Abstract
Transform faults are geological structures that interrupt the continuity of mid-ocean ridges and can act as dispersal barriers for hydrothermal vent organisms. In the equatorial Atlantic Ocean, it has been hypothesized that long transform faults impede gene flow between the northern and the southern Mid-Atlantic Ridge (MAR) and disconnect a northern from a southern biogeographic province. To test if there is a barrier effect in the equatorial Atlantic, we examined phylogenetic relationships of chemosynthetic bivalves and their bacterial symbionts from the recently discovered southern MAR hydrothermal vents at 5°S and 9°S. We examined Bathymodiolus spp. mussels and Abyssogena southwardae clams using the mitochondrial cytochrome c oxidase subunit I (COI) gene as a phylogenetic marker for the hosts and the bacterial 16S rRNA gene as a marker for the symbionts. Bathymodiolus spp. from the two southern sites were genetically divergent from the northern MAR species B. azoricus and B. puteoserpentis but all four host lineages form a monophyletic group indicating that they radiated after divergence from their northern Atlantic sister group, the B. boomerang species complex. This suggests dispersal of Bathymodiolus species from north to south across the equatorial belt. 16S rRNA genealogies of chemoautotrophic and methanotrophic symbionts of Bathymodiolus spp. were inconsistent and did not match the host COI genealogy indicating disconnected biogeography patterns. The vesicomyid clam Abyssogena southwardae from 5°S shared an identical COI haplotype with A. southwardae from the Logatchev vent field on the northern MAR and their symbionts shared identical 16S phylotypes, suggesting gene flow across the Equator. Our results indicate genetic connectivity between the northern and southern MAR and suggest that a strict dispersal barrier does not exist.
Introduction
Fracture zones on the ocean floor dissect the mid-oceanic ridges causing trench-like transform faults. They disconnect adjacent ridge segments by lateral offsets of tens to hundreds of kilometers and are thought to be geological barriers for the dispersal of hydrothermal vent organisms, thus affecting the biogeography of hydrothermal vent communities [1]. Hydrothermal vent species disperse predominantly as larvae in the water column. They rise up with the hydrothermal plume, migrate passively with currents along the ridge axes and colonize other vent sites downstream [2], [3]. Transform faults may impede species dispersal along the mid-ocean ridges by the loss of larvae from the terminal ends of ridge segments into the open ocean [4], [5]. The segregating effect of single topographic seafloor structures on species distribution has been tested recently by examining gene flow between closely related populations along ridge systems. For example, the Blanco Transform Fault in the eastern Pacific separating Juan de Fuca Ridge (JdF) and Gorda Ridge isolates mitochondrial gene haplotypes of morphologically similar limpets of the genus Lepetodrilus [6], alvinellid polychaetes display limited gene flow across fracture zones on the East Pacific Rise (EPR) [7], [8], and the Easter Island Microplate on the EPR isolates the mussel species Bathymodiolus thermophilus and B. aff. thermophilus [9]. In contrast, each of the two species “Calyptogena” magnifica and B. thermophilus share similar haplotypes along more than 4500 kilometers and across several transform faults on the EPR (“C.” magnifica: 21°N to 17°S) [10] or the EPR and the Galapagos Rift (B. thermophilus: 13°N to 17°S and Rose Garden) [9], indicating that species with sufficient dispersal potential can bridge pronounced ridge discontinuities.
In the equatorial Atlantic Ocean, a number of large transform faults interrupt the Mid-Atlantic Ridge (MAR). The largest is the Romanche Transform Fault, which reaches 7370 m depth [11] and creates an offset of 935 km in the ridge axis. These deep valleys channel Atlantic deep water across the MAR from West to East [1], [12] (Fig. 1), and it has been hypothesized that they affect the biogeography of chemosynthetic communities in two major ways. Firstly, it was suggested that the effects of large ridge offset, pronounced trench topography, and currents through the transform faults impede the dispersal of vent invertebrates between the northern and southern Mid-Atlantic Ridge (NMAR and SMAR) [13], [14]. Hydrothermal vents on the SMAR were unknown until recently and hypotheses on the barrier effect included the prospect that the equatorial belt might separate two distinct biogeographic MAR provinces, and that SMAR province communities might be most similar to Indian Ridge communities [1]. Secondly, it was hypothesized that the equatorial across-axis currents provide a west-east passage for chemosynthetic species and that the transform faults act as conduits to gene-flow across the MAR [1], [13] resulting in very similar seep communities on both sides of the Atlantic. Recent studies provide evidence for such a passage [15], [16].
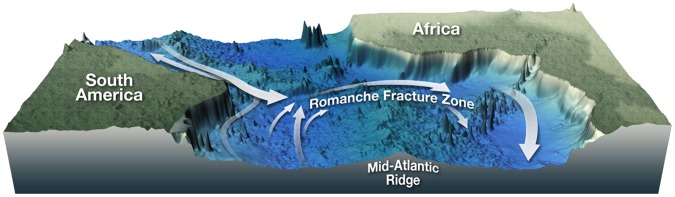
Figure 1. Deep-water currents are channeled through the equatorial transform faults crossing the Mid Atlantic Ridge.
According to current hypotheses, these currents build a conduit for dispersal of chemosynthetic organisms across the Atlantic, and together with ridge-axis offset and transform fault topography they may hinder dispersal of larvae from north to south on the MAR (after [72]).
The discovery of the hydrothermal vents and their chemosynthetic communities at 5°S and 9°S on the MAR in 2005 [17], [18], [19] (Fig. 2) provided the first opportunity to test if the Romanche Transform Fault inhibits hydrothermal vent species dispersal between NMAR and SMAR and divides these two regions into separate biogeographic provinces. The 5°S vents are located in 3000 m water depth on a 2 km long volcanically active plateau of the ridge axis and include three high-temperature vents and numerous low-temperature vent sites with diffuse fluid flow [18]. The 9°S vents lie in 1500 m water depth and consist of several low-temperature diffuse flow sites in an area of 300 m by 400 m, while hot vents have not been found [19]. Both vent fields harbor chemosynthetic communities that at first view resemble those of the NMAR. Bathymodiolus spp. are dominant at sites with diffuse venting. The mussels at 5°S are morphologically similar to the NMAR species B. puteoserpentis while 9°S mussels display more variable shell morphology and appear more similar to B. azoricus (R. von Cosel, pers. information) although their average body size was much smaller, as it was observed during repeated visits within several years [19]. Alvinocarid Rimicaris shrimp colonize the sulfide edifices of hot venting at 5°S. A vesicomyid clam found at 5°S was identified as Abyssogena southwardae based on its morphology [20], a species known from the Logatchev vent field on the NMAR [20], [21]. Vestimentiferan tubeworms typical for EPR vents or provannid gastropods characteristic for western Pacific and Indian Ridge vents were not found [18], [19].
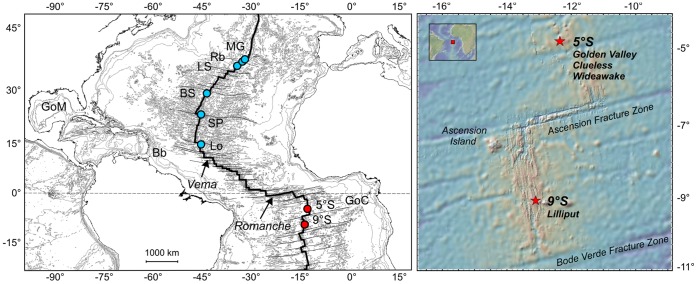
Figure 2. Hydrothermal vents on the MAR with chemosynthetic bivalves and hydrocarbon seep areas on the western and eastern Atlantic margins with the B. boomerang species complex.
Left: Overview on North Atlantic vents and seeps. Vema and Romanche are large transform faults. Right: Locations of the 5°S and 9°S vent fields on the MAR. 5°S = SMAR vents Golden Valley, Clueless and Wideawake; 9°S = SMAR vent field Lilliput; Bb = Barbados Accretionary Prism; BS = Broken Spur; GoC = Gulf of Congo; GoM = Gulf of Mexico, Lo = Logatchev Hydrothermal Vent Field; LS = Lucky Strike; MG = Menez Gwen; Rb = Rainbow; SP = Snake Pit.
In this study, we investigated the phylogenetic relationships of the chemosynthetic bivalves and their associated symbiotic bacteria from the new SMAR vent fields. We tested whether the close relationships of Bathymodiolus spp. and Abyssogena southwardae to their northern relatives based on morphology is supported by molecular data and if gene flow may have occurred across the equatorial belt. We also tested if the genetic data reveal phylogenetic relationships to chemosynthetic symbioses from the Pacific or Indian Oceans. We used the mitochondrial cytochrome c oxidase subunit I (COI) as a marker gene for the host species and 16S rRNA for their bacterial symbionts.
Materials and Methods
Animal Collection
Animal material used in this study was collected on the SMAR during RV Meteor research cruises M64/1 in May 2005 and M78/2 in May 2009 and RV l’Atalante cruise MARSUED IV in January 2008 using the ROVs Marum Quest 4000 m (Marum, Bremen) and Kiel 6000 (IFM-GEOMAR, Kiel) (Table 1). Additional NMAR material was sampled in the Logatchev hydrothermal vent field in January 2007 using ROV Jason II (Woods Hole Oceanographic Institution) during Maria S. Merian cruise MSM 04/3. For the 5°S area, 32 specimens of Bathymodiolus sp. obtained from three different diffuse vent sites were used for genetic analyses. The vent sites named Golden Valley (n = 10 specimens), Clueless (n = 10) and Wideawake (n = 12) were about 850 m from each other in 2987–2998 m water depth (Fig. 3A and B). Two Abyssogena specimens were collected from the Clueless vent site at 5°S. For the 9°S area 16 Bathymodiolus sp. specimens were used from the Lilliput vent field in 1595 m water depth (Fig. 3C and D). 21 specimens of B. puteoserpentis were obtained from the Irina II mussel bed at Logatchev in 3020 m water depth. Onboard, all specimens were kept in chilled seawater and processed as soon as possible with a maximum of 12 hours after recovery. Animals were dissected and gill tissue samples for DNA extraction were stored at −20°C until further processing.
Table 1. Material, sampling coordinates and accession numbers of investigated specimens and published sequences which were used for distance calculations (Table 2) and the reconstruction of the COI Neighbor Joining tree (Fig. 4).
| Host species | Sample | Individual ID | Sampling location | Coordinates | Depth (m) | Acc COI | Acc 16S SOX | Acc 16S MOX | Ref. |
| B. puteoserpentis | MSM04/3 244 Rov 9 | 30, 32, 34–38, 41–44, 46,48, 49, 53–59 | Logatchev, Irina II | 14°45.180′ N, 44°58.746′ W | 3022 | JQ844834 - JQ844854 | - | - | This study |
| Bathymodiolus sp. 5°S | M78/2 274 ROV 3 | 1, 2, 4–11 | Golden Valley | 4°48.162′ S, 12°21.680′ W | 2987 | JQ844798 - JQ844807 | - | - | This study |
| Bathymodiolus sp. 5°S | M78/2 302 ROV 15 | 1, 2, 3–8, 10, 11 | Clueless | 4°48.250′ S, 12°22.259′ W | 2995 | JQ844808 - JQ844817 | JQ844767 - JQ844771 | - | This study |
| Bathymodiolus sp. 5°S | ATA 37 Rov 7 | 9–11, 13–19 | Wideawake | 04°48.626′ S, 12°22.342′ W | 2998 | JQ844788 - JQ844797 | - | - | This study |
| Bathymodiolus sp. 5°S | M64/1 109 GTV-A | 7 | Wideawake | 4°29.184′ S, 12°13.416′ W | 2998 | - | JQ844772 - JQ844774 | - | This study |
| Bathymodiolus sp. 5°S | M64/1 125 Rov 7 | 4 | Wideawake | 4°48.62′ S,12°22.35′ W | 2987 | - | - | JQ844775 | This study |
| Bathymodiolus sp. 9°S | M78/2 319 ROV 10 | 1–6, 7, 8, 9, 10, 11, 26,31, 32, 36 | Lilliput | 9°32.837′ S, 13°12.549′ W | 1495 | JQ844818–JQ844832 | JQ844777JQ844778JQ844780JQ844781 | JQ844776 JQ844779 | This study |
| Bathymodiolus sp. 9°S | M64/1 200 ROV 9 | 4 | Lilliput | 9°19.716′ S, 13°07.536′ W | 1495 | JQ844833 | JQ844783 -JQ844785 | JQ844782 | This study |
| B. azoricus | n.d. | n.d. | Rainbow | 36°13′ N–33°54′ W | 2500 | FJ766849–FJ766857,FJ766870 | - | - | [40] |
| B. heckerae | n.d. | n.d. | West Florida Escarpment | 26°01.8′ N,84°54.9′ W | 3314 | AY649794 | - | - | [73] |
| B. boomerang | n.d. | n.d. | Barbados (Orenoque Dome B) | 10°19.9′ N,58°37.3′ W | 1950 | DQ513448 | - | - | [15] |
| B. aff. boomerang | n.d. | n.d. | Gulf of Congo | 5°47.8′S, 9°42.7′ E | 3170 | DQ513450 | - | - | [15] |
| A. southwardae 5°S | ATA 52 ROV 11 | 1, 2 | Clueless | 4°28.8′ S,12°22.2′ W | 2995 | JQ844786, JQ844787 | JQ844761 - JQ844766 | - | This study |
| A. southwardae Logatchev | n.d. | n.d. | Logatchev | 14°45.189′ N,44°58.829′ W;14°45.32′ N,44–58.79′ W | 3028–3038 | AF114401–AF114403 | EU403435 | - | [33], [60] |
| A. southwardae GoM | n.d. | n.d. | West Florida Escarpment | 26°01.8′ N,84°54.6′ W | 3313 | AF008280 | - | - | [44] |
| A. southwardae Barbados | n.d. | n.d. | BarbadosAccretionaryPrism | 13°50′ N, 57°45′ W | 5000 | AF008279 | - | - | [44] |
Individuals used for clone libraries of the bacterial 16S rRNA gene in bold; SOX = sulfur oxidizers; MOX = methane oxidizers, n.d. = no data.
Figure 3. Sampling locations in the SMAR vents.
A: The Golden Valley vent covers approximately 30 m of a long N-S oriented 3–5 m deep fissure at 4°48.16′S, 12°22.28′W. The walls and the bottom of the fissure are densely covered with golden-brown Bathymodiolus sp. which gave rise to the name. B: The Wideawake vent site lies in a jumbled lava sheet flow and is characterized by extensive beds of Bathymodiolus sp. and associated diverse fauna, including occasional Abyssogena southwardae clams. C: Small Bathymodiolus sp. covering pillow lava and semi-lithified Fe-oxyhydroxide crust at the Lilliput vent field at 9°32.83′S, 13°37.74′S. D: Close-up of Lilliput mussels.
DNA Extraction, PCR Amplification, Cloning and Sequencing
Genomic DNA was extracted from mussel and clam gill tissues using the DNeasy blood and tissue kit (QIAGEN, Hilden, Germany) according to the kit manual. DNA was stored in aliquots at −20°C and later used for amplification of COI genes of the host species and 16S rRNA genes of the bacterial gill symbionts.
Partial sequences (ranging from 550 to 591 nucleotides) of the COI of Bathymodiolus specimens were amplified using the primers BathCOI-F (5′-TGTGGTCTGGAATAATTGGAAC-3′) and BathCOI-R (5-ATAAAAAGATGTATTRAARTGACG-3′) [15]. The DNA amplification conditions were the same as described in Olu-Le Roy et al. (2007) [15]. Mitochondrial COI of the vesicomyid clam specimens was amplified using the primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′ TAAACTTCAGGGTGACCAAAAAATCA-3′) [22]. PCR conditions were as described in Petersen et al. (2010) [23]. PCR products were purified with the QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany) using the microcentrifuge protocol. Purified PCR products served for direct sequencing of COI using an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA, USA) with ABI BigDye and the same primers as in the amplification reaction.
16S rRNA genes of bacterial symbionts were amplified from three Bathymodiolus specimens collected at Wideawake and Clueless at 5°S, three Bathymodiolus specimens from Lilliput at 9°S and two Abyssogena specimens from 5°S using the universal bacterial primers 8F (5′-AGAGGTTGATCMTGGC-3′) and 1492R (5′-TACCTTFTTACGACTT-3′) [24]. PCR products were checked by gel electrophoresis and bands of 1500 nucleotides (nt) length were isolated and purified with the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany). Clean PCR products were cloned to separate co-occurring phylotypes in the genomic DNA. PCR amplification, cloning and sequencing of 16S rRNA genes followed the protocol in Petersen et al. (2010) [23]. All sequences are deposited at Genbank under the accession numbers provided in Table 1.
Phylogenetic Analyses
Mitochondrial COI sequences of Bathymodiolus and Abyssogena specimens were assembled using the Sequencher program (http://www.genecodes.com), imported into the ARB software package [25] and aligned against published sequences of bathymodiolin and vesicomyid species.
Pairwise genetic distances between COI sequences of Bathymodiolus from SMAR and Logatchev and published sequences of B. azoricus and the closest relatives to NMAR species from the western and eastern Atlantic margins were calculated for 519 nt based on the Kimura 2-parameter (K2P) model using MEGA version 5 [26]. K2P distances of Abyssogena spp. from 5°S, Logatchev, Gulf of Mexico (GoM) and Barbados were calculated for 513 nt. The K2P model was chosen because it allows for higher probability of transitional vs. transversional base substitution and has been used in earlier phylogenetic studies on bathymodiolins [9], [27]. Differentiation of Bathymodiolus populations was tested based on a Markov Chain method with the nonparametric exact test by Raymond and Rousset (1995) [28] using Arlequin software ver. 3.5.1.2 [29]. Nucleotide diversity within Bathymodiolus populations was calculated using DnaSP ver 5.10.01 [30]. Synonymy of Bathymodiolus COI gene nucleotide substitution was checked according to the standard invertebrate translation code (MEGA5) starting with nt 3 in the 519 nt alignment.
Phylogenetic analyses of bathymodiolins and vesicomyids were performed with the ARB software package [25] using 372 nt for bathymodiolins and 513 nt for vesicomyids. Maximum Likelihood (ML) phylogenies were calculated using the PhyML algorithm with the Generalized Time Reversible (GTR) nucleotide substitution model that was determined as the best fitting model by MODELTEST software [31]. Ratios of transitions and transversions, proportion of invariable sites and base frequencies were estimated empirically by ARB software. Bootstrapping used 1000 re-samplings.
Bacterial 16S rRNA genes were first analyzed based on partial clone sequences covering approx. 900 nt. The sequences were aligned against close relatives in ARB using the SILVA small subunit alignment [32]. Representative sequences of the major clone sequence groups were chosen for full length sequencing of the 16S rRNA gene (resulting sequences with 1323–1506 nt). 1208 nt of the full length sequences were used for further phylogenetic analyses. ML phylogenies were calculated with PhyML implemented in the ARB package using the Hasegawa, Kishino and Yano (HKY) nucleotide substitution model which is widely used for bacterial 16S rRNA analyses and allows for different base frequencies. Ratios of transitions and transversions, proportion of invariable sites and base frequencies were estimated empirically by ARB. Bootstrapping used 1000 re-samplings.
Results
Mitochondrial COI Sequence Analyses of the Hosts
Bathymodiolus spp
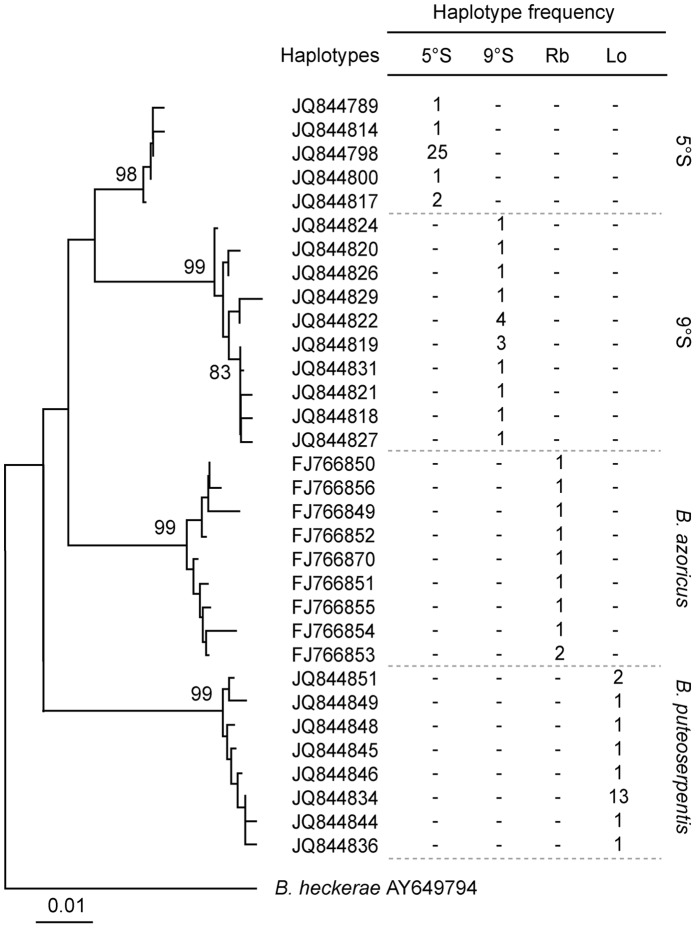
The COI sequence analysis revealed two clearly separated lineages of Bathymodiolus from 5°S and 9°S which diverged from each other as well as from the NMAR species B. azoricus and B. puteoserpentis (Fig. 4). There was no geographic overlap of haplotypes among the four lineages and differences between all of them were confirmed by the exact test for population differentiation (B. azoricus vs. 9°S p<0.014, all other p values <0.001). In 30 specimens from the three sampling sites at 5°S, we found a total of five haplotypes of which the dominant one occurred in 25 individuals (Fig. 4). This group of haplotypes included 4 polymorphic sites; the nucleotide diversity Pi was 0.00308. 15 analyzed COI sequences from the 9°S sampling site revealed 10 haplotypes with 11 polymorphic sites (Pi = 0.00612). 21 sequences of B. puteoserpentis revealed 8 haplotypes of which one was strongly dominant (8 polymorphic sites; Pi = 0.0042) while 10 published sequences of B. azoricus were much more diverse with 9 haplotypes (12 polymorphic sites; Pi = 0.00685). The 5°S and 9°S lineages were clearly separated, with a K2P distance of 0.036 and 14 fixed nucleotide substitutions (Table 2). The divergence between the two SMAR populations based on K2P distance and numbers of fixed substitutions was only about half of the divergence between the two NMAR species (Table 2). In addition to its close relationship to 9°S, the 5°S lineage was similarly closely related to B. azoricus as evidenced by 16 fixed nucleotide substitutions and a genetic distance that was only slightly larger than to 9°S (Table 2). All nucleotide substitutions were synonymous among the 32 haplotypes of all MAR populations and did not lead to amino acid substitutions according to the standard invertebrate translation code. Nonsynonymous substitutions occurred in the North Atlantic seep species B. heckerae (M → L at codon 46 and V → D at codon 132 for B. heckerae) and B. aff. boomerang (D → N at codon 171).
Figure 4. Neighbor Joining tree based on the K2P substitution model and frequencies of COI haplotypes of Bathymodiolus spp. from SMAR and NMAR vent fields and B. heckerae as an outgroup (calculated with MEGA5 [26]).
RB = Rainbow, Lo = Logatchev. Bootstrap percentage values (1000 replicates) higher 70% are marked for the relevant branches. Scale bar represents 1% estimated base substitution.
Table 2. Average pairwise nucleotide divergence (K2P; mean, standard deviation in brackets) among MAR Bathymodiolus populations and their closest relatives from the GoM, Barbados Accretionary Prism and Gulf of Congo (GoC) (above diagonal), within populations (diagonal, italics), and the number of fixed nucleotide substitutions (below diagonal) in 519 positions of COI sequences.
| B. azoricus | B. puteoserpentis | Bathymodiolus sp. 5°S | Bathymodiolus sp. 9°S | B. heckerae | B. boomerang | B. aff. boomerang | ||||||||
| B. azoricus | 0.0064 | (0.0035) | 0.0666 | (0.0023) | 0.0416 | (0.0031) | 0.0652 | (0.0036) | 0.0841 | (0.0028) | 0.0781 | (0.0033) | 0.0757 | (0.0033) |
| B. puteoserpentis | 26 | 0.0018 | (0.0019) | 0.0567 | (0.0015) | 0.0552 | (0.0016) | 0.0879 | (0.0014) | 0.0792 | (0.0014) | 0.0768 | (0.0014) | |
| Bathymodiolus sp. 5°S | 16 | 22 | 0.0006 | (0.0010) | 0.0365 | (0.0020) | 0.0721 | (0.0009) | 0.0613 | (0.0009) | 0.0634 | (0.0009) | ||
| Bathymodiolus sp. 9°S | 24 | 21 | 14 | 0.0046 | (0.0028) | 0.0862 | (0.0026) | 0.0707 | (0.0026) | 0.0726 | (0.0026) | |||
| B. heckerae | n.d. | n.d. | n.d. | n.d. | - | 0.0137 | (- ) | 0.0177 | (- ) | |||||
| B. boomerang | n.d. | n.d. | n.d. | n.d. | n.d. | - | 0.0078 | (- ) | ||||||
| B. aff. boomerang (GoC) | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | - | |||||||
Comparisons are based on 30 analyzed sequences for Bathymodiolus sp. 5°S and 15 for Bathymodiolus sp. 9°S, 21 analyzed sequences for B. puteoserpentis, 10 published sequences for B. azoricus [40], and single published sequences of B. heckerae, B. boomerang and B. aff. boomerang [15], [73] (n.d. = no data).
Maximum Likelihood phylogenetic analysis placed the SMAR lineages in a monophyletic group with B. azoricus and B. puteoserpentis confirming the close relationship of all MAR lineages (Fig. 5A). Their status as a sister group of the North Atlantic B. boomerang seep species complex, which includes B. heckerae, B. boomerang and B. aff. boomerang, is well supported by bootstrap analyses. Within the MAR species, the resolution of ML was low (bootstrap support less than 55%), and repeated ML and Maximum Parsimony analyses resulted in unstable branching topologies (data not shown).
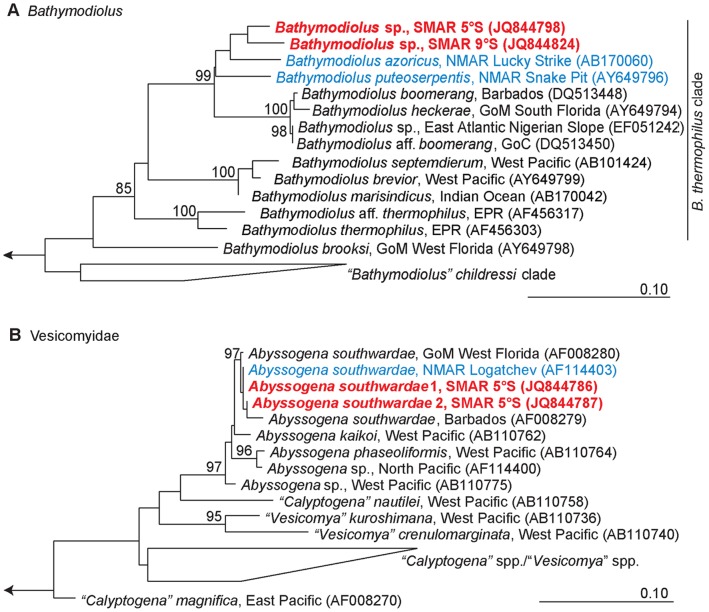
Figure 5. Phylogenetic reconstructions (Maximum Likelihood) based on COI. A: Bathymodiolins; B: Vesicomyidae.
Large chemosynthetic vesicomyids are currently under revision and generic designations will change in the future. The notation of Abyssogena species in the tree follows the preliminary nomenclature of Krylova and Sahling (2010) [47] while other vesicomyid species appear under their published names. MAR species red (SMAR) and blue (NMAR). GoC = Gulf of Congo, GoM = Gulf of Mexico, EPR = East Pacific Rise. The trees were calculated with ARB software using PhyML (GTR substitution model, estimated proportion of invariable sites, four categories of substitution rates). Bootstrap values (1000 replicates) higher 70% are marked at the relevant branches. Scale bars represent 10% estimated base substitution.
The MAR species and B. boomerang species complex together with a group of western Pacific and Indian Ocean species (B. marisindicus, B. brevior and B. septemdierum) are distinct from B. thermophilus and B. aff. thermophilus from the EPR, suggesting a common origin of species from the North Atlantic, Indian and western Pacific Oceans. However, the support for the Indian/western Pacific Ocean group was only weak (65.2%) and their positioning in this clade is uncertain.
Abyssogena southwardae
The two analyzed specimens from 5°S revealed two haplotypes that differed by a single nucleotide substitution. One of these was identical to one of three known haplotypes from Logatchev [33] and pairwise K2P distances between all SMAR and Logatchev haplotypes [33] did not exceed 0.0059 (Table 3, Fig. 5B). The largest divergence was measured between a Logatchev haplotype and Barbados. These data strongly suggest gene flow among A. southwardae populations on the MAR.
Table 3. Pairwise nucleotide divergence (K2P) of COI based on 513 nt among Abyssogena southwardae from Logatchev at the the northern Mid-Atlantic Ridge, southern Mid-Atlantic Ridge (SMAR), Florida Escarpment in the Gulf of Mexico (GoM W-Florida), and Barbados Accretionary Prism [33].
| SMAR haplotype 1 | SMAR haplotype 2 | Logatchev (3 haplotypes) | GoM W-Florida | Barbados Accretionary Prism | |
| SMAR, haplotype 1 | – | 0.0020 | 0–0.0039 | 0.0039 | 0.0138 |
| SMAR, haplotype 2 | – | 0.0020–0.0059 | 0.0059 | 0.0118 | |
| Logatchev, 3 haplotypes | 0.0020–0.0039 | 0.0039–0.0076 | 0.0138–0.0178 | ||
| GoM W-Florida | – | 0.0138 | |||
| Barbados Accretionary Prism | – |
Bathymodiolus symbionts
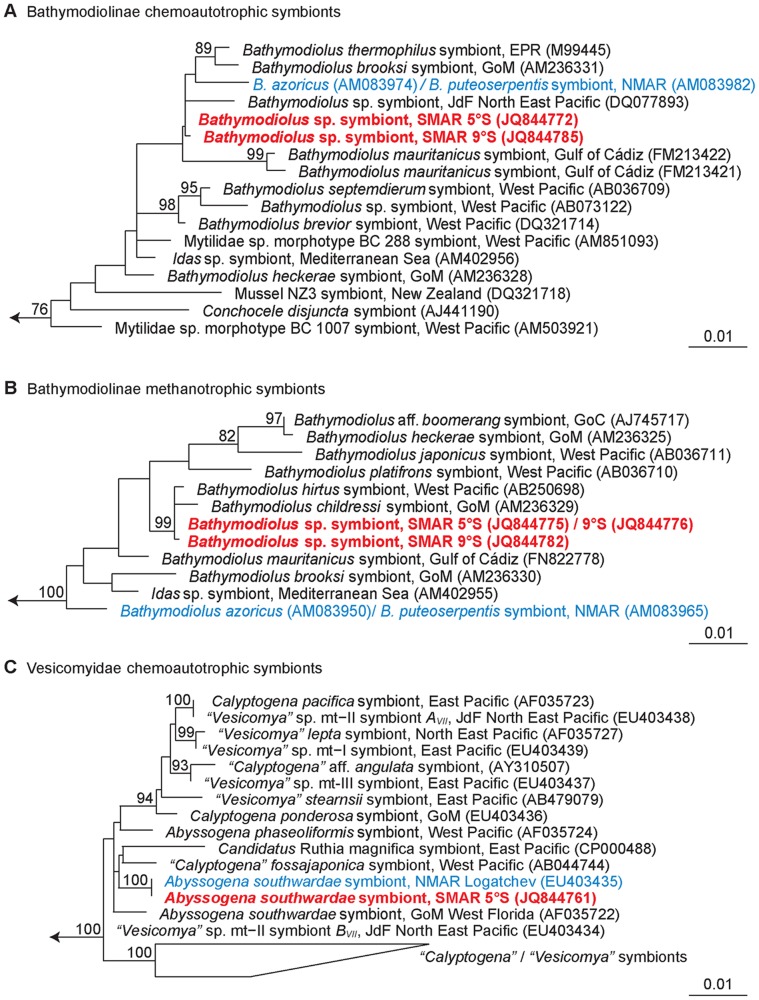
16S rRNA clone libraries constructed from 6 Bathymodiolus specimens collected at 5°S and 9°S yielded 292 partial clone sequences that fell into two groups. These clustered with γ-proteobacterial chemoautotrophic and methanotrophic symbionts from other bathymodiolin hosts, indicating that SMAR Bathymodiolus spp. also live in a dual symbiosis like their NMAR relatives.
The majority (288) of the clone sequences belonged to the chemoautotrophic group. Eight clones each of the 5°S host individuals and seven from 9°S animals were selected for full-length sequencing. These 15 full-length sequences used for further analyses shared ≥99.6% sequence similarity and the two geographical groups from 5°S and 9°S differed consistently in one substitution at E. coli position 1029 suggesting that this substitution was site-specific. Within the 5°S group, six identical sequences originated from Wideawake and Clueless hosts while the two remaining sequences differed from these by one and three substitutions (0.08–0.25% divergence). The 9°S group included four identical sequences and three other phylotypes differing by one or two substitutions (0.08–0.17%). All of these additional substitutions occurred uniquely and in moderately to highly conserved regions of the 16S rRNA gene alignment and were not specific to geographical sites. It is therefore unclear whether they were real or due to PCR or sequencing error. In contrast, the consistent presence of a site-specific substitution in all analyzed sequences suggested that Bathymodiolus hosts in 5°S and 9°S harbor very similar but distinct phylotypes of chemoautotrophic symbionts. A comparison with the chemoautotrophic symbiont of B. azoricus [34] revealed that the dominant 5°S phylotype differed by 13 substitutions (1.0% divergence) and the 9°S phylotype by 14 substitutions (1.2%).
The two dominant SMAR chemoautotrophic phylotypes were chosen for phylogenetic reconstruction (Fig. 6A). The ML analysis positioned them in a monophyletic clade together with the chemoautotrophic symbionts of B. azoricus and B. puteoserpentis from the NMAR, B. brooksi from the Gulf of Mexico, B. thermophilus from EPR, B. mauritanicus from the Gulf of Mexico and a species from the Juan de Fuca Ridge. Within this group, the chemoautotrophic SMAR symbionts appeared to be most closely related to those of Bathymodiolus sp. from JdF (0.5–0.7% nucleotide divergence), but the phylogenetic relationships were not clearly resolved.
Figure 6. Reconstruction of symbiont phylogenies (Maximum Likelihood) based on the 16S rRNA gene.
A: Chemoautotrophic symbionts of Bathymodiolus spp. B: Methanotrophic symbionts of Bathymodiolus spp. C: Chemoautotrophic symbionts of Vesicomyidae. For the notation of vesicomyid host species names, see legend for Figure 4. MAR species red (SMAR) and blue (NMAR). GoC = Gulf of Congo, JdF = Juan de Fuca Ridge. The trees were calculated with ARB software using PhyML (HKY substitution model, estimated proportion of invariable sites, four categories of substitution rates). Bootstrap values (1000 replicates) higher 70% are marked at the relevant branches. Phylotypes obtained in this study in bold red. Scale bars represent 1% estimated base substitution.
Four clone sequences (two for 5°S, two for 9°S) fell together with methanotrophic γ-proteobacterial symbionts and full sequences were obtained for all four clones. Three of the four sequences were identical revealing a common methanotrophic phylotype in 5°S and 9°S. One sequence from 9°S had two substituted positions (0.17% divergence) which were both located in moderately conserved regions. In the ML analysis, the SMAR symbionts formed a higly supported monophyletic group together with the symbionts of B. childressi from GoM and B. hirtus from the western Pacific (Fig. 6B). The methanotrophic symbionts of the two NMAR species B. azoricus and B. puteoserpentis were clearly divergent (2.5–2.7%) indicating that SMAR and NMAR Bathymodiolus spp. harbor different methanotrophic phylotypes.
Abyssogena southwardae symbionts
All 96 16S rRNA clones obtained from the two SMAR Abyssogena clams were similar to the chemoautotrophic symbiont of A. southwardae from Logatchev. Four out of six full-length sequences (1490–1506 nt) were identical while the two others had two substitutions each (0.17% divergence). These all occurred uniquely and in moderately to highly conserved regions of the 16S rRNA gene alignment, and therefore cannot be distinguished from PCR and sequencing errors. The identical sequences shared 100% identity with a sequence from Logatchev, indicating that the symbionts of A. southwardae from Logatchev and 5°S share a common phylotype (Fig. 6C). This common MAR phylotype was clearly divergent from the A. southwardae symbiont from western Florida (1.4%).
Discussion
Our analyses of Bathymodiolus and Abyssogena hosts and symbionts on the MAR revealed four main results: (i) Bathymodiolus from the SMAR and the NMAR species B. azoricus and B. puteoserpentis are monophyletic indicating that their radiation happened recently and after divergence from their sister group of the north Atlantic B. boomerang complex. (ii) Chemoautotrophic and methanotrophic symbionts of SMAR Bathymodiolus spp. are divergent from the B. azoricus/B. puteoserpentis symbionts. (iii) Bathymodiolus symbionts do not reflect the phylogenetic relationships of their host species. (iv) Identical Abyssogena southwardae COI haplotypes in NMAR and SMAR coincide with identical 16S rRNA phylotypes of their symbionts.
Phylogenetic Relationships of MAR Bathymodiolus spp
The monophyly of the SMAR and NMAR Bathymodiolus based on the COI gene indicates that all four lineages share the same evolutionary ancestor. This is in contrast to an earlier study of mitochondrial NADH dehydrogenase subunit 4 gene (ND4) suggesting that the B. boomerang complex species B. heckerae may have derived from B. azoricus [35]. That scenario was not well supported by bootstrap statistics, while a recent phylogeny based on COI [15] and our results based on the COI phylogeny, genetic distances and the distribution of non-synonymous substitutions of this gene suggest divergence of the B. boomerang complex from the MAR species before these diversified.
Bathymodiolus sp. collected at 5°S were at first glance morphologically similar to B. puteoserpentis from NMAR. Preliminary examinations of shell morphologies confirmed affinities between 5°S mussels and the Logatchev morphotype of B. puteoserpentis (R. von Cosel, pers. information) and 5°S animals were tentatively referred to as B. puteoserpentis in a first description of the 5°S vents [18] while the 9°S population shared morphological characteristics with B. azoricus (R. von Cosel, pers. information). In contrast to the similarity of the shell morphologies, we measured considerably high genetic divergence between all four MAR lineages and our results suggest a closer relationship of Bathymodiolus sp. 5°S to B. azoricus than to B. puteoserpentis. Even though the COI gene can show a high degree of polymorphism within populations of hydrothermal vent species [10] and we analyzed a limited number of specimens at each SMAR sampling site, the geographic segregation of haplotype groups, the levels of population differentiation between these groups and the measured genetic distances indicate a clear geographic separation of all four MAR lineages. The rate of nucleotide substitutions and genetic distances (Table 2) match or even exceed what was measured for the two East Pacific Rise populations of B. thermophilus and B. aff. thermophilus separated by the Easter Island Microplate, which are considered to represent a recent speciation event [9]. It is possible that the speciation process among SMAR and NMAR Bathymodiolus has reached a similar level as in EPR Bathymodiolus around the Easter Island Microplate, however, the resolution of the COI gene alone is not sufficient to clarify this (see below).
Our data clearly show a monophyletic relationship among the four MAR lineages and closest K2P-distance between 5°S and 9°S suggests most recent divergence of these two. If the equatorial belt is a dispersal barrier, we would expect a larger genetic distance between southern and northern lineages than within the north or the south. However, the largest distance was between the two NMAR species B. azoricus and B. puteoserpentis (Table 2). This could suggest that B. azoricus and B. puteoserpentis diverged before SMAR and NMAR lineages, and in this case, 5°S and 9°S lineages should share a common ancestor with one of the NMAR species. Slightly larger K2P distance and higher number of fixed substitutions between B. azoricus and B. puteoserpentis than between any of the other three lineages might suggest that B. azoricus diverged early from a common ancestor of the other three lineages. On the other hand, the real genetic divergence between pairs of lineages may be obscured by multiple substitutions of synonymous sites. Such substitutions do not affect the amino acid sequence and they can rapidly accumulate in the mitochondrial COI gene [36]. If multiple substitutions occur, the measured genetic distance underestimates the real genetic divergence, in particular if reverse substitutions lead to restorations of ancestral states, but the phylogenetic reconstruction could not resolve the history of their diversification. A reliable phylogenetic reconstruction of MAR lineages therefore requires additional markers.
The two species B. azoricus and B. puteoserpentis illustrate the complexity of MAR Bathymodiolus diversification. Allozymes and multi-locus analyses showed that these two species hybridize in a zone where their distribution ranges overlap, and that asymmetric gene flow mainly from B. azoricus to B. puteoserpentis is apparent even between geographically distant populations of the two species [37], [38], [39], [40]. Faure et al. (2009) [40] highlighted in a multi-locus population genetic study that it is difficult to decide whether gene flow between these two species happened during parapatric speciation or in the course of secondary contact after a period of allopatry. In the case of secondary contact, it could be possible that divergence of B. azoricus and B. puteoserpentis started early and that the two species entered the MAR independently at different times [40]. Because of the close relationship between all MAR Bathymodiolus lineages, their diversification and also their history of MAR colonization may only be solved by including the SMAR species in future multi-locus analyses using mitochondrial and nuclear genes.
Dispersal of Bathymodiolus spp. along the MAR
Three alternative models for the dispersal of Bathymodiolus on the MAR could explain the current species distribution: (i) The NMAR as well as both sides of the northern Atlantic Ocean may have been colonized by ancestors of the monophyletic clade of the B. boomerang complex and their MAR sister species via the southern Atlantic; (ii) ancestors of the MAR species dispersed in seep regions on the American continental margin between the GoM and off Brazil and entered the MAR independently along a longitudinal gradient north and south of the large equatorial transform faults; (iii) Bathymodiolus from hydrocarbon seeps on the northern Atlantic margins arrived on the northern MAR and dispersed southwards across the equator.
The first model assumes ancestral invasion to the southern MAR via the Indian Ridge and northwards dispersal across the equatorial belt. This is based on the monophyletic relationship of species from the MAR and the Indian and western Pacific Oceans based on mitochondrial COI and ND4 genes [41] and our own COI ML analysis (Fig. 5A). This model would match with a suggested habitat change from vents to seeps in the course of a supposed recent separation of B. heckerae from B. azoricus [35]. However, low statistical support in our analysis questions the placement of the Indo-Pacific group within the B. thermophilus clade. Moreover, if ancestral Bathymodiolus invaded the MAR from the Indian Ridge, dispersed northwards and colonized the Atlantic seeps from the MAR, we would expect that the B. boomerang complex diverged after the beginning of the MAR species radiation. However, previous analyses by Olu Le-Roy et al. (2007) [15] and our own data indicate the exact opposite, that MAR species radiated after the divergence from the B. boomerang species complex. We therefore consider this model the least likely.
The second model assumes that ancestors of the SMAR species dispersed southwards along the American continental margin. This would match a model of deep water currents presented by Van Dover et al. (2002) [1] according to which a second deep-water passage crosses the MAR south of the equatorial belt at 20°S connecting the Atlantic margins off Brazil and Namibia. Such a dispersal path is possible but appears unlikely because in that case we would expect to find species on the American continental margin that are more closely related to SMAR Bathymodiolus spp. than the B. boomerang complex. Such species have not been discovered: The seeps from the GoM, Blake Ridge and the Caribbean harbor species from the B. boomerang complex as well as the more distantly related species B. brooksi and mussels from the “B”. childressi complex [e.g. 15,16,42,43]. The genus Bathymodiolus has not yet been detected south of the Barbados seeps and, hence, there is no current support for the independent colonization of the MAR south of the equator.
The third model is the most likely explanation. It matches a current hypothesis according to which B. azoricus and B. puteoserpentis or a common ancestor entered the MAR from western Atlantic hydrocarbon seeps [37], [40] and it requires an equatorial belt open for dispersal into the southern Atlantic Ocean by west-to-east passage through the long equatorial transform faults. Faure et al. (2009) [40] estimated that B. azoricus and B. puteoserpentis diversified approximately 0.76 Ma ago, and Miyazaki et al. (2010) estimated that the monophyletic Atlantic species group including the B. boomerang complex, B. azoricus and B. puteoserpentis diversified approximately 6.2 Ma ago [41]. The entrance of Bathymodiolus from the western Atlantic margin to the MAR must therefore have happened within this time frame. The closest distance between continental margin seeps and the northern MAR today is 1400 km between the Barbados Accretionary Prism and north of the Fifteen-Twenty Fracture Zone. Assuming a constant spreading rate in this region of 23.5 km Ma-1 over the past 60 Ma since the opening of the Atlantic Ocean, the Barbados seeps and the MAR must have been separated by some 1250 km when the first Bathymodiolus species arrived. This is significantly more than the 935 km length of the Romanche Transform Fault today, and if west-to-east currents transported Bathymodiolus from the American continental margin to the MAR, passage of larvae driven by favorable currents through the Romanche Transform Fault that was even shorter in the past than today is also feasible.
Gene Flow in Abyssogena Southwardae
Our result of a common COI haplotype in Abyssogena southwardae in Logatchev and 5°S and very similar haplotypes at MAR sites and the Western Florida Escarpment in the Gulf of Mexico [33], [44] strongly suggest gene flow over large distances. Long-range dispersal capabilities of this species have also been evidenced by morphology data indicating a distribution over wide areas of the North Atlantic from offshore Virginia to the Barbados Accretionary Prism, Logatchev, Vema Transform Fault, 5°S on the SMAR and the Canary Islands [20]. All these locations are widely separated, cover an extensive depth range of 737–5107 m and include vents and seeps. This wide distribution is remarkable because the potential for dispersal in chemosynthetic vesicomyids has been considered limited as inferred from their lecitotrophic larvae which are classically considered short-lived and thus unable to reach remote areas such as widely interspersed hydrothermal vents on mid-oceanic ridges [45]. However, the dispersal capabilities of vesicomyid clams may be much greater, in particular because lecitotrophy may be an advantage for long-range dispersal in the oligotrophic deep sea waters [46]. While the majority of chemosynthetic vesicomyid species is found at continental margins where hydrocarbon seeps are abundant, only a few species occur at hydrothermal vents on the mid-oceanic ridges [47]. Among these, “Calyptogena” magnifica also shows long-range dispersal by a distribution over more than 4000 km between 21°N and 17°S on the EPR [10]. Further indications for long-range dispersal capabilities of chemosynthetic vesicomyids are given by close phylogenetic relationships based on COI between species pairs from geographically distant sites on the western Pacific and eastern Pacific margins suggesting that ancestral species migrated over large distances [48].
Biogeography of the Symbionts
Bathymodiolus symbionts
The phylogenies of MAR Bathymodiolus hosts and symbionts are not congruent. Both Bathymodiolus lineages from 5°S and 9°S share very similar chemoautotrophic and methanotrophic symbionts, and this similarity parallels previous observations in B. azoricus and B. puteoserpentis which also share highly similar to identical chemoautotrophic and methanotrophic 16S rRNA phylotypes [34]. In contrast, we observed considerable sequence divergences of 1% for the chemoautotrophs and 2.5% for the methanotrophs between NMAR and SMAR populations, which at first glance does not support genetic connectivity between NMAR and SMAR symbionts. In particular the closest phylogenetic relationship between methanotrophic symbionts of SMAR Bathymodiolus and B. childressi from the GoM suggests that transport of symbionts from the GoM to the SMAR may have occurred. This would support the hypothesis of an existing west-east passage for chemosynthetic organisms in the equatorial Atlantic [1], [13].
Bathymodiolus symbionts are most likely acquired horizontally by uptake from the environment [e.g. 49,50,51,52]. Petersen et al. (2010) [23] discussed two alternative models that could explain the geographical structuring of horizontally transmitted ectosymbionts of the MAR vent shrimp Rimicaris exoculata, which may also be valid in Bathymodiolus symbioses. In the first model (i), barriers limit the dispersal of free-living symbionts leading to geographically isolated bacterial populations. If this was the case in MAR Bathymodiolus symbioses, it would mean that SMAR hydrothermal vents provide different populations of free-living symbionts than NMAR vents. An alternative model (ii) assumes that dispersal of symbionts is not limited and the free-living populations occur ubiquitously at geographically distinct vents. In this case, geographic structuring of symbiotic associations would be due to specific bacteria-host selection from a pool of diverse free-living forms. The first model requires flexible patterns of host-symbiont recognition when host species colonize new vent sites, while the second model requires highly specific recognition mechanisms between the symbiotic partners. Such highly specific recognition patterns are known from other symbioses with horizontal symbiont transmission [53], [54] but have not yet been identified in Bathymodiolus. An additional alternative (iii) assumes that dispersing larvae may transport bacteria from their natal sites and “inoculate” new colonization sites with the symbionts of the parental host population or possibly also other bacteria from the parental habitat [51].
There are two explanations for the closer phylogenetic relationships between methanotrophic symbionts of SMAR Bathymodiolus, B. childressi from GoM and B. hirtus from the western Pacific than between SMAR and NMAR. The first is that methanotrophic bacteria closely related to NMAR symbionts were absent in SMAR vents when Bathymodiolus invaded from the north (model i), and that the hosts established a symbiotic association with a methanotroph at SMAR vents that originated from the GoM. In this first scenario we would assume stronger limits for the dispersal of bacteria along the MAR than for mussel larvae. This seems unlikely, and in fact the opposite is the case for symbionts of B. azoricus and B. puteoserpentis of the NMAR. Identical phylotypes of the two chemoautotrophic and methanotrophic symbionts co-occur in the two geographically separated mussel species, indicating that both symbionts have a wider distribution than their hosts.
Alternatively, it is possible that SMAR Bathymodiolus were initially associated with methanotrophic symbionts closely related to the NMAR symbionts, and that these hypothetical ancestors were displaced by methanotrophic symbionts that originated from the GoM. This would be consistent with larval colonization of sites that provide ubiquitously distributed symbionts (ii) and with simultaneous colonization of the sites by larvae and symbionts (iii). Displacement of one symbiont by another in marine chemosynthetic symbioses has, to our knowledge, not yet been shown. A possible scenario for how such a displacement event could occur was recently described in the hydrocarbon seep mussel Bathymodiolus heckerae that harbors two closely related γ-proteobacterial sulfur-oxidizing phylotypes [55]. Both symbionts were only rarely found in the same bacteriocyte suggesting competition between the two symbionts for the same sulfur sources [55]. If two symbionts in one host use the same source for metabolism, the less competitive one could eventually be displaced. It is possible that after the colonization of SMAR vents ancestors of methanotrophic NMAR and GoM symbionts co-occurred for a while in SMAR Bathymodiolus, and that competition between the two for methane eventually lead to the exclusion of the NMAR related symbiont.
Abyssogena southwardae symbionts
The presence of an identical symbiotic 16S rRNA phylotype in A. southwardae from Logatchev and SMAR mirrors the identical COI haplotypes of their hosts. This is in accordance with coupled dispersal of vesicomyid larvae and their symbionts in consequence of maternal (vertical) co-transmission of mitochondria and symbionts across host generations [10], [56], [57], [58], [59] and it is a strong argument for gene flow in clams and their symbionts along the MAR and across the equatorial belt. However, the presence of a divergent symbiotic phylotype in A. southwardae from western Florida is surprising. Recently discovered co-occurrence of two unrelated symbiotic phylotypes in a vesicomyid species from the Juan de Fuca Ridge indicated that vesicomyid clams can also acquire symbionts horizontally [60]. Acquisition of the divergent phylotype from the environment appeared more likely than horizontal exchange between co-occurring species or paternal transfer of symbionts associated with genetic hybridization. Divergent symbiotic phylotypes in geographically distant A. southwardae specimens could therefore suggest that populations from western Florida or the MAR acquired symbionts horizontally in the past, and would thus represent a second example of non-strictly maternal symbiont transfer in vesicomyids. If horizontal acquisition was in a MAR population, it must have happened before dispersal along the ridge axis, but more solid explanations for the divergence of symbionts in this host species requires more genetic data from these and other A. southwardae populations.
Other Indications for Gene Flow Across the Equatorial Belt
The state of taxonomic analyses of the SMAR collections does not yet allow community analyses, but a considerable degree of similarity between northern and southern MAR vent fauna is currently apparent. This suggests that along-axis faunal dispersal across the equator has happened or may still be happening. The hydrothermal vent shrimp Rimicaris exoculata is present at all 5°S hot vents [18]. Common COI haplotypes among the vent sites Rainbow, TAG, Snake Pit, Ashadze, Logatchev and 5°S indicated conspecificity for all MAR Rimicaris and recent gene flow across the equatorial belt [23], [61]. Similarly, although their γ-proteobacterial and ε-proteobacterial ectosymbionts showed geographic clustering based on 16S rRNA phylogeny, the genetic differences between these populations were correlated with distance along the MAR and did not indicate a major barrier for gene flow in the equatorial region [23]. The polychaete species Laonice athecata, Prionospio unilamellata, and Amathys lutzi found at the 5°S and 9°S vents (B. Ebbe, pers. communication) occur also at northern MAR vents. These observations suggest that the SMAR vent sites at 5°S and 9°S are not disconnected from the NMAR.
Conclusions
Our results show clearly that Bathymodiolus mussels and Abyssogena southwardae clams from the southern Mid-Atlantic Ridge at 5°S and 9°S are more closely related to species from the northern MAR than to other species: SMAR mussels form a monophyletic clade together with B. azoricus and B. puteoserpentis while A. southwardae from 5°S and from the Logatchev vent field share an identical COI haplotype. This indicates that gene flow between the northern and southern Mid-Atlantic Ridge has happened and may still continue. Our results therefore do not justify a strict dispersal barrier that disconnects northern and southern MAR communities biogeographically. Contrary to previous hypotheses, we did not observe close relationship to Indian Ocean taxa.
The dispersal of Bathymodiolus spp. most likely occurred from north to south along the MAR rather than independent colonization events from continental margin seeps north and south of the equator. The distribution pattern of the hydrothermal vent species Rimicaris exoculata across the equatorial belt testifies that such a pathway is open to species with high potential for long-range dispersal. Bathymodiolus have long-lived planktotrophic larvae that can probably spend up to a year in the water column [62] suggesting that favorable currents may transport them for several hundreds of kilometers. Vesicomyid clams may have much better dispersal capabilities than previously inferred from their mode of larval development, in particular because lecitotrophy may be of advantage for long-range dispersal in oligotrophic deep-sea waters [46]. Strong currents from west to east through the large equatorial transform faults may bridge long distances for larval transport between ridge segments and other chemosynthetic habitats including hydrocarbon seeps and possibly also sunken wood and whale carcasses [63], [64], [65] may serve as stepping stones for hydrothermal vent species. The suitability of an off-axis habitat with fluid flow induced by serpentinization processes was shown for a vent Bathymodiolus species at the Lost City site [66], [67], [68]. Other off-axis habitats may also be associated with hydrothermal fluid flow, as evidenced for example in pull-apart basins adjacent to transform faults [69], [70]. The occurrence of A. southwardae in the Vema Transform Fault [20], [71] confirmed that such stepping stones exist in the Atlantic.
Our investigations of chemosynthetic bivalves and their symbionts represent an initial test for a dispersal barrier effect of the equatorial belt based on mitochondrial COI and bacterial 16S rRNA marker genes. The results indicate that although host populations are geographically separated, a strict dispersal barrier between north and south does not exist for the species we investigated. Future multi-locus investigations of NMAR and SMAR animals using mitochondrial and nuclear phylogenetic markers will reveal if the equatorial belt exhibits a structuring effect beyond geographic distance on gene flow and speciation processes among MAR populations.
Acknowledgments
We would like to thank captains, crews and scientific parties of the cruises with RV l’Atalante MARSUED IV, RV Maria S. Merian MSM04/3, RV Meteor M64/1 and M78/2, and the teams of the ROVs Kiel 6000 (IFM-GEOMAR, Kiel) and Marum Quest 4000 m (Marum, Bremen) for their contributions to the sampling effort and data collection; Silke Wetzel, Nicole Rödiger and Lisa Drews (MPI Bremen) for their expertise in the laboratory; Nico Augustin (IfM-Geomar, Kiel) for providing a 3D reconstruction of the bathymetry of the equatorial Atlantic Ocean; and Rudo von Cosel (Muséum national d’Histoire naturelle, Paris) for comments on the manuscript. Special thanks to two anonymous reviewers who helped considerably improving the manuscript. This research contributes to the ChEss project of the Census of Marine Life and is contribution no. 67 of the DFG priority program SPP1144.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding was granted through the German Research Foundation (DFG, project no. BO2500/2-1) within the framework of the Program SPP 1144 “From Mantle to the Ocean: Energy- Material- and Life-Cycles at Spreading Axes”, the DFG Cluster of Excellence “The Ocean in the Earth System” at MARUM, Bremen, and the Max Planck Society. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Van Dover CL, German CR, Speer KG, Paarson LM, Vrijenhoek RC. Evolution and biogeography of deep-sea vent and seep invertebrates. Science. 2002;295:1253–1257. doi: 10.1126/science.1067361. [DOI] [PubMed] [Google Scholar]
- 2.Mullineaux LS, France S. Humphris SE, Zierenberg RA, Mullineaux LS, Thompson RE, editors. Dispersal mechanisms of deep-sea hydrothermal vent fauna. 1995. pp. 408–424. Seafloor hydrothermal systems: physical, chemical, biological, and geological interactions. Washington, DC: American Geophysical Union.
- 3.Marsh AG, Mullineaux LS, Young CM, Manahan DT. Larval dispersal potential of the tubeworm Riftia pachyptila at deep-sea hydrothermal vents. Nature. 2001;411:77–80. doi: 10.1038/35075063. [DOI] [PubMed] [Google Scholar]
- 4.Mullineaux LS, Speer KG, Thurnherr AM, Maltrud ME, Vangriesheim A. Implications of cross-axis flow for larval dispersal along mid-ocean ridges. Cah Biol Mar. 2002;43:281–284. [Google Scholar]
- 5.Tyler PA, Young CM. Dispersal at hydrothermal vents: a summary of recent progress. Hydrobiologia. 2003;503:9–19. [Google Scholar]
- 6.Johnson SB, Young CR, Jones WJ, Waren A, Vrijenhoek RC. Migration, isolation, and speciation of hydrothermal vent limpets (Gastropoda; Lepetodrilidae) across the Blanco Transform Fault. Biol Bull. 2006;210:140–157. doi: 10.2307/4134603. [DOI] [PubMed] [Google Scholar]
- 7.Jollivet D, Desbruyères D, Bonhomme F, Moraga D. Genetic differentiation of deep-sea hydrothermal vent alvinellid populations (Annelida: Polychaeta) along the East Pacific Rise. Heredity. 1995;74:376–391. [Google Scholar]
- 8.Plouviez S, Le Guen D, Lecompte O, Lallier FH, Jollivet D. Determining gene flow and the influence of selection across the equatorial barrier of the East Pacific Rise in the tube-dwelling polychaete Alvinella pompejana. BMC Evol Biol 10. 2010. doi: 10.1186/1471–2148–10–220. [DOI] [PMC free article] [PubMed]
- 9.Won Y, Young CR, Lutz RA, Vrijenhoek RC. Dispersal barriers and isolation among deep-sea mussel populations (Mytilidae: Bathymodiolus) from eastern Pacific hydrothermal vents. Mol Ecol. 2003;12:169–184. doi: 10.1046/j.1365-294x.2003.01726.x. [DOI] [PubMed] [Google Scholar]
- 10.Hurtado LA, Mateos M, Lutz RA, Vrijenhoek RC. Coupling of bacterial endosymbiont and host mitochondrial genomes in the hydrothermal vent clam Calyptogena magnifica. Appl Environ Microbiol. 2003;69:2058–2064. doi: 10.1128/AEM.69.4.2058-2064.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heezen BC, Bunce ET, Hersey JB, Tharp M. Chain and Romanche Fracture Zones. Deep-Sea Res. 1964;11:11–33. [Google Scholar]
- 12.Mercier H, Speer KG. Transport of bottom water in the Romanche Fracture Zone and the Chain Fracture Zone. J Phys Oceanogr. 1998;28:779–790. [Google Scholar]
- 13.Tyler PA, German CR, Ramirez-Llodra E, Van Dover CL. Understanding the biogeography of chemosynthetic ecosystems. Oceanol Acta. 2003;25:227–241. [Google Scholar]
- 14.Van Dover CL. The Ecology of deep-sea hydrothermal vents. Princeton, NJ: Princeton University Press. 424 p. 2000.
- 15.Olu-Le Roy K, von Cosel R, Hourdez S, Carney SL, Jollivet D. Amphi-Atlantic cold seep Bathymodiolus species complexes across the equatorial belt. Deep-Sea Res I. 2007;54:1890–1911. [Google Scholar]
- 16.Olu K, Cordes EE, Fisher CR, Brooks JM, Sibuet M, et al. Biogeography and potential exchanges among the Atlantic equatorial belt cold-seep faunas. PLoS ONE. 2010;5(8):e11967. doi: 10.1371/journal.pone.0011967. doi: 10.1371/journal.pone.0011967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.German CR, Bennett SA, Connelly DP, Evans AJ, Murton BJ, et al. Hydrothermal activity on the southern Mid-Atlantic Ridge: Tectonically- and volcanically-controlled venting at 4–5° S. Earth Planet Sc Lett. 2008;273:332–344. [Google Scholar]
- 18.Haase KM, Petersen S, Koschinsky A, Seifert R, Devey CW, et al. Young volcanism and related hydrothermal activity at 5° S on the slow-spreading southern Mid-Atlantic Ridge. Geochem Geophy Geosy. 2007;8:Q11002. doi: 10.1029/2006GC001509. [Google Scholar]
- 19.Haase KM, Koschinsky A, Petersen S, Devey CW, German C, et al. Diking, young volcanism and diffuse hydrothermal activity on the southern Mid-Atlantic Ridge: The Lilliput field at 9° 33′S. Mar Geol. 2009;266:52–64. [Google Scholar]
- 20.Krylova EM, Sahling H, Janssen R. Abyssogena: a new genus of the family Vesicomyidae (Bivalvia) from deep-water vents and seeps. J Mollusc Stud. 2010;76:107–132. [Google Scholar]
- 21.Gebruk AV, Chevaldonné P, Shank T, Lutz RA, Vrijenhoek RC. Deep-sea hydrothermal vent communities of the Logatchev area (14°45′N, Mid-Atlantic Ridge): Diverse biotopes and high biomass. J Mar Biol Assoc UK. 2000;80:383–393. [Google Scholar]
- 22.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek RC. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech. 1994;3:294–299. [PubMed] [Google Scholar]
- 23.Petersen JM, Ramette A, Lott C, Cambon-Bonavita MA, Zbinden M, et al. Dual symbiosis of the vent shrimp Rimicaris exoculata with filamentous gamma- and epsilonproteobacteria at four Mid-Atlantic Ridge hydrothermal vent fields. Environ Microbiol. 2010;12:2204–2218. doi: 10.1111/j.1462-2920.2009.02129.x. [DOI] [PubMed] [Google Scholar]
- 24.Muyzer G, Teske A, Wirsen CO, Jannasch HW. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig W, Strunk O, Westram R, Richter L, Meier H, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Peterson D, N P, Stecher G, Nei M, et al. MEGA5: Molecular evolutionary genetics analysis using likelihood, distance, and parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorion J, Buge B, Cruaud C, Samadi S. New insights into diversity and evolution of deep-sea Mytilidae (Mollusca: Bivalvia). Mol Phylogenet Evol. 2010;57:71–83. doi: 10.1016/j.ympev.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 28.Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 29.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 30.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 31.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 32.Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peek AS, Gaut BS, Feldman R, A., Barry JP, Kochevar RE, et al. Neutral and nonneutral mitochondrial genetic variation in deep-seaclams from the family Vesicomyidae. Mol Evol. 2000;50:141–153. doi: 10.1007/s002399910016. [DOI] [PubMed] [Google Scholar]
- 34.Duperron S, Bergin C, Zielinski F, Blazejak A, Pernthaler A, et al. A dual symbiois shared by two mussel species, Bathymodiolus azoricus and Bathymodiolus puteoserpentis (Bivalvia: Mytilidae), from hydrothermal vents along the northern Mid-Atlantic Ridge. Environ Microbiol. 2006;8:1441–1447. doi: 10.1111/j.1462-2920.2006.01038.x. [DOI] [PubMed] [Google Scholar]
- 35.Won YJ, Maas PAY, Van Dover CL, Vrijenhoek RC. Habitat reversal in vent and seep mussels: Seep species, Bathymodiolus heckerae, derived from vent ancestors. Cah Biol Mar. 2002;43:387–390. [Google Scholar]
- 36.Vrijenhoek RC. Cryptic species, phenotypic plasticity, and complex life history: Assessing deep-sea fauna diversity with molecular markers. Deep-Sea Res II. 2009;56:1713–1723. [Google Scholar]
- 37.Won Y, Hallam SJ, O’Mullan GD, Vrijenhoek RC. Cytonuclear diseqilibrium in a hybrid zone involving deep-sea hydrothermal vent mussels of the genus Bathymodiolus. Mol Ecol. 2003;12:3185–3190. doi: 10.1046/j.1365-294x.2003.01974.x. [DOI] [PubMed] [Google Scholar]
- 38.O’Mullan GD, Maas PAY, Lutz RA, Vrijenhoek RC. A hybrid zone between hydtrothermal vent musseles (Bivalvia: Mytilidae) from the Mid-Atlantic Ridge. Mol Ecol. 2001;10:2819–2831. doi: 10.1046/j.0962-1083.2001.01401.x. [DOI] [PubMed] [Google Scholar]
- 39.Maas PAY, O’Mullan GD, Lutz RA, Vrijenhoek RC. Genetic and morphological characterization of mussels (Bivalvia: Mytilidae) from Mid-Atlantic hydrothermal vents. Biol Bull. 1999;196:265–272. doi: 10.2307/1542951. [DOI] [PubMed] [Google Scholar]
- 40.Faure B, Jollivet D, Tanguy A, Bonhomme F, Bierne N. Speciation in the deep sea: Multi-locus analysis of divergence and gene flow between two hybridizing species of hydrothermal vent mussels. PloS ONE. 2009;4(8):e6485. doi: 10.1371/journal.pone.0006485. doi: 10.1371/journal.pone.0006485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyazaki J-I, de Oliveira Martins L, Fujita Y, Matsumoto H, Fujiwara Y. Evolutionary process of deep-sea Bathymodiolus mussels. PLoS ONE. 2010;5(4):e10363. doi: 10.1371/journal.pone.0010363. doi: 10.1371/journal.pone.0010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sibuet M, Olu K. Biogeography, biodiversity and fluid dependence of deep-sea cold-seep communities at active and passive margins. Deep-Sea Res II. 1998;45:517–567. [Google Scholar]
- 43.Van Dover CL, Aharon P, Bernhard JM, Caylor E, Doerries M, et al. Blake Ridge methane seeps: Characterization of a soft-sediment, chemosynthetically based ecosystem. Deep-Sea Res I. 2003;50:281–300. [Google Scholar]
- 44.Peek AS, Gustafson RG, Lutz RA, Vrijenhoek RC. Evolutionary relationships of deep-sea hydrothermal vent and cold-water seep clams (Bivalvia: Vesicomyidae): Results from mitochondrial cytochrome oxidase subunit I. Mar Biol. 1997;130:151–161. [Google Scholar]
- 45.Lutz RA, Jablonski D, Turner RD. Larval development and dispersal at deep-sea hydrothermal vents. Science. 1984;226:1451–1454. doi: 10.1126/science.226.4681.1451. [DOI] [PubMed] [Google Scholar]
- 46.Tyler PA, Young CM. Reproduction and dispersal at vents and cold seeps. J Mar Biol Assoc UK. 1999;79:193–208. [Google Scholar]
- 47.Krylova EM, Sahling H. Vesicomyidae (Bivalvia): Current taxonomy and distribution. PLoS ONE. 2010;5(4):e9957. doi: 10.1371/journal.pone.0009957. doi: 10.1371/journal.pone.0009957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kojima S, Fujikura K, Okutani T. Multiple trans-Pacific migrations of deep-sea vent/seep-endemic bivalves in the family Vesicomyidae. Mol Phylogen Evol. 2004;32:396–406. doi: 10.1016/j.ympev.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 49.Won YJ, Jones WJ, Vrijenhoek RC. Absence of cospeciation between deep-sea mytilids and their thiotrophic endosymbionts. J Shellfish Res. 2008;27:129–138. [Google Scholar]
- 50.Le Pennec M, Diouris M, Herry A. Endocytosis and lysis of bacteria in gill epithelium of Bathymodiolus thermophilus, Thyasira flexuosa and Lucinella divaricata bivalve molluscs. J Shellfish Res. 1988;7:483–490. [Google Scholar]
- 51.Won Y-J, Hallam SJ, O’Mullan GD, Pan IL, Buck CS, et al. Environmental acquisition of thiotrophic endosymbionts by deep-sea mussels of the genus Bathymodiolus. Appl Environ Microbiol. 2003;69:6785–6792. doi: 10.1128/AEM.69.11.6785-6792.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petersen JM, Dubilier N. Methanotrophic symbioses in marine invertebrates. Environ Microbiol Reports. 2009;1:319–335. doi: 10.1111/j.1758-2229.2009.00081.x. [DOI] [PubMed] [Google Scholar]
- 53.Ruby EG. Symbiotic conversations are revealed under genetic interrogation. Nat Rev Microbiol. 2008;6:752–762. doi: 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bulgheresi S, Gruber-Vodicka HR, Heindl NR, Dierks U, Kostadinova M, et al. Sequence variability of the pattern recognition receptor Mermaid mediates specificity of marine nematode symbioses. ISME J. 2011;5:986–998. doi: 10.1038/ismej.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duperron S, Sibuet M, MacGregor B, Kuypers M, Fisher C, et al. Diversity, relative abundance and metabolic potential of bacterial endosymbionts in three Bathymodiolus mussel species from cold seeps in the Gulf of Mexico. Environ Microbiol. 2007;9:1423–1438. doi: 10.1111/j.1462-2920.2007.01259.x. [DOI] [PubMed] [Google Scholar]
- 56.Endow K, Ohta S. Occurence of bacteria in the primery oocytes of vesicomyid clam Calyptogena soyoae. Mar Ecol Prog Ser. 1990;64:309–311. [Google Scholar]
- 57.Cary SC, Giovannoni SJ. Transovarial inheritance of endosymbiotic bacteria in clams inhabiting deep-sea hydrothermal vents and cold seeps. P Natl Acad Sci USA. 1993;90:5695–5699. doi: 10.1073/pnas.90.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peek AS, Vrijenhoek RC, Gaut BS. Accelerated evolutionary rate in sulfur-oxidizing endosymbiotic bacteria associated with the mode of symbiont transmission. Mol Biol Evol. 1998;15:1514–1523. doi: 10.1093/oxfordjournals.molbev.a025879. [DOI] [PubMed] [Google Scholar]
- 59.Goffredi SK, Hurtado LA, Hallam S, Vrijenhoek RC. Evolutionary relationships of deep-sea vent and cold seep clams (Mollusca: Vesicomyidae) of the “pacifica/lepta” species complex. Mar Biol. 2003;142:311–320. [Google Scholar]
- 60.Stewart FJ, Young CR, Cavanaugh CM. Lateral symbiont acquisition in a maternally transmitted chemosynthetic clam endosymbiosis. Mol Biol Evol. 2008;25:673–687. doi: 10.1093/molbev/msn010. [DOI] [PubMed] [Google Scholar]
- 61.Teixeira S, Cambon-Bonavita MA, Serrao EA, Desbruyères D, Arnaud-Haond S. Recent population expansion and connectivity in the hydrothermal shrimp Rimicaris exoculata along the Mid-Atlantic Ridge. J Biogeogr. 2011;38:564–574. [Google Scholar]
- 62.Arellano SM, Young CM. Spawning, development, and the duration of larval life in a deep-sea cold-seep mussel. Biol Bull. 2009;216:149–162. doi: 10.1086/BBLv216n2p149. [DOI] [PubMed] [Google Scholar]
- 63.Distel DL, Baco AR, Chuang E, Morril W, Cavanaugh C, et al. Do mussels take wooden steps to deep-sea vents? Nature. 2000;403:725–726. doi: 10.1038/35001667. [DOI] [PubMed] [Google Scholar]
- 64.Lorion J, Duperron S, Gros O, Cruaud C, Samadi S. Several deep-sea mussels and their associated symbionts are able to live both on wood and on whale falls. P Roy Soc Lond B-Biol. 2009;276:177–185. doi: 10.1098/rspb.2008.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith CR, Baco AR. Phylogenetic and functional affinities between whalefall, seep and vent chemoautotrophic communities. Cah Biol Mar. 1998;39:345–346. [Google Scholar]
- 66.Kelley DS, Karson JA, Fruh-Green GL, Yoerger DR, Shank TM, et al. A serpentinite-hosted ecosystem: The Lost City hydrothermal field. Science. 2005;307:1428–1434. doi: 10.1126/science.1102556. [DOI] [PubMed] [Google Scholar]
- 67.Kelley DS, Karson JA, Blackman DK, Fruh-Green GL, Butterfield DA, et al. An off-axis hydrothermal vent field near the Mid-Atlantic Ridge at 30° N. Nature. 2001;412:145–149. doi: 10.1038/35084000. [DOI] [PubMed] [Google Scholar]
- 68.DeChaine EG, Bates AE, Shank TM, Cavanaugh CM. Off-axis symbiosis found: Characterization and biogeography of bacterial symbionts of Bathymodiolus mussels from Lost City hydrothermal vents. Environ Microbiol. 2006;8:1902–1912. doi: 10.1111/j.1462-2920.2005.01113.x. [DOI] [PubMed] [Google Scholar]
- 69.Hein JR, Koski RA, Embley RW, Reid JA, Chang S-W. Diffuse-flow hydrothermal field in an oceanic fracture zone setting, Northeast Pacific: Deposit composition. Explor Min Geol. 1999;8:299–322. [Google Scholar]
- 70.Dekov V, Scholten J, Garbe-Schönberg CD, Botz R, Cuadros J, et al. Hydrothermal sediment alteration at a seafloor vent field: Grimsey Graben, Tjornes Fracture Zone, north of Iceland. J Geophys Res-Sol Ea 113. 2008.
- 71.Cannat M, Mamaloukasfrangoulis V, Auzende JM, Bideau D, Bonatti E, et al. A geological cross-section of the Vema Fracture-Zone Transverse Ridge, Atlantic-Ocean. J Geodyn. 1991;13:97–117. [Google Scholar]
- 72.Shank T. The evolutionary puzzle of seafloor life. Oceanus. 2004;42:78–85. [Google Scholar]
- 73.Jones WJ, Won Y-J, Maas PAY, Smith PJ, Lutz RA, et al. Evolution of habitat use by deep-sea mussels. Mar Biol. 2006;148:841–851. [Google Scholar]