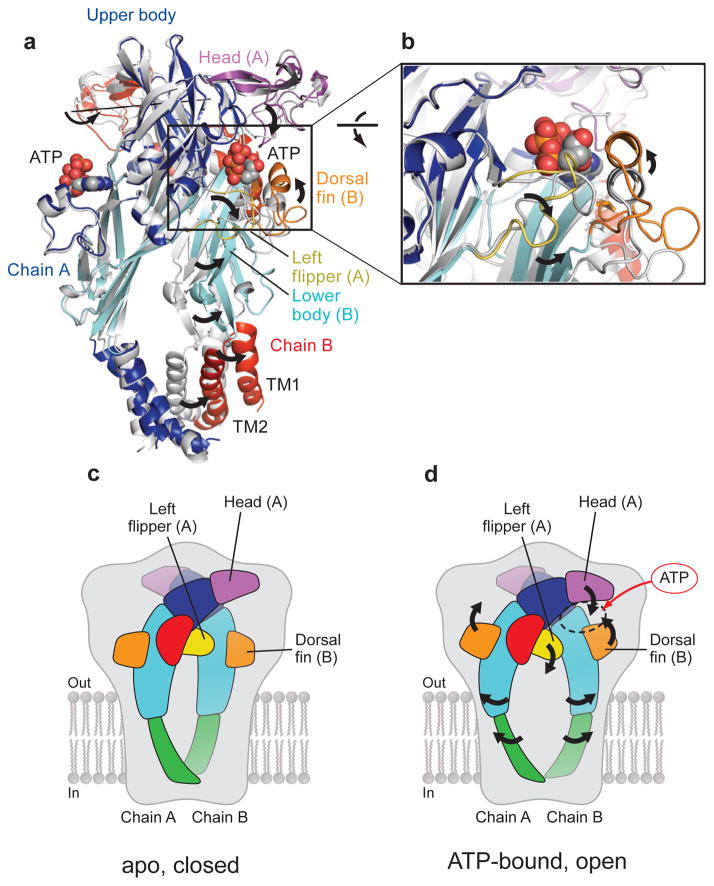
A structural comparison of the extracellular regions of the apo, closed and agonist-bound open states shows how ATP binding leads to channel activation. First, at the ATP binding site, within the intrasubunit cleft, ATP promotes cleft closure between the head and dorsal fin domains, causing the movement of the dorsal fin domain ‘up’ toward the head domain to accommodate ATP via hydrophobic interactions, whereas ATP pushes out the left flipper from the ATP binding pocket (Fig. 6, Supplementary Figs. 13, 14 and Supplementary Movie S2). Because the dorsal fin and left flipper are structurally coupled to the lower body domain (Supplementary Figs. 13, 14 and Supplementary Movie S2), there is a concomitant outward flexing of the lower body domain in the ATP bound state that substantially expands the extracellular vestibule, increasing the separation by ~10 Å (Figs. 2c, d and 6c, d). In the flexing or ‘lever arm’ motion of the lower body domains, the upper body domains of each subunit largely behave as a rigid body, or brace (Supplementary Figs. 15), and subunits rotate by ~8° around a rotation axis located in the upper body (Fig 6a). Finally, the lower body domains are directly coupled to TM1 and TM2 and thus their outward flexing directly promotes the opening of the ion channel pore by causing the TM helices to ‘expand’ in an iris-like motion (Figs. 2c, d, and 6c, d and Supplementary Movie S2).
Figure 6. Mechanism of activation.
a, A ΔP2X4-B2 subunit (grey) is superimposed on ΔP2X4-C using Cα positions of protomer A. Only two subunits in the foreground are shown. The rotation axis describes the superposition of the apo ΔP2X4-B2 B subunit onto the ATP-bound ΔP2X4-C B protomer. b, Close-up view of the conformational changes resulting from ATP binding. c, d A cartoon model of the ATP-dependent activation mechanism. The black arrows denote the movement from the apo closed state (c) to the ATP-bound open state (d).
In the context of this structure-based mechanism we speculate that competitive antagonists, such as 2′,3′-O-(2,4,6-trinitrophenyl)-adenosine-5′-triphosphate (TNP-ATP)32, antagonize the receptor by binding to the ATP site while blocking dorsal fin closure and subsequent ion channel gating because of the steric bulk of the trinitrophenyl moieties (Supplementary Fig. 16).
Conclusion
The crystal structure of the P2X4-C receptor in complex with ATP shows, to the best of our knowledge, that P2X receptors harbor an ATP binding motif not previously seen before. Agonist binding promotes cleft closure in an intersubunit binding site, resulting in the flexing of the lower body domain which in turn directly expands the region of the receptor proximal to the ion channel pore, causing an iris-like opening of an ion conductive pathway. Upon transition to the open state, potential ion pathways in the extracellular domain along the three-fold axis remain occluded by protein, and cations instead gain access to the ion channel by way of three lateral fenestrations. The open state of the receptor is characterized by few subunit-subunit contacts within the transmembrane domains and these relatively large gaps are likely occupied by lipids and allosteric modulators such as ivermectin. Sparse contacts between transmembrane domains suggest that the ion channel domain may adopt multiple conformations, a structural observation consistent with a multiplicity of pore selectivity states deduced from electrophysiological studies of P2X receptors. Taken together, this work illustrates how the binding of ATP activates P2X receptors and initiates ionotropic purinergic signaling.
Methods Summary
The zebrafish ΔP2X4-C and ΔP2X4-B proteins were expressed as N-terminal octa-histidine-enhanced green fluorescent protein (EGFP) fusions in baculovirus-infected Sf9 cells and were purified as described previously16. For samples used in crystallization, 1 mM ATP and 1 mM GdCl3 were added to purified ΔP2X4-C and ΔP2X4-B, respectively. Apo state ΔP2X4-B2 crystals were grown at 4 °C by vapour diffusion using a reservoir solution containing 18–22% PEG 3350, 100 mM MgCl2, 2M NaCl and 0.1M imidazole pH 6.5. For ATP-bound ΔP2X4-C crystals, growth occurred at 4 °C by vapour diffusion with a reservoir solution containing 20–26% PEG 2000, 300mM Mg(NO3)2 and 100mM Tris, pH8.0. Diffraction data were processed and the structures were solved by molecular replacement. The resulting models were then subjected to iterative cycles of manual adjustment and crystallographic refinement. The functional properties of the ΔP2X4-C construct were examined by two-electrode voltage clamp experiments and by [3H]-ATP saturation binding assays.
Methods
Expression and purification
The biochemically well-behaved and functionally active construct of zebrafish P2X4.1 (ΔP2X4-C) was discovered by examining C terminal deletion constructs of the zebrafish receptor as well as multiple additional P2X receptors from other organisms. These receptor candidates were screened by expression in Sf9 insect or human embryonic kidney cells and analyzed by fluorescence detection size-exclusion chromatography (FSEC)23. The ΔP2X4-C and ΔP2X4-B proteins were expressed as N-terminal EGFP fusions with an octa-histidine affinity tag in baculovirus-infected Sf9 cells and were purified as described16. For crystallization, 1 mM ATP and 1 mM GdCl3 were added to purified ΔP2X4-C and ΔP2X4-B from 100 mM stock solutions, respectively.
Crystallization
The ΔP2X4-B2 crystals were obtained at 4 °C in two weeks by vapour diffusion by mixing 1:1 or 2:1 (v/v) ratios of protein and a reservoir solution containing 18–22% PEG 3350, 100 mM MgCl2, 2M NaCl and 0.1M imidazole (pH 6.5). Crystals were collected in a harvest solution containing 20% PEG 3350, 100 mM MgCl2, 1.3M NaCl, 0.1M imidazole pH 6.5, 16% glycerol, 0.5 mM 1mM n-dodecyl β-D-maltoside (DDM) and 1 mM TNP-ATP, and incubated at 4 °C overnight, and cryoprotected by adding additional glycerol in 4% steps (final 24%). To minimize occupancy of Gd in the receptor structure, GdCl3 was excluded from harvest and cryoprotection solutions. The ΔP2X4-C crystals were grown at 4 °C in three days by vapour diffusion by mixing 1:1 or 2:1 ratios of protein and a reservoir solution containing 20–26% PEG 2000, 300 mM Mg(NO3)2 and 100 mM Tris (pH8.0). Crystals were collected in a harvest solution containing 25% PEG 2000, 300 mM Mg(NO3)2, 100 mM Tris (pH8.0), 15% glycerol, 0.5 mM DDM and 1 mM ATP, and cryoprotected by adding glycerol in 5% steps (final 25%). Crystals were flash-frozen in liquid nitrogen for x-ray diffraction experiments.
Structure determination
X-ray diffraction data sets were collected at the Advanced Photon Source (beamline 24-ID-C), and were processed using the HKL2000 suite of computer programs51. The structure of ΔP2X4-C was initially obtained by molecular replacement with the earlier ΔP2X4-B1 coordinates (PDB code 3H9V) using the program Phaser52. There is one subunit in the asymmetric unit and the entire trimeric receptor is built-up by subunits related by crystallographic symmetry. The initial model of the extracellular domain was rebuilt and refined using programs in the CCP453, COOT54 and PHENIX55 packages. The higher resolution data set of ΔP2X4-C confirmed a register shift around the residues 88–97 in the ΔP2X4-B1 structure (Supplementary Fig. 5). After iterative cycles of model building and refinement, the electron density map was recalculated using the extracellular domain of the refined ΔP2X4-C structure. The resulting electron density maps demonstrated that the TM domains associated with the ΔP2X4-C maps adopted a significantly different conformation from that of the TM domains of the ΔP2X4-B1 structure. The ΔP2X4-C TM domains were rebuilt, and further cycles of model building and refinement were performed. The final model contains residues 36–359.
The new apo structure of the zebrafish ΔP2X4-B2 construct was obtained by molecular replacement using the previous ΔP2X4-B1 crystal structure16. The new model corrected the register shift of residues 88–97 and was iteratively refined to good crystallographic residuals as described above. Although crystals were ‘back-soaked’ with a Gd3+ - free solution and soaked with TNP-ATP, we found no electron density for TNP-ATP in the ATP-binding site. Thus this structure is that of the apo conformation. The correction of the register shift was also applied to the ΔP2X4-B1 coordinates, and after refinement with the same sets of reflections as in the original article16 the Rwork and Rfree values improved with the Rfree value decreasing from 27.8% to 27.4%. All structures were validated by the computer program PROCHECK56 and MolProbity57. Pore-lining surfaces were calculated using HOLE58. The rotation axis in Fig. 6 and its angle were calculated by the Dyndom analysis59.
Electrophysiology
RNA encoding the full-length of zebrafish P2X4, ΔP2X4-C and the N-terminal EGFP fusion of the ΔP2X4-C construct was transcribed from pCDNA3.1x plasmids using the mMessage mMachine T7 Ultra kit (Ambion). 2.5–5 ng of RNA was injected into Xenopus oocytes. The recording solution was composed of 100 mM NaCl, 5 mM HEPES, 1 mM MgCl2, and 0.3 mM CaCl2 (pH 7.6)49. Test solutions containing ATP were freshly prepared each day. Recording electrode pipettes (0.5–2M Ω) were filled with 3M KCl. The holding potential was at −80mV. Currents under two-electrode voltage clamp were recorded using a Axoclamp 2B and 900A, GeneClamp 500 amplifiers (Axon Instruments) and digitized using a Digidata 1440A and pClamp 10 (Molecular Devices). No significant currents to test solutions were observed from uninjected oocytes.
The dose-response relationship for ATP activation was obtained by measuring peak current amplitudes in response to ATP application. The peak current from the test solution at the reference concentration of ATP (6.5 μM) was measured first and the peak current for each ATP test solution was measured 4 minutes later and normalized to the peak current evoked by the reference solution. Each ATP concentration was tested on four oocytes. The data were fit to the Hill equation using the Graphpad Prism4 program.
Radioligand binding experiments
The N-terminal, GFP-fusion ΔP2X4-C construct was expressed and purified as described above, concentrated and dialysed overnight at 4 °C against three changes of a dialysis buffer containing 20 mM HEPES, pH7.0, 80 mM NaCl, 20 mM KCl, 15% glycerol and 0.5 mM DDM, and stored at −80 °C before use. Measurements of total ATP binding were obtained by adding GFP-fusion ΔP2X4-C to a final concentration of 15 nM in 250 μl dialysis buffer containing 0 to 293 nM 3H-ATP (Perkin Elmer) where the hot ATP was diluted with cold ATP in a ratio of 1:4, yielding a final specific activity of 7.5 Ci/mmol. Samples were incubated at 4 °C overnight and then binding was terminated by filtering through GSWP 02500 nitrocellulose membranes (Millipore) pre-equilibrated with dialysis buffer containing 100 μM cold ATP. The membranes were subsequently washed three times with 2 ml buffer I, transferred to scintillation vials containing 6 ml of Ultima Gold scintillation cocktail (Perkin Elmer), and counted. Estimates of nonspecific binding were obtained by reactions carried out in the presence of 100 μM cold ATP. The determination of specific binding was derived by subtraction of the nonspecific binding from total binding. The entire experiment was performed in triplicate. The data were fit to a rectangular hyperbola using the Graphpad Prism4 program. For [3H]-ATP competition assay, measurements of total ATP binding were obtained by adding 10 nM of GFP-fusion ΔP2X4-C, 10 nM of 3H-ATP (7.5 Ci/mmol) at the final concentration in 250 μl dialysis buffer containing cold nucleotides. Estimates of nonspecific binding were obtained by reactions carried out in the presence of 100 μM cold ATP. Reactions were terminated by filtering, as described above. The entire experiment was performed in triplicate. Data were fit to a sigmoidal dose response equation using the Graphpad Prism4 program.
Supplementary Material
Acknowledgments
We thank C. Alexander and D. C. Dawson for providing Xenopus oocytes, K. L. Dürr and T. Friedrich for providing the pCNA3.1x vector, L. Vaskalis for assistance with figure illustrations, K. J. Swartz and M. P. Kavanaugh for advice related to the oocyte experiments, and Gouaux lab members for helpful discussions. We are also grateful to the staff at the Advanced Photon Source beamline 24-ID-C for help with X-ray data collection. This work was supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad (M.H.) and by the American Asthma Foundation (E.G.). E.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Author Contributions
M.H. and E.G. contributed to all aspects of the project.
The coordinates and structure factors for the zebrafish apo ΔP2X4-B2 and ATP-bound ΔP2X4-C have been deposited in the Protein Data Bank under the accession codes 4DW0 and 4DW1, respectively. Reprints and permissions information is available at www.nature.com/reprints
The authors declare no competing financial interests
Readers are welcome to comment on the online version of this article at www.nature.com/nature
References
- 1.Burnstock G. Purinergic nerves. Pharmacol Rev. 1972;24:509–581. [PubMed] [Google Scholar]
- 2.Valera S, et al. A new class of ligand-gated ion channel defined by P2x receptor for extracellular ATP. Nature. 1994;371:516–519. doi: 10.1038/371516a0. [DOI] [PubMed] [Google Scholar]
- 3.Webb TE, et al. Cloning and functional expression of a brain G-protein-coupled ATP receptor. FEBS Lett. 1993;324:219–225. doi: 10.1016/0014-5793(93)81397-i. [DOI] [PubMed] [Google Scholar]
- 4.Surprenant A, North RA. Signaling at purinergic P2X receptors. Annu Rev Physiol. 2009;71:333–359. doi: 10.1146/annurev.physiol.70.113006.100630. [DOI] [PubMed] [Google Scholar]
- 5.Burnstock G, Kennedy C. P2X receptors in health and disease. Adv Pharmacol. 2011;61:333–372. doi: 10.1016/B978-0-12-385526-8.00011-4. [DOI] [PubMed] [Google Scholar]
- 6.Coddou C, Yan Z, Obsil T, Huidobro-Toro JP, Stojikovic SS. Activation and regulation of purinergic P2X receptor channels. Pharmacol Rev. 2011;63:641–683. doi: 10.1124/pr.110.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 8.Nicke A, et al. P2X1 and P2X3 receptors form stable trimers: a novel structural motif of ligand-gated ion channels. EMBO J. 1998;17:3016–3028. doi: 10.1093/emboj/17.11.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aschrafi A, Sadtler S, Niculescu C, Rettinger J, Schmalzing G. Trimeric architecture of homomeric P2X2 and heteromeric P2X1+2 receptor subtypes. J Mol Biol. 2004;342:333–343. doi: 10.1016/j.jmb.2004.06.092. [DOI] [PubMed] [Google Scholar]
- 10.Brake AJ, Wagenbach MJ, Julius D. New structural motif for ligand-gated ion channels defined by an ionotropic ATP receptor. Nature. 1994;371:519–523. doi: 10.1038/371519a0. [DOI] [PubMed] [Google Scholar]
- 11.Browne LE, Jiang LH, North RA. New structure enlivens interest in P2X receptors. Trends Pharmacol Sci. 2010;31:229–237. doi: 10.1016/j.tips.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khakh BS, Bao XR, Labarca C, Lester HA. Neuronal P2X transmitter-gated cation channels change their ion selectivity in seconds. Nature Neurosci. 1999;2:322–330. doi: 10.1038/7233. [DOI] [PubMed] [Google Scholar]
- 13.Virginio C, MacKenzie A, Rassendren FA, North RA, Surprenant A. Pore dilation of neuronal P2X receptor channels. Nat Neurosci. 1999;2:315–321. doi: 10.1038/7225. [DOI] [PubMed] [Google Scholar]
- 14.Jarvis MF, Khakh BS. ATP-gated P2X cation-channels. Neuropharmacology. 2009;56:208–215. doi: 10.1016/j.neuropharm.2008.06.067. [DOI] [PubMed] [Google Scholar]
- 15.Kucenas S, Li Z, Cox JA, Egan TM, Voigt MM. Molecular characterization of the zebrafish P2X receptor subunit gene family. Neuroscience. 2003;121:934–945. doi: 10.1016/s0306-4522(03)00566-9. [DOI] [PubMed] [Google Scholar]
- 16.Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X4 ion channel in the closed state. Nature. 2009;460:592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang LH, Rassendren F, Surprenant A, North RA. Identification of amino acid reisdues contributing to the ATP-binding site of a purinergic P2X receptor. J Biol Chem. 2000;275:34190–34196. doi: 10.1074/jbc.M005481200. [DOI] [PubMed] [Google Scholar]
- 18.Ennion S, Hagan S, Evans RJ. The role of positively charged amino acids in ATP recognition by human P2X(1) receptors. J Biol Chem. 2000;275:29361–29367. doi: 10.1074/jbc.M003637200. [DOI] [PubMed] [Google Scholar]
- 19.Marquez-Klaka B, Rettinger J, Bhargava Y, Eisele T, Nicke A. Identification of an intersubunit cross-link between substituted cysteine residues located in the putative ATP binding site of the P2X1 receptor. J Neurosci. 2007:1456–1466. doi: 10.1523/JNEUROSCI.3105-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts JA, Evans RJ. Cysteine substitution mutants give structural insight and identify ATP binding and activation sites at P2X receptors. J Neurosci. 2007;27:4072–4082. doi: 10.1523/JNEUROSCI.2310-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts JA, et al. Cysteine substitution mutagenesis and the effects of methanethiosulfonate reagents at P2X2 and P2X4 receptors support a core common mode of ATP action at P2X receptors. J Biol Chem. 2008;283:20126–20136. doi: 10.1074/jbc.M800294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodnar M, et al. Amino acid residues constituting the agonist binding site of the human P2X3 receptor. J Biol Chem. 2011;286:2739–2749. doi: 10.1074/jbc.M110.167437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson WJ, Jiang LH, Surprenant A, North RA. Role of ectodomain lysines in the subunits of the heteromeric P2X2/3 receptor. Mol Pharmacol. 2006;70:1159–1163. doi: 10.1124/mol.106.026658. [DOI] [PubMed] [Google Scholar]
- 25.Cavarelli J, et al. The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J. 1994;13:327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gever JR, Cockayne DA, Dillon MP, Burnstock G, Ford APDW. Pharmacology of P2X channels. Pflugers Arch. 2006;452:513–537. doi: 10.1007/s00424-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 27.Evans RJ, et al. Pharmacological characterization of heterologously expressed ATP-gated cation channels (P2x purinoceptors) Mol Pharmacol. 1995;48:178–183. [PubMed] [Google Scholar]
- 28.Wildman SS, Brown SG, King BF, Burnstock G. Selectivity of diadenosine polyphosphates for rat P2X receptor subunits. Eur J Pharmacol. 1999;367:119–123. doi: 10.1016/s0014-2999(98)00976-5. [DOI] [PubMed] [Google Scholar]
- 29.Roberts JA, Valente M, Allsopp RC, Watt D, Evans RJ. Contribution of the region Glu181 to Val200 of the extracellular loop of the human P2X1 receptor to agonist binding and gating revealed using cysteine scanning mutagenesis. J Neurochem. 2009;109:1042–1052. doi: 10.1111/j.1471-4159.2009.06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang R, et al. Agonist trapped in ATP-binding sites of the P2X2 receptor. Proc Natl Acad Sci USA. 2011;108:9066–9071. doi: 10.1073/pnas.1102170108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bianchi BR, et al. Pharmacological characterization of recombinant human and rat P2X receptor subtypes. Eur J Pharmacol. 1999;376:127–138. doi: 10.1016/s0014-2999(99)00350-7. [DOI] [PubMed] [Google Scholar]
- 32.Virginio C, Robertson G, Surprenant A, North RA. Trinitrophenyl-substituted nucleotides are potent antagonists selective for P2X1, P2X3, and heteromeric P2X2/3 receptors. Mol Pharmacol. 1998;53:969–973. [PubMed] [Google Scholar]
- 33.Soto F, et al. P2X4: an ATP-activated ionotropic receptor cloned from rat brain. Proc Natl Acad Sci USA. 1996;93:3684–3688. doi: 10.1073/pnas.93.8.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Guzman M, Soto F, Gomez-Hernandez JM, Lund PE, Stühmer W. Characterization of recombinant human P2X4 receptor reveals pharmacological differences to the rat homologue. Mol Pharmacol. 1997;51:109–118. doi: 10.1124/mol.51.1.109. [DOI] [PubMed] [Google Scholar]
- 35.Egan TM, Haines WR, Voigt MM. A domain contributing to the ion channel of ATP-gated P2X2 receptors identified by the substituted cysteine accessibility method. J Neurosci. 1998;18:2350–2359. doi: 10.1523/JNEUROSCI.18-07-02350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rassendren F, Buell G, Newbolt A, North RA, Surprenant A. Identification of amino acid residues contributing to the pore of a P2X receptor. EMBO J. 1997;16:3446–3454. doi: 10.1093/emboj/16.12.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, Chang TH, Silberberg SD, Swartz KJ. Gating the pore of P2X receptor channels. Nat Neurosci. 2008;11:883–887. doi: 10.1038/nn.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, Kawate T, Silberberg SD, Swartz KJ. Pore-opening mechanism in trimeric P2X receptor channels. Nat Commun. 2010;1:1–7. doi: 10.1038/ncomms1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kracun S, Chaptal V, Abramson J, Khakh BS. Gated access to the pore of a P2X receptor: structural implications for closed-open transitions. J Biol Chem. 2010;285:10110–10121. doi: 10.1074/jbc.M109.089185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans RJ, et al. Ionic permeability of, and divalent cation effects on, two ATP-gated cation channels (P2X receptors) expressed in mammalian cells. J Physiol. 1996;497:413–422. doi: 10.1113/jphysiol.1996.sp021777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu DM, Adams DJ. Ionic selectivity of native ATP-activated (P2X) receptor channels in dissociated neurones from rat parasympathetic ganglia. J Physiol. 2001;534:423–435. doi: 10.1111/j.1469-7793.2001.00423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villarroel A, Burnashev N, Sakmann B. Dimensions of the narrow portion of a recombinant NMDA receptor channel. Biophys J. 1995;68:866–875. doi: 10.1016/S0006-3495(95)80263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Migita K, Haines WR, Voigt MM, Egan TM. Polar residues of the second transmembrane domain influence cation permeability of the ATP-gated P2X(2) receptor. J Biol Chem. 2001;276:30934–30941. doi: 10.1074/jbc.M103366200. [DOI] [PubMed] [Google Scholar]
- 44.Browne LE, et al. P2X receptor channels show threefold symmetry in ionic charge selectivity and unitary conductance. Nat Neurosci. 2011;14:17–18. doi: 10.1038/nn.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawate T, Robertson JL, Li M, Silberberg SD, Swartz KJ. Ion access pathway to the transmembrane pore in P2X receptor channels. J Gen Physiol. 2011;137:579–590. doi: 10.1085/jgp.201010593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samways DS, Khakh BS, Dutertre S, Egan TM. Preferential use of unobstructed lateral portals as the access route to the pore of human ATP-gated ion channels (P2X receptors) Proc Natl Acad Sci USA. 2011;108:13800–13805. doi: 10.1073/pnas.1017550108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujiwara Y, Keceli B, Nakajo K, Kubo Y. Voltage- and [ATP]-dependent gating of the P2X(2) ATP receptor channel. J Gen Physiol. 2009;133:93–109. doi: 10.1085/jgp.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jelinkova I, et al. Identification of P2X4 receptor-specific residues contributing to the ivermectin effects on channel activation. Biochem Biophys Res Comm. 2006;349:619–625. doi: 10.1016/j.bbrc.2006.08.084. [DOI] [PubMed] [Google Scholar]
- 49.Silberberg SD, Li M, Swartz KJ. Ivermectin interaction with transmembrane helices reveals widespread rearrangements during opening of P2X receptor channels. Neuron. 2007;54:263–274. doi: 10.1016/j.neuron.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 50.Khakh BS, Proctor WR, Dunwiddie TV, Labarca C, Lester HA. Allosteric control of gating and kinetics at P2X(4) receptor channels. J Neurosci. 1999;19:7289–7299. doi: 10.1523/JNEUROSCI.19-17-07289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 52.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.CCP4 Project 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 56.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 57.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smart OS, Neduvelil JG, Wang X, Wallace BA, Samsom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 59.Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model. 2002;21:181–183. doi: 10.1016/s1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.