Abstract
Pathogenic protozoa threaten lives of several hundred million people throughout the world and are responsible for large numbers of deaths globally. The parasites are transmitted to humans by insect vectors, more than a hundred of infected mammalian species forming reservoir. With human migrations, HIV-coinfections, and blood bank contamination the diseases are now spreading beyond the endemic tropical countries, being found in all parts of the world including the USA, Canada and Europe. In spite of the widely appreciated magnitude of this health problem, current treatment for sleeping sickness (Trypanosoma brucei), Chagas disease (Trypanosoma cruzi) and leishmaniasis (Leishmania spp.) remains unsatisfactory. The drugs are decades old, their efficacy and safety profiles are unacceptable. This review describes sterol 14α-demethylase, an essential enzyme in sterol biosynthesis in eukaryotes and clinical target for antifungal azoles, as a promising target for antiprotozoan chemotherapy. While several antifungal azoles have been proven active against Trypanosomatidae and are under consideration as antiprotozoan agents, crystal structures of sterol 14α-demethylases from three protozoan pathogens, Trypanosoma brucei, Trypanosoma cruzi and Leishmania infantum provide the basis for the development of new, highly potent and pathogen-specific drugs with rationally optimized pharmacological properties.
Keywords: Antiprotozoan chemotherapy, crystal structure, enzyme inhibitors, leishmaniasis, sterol 14alpha-demethylase (CYP51), sterol biosynthesis, trypanosomiasis
INTRODUCTION
The Pathogens, Diseases and Geographic Distribution
Trypanosomatidae form a family of unicellular eukaryotic parasites from the order Kinetoplastida, phylum Euglenozoa, supergroup Excavates [1]. The major human pathogens include a number of species in the genera Leishmania and Trypanosoma. All these organisms have complex life-cycles shuttling between insect and mammalian hosts [2].
Trypanosoma cruzi causes Chagas disease, or American trypanosomiasis [3]. It is transmitted by Triatomine vectors (kissing bugs). In mammals T. cruzi resides both extra- and intra-cellularly, as bloodstream trypomastigotes and proliferative amastigotes, respectively. The severity of the acute stage of infection varies from non-symptomatic to fatal (up to 2%) cases, depending on the parasite burden and strain, host immunosystem, etc. Chronic form of the disease develops in 30–40% infected, often 10 to 20 years later. It predominantly affects the heart and gastrointestinal tract, though the parasite is also found in other organs and tissues. The disease is endemic in 18 countries in South and Central America. 16 to 18 million people are infected, ~50,000 die each year and more than 100 million people are at risk.
Two morphologically indistinguishable species of Trypanosoma brucei, transmitted by tsetse fly (Glossina), cause sleeping sickness: West African trypanosomiasis (T. brucei gambiense) or East African trypanosomiasis (T. brucei rhodesiense) [4]. This extracellular parasite first multiplies in the bloodstream; later it crosses the blood-brain barrier and migrates to the central nervous system, invading cerebrospinal fluid. The symptoms include psychiatric disorders, seizures, coma and ultimately death. Sleeping sickness is endemic in 36 Sub-Saharan African countries, with an estimated 300,000 new cases and ~30,000 deaths per year, >60 million people are at risk.
Leishmania is transmitted by sand fly (Plebotomine) [5]. In mammals the parasite exists intracellularly, multiplying within host macrophages. About 30 pathogenic species are morphologically indistinguishable but cause different forms of the disease. The cutaneous forms, localized and diffuse (L. major, L. tropica, L. mexicana, L. aethiopia), appear as obvious skin reactions. In mucocutaneous forms (L. braziliensis, L. panamensis) the lesions destroy the mucous membranes of the nose, mouth and throat cavities and surrounding tissues. Visceral leishmaniasis (L. donovani, L. infantum/L. chagasi), also known as kala-azar, is characterized by high fever, substantial weight loss, swelling of the spleen and liver, and anemia. Post-kala-azar dermal leishmaniasis may follow successful treatment of visceral leishmaniasis. Leishmaniasis is endemic in 88 countries in Africa, America, Asia and Europe, with ~12 million people infected, ~2 million new cases per year and 1/10th of the world population at risk of infection.
In addition to being transmitted by insects, the diseases are also spread by blood transfusion, organ transplantation, from mother to child. Sleeping sickness, visceral leishmaniasis and chronic form of Chagas disease are invariably fatal when untreated.
CURRENT TREATMENT AND FUTURE PROSPECTS
Though parasitic protozoan diseases constitute the world’s most widely spread human health problem since they are concentrated in the poorest parts of the world, they remain neglected and receive little attention from the pharmaceutical industry and scientific funding agencies [6]. There are currently no vaccines [2, 7], and therefore chemotherapy remains the only option.
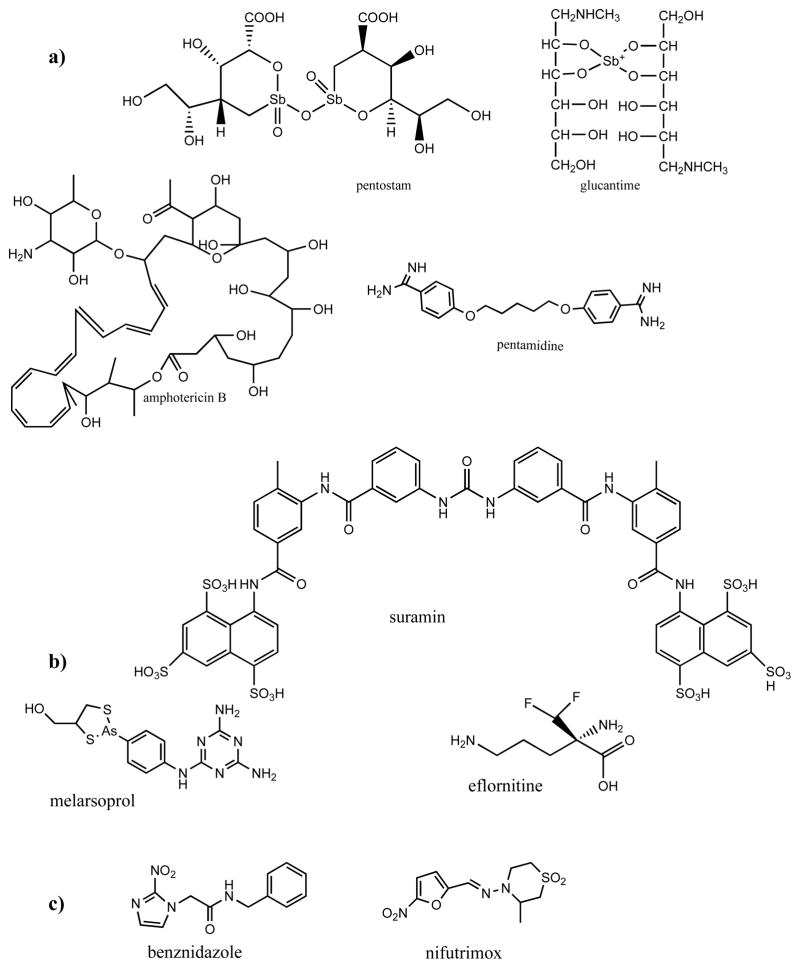
Four major clinical drugs used worldwide for treatment of leishmaniasis Fig. (1) are: two pentavalent antimonials including meglumine antimoniate (pentostam, since 1947) and sodium stibogluconate (glucantime, since 1950), pentamidine (lomidine, since 1940) and amphotericin B (fungizone, since1959), the incidence of resistance being 10–25% of cases. Recently, a new oral chemotherapeutical agent miltefosine (not shown) has become available for treatment of cutaneous and visceral leishmaniasis in India, Colombia and Germany [http://en.wikipedia.org/wiki/Miltefosine]. Amongst the four drugs used against sleeping sickness, suramin (since 1921) and pentamidine are only effective at the first, acute stage of infection since they do not cross the blood brain barrier. Melarsoprol (since 1949) is extremely toxic causing death in up to 10% of patients and eflornithine (developed in 1990) generally does not cure East African trypanosomiasis. Two clinical drugs against Chagas disease, benznidazole and nifurtimox (both introduced in the late 1970s) are effective only for the acute stage of infection but do not treat the symptomatic, chronic form of the disease.
Fig. 1.
Clinical drugs used for treatment of (a) leishmaniasis, (b) sleeping sickness and (c) Chagas disease.
Except for the polyene antibiotic amphotericin B, which depletes ergosterol from the parasite membranes, the drugs are non-specific, the mechanisms of their action remain unclear. The disadvantages include high toxicity, severe side effects and low efficacy. Safer and more efficient drugs are badly needed. Major current approaches for the development of new therapies for human trypanosomiasis and leishmaniasis include blind screening for compounds having expressed antiparasitic effect in cellular experiments, searches for novel targets in the parasite genomes as well as searches for new effective compounds to act on potential drug-targets known to be essential for parasite biology [8–17].
1. Sterol Biosynthesis in Trypanosomatidae
Sterol biosynthesis is an ancient metabolic pathway which is believed to play a key role in the diversification and evolution of eukaryotic organisms [18, 19]. Sterols (cholesterol in vertebrates, ergosterol in fungi and sitosterol in plants) are required for the formation of viable eukaryotic membranes [20]. They define the membrane fluidity and permeability; modulate activity of membrane-bound proteins and ion channels. In addition, sterols serve as precursors for biologically active molecules that act at a nanomolar concentration (hormonal) level and regulate growth and development processes [21, 22]. Essential roles of steroid hormones in animals and plants are well established. Several reports indicate that functional (regulatory) sterols must be important in fungi [23–26] and Trypanosomatidae as well [27–29].
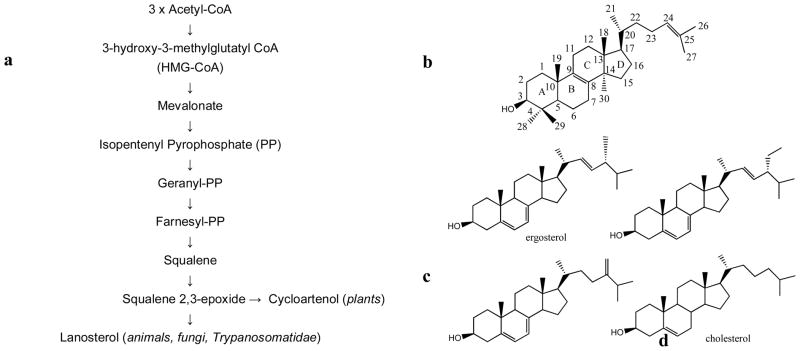
Though some protozoa (e.g. Plasmodium) do not have a sterol biosynthetic pathway in their genome, sequencing of nine genomes of Trypanosomatidae (T. brucei brucei, T. brucei gambiense, T. congolense, T. vivax, T. cruzi, L. major, L. infantum, L. brasiliensis, L. amazonensis) established that all the enzymes from this pathway are present in these parasites. Sterol biosynthesis in Trypanosomatidae begins with acetoacetyl-CoA condensation, and proceeds via mevalonate and farnesyl-pyrophosphate to produce squalene and then squalene 2,3-epoxide Fig. (2a). Similar to fungi and animals and opposite to plants and algae, where squalene 2,3-epoxide is converted into cycloartenol, Trypanosomatidae cyclyze it into lanosterol Fig. (2b). This is in agreement with other genomic information supporting divergence of Trypanosomatidae from an evolutionary ancestor other than photosynthetic organisms [1, 30]. Further reactions on the lanostan skeleton comprise demethylations of the sterol core and modification of the sterol rings and its C20–C27 arm. Although the sequence of these postlanosterol steps in the pathway(s) remains to be clarified, analysis of Trypanosomatidae sterol composition indicates that sterol biosynthesis in these organisms leads to formation of multiple ergostane-based products, the major sterol component in T. cruzi and Leishmania spp. being represented by ergosterol and its 24-methylated and alkylated derivatives Fig. (2c) [27, 28, 31–33]. Contrary to other Trypanosomatidae, bloodstream forms of T. brucei are known to build their membranes using host cholesterol from the human plasma. Therefore exogenous cholesterol Fig. (2d) is the major sterol in bloodstream forms of T. brucei. However, it has been shown that formation of trace amounts of ergosterol derivatives is still required for the parasite growth, indicating that in the conditions of cholesterol abundance, the endogenous sterol biosynthesis is down-regulated but not eliminated completely [28].
Fig. 2.
Sterol biosynthesis in Trypanosomatidae. (a) Formatin of the first cyclzed precursor, lanosterol. (b) Sterol core rings and carbon atoms and nomenclature. Major (c) endogenous and (d) exogenous sterols in the parasites.
Interestingly, there are several publications that report a possibility of multi-organelle location of the sterol biosynthetic pathway in Trypanosomatidae. In addition to endoplasmic reticulum, the sterol biosynthetic enzymes have been found in the parasite mitochondrion and glycosomes [34–36]. The reason for the possible broader subcellular distribution of the pathway is not quite clear, yet it might support multiple regulatory functions of endogenous sterols in Trypanosomatidae.
2. Sterol Biosynthetic Enzymes as Potential Drug Targets for Trypanosomatidae
There are several enzymes on the sterol biosynthetic pathway that have potential to serve as future targets for anti-trypanosomal chemotherapy. Extensive reviews of the details can be found in [36–38]. The most apparent examples on the list comprise HMG-CoA reductase, which in humans serves as clinical target for statins [39], farnesyl diphosphate synthase, the target for bisphosphonates [40], squalene synthase, which is inhibited by quinuclidine derivatives [41], sterol 24-methyl transferase inhibited by azasterols [42, 43] (this enzyme is unique for human pathogens and not present in humans [44]) and sterol 14α-demethylsae.
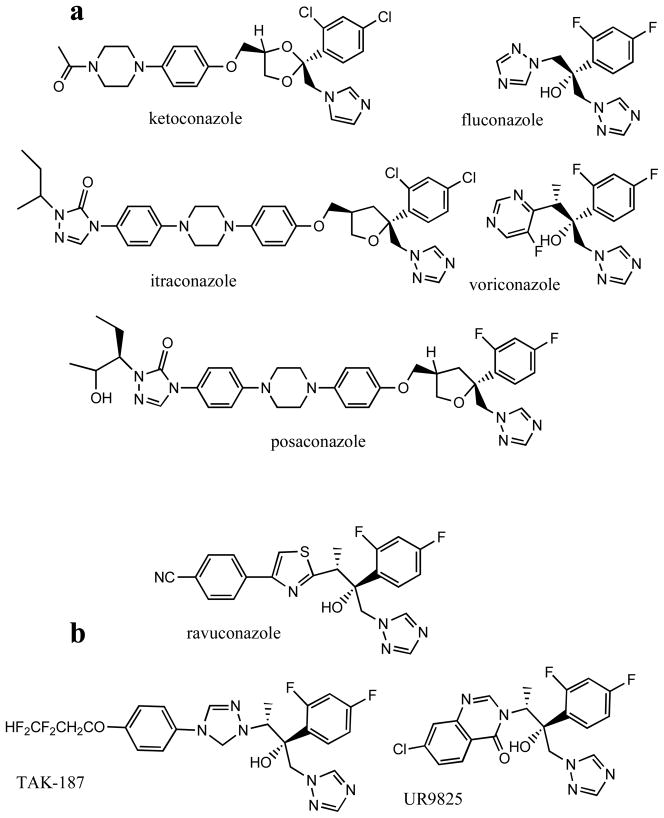
The major advantage of sterol 14α-demethylase as an antiprotozoan drug target is connected with the fact that inhibitors of this enzyme imidazole and triazole derivatives Fig. (3a) are already efficiently used as antifungal agents, in clinical medicine and in agriculture [45, 46]. In addition to blocking sterol biosynthesis, the potency of azoles is enhanced by accumulation of toxic methylated sterols that lead to fungal growth arrest and cell death. An antiparasitic effect of antifungal azoles in Trypanosomatidae cells has been observed by multiple investigators, the first reports being published in 1981 [47–53]. Azoles damage the parasite membranes, affect division, multiplication and dramatically alter sterol composition [31–33, 54]. Antifungal drugs ketoconazole and fluconazole were shown to be effective in vivo for treatment of leishmaniasis [e.g 55, 56]. The newest clinical antifungal drug, posaconazole, approved in 2006 by FDA as a salvage therapy for treatment of opportunistic fungal infections in immumocompromised patients [57] was shown to produce a curative effect in the acute and chronic forms of Chagas disease [58–60] and has been reported to enter clinical trials in 2010 [37, 61]. Other azoles obtained from anti-fungal drug development programs Fig. (3b) were also proven to have trypanocidal activity, both in vitro and in vivo [62–64].
Fig. 3.
Antifungal azoles. (a) Five drugs clinically available for systemic treatment of fungal infections. (b) Antifungal agents that are under trials.
3. Sterol 14α-Demethylase: Catalytic Reaction, Substrate Specificity and the basis for Inhibition and Screening for New Binding Ligands
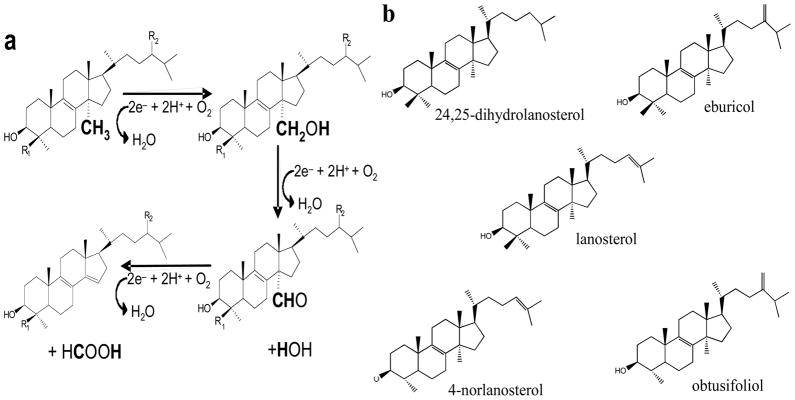
Sterol 14α-demethylase is a cytochrome P450 monooxynenase (CYP51 gene family), the heme-thiolate protein that catalyses a unique three-step reaction of oxidative removal of the 14α-methyl group from the sterol core Fig. (4a). The reaction involves three sequential cytochrome P450 catalytic cycles [http://en.wikipedia.org/wiki/Cytochrome_P450] each requiring delivery of two electrons and two protons. When the sterol substrate binds in the enzyme active site in such a way that the 14α-methyl group is positioned about 5 Å above the heme iron plane, the P450 accepts the first electron from the electron transfer protein, cytochrome P450 reductase (CPR), and its heme iron is reduced from the resting ferric (Fe3+) to the active ferrous (Fe2+) state [65]. Delivery of the first electron enables binding of an oxygen molecule in the distal axial position of the reduced iron, in close proximity to the sterol 14α-methyl group. After this, the second electron is transferred by CPR, reducing the heme bound oxygen. Then two catalytic protons arrive from the protein surface causing the molecular oxygen scission and release of one water molecule while introducing the other atomic oxygen into the methyl group of the substrate (-C-H →-C-O-H). The second cycle of catalysis converts the 14α-alcohol group into the 14α-aldehyde; the third cycle results in release of formic acid and introduction of the Δ14–15-double bond into the sterol ring D.
Fig. 4.
Sterol 14α-demethylase catalysis. (a) The tree step reaction, each step requires two reducing equivalents, two protons and one molecular of oxygen. The electrons are delivered from NADPH by cytochrome P450 reductase (CPR). CPR uses FAD and FMN as cofactors, and the electron flow occurs from NADPH to EAD and then to FMN. Proton delivery route in sterol 14a-demethylase most likely begins from the cytoplasm-exposed surface of the molecule and involves conserved E205 (Helix F′, T. brucei numbering)), the I-helix CYP51 signature residues H297, T295, A291 and the active site water molecule [29]. (b) Five substrates of sterol 14a-demethylases.
There are only five naturally occurring compounds that can be metabolized by sterol 14α-demethylases Fig. (4b). Lanosterol and 24,25-dihydrolanosterol are their natural substrates in animals. Fungal sterol 14α-demethylases metabolize lanosterol and/or 24-methylene-24,25-dihydrolanosterol (eburicol). Obtusifoliol is the substrate for sterol 14α-demethylase from plants. Amongst Trypanosomatidae, sterol 14α-demethylase in T. cruzi so far represents the only exception in terms of its substrate preferences. In vitro, the enzyme clearly prefers eburicol [66], suggesting that this parasite uses the postlanosterol portion of the sterol biosynthetic pathway similar to that in filamentous fungi. Accumulation of eburicol in T. cruzi upon treatment of the parasite with antifungal azoles [31–33, 54] supports this suggestion. We have found that this preference toward the C4-doublemethylated sterol substrates in T. cruzi sterol 14α-demethylase is connected with one amino acid, I105, in the B′-helix [66]. Sterol 14α-demethylases from other Trypanosomatidae have plant-specific phenylalanine in this position (F105). Accordingly, similar to the plant orthologs, they reveal strict specificity to C4-monomethylated sterols (obtusifoliol and 4-norlanosterol) [67, 68]. Although both obtusifoliol and norlanosterol have been identified in Leishmania spp. upon their inhibition with azoles [27, 69], it remains to be clarified which of them or both serve as the natural sterol 14α-demethylase substrate in vivo. While the differentiation between the C4-double versus C4-monomethylated sterol substrates is defined by a single amino acid residue, general sequence identity amongst sterol 14α-demethylases across phyla varies between 22 and 30%. The question how these enzymes maintain their strict functional conservation at such a low amino acid sequence identity has remained a major puzzle in the CYP51 family for many years.
In the substrate-free state the active site cavity of sterol 14α-demethylase contains the water molecule that is coordinated to the heme iron. Apparently, this water is rather strongly bound to the iron and only partially displaced by the substrate, remaining in the iron proximity and participating in the catalytic proton delivery [29]. Heterocyclic compounds, such as imidazole, triazole, pyridine and pyrimidine derivatives, which contain a basic atom, serve as stronger ligands for the heme iron. They coordinate to the heme easily replacing the water from the iron coordination sphere and affect substrate binding and metabolism.
Alterations in the heme iron environment upon ligand binding are reflected in the P450 absorbance spectrum. Thus, the five-coordinated high spin form of the protein, e.g.: if the water is expelled from the iron coordination sphere by a substrate analogous compound, reveals a blue shift in the Soret band maximum (from 417 to 390 nm). Replacement of the water by another ligand that directly coordinates to the heme iron causes a red shift in the maximum (from 417 to 420–427 nm, depending on the compound). This feature of the enzyme can serve as a basis for optical screening for new potential inhibitors [31].
4. Low Sequence Similarity of Sterol 14α-Demethylases Across Biological Kingdoms Predicts a Possibility their Selective Inhibition
It is well known that inhibitory effects of azoles drastically vary depending on the composition of the non-ligated portion of their molecule [70]. Thus, small compounds (e.g. imidazole) have very weak influence on the enzyme activity, while inhibitory potencies of the larger structures e.g. in Fig. (3) are sufficient to cure fungal infections. This implied an important role of additional interactions that must be formed between the inhibitor and amino acid residues inside the protein globule.
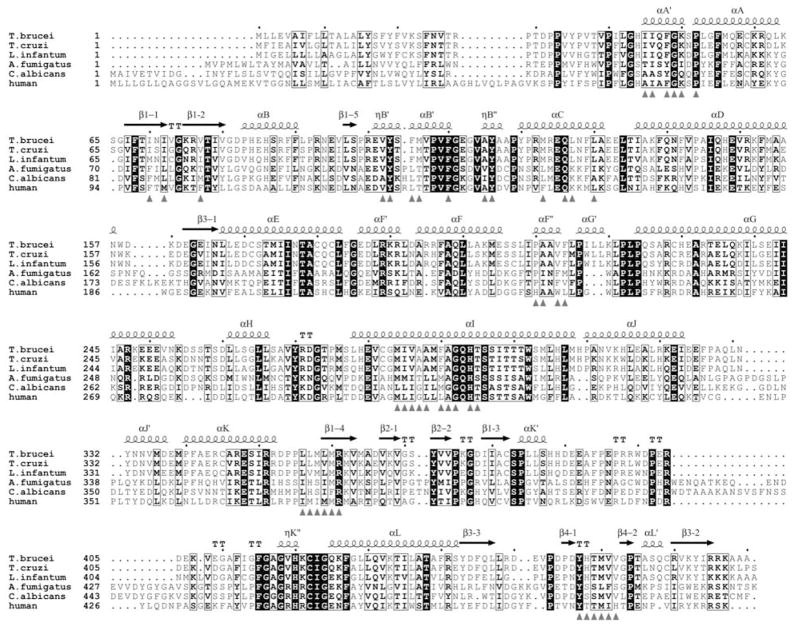
Accordingly, low amino acid sequence identities between the sterol 14α-demethylase orthologs from fungal pathogens and human host suggested that azoles with high potencies to inhibit fungal cell growth and low toxicity in human cells are likely to act as selective inhibitors of fungal enzymes [70]. Later, when heterologously expressed and purified eukaryotic sterol 14α-demethylases became available for reconstitution of their activity in vitro [21], high selectivity of fluconazole towards the enzyme from C. albicans with no inhibitory effect on the human ortholog has been proven [71]. Sterol 14α-demethylases in Trypanosomatidae also have low amino acid sequence identity to the human enzyme (25–26%). On the other hand, their identities to fungal sterol 14α-demethylases are even lower, only 22–24% Fig. (5). Therefore, azoles that serve as potent antifungal drugs are not necessarily the best inhibitors for Trypanosomatidae.
Fig. 5.
Structure-based alignment of sterol 14α-demethylases from Trypanosomatidae, fungi and human. Amino acid residues forming substrate-binding cleft surface are marked with grey triangles.
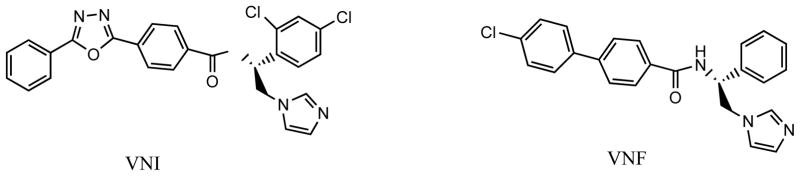
We have cloned, expressed, purified and reconstituted enzymatic activities of sterol 14α-demethylases from T. brucei [67], T. cruzi [66] and L. infantum [72]. Including the direct testing of the inhibitory effects on the enzyme activity into the search for new inhibitors allowed us to identify several novel highly potent compounds. Two carboxamide-containing imidazole derivatives in Fig. (6) are amongst the most promising leads. We have found that completely inhibiting activity of sterol 14α-demethylases from Trypanosomatidae at equimolar ratio to the enzyme [e.g. 31], these compounds, unlike ketoconazole or posaconazole, do not have any inhibitory effect on human sterol 14α-demeyhlase, molar excess of the inhibitor over the human enzyme up to 200-fold has been tested [29, 73]. Their direct targeting of the parasite sterol 14a-demethylase in cellular experiments has been confirmed by T. cruzi sterol analysis [31, 54]. These inhibitors have low general cytotoxicity (EC50 >50 μM in the human leukemia cell line HL60 [31]) and produce antiparasitic effects in trypanosomal cells comparable with that of posaconazole (EC99<1μM) [31, 73]. However, contrary to posaconazole or fluconazole, they do not enhance the T. cruzi CYP51 gene expression and do not require increase in the dosage to maintain constant cellular growth inhibition over time [73], which we believe suggests their weaker propensity to induce resistance in the parasite. Evaluation of these inhibitors in vivo, in animal model of Chagas disease as well as analysis of their pharmacokinetic properties is currently underway.
Fig. 6.
Two potent carboxamide-containing imidazole inhibitors of protozoan sterol 14α-demethylases. VNI [29] and VNF [72] are the Protein Data Bank codes of the compounds.
Two other highly potent inhibitory scaffolds, disubstituted imidazoles and derivatives of anticancer drug tipifarnib (triazole-based) were identified by Buckner et al. [74, 75]. In this case the investigators originally observed strong anti-parasitic effects the compounds T. cruzi and then analysis of the parasite sterols has shown that they actually inhibit sterol 14α-demethylase. Testing these inhibitors in the reconstituted reaction confirmed their high inhibitory potencies and selectivity to the enzyme from T. cruzi [76]. Curative effects of these compounds on Chagas disease have already been proven in murine models.
5. Structural basis for the Development of Selective Anti-protozoan Sterol 14α-Demethylase Inhibitors
Though humans can consume cholesterol from the diet, inhibitors selective for pathogenic sterol 14α-demethylases are still highly desirable, especially in the cases of extended treatment, to prevent formation of harmful methylated sterols in human body and also to avoid potentially negative effects, e.g. on steroid hormone biosynthesis. Crystal structures of protozoan sterol 14α-demethylases provide an opportunity for structure-directed development of such inhibitors. We have determined structures of the enzymes from T. brucei, ligand-free (3g1q) and VNI-bound (3gw9) [29], T. cruzi, bound to posaconazole (3k1o), fluconazole (3khm) and VNF (3ksw) [73] and L. infantum, bound to fluconazole (3l4d) [72].
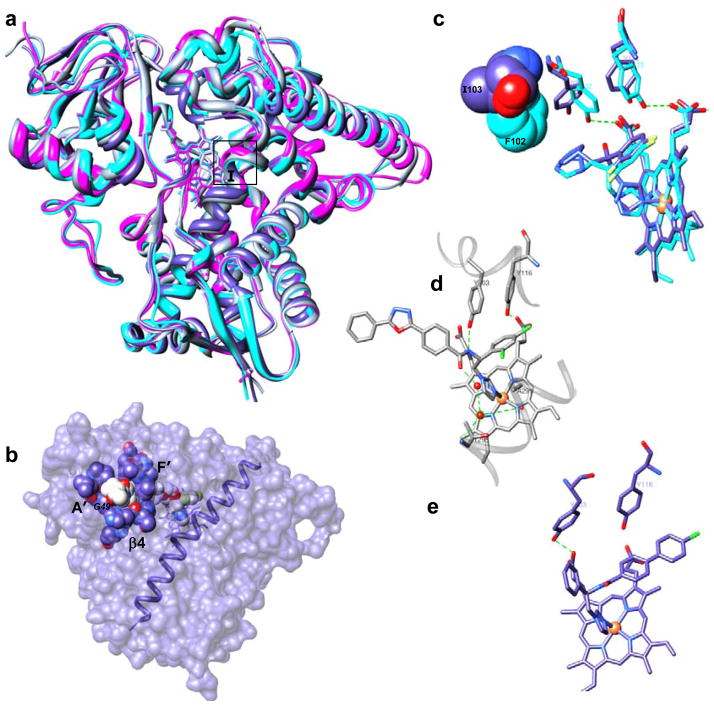
We have found that unlike many (especially drug-metabolizing) cytochromes P450, which are know to exhibit high conformational flexibility, sterol 14α-demethylases have a very rigid active site cavity. This feature must be essential for the enzyme ability to maintain proper orientation of the substrate during the three steps of catalysis and preserve their functional conservation across phylogeny. Binding of different ligands do not cause significant rearrangements within the cavity, except for the subtle movement in the closest to the heme part of the I-helix (A291-region). In all inhibitor-bound sterol 14α-demethylases this part of the helix shifts about 1.5 Å away from the heme iron providing space for the azole ring coordination Fig. (7a).
Fig. 7.
Crystal structures of protozoan sterol 14a-demethylases. (a) Superimposition of ligand-free (magenta, T. brucei) and inhibitor-bound structures: VNI-bound T. brucei (gray), posaconazole-bound T. cruzi (blue) and fluconazole-bound L. infantum (cyan) orthologs, ribbon representation. Middle part of the I-helix is framed. (b) Surface binding subsite in posaconazole-bound T. cruzi sterol 14α-demethylase (surface representation, I-helix is shown). (c) Fluconazole bound to sterol 14a-demethylases from L. infantum and T. cruzi. Substrate preference defining residues (F104 in L. infantum (cyan) vs I105 in T. cruzi (blue)) are shown as Van der Waals spheres. (d) VNI bound in the active center of sterol 14a-demethylase from T. brucei, hydrogen bond network around the inhibitor carboxamide fragment is shown as green dotted lines. (e) VNF bound in the active center of sterol 14a-demethylase from T. cruzi. Protein is in the same orientation as in d.
The structures uncover specific details essential for the potency of each particular inhibitor. Thus, the complex of T. cruzi sterol 14α-demethylase with posaconazole [73] depicts an additional binding subsite on the protein surface, around the substrate access channel. This subsite is formed about 25 Å away from the heme iron by 12 amino acid residues from helices A′, F″ and β4-hairpin. The residues surround the posaconazole long arm which extends outside the protein globule Fig. (7b). As a result, in addition to the azole ring coordination to the heme iron, 25 protein residues (13 of them are from the active site cavity) tightly wrap the inhibitor and position it within the enzyme geometry. The surface interactions must be importantfor the potency of posaconazole to inhibit fungal sterol 14α-demethylase as wells, since single amino acid substitutions of G54 (that align with G49 in Trypanosomatidae) by the bulky and less flexible arginine or tryptophan are found in the sequences of sterol 14α-demethylase from posaconazole-resistant strains of Aspergillus fumigatus [77].
Another example, stronger inhibitory effect of fluconazole on sterol 14α-demethylase from T. cruzi versus L. infantum and T. brucei can be due to disruption of the heme support in the T. cruzi enzyme as a result of repositioning of two conserved heme-contacting tyrosines (Y103 and Y116) upon fluconazole binding Fig. (7c). No such repositioning is seen in the L. infantum enzyme (Y102 and Y115), most likely because of neighboring F104 (substrate preference defining I105 in T. cruzi sterol 14α-demethylase). This bulky residue pushes fluconazole back to the I-helix, restraining it from intercalation between the heme plane and the heme propionate supporting tyrosines.
The complex of T. brucei sterol 14α-demethylase with VNI (1.9 Å resolution), in addition to the heme iron coordination with the imidazole nitrogen and multiple van der Waals contacts within the active site cavity (at the distance of 5 Å VNI is surrounded by 25 amino acid residues, 15 of them being located within 4 Å), reveals formation of a hydrogen bond network not seen before in other cytochrome P450-inhibitor complexes. This network Fig. (7d) connects, via the inhibitor carboxamide group fragment, unique for this inhibitory scaffold, two functionally essential CYP51 family signature [21] regions, helices B′ and I, strengthening the binding and altering the heme environment. We believe that this hydrogen bond network adds to the VNI inhibitory potency and is likely to be a reason for its high selectivity to the protozoan sterol 14α-demethylases [29]. Quite unexpected, VNF, which is structurally highly similar to VNI binds to T. cruzi sterol 14α-demethylase in an opposite orientation Fig. (7e), including 180° rotation of its carboxamide group. Although the electron density for the hydrogen bond network around VNF can not be clearly seen at medium resolution (3.05 Å), it is quite likely that the comparably strong inhibitory potencies and antiprotozoan selectivity of these two compounds might have the same origin [73].
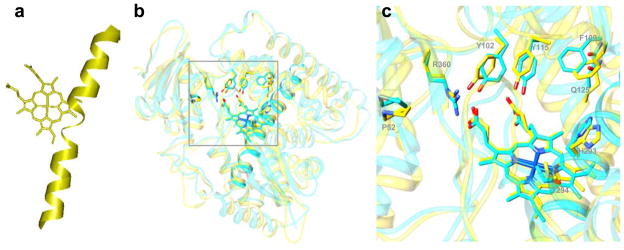
Structural information provides important insights into general aspects of sterol 14α-demethylase inhibition. Thus, elevated susceptibility of the enzyme to the heme-coordinated inhibitors might be connected with the high proximity of the middle part of their I-helix to the heme. The I-helix potentially can serve as a “seat-belt” holding the inhibitors and preventing their replacement in the active site by the substrate. In this regard, it is not excluded that weaker inhibition of the human sterol 14α-demethylase by azoles might be its “lucky” specific feature: while in the sterol 14α-demethylases from protozoan pathogens the I-helix is whole, there appear to be a low energy loop-like interruption in the middle part of the I-helix in the human ortholog Fig. (8a).
Fig. 8.
Sterol 14α-demethylases from protozoa and human reveal minor structural differences that allow selective inhibition of the enzyme from pathogens. (a) The I-helix in human sterol 14α-demethylase (Accelrys). (b) Superimposition of the orthologs from L. infantum and human, the protein backbone is displayed as ribbon diagram (rms deviation for the Ca atoms 1.3 Å), the side chains of the residues in the active site cleft that are identical in both enzymes are shown. (c) Enlarged view of the active site area (L. infantum numbering). I102, Y115 and R360 provide hydrogen bonds to the heme propionates in all sterol 14α-demethylases, H293 and T294 are likely to participate in the proton delivery route [29]. Variations in the amino acid composition of the rest of the cavity can be used as a basis for the development of highly specific inhibitors.
Moreover, although eukaryotic sterol 14α-demethylases reveal high similarity in the location of the backbone of their major structural elements across species Fig. (8b), amongst 47 residues that form their access channel and active site cavity (the residues are marked with black triangles in Fig. (5)) only 9 (G49, P52, Y103, F110, Y116, G292, H294, T/s295 and R361 (T. brucei numbering)) are conserved amongst protozoan, fungal and human orthologs Fig. (8c); while the majority of the other residues disclose a high tendency to be phyla-specific [78]. Differences in the topology of the area potentially available for ligand binding can be used for structure-directed development of highly selective, ideally species-specific sterol 14α-demethylase inhibitors.
6. Non-Azole Structures as Potential Alternative Inhibitors of Sterol 14α-Demethylase
Acquired azole resistance is a major problem in current antifungal chemotherapy [79], four different mechanisms being discussed as its possible causes (mutations in sterol 14α-demethylase, the CYP51 gene amplification, development of active azole efflux pumps or activation of alternative routes for sterol biosynthesis). Development of resistance to fluconazole was demonstrated in the laboratory strains of T. cruzi [80]. Although highly effective inhibitors potentially have lower propensity to induce resistance, search for non-azole inhibitory structures can be a very helpful alternative.
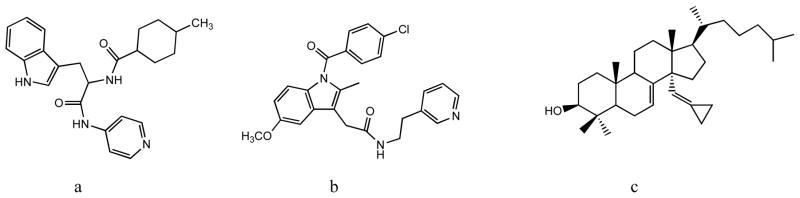
Using optical high-throughput screening for new binding ligands followed by web search for structurally similar compounds, we identified several non-azole inhibitors of protozoan sterol 14α-deethylases [54, 81]. Though their inhibitory potencies are so far relatively lower than those of the best azoles, we have found that they produce strong a antiparasitic effect in Trypanosomatidae: e.g. compounds a and b in Fig. (9) at 20 μM concentration being able to completely clear the parasite amastigotes from T. cruzi infected cardiomyocytes [81]. We believe that structure-directed modification of these compounds will allow us to further increase their inhibitory potencies.
Fig. 9.
Non-azole inhibitors of protozoan sterol 14α-demethylases. HTS-derived (a) N-(3-(1H-indol-3-yl)-1-oxo-1(pyridin-4-ylamino)propan-2-yl)-4-methylcyclohexanecarboxamide and (b) 2-(1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl)-N-(2-(pyridin-3-yl)ethyl)acetamide and (c) substrate analog delta7-14amethylenecyclopropyl-dihydrolanosterol.
The substrate analog Δ7-14α-methylenecyclopropyl-dihydrolanosterol (MCP) is another promising example of a non-azole CYP51 inhibitor Fig. (9c), EC50 in T. cruzi cells being ~5 μM [54]. Interestingly, we found that while MCP can be easily replaced by VNI in the oxidized (ferric) form of the enzyme, the inhibitory effects of MCP and VNI in the reaction (ferrous form) are comparable. It suggests that MCP may work as a mechanism-based inhibitor [54], which would be the first example of this type of inhibitor for sterol 14α-demethylase.
CONCLUSIONS
Current lack of efficient treatment for human trypanosomiasis and leishmaniasis is largely related to the lack of attention to this group of neglected tropical diseases. Multiple searches for new antiprotozoan drugs and drug targets must eventually change the situation. Identification of more than 8,000 new protein coding genes in the pathogen genomes [82] vastly expands potential options for investigations aimed at development of novel highly specific agents. However, this requires time, while millions of people are suffering presently. Inhibitors of sterol 14α-demethylase obtained from antifungal drug development programs can be used in antitrypamosomal chemotherapy almost immediately. Their combination with the currently available clinical antiprotozoan drugs should allow decreasing the drug dosages thus minimizing their side-effects, toxicity and shortening the treatment time. High resolution three-dimensional structures of Trypanosomatidae CYP51s open an excellent opportunity for rational development of new inhibitors, highly potent and specific for the protozoan sterol 14α-demethylases which should allow achieving much stronger antiparasitic efficiency and therefore a weaker tendency to cause resistance. As a future prospective, combinatory chemotherapy, which would concomitantly target several sterol biosynthetic enzymes, appears to be especially advantageous since it increases the chances of complete blockage of the endogenous sterol flow in the parasites.
Acknowledgments
The authors were supported by the National Institute of Health grant GM067871.
References
- 1.Koonin EV. The incredible expanding ancestor of eukaryotes. Cell. 2010;140(5):606–608. doi: 10.1016/j.cell.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kreier T, Julius P. Parasitic Protozoa. 2. Academic Press; San Diego, CA: 1991. [Google Scholar]
- 3.World Health Organization. www.who.int/neglected_diseases/diseases/chagas/en/index.html.
- 4.World Health Organization. www.who.int/trypanosomiasis_african/en/index.html.
- 5.World Health Organization. www.who.int/leishmaniasis/en/
- 6.Chaudhary K, Ross D. Protozoan genomics for drug discovery. Nat Biotechnol. 2005;23:1089–1091. doi: 10.1038/nbt0905-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavalli A, Bolognesi ML. Neglected Tropical diseases: Multi-target directed Ligands in the Search for novel Lead Candidates against Trypanosoma and Leishmania. J Med Chem. 2009;52:7339–7359. doi: 10.1021/jm9004835. [DOI] [PubMed] [Google Scholar]
- 8.Hoet S, Opperdoes F, Brun R, Quetin-Leclercq J. Natural products active against African trypanosomes: a step towards new drugs. Nat Prod Rep. 2004;21(3):353–364. doi: 10.1039/b311021b. [DOI] [PubMed] [Google Scholar]
- 9.Barret MP, Burchmore RJB. The Trypanosomiases. Lancet. 2003;362:1469–1480. doi: 10.1016/S0140-6736(03)14694-6. [DOI] [PubMed] [Google Scholar]
- 10.Mishra J, Saxena A, Singh S. Chemotherapy of leishmaniasis: past, present and future. Curr Med Chem. 2007;14(10):1153–1169. doi: 10.2174/092986707780362862. [DOI] [PubMed] [Google Scholar]
- 11.Myler PJ. Searching the Tritryp genomes for drug targets. Adv Exp Med Biol. 2008;625:133–140. doi: 10.1007/978-0-387-77570-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soeiro MN, de Castro SL. Trypanosoma cruzi targets for new chemotherapeutic approaches. Expert Opin Ther Targets. 2009;13(1):105–121. doi: 10.1517/14728220802623881. [DOI] [PubMed] [Google Scholar]
- 13.Cruz AK, de Toledo JS, Falade M, Terrão MC, Kamchonwongpaisan S, Kyle DE, Uthaipibull C. Current treatment and drug discovery against Leishmania spp. and Plasmodium spp.: a review. Curr Drug Targets. 2009;10(3):178–192. doi: 10.2174/138945009787581177. [DOI] [PubMed] [Google Scholar]
- 14.Lüscher A, de Koning HP, Mäser P. Chemotherapeutic strategies against Trypanosoma brucei: drug targets vs. drug targeting. Curr Pharm Des. 2007;13(6):555–567. doi: 10.2174/138161207780162809. [DOI] [PubMed] [Google Scholar]
- 15.Datta AK, Datta R, Sen B. Antiparasitic chemotherapy: tinkering with the purine salvage pathway. Adv Exp Med Biol. 2008;625:116–132. doi: 10.1007/978-0-387-77570-8_10. [DOI] [PubMed] [Google Scholar]
- 16.Landfear SM. Drugs and transporters in kinetoplastid protozoa. Adv Exp Med Biol. 2008;625:22–32. doi: 10.1007/978-0-387-77570-8_3. [DOI] [PubMed] [Google Scholar]
- 17.Frearson JA, Brand S, McElroy SP, Cleghorn LA, Smid O, Stojanovski L, Price HP, Guther ML, Torrie LS, Robinson DA, Hallyburton I, Mpamhanga CP, Brannigan JA, Wilkinson AJ, Hodgkinson M, Hui R, Qiu W, Raimi OG, van Aalten DM, Brenk R, Gilbert IH, Read KD, Fairlamb AH, Ferguson MA, Smith DF, Wyatt PG. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature. 2010;464(7289):728–732. doi: 10.1038/nature08893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohmer M, Bouvier P, Ourisson G. Molecular evolution of biomembranes: structural equivalents and phylogenetic precursors of sterols. Proc Natl Acad Sci U S A. 1979;76(2):847–851. doi: 10.1073/pnas.76.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volkman JK. Sterols and other triterpenoids: source specificity and evolution of biosynthetic pathways. Org Geochem. 2005;36(2):139–159. [Google Scholar]
- 20.Nes WR, McKean ML. Biochemistry of Steroids and Other Isoprenoids. University Park Press; Baltimore: 1977. [Google Scholar]
- 21.Lepesheva GI, Waterman MR. Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim Biophys Acta. 2007;1770(3):467–477. doi: 10.1016/j.bbagen.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards PA, Ericsson J. Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu Rev Biochem. 1999;68:157–185. doi: 10.1146/annurev.biochem.68.1.157. [DOI] [PubMed] [Google Scholar]
- 23.Dahl C, Biemann HP, Dahl J. A protein kinase antigenically related to pp60v-src possibly involved in yeast cell cycle control: positive in vivo regulation by sterol. Proc Natl Acad Sci USA. 1987;84(12):4012–4016. doi: 10.1073/pnas.84.12.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parks LW, Casey WM. Physiological implications of sterol biosynthesis in yeast. Annu Rev Microbiol. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- 25.Vanden Bossche H, Koymans L, Moereels H. P450 inhibitors of use in medical treatment: focus on mechanisms of action. Pharmacol Ther. 1995;67:79–100. doi: 10.1016/0163-7258(95)00011-5. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 27.Roberts CW, McLeodm R, Rice DW, Ginger M, Chance ML, Goad LJ. Fatty acid and sterol metabolism: potential antimicrobial targets in apicomplexan and trypanosomatid parasitic protozoa. Mol Biochem Parasitol. 2003;126:129–142. doi: 10.1016/s0166-6851(02)00280-3. [DOI] [PubMed] [Google Scholar]
- 28.Zhou W, Cross GA, Nes WD. Cholesterol import fails to prevent catalyst-based inhibition of ergosterol synthesis and cell proliferation of Trypanosoma brucei. J Lipid Res. 2007;48(3):665–673. doi: 10.1194/jlr.M600404-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Lepesheva GI, Park HW, Hargrove TY, Vanhollebeke B, Wawrzak Z, Harp JM, Sundaramoorthy M, Nes WD, Pays E, Chaudhuri M, Villalta F, Waterman MR. Crystal structures of Trypanosoma brucei sterol 14alpha-demethylase and implications for selective treatment of human infections. J Biol Chem. 2010;285(3):1773–1780. doi: 10.1074/jbc.M109.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, Hertz-Fowler C, Ghedin E, Peacock C, Bartholomeu DC, Haas BJ, Tran AN, Wortman JR, Alsmark UC, Angiuoli S, Anupama A, Badger J, Bringaud F, Cadag E, Carlton JM, Cerqueira GC, Creasy T, Delcher AL, Djikeng A, Embley TM, Hauser C, Ivens AC, Kummerfeld SK, Pereira-Leal JB, Nilsson D, Peterson J, Salzberg SL, Shallom J, Silva JC, Sundaram J, Westenberger S, White O, Melville SE, Donelson JE, Andersson B, Stuart KD, Hall N. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309(5733):404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 31.Lepesheva GI, Ott RD, Hargrove TY, Kleshchenko YY, Schuster I, Nes WD, Hill GC, Villalta F, Waterman MR. Sterol 14alpha-demethylase as a potential target for antitrypanosomal therapy: enzyme inhibition and parasite cell growth. Chem Biol. 2007;14(11):1283–1293. doi: 10.1016/j.chembiol.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urbina JA, Payares G, Sanoja C, Molina J, Lira R, Brener Z, Romanha AJ. Parasitological cure of acute and chronic experimental Chagas disease using the long-acting experimental triazole TAK-187. Activity against drug-resistant Trypanosoma cruzi strains. Int J Antimicrob Agents. 2003;1:39–48. doi: 10.1016/s0924-8579(02)00274-1. [DOI] [PubMed] [Google Scholar]
- 33.Urbina JA, Payares G, Contreras LM, Liendo A, Sanoja C, Molina J, Piras M, Piras R, Perez N, Wincker P, Loebenberg D. Antiproliferative effects and mechanism of action of SCH 56592 against Trypanosoma (Schizotrypanum) cruzi: in vitro and in vivo studies. Antimicrob Agents Chemother. 1998;42(7):1771–1777. doi: 10.1128/aac.42.7.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinones W, Urbina JA, Dubourdieu M, Concepcion JL. The glycosome membrane of Trypanosoma cruzi epimastigotes: protein and lipid composition. Exper Parasitol. 2004;106:135–149. doi: 10.1016/j.exppara.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Pena-Diaz J, Montalvetti A, Flores CL, Constán A, Hurtado-Guerrero R, De Souza W, Gancedo C, Ruiz-Perez LM, Gonzalez-Pacanowska D. Mitochondrial localization of the mevalonate pathway enzyme 3-hydroxy-3-methyl-glutaryl-CoA reductase in the Trypanosomatidae. Mol Biol Cell. 2004;15:1356–1363. doi: 10.1091/mbc.E03-10-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Souza W, Rodrigues JC. Sterol biosynthesis pathway as target for anti-trypanosomatid drugs. Interdiscip. Perspect Infect Dis. 2009:642502. doi: 10.1155/2009/642502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbina JA. Ergosterol biosynthesis and drug development for Chagas disease. Mem Inst Oswaldo Cruz. 2009;104:311–318. doi: 10.1590/s0074-02762009000900041. [DOI] [PubMed] [Google Scholar]
- 38.Gebre-Hiwot A, Frommel D. The in vitro anti-leishmania activity of inhibitors of ergosterol biosynthesis. J Antimicrob Chemother. 1993;32:837–842. doi: 10.1093/jac/32.6.837. [DOI] [PubMed] [Google Scholar]
- 39.Puccetti L, Acampa M, Auteri A. Pharmacogenetics of statins therapy. Recent Pat Cardiovasc Drug Discov. 2007;2(3):228–236. doi: 10.2174/157489007782418982. [DOI] [PubMed] [Google Scholar]
- 40.Kavanagh KL, Guo K, Dunford JE, Wu X, Knapp S, Ebetino FH, Rogers MJ, Russell RG, Oppermann U. The molecular mechanism of nitrogen-containing bisphosphonates as antiosteoporosis drugs. Proc Natl Acad Sci U S A. 2006;103(20):7829–3784. doi: 10.1073/pnas.0601643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cammerer SB, Jimenez C, Jones S, Gros L, Lorente SO, Rodrigues C, Rodrigues JC, Caldera A, Ruiz Perez LM, da Souza W, Kaiser M, Brun R, Urbina JA, Gonzalez Pacanowska D, Gilbert IH. Quinuclidine derivatives as potential antiparasitics. Antimicrob Agents Chemother. 2007;51(11):4049–4061. doi: 10.1128/AAC.00205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magaraci F, Jimenez CJ, Rodrigues C, Rodrigues JC, Braga MV, Yardley V, de Luca-Fradley K, Croft SL, de Souza W, Ruiz-Perez LM, Urbina J, Gonzalez Pacanowska D, Gilbert IH. Azasterols as inhibitors of sterol 24-methyltransferase in Leishmania species and Trypanosoma cruzi. J Med Chem. 2003;46(22):4714–4727. doi: 10.1021/jm021114j. [DOI] [PubMed] [Google Scholar]
- 43.Gros L, Castillo-Acosta VM, Jiménez Jiménez C, Sealey-Cardona M, Vargas S, Manuel Estévez A, Yardley V, Rattray L, Croft SL, Ruiz-Perez LM, Urbina JA, Gilbert IH, González-Pacanowska D. New azasterols against Trypanosoma brucei: role of 24-sterol methyltransferase in inhibitor action. Antimicrob, Agents Chemother. 2006;50(8):2595–2601. doi: 10.1128/AAC.01508-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nes WD. Enzyme mechanisms for sterol C-methylations. Phytochemistry. 2003;64(1):75–95. doi: 10.1016/s0031-9422(03)00349-2. [DOI] [PubMed] [Google Scholar]
- 45.Petrikkos G, Skiada A. Recent advances in antifungal chemotherapy. Int J Antimicrob Agents. 2007;30:108–117. doi: 10.1016/j.ijantimicag.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Zonios DI, Bennet JE. Update on azole antifungals. Semin Respir Crit Care Med. 2008;29:198–210. doi: 10.1055/s-2008-1063858. [DOI] [PubMed] [Google Scholar]
- 47.Docampo R, Moreno SN, Turrens JF, Katzin AM, Gonzalez-Cappa SM, Stoppani AO. Biochemical and ultrastructural alterations produced by miconazole and econazole in Trypanosoma cruzi. Mol Biochem Parasitol. 1981;3:169–180. doi: 10.1016/0166-6851(81)90047-5. [DOI] [PubMed] [Google Scholar]
- 48.Beach DH, Goad LJ, Holz GG. Effects of ketoconazole on sterol biosynthesis by Trypanosoma cruzi epimastigotes. Biochem Biophys Res Commun. 1986;136:851–856. doi: 10.1016/0006-291x(86)90410-9. [DOI] [PubMed] [Google Scholar]
- 49.Urbina JA, Lazardi K, Aguirre T, Piras MM, Piras R. Antiproliferative synergism of the allylamine SF 86–327 and ketoconazole on epimastigotes and amastigotes of Trypanosoma (Schizotrypanum) cruzi. Antimicrob Agents Chemother. 1988;32:1237–1242. doi: 10.1128/aac.32.8.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Apt W, Aguilera X, Arribada A, Pérez C, Miranda C, Sánchez G, Zulantay I, Cortés P, Rodriguez J, Juri D. Treatment of chronic Chagas’ disease with itraconazole and allopurinol. Am J Trop Med Hyg. 1998;59:133–138. doi: 10.4269/ajtmh.1998.59.133. [DOI] [PubMed] [Google Scholar]
- 51.Araújo MS, Martins-Filho OA, Pereira ME, Brener ZJ. A combination of benznidazole and ketoconazole enhances efficacy of chemotherapy of experimental Chagas' disease. Antimicrob Chemother. 2000;45:819–824. doi: 10.1093/jac/45.6.819. [DOI] [PubMed] [Google Scholar]
- 52.Molina J, Martins-Filho O, Brener Z, Romanha AJ, Loebenberg D, Urbina JA. Activities of the triazole derivative SCH 56592 (posaconazole) against drug-resistant strains of the protozoan parasite Trypanosoma (Schizotrypanum) cruzi in immunocompetent and immunosuppressed murine hosts. Antimicrob Agents Chemother. 2000;44:150–155. doi: 10.1128/aac.44.1.150-155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buckner FS. Sterol 14-demethylase inhibitors for Trypanosoma cruzi infections. Adv Exp Med Biol. 2008;625:61–80. doi: 10.1007/978-0-387-77570-8_6. [DOI] [PubMed] [Google Scholar]
- 54.Lepesheva GI, Hargrove TY, Kleshchenko Y, Nes WD, Villalta F, Waterman MR. CYP51: A major drug target in the cytochrome P450 superfamily. Lipids. 2008;43:1117–1125. doi: 10.1007/s11745-008-3225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinrauch L, Livshin R, el-On J. Ketoconazole in cutaneous leishmaniasis. Br J Dermatol. 1987;117(5):666–668. doi: 10.1111/j.1365-2133.1987.tb07504.x. [DOI] [PubMed] [Google Scholar]
- 56.Alrajhi AA, Ibrahim EA, De Vol EB, Khairat M, Faris RM, Maguire JH. Fluconazole for the Treatment of Cutaneous Leishmaniasis Caused by Leishmania major. N Engl J Med. 2002;346(12):891–895. doi: 10.1056/NEJMoa011882. [DOI] [PubMed] [Google Scholar]
- 57.Nagappan V, Deresinski S. Reviews of anti-infective agents: posaconazole: a broad-spectrum triazole antifungal agent. Clin Infect Dis. 2007;45:1610–1617. doi: 10.1086/523576. [DOI] [PubMed] [Google Scholar]
- 58.Urbina JA, Docampo R. Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol. 2003;19 (11):495–501. doi: 10.1016/j.pt.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 59.Urbina JA. Specific chemotherapy of Chagas disease: relevance, current limitations and new approaches. Acta Trop. 2010;115(1–2):55–68. doi: 10.1016/j.actatropica.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 60.Pinazo MJ, Espinosa G, Gállego M, López-Chejade PL, Urbina JA, Gascón J. Successful treatment with posaconazole of a patient with chronic Chagas disease and systemic lupus erythematosus. Am J Trop Med Hyg. 2010;82(4):583–587. doi: 10.4269/ajtmh.2010.09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olivieri BP, Molina JT, de Castro SL, Pereira MC, Calvet CM, Urbina JA, Araújo-Jorge TC. A comparative study of posaconazole and benznidazole in the prevention of heart damage and promotion of trypanocidal immune response in a murine model of Chagas disease. Int J Antimicrob Agents. 2010;36(1):79–83. doi: 10.1016/j.ijantimicag.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 62.Urbina JA, Payares G, Sanoja C, Molina J, Lira R, Brener Z, Romanha AJ. Parasitological cure of acute and chronic experimental Chagas disease using the long-acting experimental triazole TAK-187. Activity against drug-resistant Trypanosoma cruzi strains. Int J Antimicrob Agents. 2003;21(1):39–48. doi: 10.1016/s0924-8579(02)00274-1. [DOI] [PubMed] [Google Scholar]
- 63.Urbina JA, Payares G, Sanoja C, Lira R, Romanha AJ. In vitro and in vivo activities of ravuconazole on Trypanosoma cruzi, the causative agent of Chagas disease. Int J Antimicrob Agents. 2003;21:27–38. doi: 10.1016/s0924-8579(02)00273-x. [DOI] [PubMed] [Google Scholar]
- 64.Urbina JA, Lira R, Visbal G, Bartrolí J. In vitro antiproliferative effects and mechanism of action of the new triazole derivative UR-9825 against the protozoan parasite Trypanosoma (Schizotrypanum) cruzi. Antimicrob Agents Chemother. 2000;44:2498–2502. doi: 10.1128/aac.44.9.2498-2502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murataliev MB, Feyereisen R, Walker FA. Electron transfer by diflavin reductases. Biochim Biophys Acta. 2004;1698:1–26. doi: 10.1016/j.bbapap.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 66.Lepesheva GI, Zaitseva NG, Nes WD, Zhou W, Arase M, Liu J, Hill GC, Waterman MR. CYP51 from Trypanosoma cruzi: a phyla-specific residue in the B′ helix defines substrate preferences of sterol 14alpha-demethylase. J Biol Chem. 2006;281(6):3577–3585. doi: 10.1074/jbc.M510317200. [DOI] [PubMed] [Google Scholar]
- 67.Lepesheva GI, Nes WD, Zhou W, Hill GC, Waterman MR. CYP51 from Trypanosoma brucei is obtusifoliol-specific. Biochemistry. 2004;43(33):10789–10799. doi: 10.1021/bi048967t. [DOI] [PubMed] [Google Scholar]
- 68.Lepesheva GI, Hargrove TY, Ott RD, Nes WD, Waterman MR. Biodiversity of CYP51 in trypanosomes. Biochem Soc Trans. 2006;34(Pt 6):1161–1164. doi: 10.1042/BST0341161. [DOI] [PubMed] [Google Scholar]
- 69.Haughan PA, Goad LJ. Lipid Biochemistry of Trypanosomatids. In: Coombs G, North M, editors. Biochemical Protozoology. Taylor & Francis; London: 1991. pp. 312–328. [Google Scholar]
- 70.Ortiz de Montellano PR, Correia MA. Inhibition of cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. Ch 3. Plenum Publishing Corp; New York: 1995. pp. 305–364. [Google Scholar]
- 71.Bellamine A, Lepesheva GI, Waterman MR. Fluconazole binding and sterol demethylation in three CYP51 isoforms indicate differences in active site topology. J Lipid Res. 2004;45(11):2000–2007. doi: 10.1194/jlr.M400239-JLR200. [DOI] [PubMed] [Google Scholar]
- 72.Hargrove TY, Wawrzak Z, Liu J, Nes WD, Waterman MR, Lepesheva GI. Substrate preferences and catalytic parameters determined by structural characteristics of Sterol 14{alpha}-demethylase (CYP51) from Leishmania infantum. J Biol Chem. 2011 May 31; doi: 10.1074/jbc.M111.237099. < http://www.ncbi.nlm.nih.gov/pubmed/21632531>. [Epub ahead of print] PMID: 21632531. [DOI] [PMC free article] [PubMed]
- 73.Lepesheva GI, Hargrove TY, Anderson S, Kleshchenko Y, Furtak V, Wawrzak Z, Villalta F, Waterman MR. Structural insights into inhibition of sterol 14α-demethylase in the human pathogen Trypanosoma cruzi. J Biol Chem. 2010;285(33):25582–25590. doi: 10.1074/jbc.M110.133215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buckner F, Yokoyama K, Lockman J, Aikenhead K, Ohkanda J, Sadilek M, Sebti S, Van Voorhis W, Hamilton A, Gelb MH. A class of sterol 14-demethylase inhibitors as anti-Trypanosoma cruzi agents. Proc Natl Acad Sci U S A. 2003;100(25):15149–15153. doi: 10.1073/pnas.2535442100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hucke O, Gelb MH, Verlinde CL, Buckner FS. The protein farnesyltransferase inhibitor Tipifarnib as a new lead for the development of drugs against Chagas disease. J Med Chem. 2005;48(17):5415–5418. doi: 10.1021/jm050441z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kraus JM, Verlinde CL, Karimi M, Lepesheva GI, Gelb MH, Buckner FS. Rational modification of a candidate cancer drug for use against Chagas disease. J Med Chem. 2009;52(6):1639–1647. doi: 10.1021/jm801313t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mann PA, Parmegiani RM, Wei SQ, Mendrick CA, Li X, Loebenberg D, DiDomenico B, Hare RS, Walker SS, McNicholas PM. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14alpha-demethylase. Antimicrob Agents Chemother. 2003;47:577–581. doi: 10.1128/AAC.47.2.577-581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lepesheva GI, Waterman MR. Structural basis for conservation in the CYP51 family. Biochim Biophys Acta. 2010;1814(1):88–93. doi: 10.1016/j.bbapap.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanafani ZA, Perfect JR. Antimicrobial resistance: resistance to antifungal agents: mechanisms and clinical impact. Clin Infect Dis. 2008;46:120–128. doi: 10.1086/524071. [DOI] [PubMed] [Google Scholar]
- 80.Buckner FS, Wilson AJ, White TC, Van Voorhis WC. Induction of resistance to azole drugs in Trypanosoma cruzi. Antimicrob Agents Chemother. 1998;42(12):3245–50. doi: 10.1128/aac.42.12.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Konkle ME, Hargrove TY, Kleshchenko YY, von Kries JP, Ridenour W, Uddin MJ, Caprioli RM, Marnett LJ, Nes WD, Villalta F, Waterman MR, Lepesheva GI. Indomethacin amides as a novel molecular scaffold for targeting Trypanosoma cruzi sterol 14alpha-demethylase. J Med Chem. 2009;52(9):2846–2853. doi: 10.1021/jm801643b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, Kerhornou A, Ivens A, Fraser A, Rajandream MA, Carver T, Norbertczak H, Chillingworth T, Hance Z, Jagels K, Moule S, Ormond D, Rutter S, Squares R, Whitehead S, Rabbinowitsch E, Arrowsmith C, White B, Thurston S, Bringaud F, Baldauf SL, Faulconbridge A, Jeffares D, Depledge DP, Oyola SO, Hilley JD, Brito LO, Tosi LR, Barrell B, Cruz AK, Mottram JC, Smith DF, Berriman M. Comparative genomis analysis of three Leishmania species that cause diverse human disease. Nat Gen. 2007;39(7):839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]