Abstract
We recently reported that mutations in the VCP gene are a cause of 1–2% of familial amyotrophic lateral sclerosis (ALS) cases, but their role in the pathogenesis of sporadic ALS is unclear. We undertook mutational screening of VCP in 701 sporadic ALS cases. Three pathogenic variants (p.Arg159Cys, p.Asn387Thr, and p.R662C) were found in three US cases, each of whom presented with progressive upper and lower motor neuron signs consistent with definite ALS by El Escorial diagnostic criteria. Our data indicate that VCP mutations may underlie apparently sporadic ALS, but account for less than 1% of this form of disease.
Keywords: Amyotrophic lateral sclerosis, valosin-containing protein, mutations, sporadic disease
1. Introduction
Using an exome sequencing approach, we recently found that mutations in the Valosin containing protein (VCP) gene on the short arm of chromosome 9 are a cause of familial amyotrophic lateral sclerosis (ALS) (Johnson, et al., 2010). Mutations in this gene were previously known to cause an unusual clinical syndrome characterized by inclusion body myopathy with early-onset Paget’s disease and fronto-temporal dementia (IBMPFD, OMIM# 167320) (Watts, et al., 2004). Our data extend the phenotype associated with VCP mutations to include progressive upper and lower motor neuron degeneration consistent with ALS (without concomitant muscle or bone involvement), and provide fundamental insight into the pathogenesis of motor neuron degeneration.
Approximately 5% of ALS cases are familial in nature, whereas the bulk of patients have no family history of ALS and are presumed to represent sporadic disease (Chiò, et al., 2008). Although VCP mutations account for 1–2% of familial cases, the role of this gene in the more common sporadic form of ALS remains unclear. To address this gap in our knowledge, we undertook mutational screening of VCP in a large cohort of patients diagnosed with sporadic ALS.
2. Methods
2.1 Study population
Case samples consisted of 459 US and 242 Italian individuals diagnosed with ALS based on El Escorial criteria (Brooks, 1994). The US samples were obtained from the NINDS Human Genetics DNA and Cell Line Repository at Coriell (catalog numbers for the precompiled panels of DNA samples used in this study: NDPT025, NDPT026, NDPT100, NDPT103 and NDPT106, see www.coriell.org). The Italian samples were collected from the Piedmont region having been identified by the population-based ALS Registry that has been operating in that region since 1995 (Chiò, et al., 2009a). Control samples consisted of neurologically normal individuals obtained from Coriell (US, n = 569) and Italy (n = 636). An additional 364 samples that are part of the Human Genome Diversity Panel (HGDP) (Cann, et al., 2002) were included in the mutational analysis as controls to evaluate the genetic variability of VCP in non-Caucasian populations (Johnson, et al, 2010). Demographics and clinical features of these samples are summarized in Supplementary Table 1, whereas the frequencies of SOD1, TDP-32, and FUS mutations, as well as the C9ORF72 hexanucleotide repeat expansion in the Italian and US patients are shown in Supplemental Table 2.
2.2 Mutational screening
All patients underwent mutational analysis for VCP mutations. Specifically, all the coding exons and 30bp of the flanking intron-exon boundaries of VCP (NM_007126.3) were PCR amplified, sequenced using the Big-Dye Terminator v3.1 sequencing kit, run on an ABI 3730xl genetic analyzer (Applied Biosystems Inc.), and analyzed using Sequencer software version 4.2 (Gene Codes Corp., Ann Arbor, MI, USA).
3. Results
We performed sequence analysis of the VCP gene in 701 Caucasian cases diagnosed with sporadic ALS. This identified three missense mutations, namely a p.Arg159Cys (c.864C>T) mutation, a p.Asn387Thr (c.1549A>C) mutation and a p.Arg662Cys (c.2373C>T) mutation, each of which were observed in single cases (Table 1). The three cases with these VCP mutations did not carry a FUS mutation or the pathogenic hexanucleotide expansion of the C9ORF72 gene (Lai, et al. 2011; Majounie, et al., 2012). The p.Arg159Cys variant is known to be pathogenic, in that it has been previously found in patients with IBMPFD (Bersano, et al., 2009, Spina, et al., 2008). The p.Asn387Thr variant represents a novel amino acid shift, though a different mutation (p.Asn387His) involving the same codon has also been described as causative for IBMPFD (Watts, et al., 2007). The p.Arg662Cys variant also represents a novel mutation. None of the variants were found in control samples screened in our own laboratory (n = 1,569 individuals, equating to 3,138 chromosomes), or in the dbSNP (Build 133, http://www.ncbi.nlm.nih.gov/projects/SNP/) and the 1000 genomes online databases (accessed 11th April 2011, n = 629 individuals, www.1000genomes.org) of human population polymorphisms.
Table 1.
VCP mutations in sporadic ALS cases with clinical data
| Coriell Sample ID | Mutation | Age at Onset | Gender | Race (Origin) | Site of Onset | Cognitive Impairment | ALSFRS-R | Neurological Examination | Duration from Onset |
|---|---|---|---|---|---|---|---|---|---|
| ND11807 | p.Arg159Cys (c.864C>T) | 68 | Female | Caucasian (US) | Lower limb | No impairment reported | 40/48 | UMN and LMN signs in 4 limbs | Alive at 5 years |
| ND12329 | p.Asn387Thr (c.1549A>C) | 57 | Male | Caucasian (US) | Lower limb | No impairment reported | 38/48 | UMN and LMN signs in 4 limbs | Alive at 5 years |
| ND10069 | p.Arg662Cys (c.2373C>T) | 67 | Male | Caucasian (US) | Lower limb | No impairment reported | 38/42* | UMN and LMN signs in 4 limbs; Bulbar LMN signs | Alive at 2 years |
Additional phenotype data is available for these samples at www.coriell.org; ALSFRS-R, Revised ALS Functional Rating Scale;
this value represents the ALSFRS score (maximum value = 42);
UMN, upper motor neuron; LMN, lower motor neuron; base pair coordinates are based on VCP transcript NM_007126.3.
In addition to the three missense mutations, we also found variants for which the pathogenicity is less clear. These are listed in Supplementary Table 3, and consist of the synonymous variant p.Tyr755 (c.2654T>C, n = 1), and intronic variants IVS12+9 T>C (n = 1) and 3′UTR *12 C>T (n = 1). Again, none of these variants were found in controls sequenced in our own laboratory, or in the dbSNP or 1000 genomes databases of human population polymorphisms.
All of the patients carrying VCP mutations developed lower limb weakness in middle to late age, which progressed to involve all four limbs over a number of years. Apart from the mother of ND12329, who was diagnosed with unspecified dementia, there was no reported personal or family history of muscle disease, frontotemporal dementia or bone disease. Examination at the time of sampling revealed upper and lower motor neuron signs in all four limbs, and widespread ongoing denervation and chronic reinnervation changes on EMG. One of the patients had evidence of bulbar involvement at the time of sampling. Revised ALS Functional Rating Scale (ALSFRS-R) scores revealed moderate disability. The clinical pictures of the patients carrying mutations of the VCP gene were consistent with definite ALS by El Escorial diagnostic criteria, and they had been treated with Riluzole.
4. Discussion
By definition, it is not possible to prove causation by showing segregation of a mutation with disease in sporadic ALS cases. Despite this, the three missense mutations of VCP identified in our cohort are likely to be pathogenic for several reasons: first, one of them has previously been described in patients with IBMPFD (Kimonis, et al., 2008), and another involves the same codon as another mutation known to be pathogenic; none of the variants were found in large numbers of controls nor have been described as a population polymorphism; furthermore, the p.Arg159Cys variant lies in the known mutational hotspot of the gene, a region of the VCP protein that is thought to be essential to its proper cellular function (Weihl, et al., 2009).
We found that mutations of the VCP gene account for less than 1% of sporadic cases in our cohort (3 mutations out of 701 cases screened = 0.43%). Thus, mutations in this gene account for a smaller percentage of sporadic cases compared to the 1–2% rate of VCP mutations seen in familial ALS (Johnson, et al., 2010). A similar pattern has been observed with other familial ALS genes: SOD1 mutations accounts for ~13% of familial ALS cases in Italy, but are found in less than 1% of sporadic cases (Chiò, et al., 2008); TDP-43 and FUS mutations are each found in ~3–4% of familial cases, but have been described in less than 1% of sporadic cases in the general European population (Chiò, et al., 2009b, Guerreiro, et al., 2008, Kabashi, et al., 2008, Lai, et al., 2011, Mackenzie, et al., 2010).
The question arises as to whether the sporadic cases with mutations in VCP or any other familial ALS gene are truly sporadic cases or whether they represent cryptically-related cases. This scenario may occur for many reasons: lack of knowledge of the pedigree on the part of the patient or neurologist; previous generations dying at a young age prior to the onset of neurological symptoms; decreased penetrance of genes where not all individuals carrying the mutation manifest a clinical phenotype, which may be particularly relevant in any late-onset disease, such as ALS (Lai, et al., 2011). Indeed, VCP mutations are known to be highly variable with respect to both penetrance and phenotype expressivity: 90% developing weakness, 51% having osteolytic lesions, but only one third of cases manifesting FTD, and a smaller percentage developing ALS (Johnson, et al., 2010, Weihl, et al., 2009). Despite these caveats, it is clear that mutations of SOD1 and FUS can underlie truly sporadic ALS, as there are documented de novo mutations in both these genes (Alexander, et al., 2002, Chiò, et al., 2011, DeJesus-Hernandez, et al., 2010). Furthermore, recent data has eroded the artificial barrier between familial and sporadic disease, and suggest that the definition of sporadic disease should be considered operational, rather than definitive (Majounie, et al., 2012).
We found a number of variants that were present only in cases, which did not obviously alter amino acid structure of the VCP protein, or were located in the intron close to the splice site. Both types of mutations can occasionally cause disease by altering splice patterns of genes. For example, a synonymous mutation (p.G608) that introduces a cryptic splice site in the LMNA gene is responsible for a large proportion of Hutchinson-Gilford progeria cases (Eriksson, et al., 2003). Furthermore, mutations in the 5′-splice site of exon 10 of the MAPT gene are known to be pathogenic in families with frontotemporal dementia with parkinsonism (Hutton, et al., 1998). Although these variants were not found in controls, it would be inappropriate to label them as pathogenic at this stage, especially as RNA is not available from these cases to experimentally confirm aberrant splicing. It will be interesting to see if other groups find these variants in their cohorts of familial and sporadic cases, thus providing additional evidence pointing toward their pathogenicity.
In summary, our data indicate that mutations of VCP can be responsible for occasional cases of sporadic ALS, but their low frequency means that there is little need to routinely screen the gene in such cases in the absence of additional clinical features or family history of concomitant bone disease, muscle disease or frontotemporal dementia.
Supplementary Material
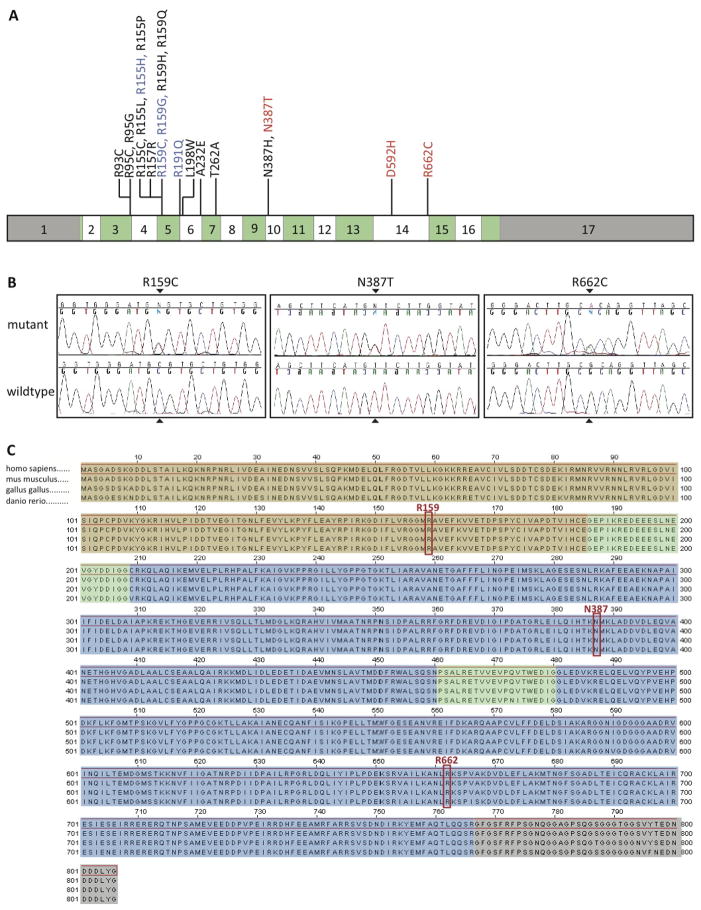
Figure 1.
(A) Graphical representation of the VCP gene showing its 17 exons. Coding exons are colored in alternating green and white, and non-coding regions are shown in grey. Mutations known to cause IBMPFD are shown in black, mutations detected in both IBMPFD and ALS cases are shown in blue, and mutations that have been found in only ALS cases are displayed in red. (B) Chromatograms showing mutant and wild-type alleles of the three variants found in sporadic ALS cases. (C) Sequence alignment demonstrates near complete protein conservation across species (brown shading, N-terminal domain; green, linker domains; blue, AAA-domains; grey, C-terminal domain). Mutated residues are highlighted in red.
Acknowledgments
This work was supported in part by the Intramural Research Programs of the NIH, National Institute on Aging (Z01-AG000949-02). The work was also funded by the Packard Center for ALS Research at Hopkins, the ALS Association, the Fondazione Vialli e Mauro for ALS Research Onlus, Federazione Italiana Giuoco Calcio (FICG), and the Ministero della Salute (Ricerca Sanitaria Finalizzata 2007). DNA samples for this study were obtained in part from the NINDS Human Genetics DNA and Cell Line Repository at Coriell (www.coriell.org).
Footnotes
Disclosure statement
None of the other authors report any conflicts of interest. Informed consent for genetic analysis was obtained from each individual, and appropriate institutional review boards approved the study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander MD, Traynor BJ, Miller N, Corr B, Frost E, McQuaid S, Brett FM, Green A, Hardiman O. “True” sporadic ALS associated with a novel SOD-1 mutation. Ann Neurol. 2002;52:680–683. doi: 10.1002/ana.10369. [DOI] [PubMed] [Google Scholar]
- Bersano A, Del Bo R, Lamperti C, Ghezzi S, Fagiolari G, Fortunato F, Ballabio E, Moggio M, Candelise L, Galimberti D, Virgilio R, Lanfranconi S, Torrente Y, Carpo M, Bresolin N, Comi GP, Corti S. Inclusion body myopathy and frontotemporal dementia caused by a novel VCP mutation. Neurobiol Aging. 2009;30:752–758. doi: 10.1016/j.neurobiolaging.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci. 1994;124(Suppl):96–107. doi: 10.1016/0022-510x(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Chiò A, Calvo A, Moglia C, Ossola I, Brunetti M, Sbaiz L, Lai SL, Abramzon Y, Traynor BJ, Restagno G. A de novo missense mutation of the FUS gene in a “true” sporadic ALS case. Neurobiol Aging. 2011;32:553.e523–553.e526. doi: 10.1016/j.neurobiolaging.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Mora G, Calvo A, Mazzini L, Bottacchi E, Mutani R. Epidemiology of ALS in Italy: a 10-year prospective population-based study. Neurology. 2009a;72:725–731. doi: 10.1212/01.wnl.0000343008.26874.d1. [DOI] [PubMed] [Google Scholar]
- Chiò A, Restagno G, Brunetti M, Ossola I, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Mandrioli J, Salvi F, Spataro R, Schymick J, Traynor BJ, La Bella V. Two Italian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation. Neurobiol Aging. 2009b;30:1272–1275. doi: 10.1016/j.neurobiolaging.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Traynor BJ, Lombardo F, Fimognari M, Calvo A, Ghiglione P, Mutani R, Restagno G. Prevalence of SOD1 mutations in the Italian ALS population. Neurology. 2008;70:533–537. doi: 10.1212/01.wnl.0000299187.90432.3f. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Kocerha J, Finch N, Crook R, Baker M, Desaro P, Johnston A, Rutherford N, Wojtas A, Kennelly K, Wszolek ZK, Graff-Radford N, Boylan K, Rademakers R. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum Mutat. 2010;31:E1377–1389. doi: 10.1002/humu.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Schymick JC, Crews C, Singleton A, Hardy J, Traynor BJ. TDP-43 is not a common cause of sporadic amyotrophic lateral sclerosis. PLoS One. 2008;3:e2450. doi: 10.1371/journal.pone.0002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, Galassi G, Scholz SW, Taylor JP, Restagno G, Chio A, Traynor BJ. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kimonis VE, Fulchiero E, Vesa J, Watts G. VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys Acta. 2008;1782:744–748. doi: 10.1016/j.bbadis.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Lai SL, Abramzon Y, Schymick JC, Stephan DA, Dunckley T, Dillman A, Cookson M, Calvo A, Battistini S, Giannini F, Caponnetto C, Mancardi GL, Spataro R, Monsurro MR, Tedeschi G, Marinou K, Sabatelli M, Conte A, Mandrioli J, Sola P, Salvi F, Bartolomei I, Lombardo F, Mora G, Restagno G, Chio A, Traynor BJ. FUS mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2011;32:550, e551–550, e554. doi: 10.1016/j.neurobiolaging.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chiò A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ The Chromosome 9-ALS/FTD Consortium; The French research network on FTLD/FTLD/ALS; The ITALSGEN Consortium. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina S, Van Laar AD, Murrell JR, de Courten-Myers G, Hamilton RL, Farlow MR, Quinlan J, DeKosky ST, Ghetti B. Frontotemporal dementia associated with a Valosin-Containing Protein mutation: report of three families. FASEBJ. 2008:22. [Google Scholar]
- Watts GD, Thomasova D, Ramdeen SK, Fulchiero EC, Mehta SG, Drachman DA, Weihl CC, Jamrozik Z, Kwiecinski H, Kaminska A, Kimonis VE. Novel VCP mutations in inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Clin Genet. 2007;72:420–426. doi: 10.1111/j.1399-0004.2007.00887.x. [DOI] [PubMed] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Weihl CC, Pestronk A, Kimonis VE. Valosin-containing protein disease: inclusion body myopathy with Paget’s disease of the bone and fronto-temporal dementia. Neuromuscul Disord. 2009;19:308–315. doi: 10.1016/j.nmd.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.