Abstract
The prevalence of diabetes mellitus is increasing dramatically throughout the world, and the disease has become a major public health issue. The most common form of the disease, type 2 diabetes, is characterized by insulin resistance and insufficient insulin production from the pancreatic beta-cell. Since glucose is the most potent regulator of beta-cell function under physiological conditions, identification of the insulin secretory defect underlying type 2 diabetes requires a better understanding of glucose regulation of human beta-cell function. To this aim, a bottom-up LC-MS/MS-based proteomics approach was used to profile pooled islets from multiple donors under basal (5 mM) or high (15 mM) glucose conditions. Our analysis discovered 256 differentially abundant proteins (~p<0.05) after 24 h of high glucose exposure from more than 4500 identified in total. Several novel glucose-regulated proteins were elevated under high glucose conditions, including regulators of mRNA splicing (Pleiotropic regulator 1), processing (Retinoblastoma binding protein 6), and function (Nuclear RNA export factor 1), in addition to Neuron navigator 1 and Plasminogen activator inhibitor 1. Proteins whose abundances markedly decreased during incubation at 15 mM glucose included Bax inhibitor 1 and Synaptotagmin-17. Up-regulation of Dicer 1 and SLC27A2 and down-regulation of Phospholipase Cβ4 were confirmed by Western blots. Many proteins found to be differentially abundant after high glucose stimulation are annotated as uncharacterized or hypothetical. These findings expand our knowledge of glucose regulation of the human islet proteome and suggest many hitherto unknown responses to glucose that require additional studies to explore novel functional roles.
Keywords: human, pancreatic islet, glucose, type 2 diabetes, proteomics, mass spectrometry, LC-MS/MS
INTRODUCTION
The application of proteomics and transcriptomics to characterize protein and gene expression on a global scale has considerable potential for understanding disease processes, including the pathogenesis of diabetes mellitus. Type 1 and type 2 diabetes are characterized by absolute or relative insulin deficiency, respectively. Insulin secretion from pancreatic beta cells of the islets of Langerhans plays a crucial role in glucose homeostasis, and impaired pancreatic islet function is key to the development of both forms of diabetes [1]. Under normal circumstances, the maintenance of blood glucose levels within a narrow physiological range relies on coordinated regulation of insulin secretion through nutrient availability, hormones, and neural inputs. Amongst these factors, glucose is by far the most potent and physiologically important regulator of beta-cell function through coordinate stimulation of insulin gene transcription, proinsulin biosynthesis, and insulin secretion from pancreatic beta-cells [2, 3]. Since type 2 diabetes is characterized by a loss of first-phase insulin secretion in response to glucose [4], identification of the molecular defects underlying beta-cell failure in type 2 diabetes first requires a better understanding of the normal regulation of beta-cell function by glucose. The advent of global ‘-omics’ technologies has enabled researchers to address this issue on a much larger scale than traditional approaches and has resulted in a number of studies examining pancreatic islet mRNA and protein content. Most of these studies have investigated basal -omics expression [5–10]; however, the effect of elevated glucose on rodent beta-cells has also been examined [11–14]. Two-dimensional (2-D) gel-based proteomics approaches have been used to identify proteins altered in response to high glucose exposure in mouse islets [11] and the rat insulin-producing INS-1E cell line [15]. These efforts identified a number of significant proteins illustrative of known biology and suggestive of new perspectives, but were limited by an inherent modest dynamic range. Advanced multi-dimensional liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomics technologies have demonstrated a wider dynamic range, higher sensitivity, and higher throughput for islet samples relative to 2-D gel-based methods [16, 17]. LC-MS/MS data from Waanders et al. [13] and Lu et al. [8] provided increased coverage of the mouse islet proteome during glucose stimulation, although extrapolation to human pancreatic islet biology is still limited. These comparative, glucose-stimulated islet proteomics studies revealed the need for a better understanding of glucose regulation at the molecular level and have prompted their application to the examination of protein abundances from human pancreatic donor islets under hyperglycemic conditions.
This work was undertaken to determine the effects of elevated glucose levels on the proteome of isolated human islets. To this aim, islets from multiple human pancreatic donors were incubated for 24 h at basal or high glucose levels and pooled into two groups representing each glucose concentration. Extracted protein samples were then fractionated in two dimensions (based upon solubility and charge) to increase the depth of proteome coverage, and peptides were analyzed by reversed-phase LC-MS/MS. The islet proteome was characterized for each culture condition, and spectral count data [18] was used to identify proteins differentially regulated by glucose. Over 40,000 unique peptides corresponding to more than 4500 proteins (≥2 unique peptide identifications per protein) were detected in these analyses; 256 were found differentially abundant (~p<0.05 by the G test statistic) [19]. Of these, the regulation of selected proteins was confirmed by Western blot. These findings revealed several novel glucose-regulated proteins and expand our understanding of glucose regulation of human islets.
MATERIALS AND METHODS
Reagents
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless stated otherwise. Ammonium bicarbonate and acetonitrile were purchased from Fisher Scientific (Pittsburgh, PA), and sequencing grade modified trypsin was purchased from Promega (Madison, WI). The Bicinchoninic acid (BCA) protein assay was purchased from Pierce (Rockford, IL), and purified, deionized water, >18 MΩ, (Nanopure Infinity ultrapure water system, Barnstead, Newton, WA) was used to make all aqueous biological and HPLC buffers.
Human pancreatic islets
Approval for the conduct of this programmatic research was obtained from the Institutional Review Boards of the participating institutions. Twenty-one batches of pancreatic human islets from non-diabetic donors were obtained through the NIH-supported Integrated Islet Distribution Program (http://www.iidp.coh.org), as well as from the Islet Transplant Program at the University of Alberta. The average age of the donors (13 females, 8 males) was 42 ± 3 years (range: 20–65 years), and their body mass index was 27.9 ± 1.6 (range: 20.2–48.2). The initial purity of the preparations was 77 ± 3 % and the viability was 84 ± 2 %, as assessed by the centers prior to shipment. Upon arrival, human islets from individual donor preparations were divided into two equivalent batches, washed with 1X with PBS, hand-picked, and cultured for 24 h at 37°C in RPMI-1640 (Invitrogen, Burlington, ON) containing 1% penicillin/streptomycin (Wisent, Saint Bruno, QC) and 1% human serum albumin (Hema-Québec, Saint Laurent, QC), in the presence of either 5 mM or 15 mM glucose (Sigma-Aldrich Canada Ltd., Oakville, ON) to examine the effects of glucose on protein expression (Figure 1). At the end of 24-h incubation, islets were centrifuged at 340 g for 2 min at 4°C and washed once with ice-cold 5 mM glucose in Optima water (Optima, Fisher Scientific, Pittsburgh, PA), transferred to siliconized tubes, and centrifuged at 3409 g for 2 min at 4°C. Pellets were snap-frozen and kept at −80 °C pending membrane and soluble protein extraction as described below.
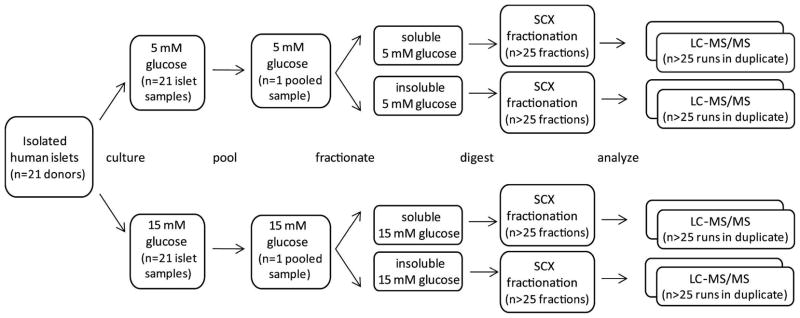
Figure 1.
Schematic showing the approach used to characterize the islet proteome under normal and high glucose conditions. Isolated human islets (handpicked from 21 pancreatic donors) were divided in half (by donor) and incubated for 24 h with either 5 mM or 15 mM glucose. Individual samples were pooled based on glucose concentration, cells were lysed, and proteins were separated based on solubility. Each of the resulting four samples was digested, and peptides were fractionated by strong cation exchange chromatography (SCX). All SCX fractions were analyzed in duplicate by reversed-phase LC-MS/MS (n = 217).
Protein extraction and enzymatic digestion
Thawed islets from 21 different donors were pooled to create uniform samples of ~13,000 total islet equivalents for each glucose concentration. Pooled islets from each glucose concentration were lysed for 30 min on ice in 5 mM sodium phosphate, pH 8.0, containing 1 mM EDTA, and soluble proteins were then isolated from membrane proteins by centrifugation at 16,000×g for 60 min at 4°C. The soluble proteins were transferred into a fresh tube, and the membrane proteins were reconstituted in 50 mM ammonium bicarbonate, pH 7.8. Protein concentrations for both soluble and membrane protein fractions were calculated by the BCA protein assay. Soluble and membrane proteins were next denatured by incubation with 50% 2,2,2-trifluoroethanol [20] at 60°C for 2 h with constant shaking at 300 rpm (Thermomixer R, Eppendorf, Hamburg, Germany). Sample reduction was achieved by 1 h incubation in 2 mM dithiothreitol at 37°C with constant shaking at 300 rpm. Denatured and reduced samples were then diluted 5-fold with 50 mM ammonium bicarbonate, pH 7.8, before tryptic digestion. Activated sequencing grade-modified trypsin was prepared by adding 20 μL of 50 mM ammonium bicarbonate, pH 7.8, to a vial containing 20 μg of lyophilized trypsin, followed by incubation for 10 min at 37°C. Activated trypsin was next added to the samples at 1:50 (w/w) trypsin-to-protein ratio, and tryptic digestion was carried out at 37°C for 3 h. Tryptic digestion was quenched by rapid freezing of the samples in liquid nitrogen. The samples were concentrated to 50 μL (Speed-Vac SC 250 Express, Thermo Savant, Holbrook, NY), then centrifuged at 88,760×g for 5 min at 4°C. The supernatants were placed into new tubes, and the BCA protein assay was performed to estimate final concentrations of peptide solutions.
Strong-cation exchange fractionation of enzymatic digests
Strong-cation exchange (SCX) fractionation of enzymatic digests was performed as previously described [16, 17]. Briefly, human pancreatic islet peptides were diluted with 850 μL of 10 mM ammonium formate (pH 3.0) in water containing 25% acetonitrile and fractionated by SCX chromatography on a Polysulfoethyl A 200 mm × 2.1 mm column (PolyLC, Columbia, MD) that was preceded by a 10 mm × 2.1 mm guard column of the same material. The separations were performed at a flow rate of 0.2 mL/min using an Agilent 1100 series HPLC system (Agilent Technologies, Santa Clara, CA), with mobile phases consisting of 10 mM ammonium formate, pH 3.0, in water containing 25% acetonitrile (A), and 500 mM ammonium formate, pH 6.8, in water containing 25% acetonitrile (B). After loading 350 μg of peptides onto the column, the gradient was maintained at 100% A for 10 min. Peptides were then separated using a gradient from 0 to 50% B over 40 min, followed by a gradient of 50–100% B over 10 min. The gradient was then held at 100% B for 20 min. A total of 30 fractions were collected from 30 to 65 min of the separation for pooled islets cultured in both normal and high glucose. All fractions were dried under vacuum and stored at −80°C until LC-MS/MS analysis.
Reversed-phase nanocapillary LC-MS/MS analyses
Dried peptide fractions were reconstituted in 30 μL of 25 mM ammonium bicarbonate, pH 7.8, and analyzed in duplicate and random order using a custom-built 4-column nanocapillary LC system coupled online to a linear ion trap mass spectrometer (LTQ; ThermoElectron, Waltham, MA) by way of an in-house manufactured electrospray ionization interface [21]. Electrospray emitters were custom made using 150 μm o.d. × 20 μm i.d. chemically etched fused silica, as previously described [22]. The reversed-phase capillary columns were prepared by slurry packing 3-μm Jupiter C18 bonded particles (Phenomenex, Torrence, CA) into 75 μm × 65 cm fused silica capillaries (Polymicro Technologies, Phoenix, AZ) using 0.5 cm sol-gel plugs for particle retention [23]. The mobile phase consisted of 0.2% acetic acid and 0.05% TFA in water (A) and 0.1% TFA in 90% acetonitrile/10% water (B). Mobile phases were degassed on-line using a Degassex Model DG4400 vacuum degasser (Phenomenex, Torrence, CA), and the HPLC system was equilibrated at 10,000 psi with 100% mobile phase A for initial starting conditions. After loading 2.5 μg of peptides onto the column, the mobile phase was held at 100% A for 50 min. Exponential gradient elution was performed by increasing the mobile-phase composition from 0 to 55% B over 100 min, using a 2.5 mL stainless steel mixing chamber, followed by a rapid increase to ~100% B for 10 min to wash the column. To identify the eluting peptides, the LTQ was operated in a data-dependent MS/MS mode (m/z 400–2,000), in which a full MS scan was followed by ten MS/MS scans using a normalized collision energy of 35%. A dynamic exclusion window of 1 min was used to discriminate against previously analyzed ions. The temperature of the heated capillary and the ESI voltage were 200 °C and 2.2 kV, respectively.
Peptide identification and protein categorization
RAW files for all datasets may be downloaded at http://omics.pnl.gov. Peak lists were generated using Extract MSN and SEQUEST peptide identification software [24] was used to match the MS/MS fragmentation spectra with sequences from the August 2006 IPI human database (version 3.20), containing 61,231 entries. The parent mass tolerance window used for matching was set to ±3 Da, and the fragment ion window was set to ±1 amu. A maximum of three missed cleavages were allowed. Protein identifications were retained if their identified peptide sequence met the following criteria: 1) SEQUEST DelCn2 value (normalized Xcorr difference between top scoring peptide and second highest scoring peptide in each MS/MS spectrum) of ≥ 0.10 and 2) SEQUEST correlation score (Xcorr) ≥ 2.4 for charge state 1+ for fully tryptic peptides and Xcorr ≥ 2.5 for 1+ for partially tryptic peptides; Xcorr ≥ 2.7 for charge state 2+ and fully tryptic peptides and Xcorr ≥ 3.7 for charge state 2+ and partially tryptic peptides; Xcorr ≥ 3.4 for charge state 3+ and fully tryptic peptides and Xcorr ≥ 4.2 for charge state 3+ and partially tryptic peptides. Proteins used in the analysis were required to have ≥ 2 unique peptides for identification. Using the reverse database approach [25], the false discovery rate (FDR) was estimated to be 1.7% at the spectra level and 0.2% at the protein level.
ProteinProphet [26] was used as previously described [16] to generate a set of proteins and protein groups from all peptide observations searched against the January 2009 IPI human database (version 3.54) in order to remove deleted entries. Protein ‘groups’ were assigned when peptides were degenerate (i.e., peptide sequences were present in homologous/redundant proteins within the protein sequence database). This analysis reduced the list of 20,829 possible proteins mapped to identified peptides to 4594 confident proteins/protein groups with two or more peptides per protein (Supplemental Table S1).
In cases where multiple proteins mapped to a single group, the Entrez ID corresponding to the protein with the largest number of unique peptides was chosen to be representative of the group for downstream analyses. An additional 1592 proteins/protein ‘groups’ were identified by only one unique peptide (Supplemental Table S2). These proteins were not included in any lists of detected genes for analysis (GO clustering or chart, or G-test), but were used in DAVID to generate background lists of possible genes for statistical calculations of GO terms (as described in Data Analysis below).
Western blots
At the end of 24-h incubation, batches of 200 islets were centrifuged at 1200 rpm for 2 min at 4°C and proteins were extracted as previously described [27]. Proteins (30 μg) were resolved by 7.5 or 10% SDS-PAGE and electrotransferred to nitrocellulose membrane (BioRad, Hercules, CA). After blocking with 5% nonfat milk TBS-T (10 mM Tris, pH 7.5, 150 mM NaCl, and 0.1% Tween 20) for 1h at room temperature, membranes were blotted overnight at 4°C with antibodies against Phospholipase Cβ4 (BD Transduction Laboratories, Mississauga, ON), SLC27A2 (Abcam Inc., Cambridge, MA), Dicer 1 (Abcam), or alpha Tubulin (Abcam). Signals were detected using a horseradish peroxidase-labeled anti-rabbit or anti-mouse IgG (BioRad) and enhanced chemiluminescence (ECL, PerkinElmer Las Canada Inc., Woodbridge, ON) on Kodak BioMax XAR films (Kodak, Rochester, NY). Molecular mass was estimated using protein molecular standards (Invitrogen).
Data analysis
Spectral counts were calculated as the sum of all MS/MS spectra for all peptides that map to each protein [18]. Percent coefficient of variance (CV) (%CV=σ/μ*100) values were calculated for replicates (e.g., total spectral counts for all SCX fractions for both membrane and soluble preparations for 15 mM (or 5 mM) replicate one versus replicate two). CV values of 0.00 occurred in cases where SCs for both replicates were identical nonzero values, and CV values reported as NA occurred when no spectra were identified in either replicate of either condition. Estimates of relative protein abundance were made by normalizing spectral counts by protein sequence length on an individual protein basis. For groups that included multiple proteins, the protein with the largest number of unique peptides was used in the calculation.
A G-test (likelihood ratio test for goodness-of-fit) [19, 28] was performed to determine proteins whose abundances changed with glucose concentration. Protein spectral counts were calculated as the sum of all observations for all peptides mapping to an individual protein for each glucose concentration. A global normalization was applied to equate the total spectral counts for all proteins for each sample, and a Yate’s correction was incorporated to generate the final values for normalized spectral counts [29]. Differentially expressed proteins were considered significant based on the G-value relative to the Chi-square distribution table with one degree of freedom (G>3.84 approximates p<0.05).
Proteins were classified with general Gene Ontology (GO) biological process terms using the GO Slim tool at the UniProtKB-GOA [30]. Entrez gene IDs were extracted for all proteins and a non-redundant list was uploaded for each analysis. The Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resource 6.7 (National Institute of Allergy and Infectious Diseases (NIAID), NIH) [31, 32] was used to classify and annotate proteins. For global KEGG pathway coverage, confidently identified proteins (≥2 unique peptides per protein) and the human genome were used as the gene list and background, respectively. The Functional Annotation Chart tool was used to determine KEGG pathways associated with imported identifiers. For clustering and chart analysis of proteins regulated by glucose, significant proteins (G>3.84) and all identified proteins (≥1 unique peptide per protein) were used as the gene list and background, respectively.
Clustering and visualization
Spectral count data were imported into OmniViz 6.0 (OmniViz, Maynard, MA). Counts were log2 transformed and normalized to the sum of the row (i.e., sum of spectral counts for all four columns, which included two replicates of each glucose concentration). Data were clustered using Hierarchical-standard clustering by magnitude and shape (Euclidean) with 18 clusters.
RESULTS
Proteome Coverage
Previous studies have defined comprehensive islet gene [6, 7, 9, 12] and protein [5, 8, 16, 17] expression patterns in both murine and human samples, yet the effect of elevated glucose on islet protein expression has, to date, been thoroughly examined only in mouse models [11, 13]. In the present study, a semi-quantitative proteomics approach was used to determine the human islet proteome under basal (5 mM) or elevated (15 mM) glucose conditions. Islet preparations from each of the 21 donors were divided in half, incubated with either 5 or 15 mM glucose for 24 h at 37 °C, and pooled into samples comprised of approximately 13,000 islets for each condition (summarized in Figure 1). Because islet proteins have been shown to have a large dynamic range [17], two levels of fractionation were used to increase the depth of proteome coverage. Proteins were separated into soluble and insoluble preparations and digested, and subsequent peptides were then fractionated using strong cation exchange (SCX). Duplicate LC-MS/MS analyses of SCX-fractionated samples resulted in the detection of 41,784 unique peptides corresponding to 4594 confident protein identifications having at least 2 unique peptides per protein. An additional 1592 proteins were identified by single peptides but were not included in any subsequent comparative data analysis. Detailed lists of protein identifications are provided as Supplemental Tables S1 and S2.
During LC-MS/MS analyses of complex protein mixtures, peptides derived from proteins of higher abundance are sampled more often than those from proteins of lower abundance [18]. Consequently, estimates of relative protein abundance can be made using spectral count information. Spectral counts refer to the sum of all MS/MS spectra for all peptides that map to individual proteins. Since higher counts would be expected for larger proteins, counts were normalized by protein sequence length on an individual protein basis. This approach was used to order the set of proteins found for the 5 mM glucose-exposed islet proteome by relative abundance (Supplemental Table S3). The most abundant proteins detected include histone 2A, glucagon, transthyretin, actin, and annexin A2.
Protein Classification
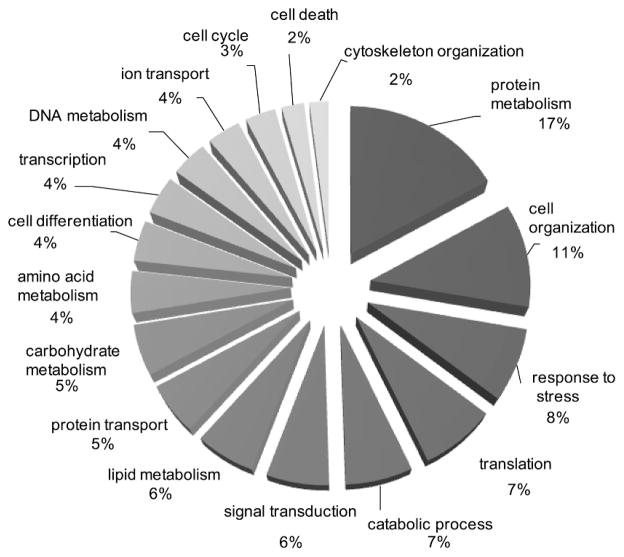
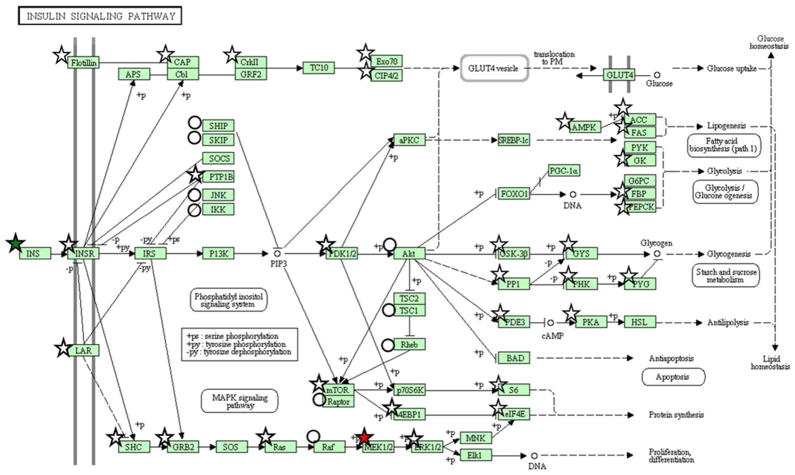
In order to extract biological processes associated with the islet proteome, proteins were classified using gene ontology (GO) categories. GO Slim [30] was used to group GO categories into general functional annotations as shown in Figure 2. Nearly one-third of the proteins play a role in protein (15%), lipid (5%), carbohydrate (5%), amino acid (5%), and DNA (3 %) metabolic processes, respectively. Proteins were also mapped to KEGG pathways to examine pathway coverage. Fifty-two pathways exhibited high coverage (≥10 proteins), including 10 of the 15 pathways associated with carbohydrate metabolism (Supplemental Table S4). Several members of the insulin signaling pathway were detected (Figure 3) and this pathway, interestingly, was modestly regulated by glucose. Other pathways exhibiting high coverage were associated with the processing of genetic information and included ‘transcription’ (e.g., spliceosome), ‘translation’ (e.g., ribosome and aminoacyl-tRNA biosynthesis), and ‘folding, sorting, and degradation’ (e.g., RNA degradation and proteasome).
Figure 2.
Functional classification of the human islet proteome. GO slim was used to group confidently identified proteins (≥2 peptides per protein) into general gene ontology biological process terms (http://www.ebi.ac.uk/QuickGO/).
Figure 3.
Proteome coverage of the KEGG ‘Insulin Signaling’ pathway. Proteins marked by stars and circles indicate identification by two or more unique peptides or a single unique peptide, respectively. Symbols are colored to indicate direction of observed abundance change (15 mM versus 5 mM) after 24 h of incubation based upon significance by the G-test statistic: red (up-regulation), green (down-regulation), or white (no change).
Differential Abundance
As an indication of the reproducibility of the LC-MS/MS analyses, peptides from 91% and 89% of all proteins identified in their respective 15 mM- and 5 mM-glucose-treated samples were detected in both replicates. The median coefficients of variance in spectral counts for the 4594 proteins identified by at least 2 unique peptides were 17% and 19% for proteins identified in the 15mM- and 5mM samples, respectively (Supplemental Table S5). A G-test [29] was performed on pairwise comparisons of normalized spectral counts from islets cultured in 5 mM glucose versus islets cultured in 15 mM glucose to define proteins with significant changes in protein abundance (Supplemental Table S5). Of the 4594 proteins identified, 256 proteins were determined to be differentially abundant (G> 3.84 ≈ p< 0.05) with 138 proteins exhibiting higher abundance in 15 mM glucose and 118 proteins showing lower abundance. Fold changes were calculated from the normalized spectral count data and indicated that the abundances of 14 and 33 of the significant proteins were changed by at least 10- and 5-fold, respectively, when the concentration of glucose was increased from 5 mM to 15 mM. Table 1 shows that while the numbers of spectral counts for these proteins are relatively low, they represent mostly qualitative differences in protein expression between the two conditions, rather than true quantitative differences. Many of the proteins detected in one condition were not detected at all (0 to 2 total spectral counts at most) in the other condition. The lone exception to this observation is 40s Ribosomal Protein s15, with 17 spectral counts for islets cultured in 5 mM glucose versus 142 spectral counts for islets cultured in 15 mM glucose.
Table 1.
Human islet proteome proteins with significant differential abundances (G>3.84) greater than 5-fold up- or down-regulated after 24h culture in 15 vs 5 mM glucose.
| Description (Protein IPI) | Entrez Gene Symbol | Raw Spectral Counts (5 mM) a | Raw Spectral Counts (15 mM)a | Biological Relevanceb | Function in Beta Cellsc |
|---|---|---|---|---|---|
| Isoform 1 of pleiotropic regulator 1 (IPI00002624.1) | PLRG1 | 0 | 12 | Regulator of cell development and apoptosis [48] | Unknown |
| Plasminogen activator inhibitor 1 (IPI00007118.1) | SERPINE1 | 0 | 9 | Pro-inflammatory cytokine. Knock-out mice are protected from high-fat diet-induced insulin resistance [49] | Unknown |
| Isoform 1 of retinoblastoma-binding protein 6 (IPI00337315.1) | RBBP6 | 0 | 8 | E3 ubiquitin-protein ligase; may function as negative regulator of p53/TP53 leading to both apoptosis and cell growth retardation [50] | Unknown |
| Nuclear RNA export factor 1 (IPI00033153.1) | NXF1 | 0 | 8 | Protein involved in nuclear RNA export [51] | Unknown |
| Isoform 3 of sodium bicarbonate cotransporter 3 (IPI00021058.1) | SLC4A7 | 0 | 8 | Regulates intracellular pH and may play a role in bicarbonate salvage in secretory epithelia. May also have an associated sodium channel activity [52, 53] | Unknown |
| Very long-chain acyl-coa synthetase (IPI00024787.1) | SLC27A2 | 0 | 7 | Plays a key role in lipid biosynthesis and fatty acid metabolism [54] | Expressed in pancreatic endocrine tumors [45] |
| Dicer1 (IPI00219036.5) | DICER1 | 0 | 7 | Ribonuclease that is required by the RNA interference pathway for repression of gene expression through destruction of complementary RNAs [55] | Knock-out in the mouse leads to agenesis of beta-cells [44] |
| Isoform 1 of craniofacial development protein 1 (IPI00007306.1) | CFDP1 | 0 | 7 | May play a role during embryogenesis [56]; SNP associated with type 1 diabetes [57, 58] | Unknown |
| Laminin alpha 2 subunit isoform b precursor (IPI00218725.3) | LAMA2 | 0 | 7 | Major component of the basement membrane [59]; SNP associated with type 1 and type 2 diabetes [57, 58, 60] | Unknown |
| Isoform 1 of huntingtin-interacting protein k (IPI00335001.2) | HYPK | 0 | 7 | Binds to huntingtin and prevents huntingtin-mediated apoptosis in neuronal cells [61] | Unknown |
| 28s ribosomal protein s32, mitochondrial (IPI00022320.1) | MRPL42 | 0 | 6 | Belongs to both the 28S and the 39S ribosomal subunits [62, 63] | Unknown |
| Gamma-aminobutyric acid receptor-associated protein-like 1 (IPI00027733.1) | GABARAPL1 | 0 | 6 | Increases cell-surface expression of kappa-type opioid receptor [64] | Unknown |
| Isoform 3 of cd99 antigen-like protein 2 (IPI00152488.1) | CD99L2 | 1 | 13 | Endothelial surface protein involved in neutrophil extravasation [65] | Unknown |
| 40s ribosomal protein s15 (IPI00479058.2) | RPS15 | 17 | 142 | Belongs to the 40S ribosomal subunit | Upregulated in insulinomas [66] |
| Isoform 4 of neuron navigator 1 (IPI00550839.1) | NAV1 | 1 | 12 | Sodium channel, thought to play a role in neuronal development and regeneration [42] | Unknown |
| Charged multivesicular body protein 7 (IPI00395463.3) | CHMP7 | 1 | 10 | Plays a role in the endosomal sorting pathway [67] | Unknown |
| NADP-dependent malic enzyme, mitochondrial (IPI00003970.2) | ME3 | 1 | 10 | Catalyzes the oxidative decarboxylation of malate to pyruvate [68]; SNP associated with type 1 diabetes [57, 58] | Decreased malic enzyme activity in type 2 diabetic islets [69] |
| Scy1-like protein 2 (IPI00396218.2) | SCYL2 | 1 | 10 | Component of AP2-containing clathrin coated structures at the plasma membrane or of endocytic coated vesicles [70] | Unknown |
| Aldehyde oxidase (IPI00029715.4) | AOX1 | 1 | 10 | Produces hydrogen peroxide and can catalyze the formation of superoxide | Unknown |
| Mesoderm development candidate 2 (IPI00399089.4) | MESDC2 | 1 | 9 | Chaperone specifically assisting the folding of beta-propeller/EGF modules within the family of low-density lipoprotein receptors [71, 72] | Unknown |
| Potassium channel tetramerisation domain containing 14 (IPI00181836.7) | KCTD14 | 1 | 9 | Inferred voltage-gated potassium channel activity | Unknown |
| Isoform 1 of regulator of microtubule dynamics protein 3 (IPI00410079.3) | FAM82A2 | 1 | 9 | Also known as PTPIP51. Interacts with PTP1B. May participate in differentiation and apoptosis of keratinocytes [72] | Unknown |
| Zinc finger protein 622 (IPI00056499.1) | ZNF622 | 1 | 9 | May behave as an activator of the bound transcription factor, MYBL2 [73]; SNP associated with type 2 diabetes [57, 58] | Unknown |
| Trafficking protein particle complex subunit 10 (IPI00298870.1) | TRAPPC10 | 8 | 1 | Part of the multi-subunit transport protein particle (TRAPP) complex; may be involved in vesicular transport from the endoplasmic reticulum to the Golgi [74] | Unknown |
| Isoform 1 of dnaj homolog subfamily c member 25 (IPI00027909.1) | DNAJC25 | 13 | 2 | Heat shock protein | Unknown |
| Comm domain-containing protein 8 (IPI00016447.5) | COMMD8 | 9 | 1 | Functional homologue of MURR1; Inhibits the association of NFκB with chromatin [75] | Unknown |
| Protein yipf4 (IPI00031127.1) | YIPF4 | 9 | 1 | Unknown | Unknown |
| Isoform 1 of uncharacterized protein c1orf71 (IPI00465154.4) | C1orf71 | 9 | 1 | Integral membrane protein that acts as a binding partner of connexins [76]; SNP associated with type 1 diabetes [57, 58] | Unknown |
| Phospholipase c beta 4 isoform a (IPI00783004.2) | PLCB4 | 10 | 1 | Catalyzes the formation of inositol 1,4,5-trisphosphate and diacylglycerol from phosphatidylinositol 4,5-bisphosphate; SNP associated with type 2 diabetes [57, 58] | Member of the phospholipase C beta family, involved in Gq-coupled GPCR signaling [77] |
| Bax inhibitor 1 (IPI00022748.4) | TMBIM6 | 6 | 0 | Suppressor of apoptosis [39] | Unknown |
| Proteasome subunit beta type-10 (IPI00027933.3) | PSMB10 | 6 | 0 | Member of the proteasome B-type family; a 20S core beta subunit | Unknown |
| Isoform 1 of tetratricopeptide repeat protein 21b (IPI00178386.4) | TTC21B | 6 | 0 | May negatively modulate sonic hedgehog signal transduction; may play a role in retrograde intraflagellar transport in cilia [78] | Unknown |
| Synaptotagmin-17 (IPI00305536.3) | SYT17 | 8 | 0 | Calcium binding protein. Plays a role in exocytosis by interacting with the SNARE complex [37] | Unknown |
Raw spectral count values. See Supplemental Table S5 for normalized spectral counts;
Biological relevancies obtained from UniProtKB summaries;
Function in beta cells deduced from a PubMed search using the terms “Gene and Islet” or “Gene and Beta-cell”
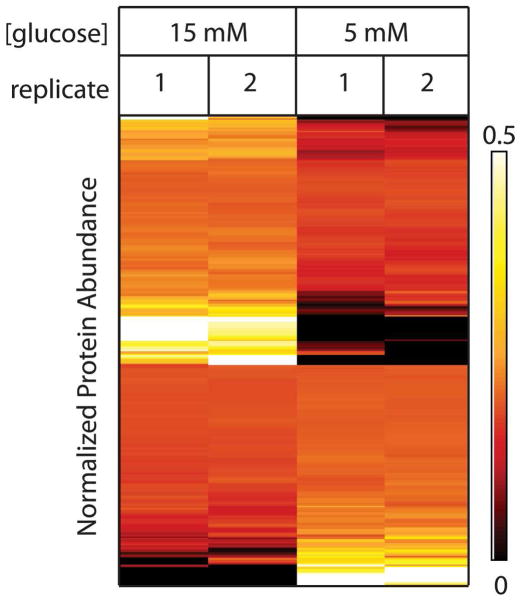
Figure 4 reflects the reproducibility of the measurements for the differentially abundant proteins with a heat map view showing proteins on rows and samples in columns. To remove the visual bias that occurs due to the large dynamic range within the proteome, protein abundance values were normalized for each protein individually by the sum of spectral counts for all samples and colored from black (protein was not observed or had a low abundance relative to the total abundance across all samples) to red to yellow to white (increasingly higher abundance relative to the total across all samples). Examples of proteins whose abundances were most affected by high glucose exposure included regulators of mRNA splicing (Pleiotropic Regulator 1), processing (Retinoblastoma Binding Protein 6) and function (Nuclear RNA Export Factor 1), in addition to Neuron Navigator 1 and Plasminogen Activator Inhibitor 1. Proteins whose abundances markedly decreased at 15 mM included Bax inhibitor 1 (BI-1) and Synaptotagmin-17.
Figure 4.
Clustering analysis of proteins with differential abundance shows reproducibility. Heat map representation of the data shows relative levels of differentially abundant proteins on each row with the cell color representing relative protein abundance [black (low abundance) to red to yellow to white (increasingly higher abundance)].
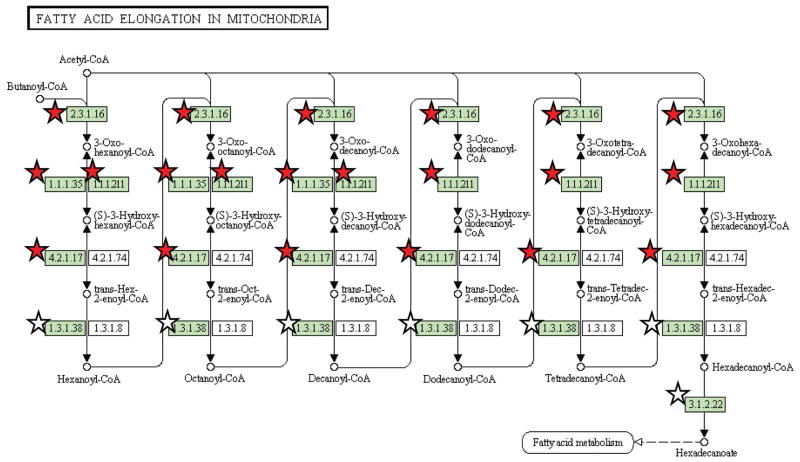
Similar to the functional analysis approach used for all confidently identified proteins, GO categories and KEGG pathway coverage were determined for all differentially abundant proteins. The web-based DAVID bioinformatics annotation tool [31, 32] was used to group the set of significant proteins into functional categories that are statistically over-represented relative to all islet proteins identified in the analysis. The clustering feature was used to combine functionally related annotation terms into clusters. Table 2 highlights a representative category from each of the significantly overrepresented clusters (Enrichment Score > 1.3 ≈ p < 0.05). Regulation of glucose metabolism, apoptosis, and macromolecular complex assembly are among the subset of the islet proteome most altered by 24 h of high glucose exposure. Full functional clustering is illustrated in more detail in Supplemental Table S6. DAVID was also used to examine the coverage of KEGG biological pathways by differentially abundant proteins. ‘Fatty acid elongation in mitochondria’ was identified as having significant coverage with three of the eight proteins associated with this pathway (Figure 5) having higher abundances at the elevated glucose level.
Table 2.
Significant gene ontology (GO) biological process and molecular function terms associated with differentially abundant proteins after 24h culture in 15 versus 5 mM glucose.
| GO Term | Enrichment Score | Protein Count | % of All Differentially Abundant Proteins Within GO Term | |
|---|---|---|---|---|
| GO:005125 | protein polymerization | 2.98 | 9 | 3.7 |
| GO:003462 | cellular macromolecular complex | 2.19 | 16 | 6.5 |
| GO:001646 | pyrophosphatase activity | 1.85 | 29 | 11.8 |
| GO:004616 | alcohol catabolic process | 1.77 | 8 | 3.3 |
| GO:002200 | neurogenesis | 1.68 | 15 | 6.1 |
| GO:004298 | regulation of apoptosis | 1.55 | 23 | 9.4 |
| GO:000600 | glucose metabolic process | 1.40 | 10 | 4.1 |
Figure 5.
Proteome coverage of the KEGG ‘Fatty acid elongation in mitochondria’ pathway. Stars represent proteins identified by two or more unique peptides. Symbols colored red indicate proteins deemed to have significant differential abundance (by G test statistic) after 24 h of high glucose incubation.
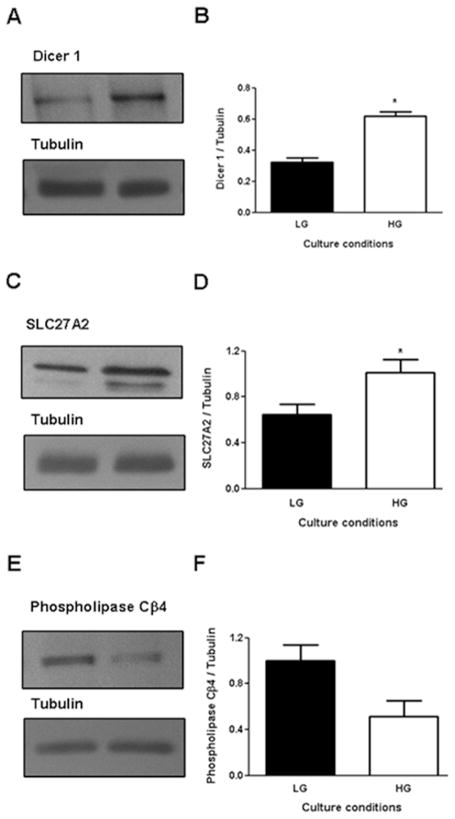
The regulation of expression by glucose was examined using Western blots for selected proteins (Figure 6). We confirmed that a 24-h exposure of human islets to 15 mM glucose increases protein expression of Dicer 1 (Figure 6A&B) and SLC27A2 (Figure 6C&D) and decreases expression of Phospholipase Cβ4 (Figure 6D&F).
Figure 6.
Glucose-regulated expression of selected proteins in human islets by Western blot. Human islets from individual donors were cultured at 5 mM (low glucose, LG) or 15 mM (high glucose, HG) glucose for 24h and protein extracts were examined by immunoblotting using specific antibodies. Representative immunoblots (A, C, E) and quantification of 3–4 replicate experiments (B, D, F) of Dicer 1 (A, B), SLC27A2 (C, D), Phospholipase Cb4 (E, F) are shown. Tubulin was used as a control for protein loading.
DISCUSSION
This study was initiated on the premise that a better understanding of glucose regulation can be elucidated from the transcriptional and translational responses of islets to glucose levels, and that this knowledge can be applied to benefit islet transplantation and novel diabetes therapies. The highly coordinated regulation of gene and protein expression in response to glucose stimulation is responsible for many established cellular functions such as glycolysis and insulin biosynthesis/secretion, but also for many ambiguous or unknown responses. A number of studies have profiled the comprehensive pancreatic islet transcriptome and proteome, yet the effect of elevated glucose has not been examined in proteomics analyses of human islets. The current study highlights changes in proteomic abundance after 24 h of 5 or 15 mM glucose incubation. To augment protein identification across the large dynamic range of the islet proteome [17], extensive separation was performed at both the protein and subsequent peptide levels. This approach resulted in the confident identification of nearly 4600 proteins/protein groups, of which 256 exhibited altered protein abundance with glucose stimulation.
A majority of proteins identified in this study have also been characterized in previously published islet proteomic profiles [13, 16, 17]. These include the islet hormones insulin, glucagon, somatostatin, and pancreatic polypeptide, in addition to most glycolytic proteins (Supplemental Figure S1). Transthyretin, glucagon, and annexin A2, some of the most abundant proteins identified in this study, were also found to be abundantly expressed at the mRNA level in several islet gene expression datasets available at http://T1Dbase.org [33].
Of the 4594 proteins identified in the current study, 81% were identified in at least one other previous LC-MS/MS study [13, 16, 17]. Comparison of protein abundance levels (based on spectral counts or peptide signal intensities) from the current study with murine islet protein expression data [13, 17] suggests a correlation of protein expression between species. We refer here to relative, not absolute, abundance; however, measures of high or low abundance proteins can still be inferred. Of the top 10% most abundant proteins identified after 24 h of 5 mM glucose incubation, 85% were identified in at least one murine study. In all three studies, glucagon (GCG), ATP synthase subunit beta, mitochondrial (ATP5B), protein disulfide isomerase A3 (PDIA3), protein disulfide-isomerase (P4HB), 78 kDa glucose-regulated protein precursor (HSPA5), elongation factor 1-alpha 1 (EEF1A1), chromogranin A (CHGA), and 14-3-3 protein epsilon (YWHAE) were among the most abundant proteins detected.
Table 1 summarizes the proteins that were observed to be regulated greater than five-fold in this study. The relatively low spectral counts for these proteins are likely a reflection of their abundance within the islets. Two previously published papers describing multi-dimensional LC-MS/MS analyses of islets, Metz et al. [16] and Petyuk et al. [17], reported the identification of 29,021 (3365 unique proteins) and 17,350 (2612 unique proteins) unique peptides for human and mouse islets, respectively. Of the differentially expressed proteins shown in Table 1, Metz et al. identified fourteen and Petyuk et al identified twelve, with 1 to 2 spectral counts of up to 3 unique peptides per protein observed in either study, indicating that these proteins are present in relatively low abundance in both human and mouse islets. Importantly, the studies reported by Metz et al. and Petyuk et al. used identical protein digestion, SCX fractionation, and capillary LC-MS/MS analysis protocols to the study reported here, enabling direct comparison of the results. The current study likely achieved higher coverage (in terms of both numbers of unique peptides and spectral counts) of these proteins due to the additional fractionation of the islet proteome into soluble and membrane proteins.
Examination of the annotations for the differentially regulated proteins and functional clusters identified mRNA processing and function, macromolecular complex assembly, and protein polymerization as being highly responsive to glucose. These findings are consistent with the notion that glucose is a major regulator of transcription and translation in pancreatic beta-cells, an effect that is necessary for the long-term maintenance of the highly differentiated state of the cell and the secretory requirements imposed by prolonged elevations of glucose levels [34]. In addition, considering that the beta-cell is highly metabolically active and that insulin secretion is tightly coupled to glucose metabolism, it is not surprising that the most highly glucose-regulated proteins in this study are implicated in glucose metabolism. Consistent with this notion, the data suggest that glucose promotes the coordinated expression of enzymes involved in fatty-acid synthesis and, more specifically, fatty-acid elongation. Although glucose is known to promote fatty-acid esterification and triglyceride synthesis in beta-cells [35], its effect on de novo fatty acid synthesis is thought to be minimal, at least in rodent beta-cells, due to low levels of expression of fatty-acid synthase [36]. The indication that glucose stimulates the expression of enzymes involved in fatty-acid elongation suggests that perhaps the rate of de novo fatty-acid synthesis might be higher in human than rodent beta-cells, although this would need to be directly measured. Importantly, the effect of glucose on protein expression was relatively selective and did not reflect a global stimulation of translation, as illustrated by the fact that most of the proteins in the insulin signaling pathway were not differentially regulated (Figure 3).
Perhaps the most important observation of this study is the identification of several novel glucose-regulated proteins. Amongst those that were most profoundly reduced in expression in the presence of high glucose was synaptotagmin 17. Synaptotagmins are calcium-binding proteins that interact with proteins of the SNARE (soluble N-ethylmaleimide–sensitive factor attachment protein receptor) complex, and this interaction plays an essential role in secretory granule membrane fusion [37]. Although synaptotagmin 17 protein expression has not been reported in beta-cells, it is noteworthy that mRNA expression of another gene from the synaptotagmin family, synaptotagmin V, is down-regulated in islets from type 2 diabetic patients [38]. Consistent with this observation is the finding that insulin was expressed at lower levels in islets cultured in high glucose, as were several members of the tubulin family (tubulins alpha 1a, alpha 4a, alpha 8, beta 2a, beta 2b, beta 3, beta 5, and beta 6; Supplemental Table S5). These results correlate with findings in islets [8, 13] and insulin-secreting INS1 cells [15] that show decreases of both insulin and vesicle-transport related protein levels [8, 13] suggesting that exposure of islets to elevated glucose for even a relatively short period of time in culture (24 h) might down-regulate the expression of proteins involved in insulin secretion. Interestingly, altered regulation of cytoskeletal proteins have also been observed in non-human species: tubulin alpha 1c [11] and beta 5 [8, 11] were up-regulated and tubulin 2a was down-regulated by glucose in mouse islets, and, in a recent report in INS1 cells, tubulin beta 2c was down-regulated by glucose [15], suggesting potentially important differences in tubulin chains among species.
Another protein whose expression was markedly decreased by glucose is BI-1. BI-1 is an evolutionarily conserved endoplasmic reticulum-associated protein which protects against endoplasmic reticulum stress and apoptosis [39], a recognized mechanism of glucose-mediated beta cell dysfunction. Although the function of BI-1 in the beta-cell is, to our knowledge, unknown, a decrease in expression upon culture at 15 mM glucose would be anticipated to lead to increased apoptosis. We did not, however, observe any significant differences in cell viability between the culture conditions (data not shown).
Amongst the proteins whose expression were the most significantly augmented in the presence of glucose but which, to our knowledge, have not been described in islets are: PLRG1, a protein involved in the regulation of mRNA splicing [40]; the retinoblastoma tumor suppressor RBP6; NXF1, a regulator of mRNA nuclear export [41]; NAV1, a protein of the neuron navigator family expressed predominantly in the nervous system [42]; the sodium bicarbonate cotransporter SLC4A7; laminin alpha 2 (LAMA2); and protein tyrosine phosphatase interacting protein 51 (PTPIP51), which interacts with PTP1B and plays a role in cell differentiation and apoptosis [43]. Also strongly stimulated by glucose (and confirmed by Western blot, Figure 6) are Dicer 1, an enzyme involved in microRNA processing, whose gene deletion leads to beta-cell agenesis [44], and the fatty-acid transporter SLC27A2, which is overexpressed in pancreatic endocrine neoplasms [45].
Dicer 1 is an RNA-specific endonuclease which is required for the synthesis of micro RNAs (miRNAs). Conditional deletion of Dicer 1 in the pancreas profoundly alters endocrine cell development, particularly beta-cells [44]. While to our knowledge the role of Dicer 1 in adult beta-cell function has not been directly examined, miRNAs such as miR-375 have been shown to play a role in insulin secretion [46]. Therefore, it is conceivable that glucose upregulation of Dicer 1 expression in human islets contributes to the biogenesis of miRNAs that are important to enhance insulin secretion in response to elevated glucose levels.
This study has identified a number of glucose-regulated proteins whose functions are presently unknown in beta-cells, and paves the way for additional studies to explore their functional roles. For example, the marked reduction in synaptotagmin 17, as well as the coordinated reduction in several members of the tubulin family in response to glucose, might suggest an additional mechanism by which glucose toxicity impairs insulin exocytosis in human islets [47]. Also, confirmation of the functional importance of the fatty-acid transporter SLC27A2, which was strongly upregulated by glucose in this study, would shed light on the debated mechanisms of fatty-acid uptake and transport in beta cells.
Global identification and quantification of human islet proteins by two-dimensional fractionation and reversed phase LC-MS/MS has enabled deep coverage of the islet proteome and identification of novel glucose-regulated proteins. Since the LC-MS/MS analyses were conducted on peptides from pooled islet samples, the identified glucose-regulated proteins likely represent a conservative group and the reproducibility of the findings across individual islet preparations remains to be confirmed. Nevertheless, these results suggest several targets for future efforts focused on a better understanding of the glucose regulation of human beta-cell function and the molecular defects underlying beta-cell dysfunction in type 2 diabetes mellitus.
Supplementary Material
Acknowledgments
The authors wish to thank Ms. Melissa M. Matzke and Dr. Samuel H. Payne of Pacific Northwest National Laboratory (PNNL) for helpful discussions. This research was supported by NIDDK grant DK070146. Portions of this research were supported by the National Institute of Allergy and Infectious Diseases NIH/DHHS through Interagency agreement Y1-AI-8401. The work utilized proteomics capabilities developed under support from the U.S. Department of Energy (DOE) Office of Biological and Environmental Research and the NIH National Center for Research Resources (grant RR018522), and was performed in the Environmental Molecular Sciences Laboratory, a DOE national scientific user facility at PNNL. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under contract DE-AC05-76RLO 1830. Isolated human islets were provided by the NIH-supported Integrated Islet Distribution Program and by the Islet Transplant Program at the University of Alberta. VP holds the Canada Research Chair in Diabetes and Pancreatic Beta-Cell Function. GF was supported by a post-doctoral fellowship from the Canadian Diabetes Association.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supplemental Tables (S1–S6) are available showing protein identifications having at least two peptides per protein (Table S1) or one peptide per protein (Table S2), and illustrating relative protein abundances (Table S3), significant KEGG pathways (Table S4), calculated G-test statistics (Table S5), and gene ontology (GO) functional clustering (Table S6). Supplemental Figure S1 shows protein identifications (>= 2 unique peptides/protein) mapped to the KEGG glycolysis pathway using the web-based DAVID bioinformatics tool.
References
- 1.Prentki M, Nolan CJ. Islet beta cell failure in type 2 diabetes. J Clin Invest. 2006;116(7):1802–12. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poitout V, Stein R, Rhodes CJ. Insulin Gene Expression and Biosynthesis. In: DeFronzo R, et al., editors. International Textbook of Diabetes Mellitus. John Wiley & Sons; Chichester: 2004. pp. 98–123. [Google Scholar]
- 3.Henquin JC, Dufrane D, Nenquin M. Nutrient control of insulin secretion in isolated normal human islets. Diabetes. 2006;55(12):3470–7. doi: 10.2337/db06-0868. [DOI] [PubMed] [Google Scholar]
- 4.Brunzell JD, et al. Relationships between fasting plasma glucose levels and insulin secretion during intravenous glucose tolerance tests. J Clin Endocrinol Metab. 1976;42:222–229. doi: 10.1210/jcem-42-2-222. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed M, Forsberg J, Bergsten P. Protein profiling of human pancreatic islets by two-dimensional gel electrophoresis and mass spectrometry. J Proteome Res. 2005;4(3):931–40. doi: 10.1021/pr050024a. [DOI] [PubMed] [Google Scholar]
- 6.Cras-Meneur C, et al. An expression profile of human pancreatic islet mRNAs by Serial Analysis of Gene Expression (SAGE) Diabetologia. 2004;47(2):284–99. doi: 10.1007/s00125-003-1300-8. [DOI] [PubMed] [Google Scholar]
- 7.Hui H, et al. Gene expression profiling of cultured human islet preparations. Diabetes Technol Ther. 2004;6(4):481–92. doi: 10.1089/1520915041705866. [DOI] [PubMed] [Google Scholar]
- 8.Lu H, et al. The identification of potential factors associated with the development of type 2 diabetes: a quantitative proteomics approach. Mol Cell Proteomics. 2008;7(8):1434–51. doi: 10.1074/mcp.M700478-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marselli L, et al. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab. 2008;93(3):1046–53. doi: 10.1210/jc.2007-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolls MR, et al. Proteomics as a tool for discovery: proteins implicated in Alzheimer’s disease are highly expressed in normal pancreatic islets. J Proteome Res. 2003;2(2):199–205. doi: 10.1021/pr025576x. [DOI] [PubMed] [Google Scholar]
- 11.Ahmed M, Bergsten P. Glucose-induced changes of multiple mouse islet proteins analysed by two-dimensional gel electrophoresis and mass spectrometry. Diabetologia. 2005;48(3):477–85. doi: 10.1007/s00125-004-1661-7. [DOI] [PubMed] [Google Scholar]
- 12.Shalev A, et al. Oligonucleotide Microarray Analysis of Intact Human Pancreatic Islets: Identification of Glucose-Responsive Genes and a Highly Regulated TGF{beta} Signaling Pathway. Endocrinology. 2002;143(9):3695–8. doi: 10.1210/en.2002-220564. [DOI] [PubMed] [Google Scholar]
- 13.Waanders LF, et al. Quantitative proteomic analysis of single pancreatic islets. Proc Natl Acad Sci U S A. 2009;106(45):18902–7. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bensellam M, et al. Cluster analysis of rat pancreatic islet gene mRNA levels after culture in low-, intermediate- and high-glucose concentrations. Diabetologia. 2009;52(3):463–76. doi: 10.1007/s00125-008-1245-z. [DOI] [PubMed] [Google Scholar]
- 15.Maris M, et al. High Glucose Induces Dysfunction in Insulin Secretory Cells by Different Pathways: A Proteomic Approach. J Proteome Res. 2010;9(12):6274–6287. doi: 10.1021/pr100557w. [DOI] [PubMed] [Google Scholar]
- 16.Metz TO, et al. Characterization of the human pancreatic islet proteome by two-dimensional LC/MS/MS. J Proteome Res. 2006;5(12):3345–54. doi: 10.1021/pr060322n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petyuk VA, et al. Characterization of the mouse pancreatic islet proteome and comparative analysis with other mouse tissues. J Proteome Res. 2008;7(8):3114–26. doi: 10.1021/pr800205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Sadygov RG, Yates JR. A Model for Random Sampling and Estimation of Relative Protein Abundance in Shotgun Proteomics. Analytical Chemistry. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 19.Sokal RR, Rohlf FJ. Biometry: the Principles and Practice of Statistics in Biological Research. 3. New York: W. H. Freeman and Co; 1995. [Google Scholar]
- 20.Wang H, et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4(6):2397–403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livesay EA, et al. Fully automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal Chem. 2008;80(1):294–302. doi: 10.1021/ac701727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly RT, et al. Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal Chem. 2006;78(22):7796–801. doi: 10.1021/ac061133r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiolica A, Borsotti D, Rappsilber J. Self-made frits for nanoscale columns in proteomics. Proteomics. 2005;5(15):3847–50. doi: 10.1002/pmic.200402010. [DOI] [PubMed] [Google Scholar]
- 24.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 25.Qian WJ, et al. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2005;4(1):53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 26.Nesvizhskii AI, et al. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 27.Hagman DK, et al. Palmitate inhibits insulin gene expression by altering PDX-1 nuclear localization and reducing MafA expression in isolated rat islets of Langerhans. J Biol Chem. 2005;280(37):32413–8. doi: 10.1074/jbc.M506000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Old WM, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4(10):1487–502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Ambatipudi KS, et al. Quantitative analysis of age specific variation in the abundance of human female parotid salivary proteins. J Proteome Res. 2009;8(11):5093–102. doi: 10.1021/pr900478h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrell D, et al. The GOA database in 2009--an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37(Database issue):D396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Kutlu B, et al. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuit F, et al. Glucose-regulated gene expression maintaining the glucose-responsive state of beta-cells. Diabetes. 2002;51(Suppl 3):S326–32. doi: 10.2337/diabetes.51.2007.s326. [DOI] [PubMed] [Google Scholar]
- 35.Briaud I, et al. Lipotoxicity of the pancreatic beta-cell is associated with glucose-dependent esterification of fatty acids into neutral lipids. Diabetes. 2001;50(2):315–21. doi: 10.2337/diabetes.50.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brun T, et al. Evidence for an anaplerotic/malonyl-CoA pathway in pancreatic beta-cell nutrient signaling. Diabetes. 1996;45(2):190–8. doi: 10.2337/diab.45.2.190. [DOI] [PubMed] [Google Scholar]
- 37.Gauthier BR, Wollheim CB. Synaptotagmins bind calcium to release insulin. Am J Physiol Endocrinol Metab. 2008;295(6):E1279–86. doi: 10.1152/ajpendo.90568.2008. [DOI] [PubMed] [Google Scholar]
- 38.Ostenson CG, et al. Impaired gene and protein expression of exocytotic soluble N-ethylmaleimide attachment protein receptor complex proteins in pancreatic islets of type 2 diabetic patients. Diabetes. 2006;55(2):435–40. doi: 10.2337/diabetes.55.02.06.db04-1575. [DOI] [PubMed] [Google Scholar]
- 39.Chae HJ, et al. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol Cell. 2004;15(3):355–66. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 40.Lleres D, et al. Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice. EMBO Rep. 2010;11(6):445–51. doi: 10.1038/embor.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erkmann JA, Kutay U. Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp Cell Res. 2004;296(1):12–20. doi: 10.1016/j.yexcr.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 42.Maes T, Barcelo A, Buesa C. Neuron navigator: a human gene family with homology to unc-53, a cell guidance gene from Caenorhabditis elegans. Genomics. 2002;80(1):21–30. doi: 10.1006/geno.2002.6799. [DOI] [PubMed] [Google Scholar]
- 43.Stenzinger A, et al. Cell and molecular biology of the novel protein tyrosine-phosphatase-interacting protein 51. Int Rev Cell Mol Biol. 2009;275:183–246. doi: 10.1016/S1937-6448(09)75006-3. [DOI] [PubMed] [Google Scholar]
- 44.Lynn FC, et al. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56(12):2938–45. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 45.Hansel DE, et al. Met proto-oncogene and insulin-like growth factor binding protein 3 overexpression correlates with metastatic ability in well-differentiated pancreatic endocrine neoplasms. Clin Cancer Res. 2004;10(18 Pt 1):6152–8. doi: 10.1158/1078-0432.CCR-04-0285. [DOI] [PubMed] [Google Scholar]
- 46.Poy MN, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–30. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 47.Dubois M, et al. Glucotoxicity inhibits late steps of insulin exocytosis. Endocrinology. 2007;148(4):1605–14. doi: 10.1210/en.2006-1022. [DOI] [PubMed] [Google Scholar]
- 48.Kleinridders A, et al. PLRG1 is an essential regulator of cell proliferation and apoptosis during vertebrate development and tissue homeostasis. Mol Cell Biol. 2009;29(11):3173–85. doi: 10.1128/MCB.01807-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma LJ, et al. Prevention of obesity and insulin resistance in mice lacking plasminogen activator inhibitor 1. Diabetes. 2004;53(2):336–46. doi: 10.2337/diabetes.53.2.336. [DOI] [PubMed] [Google Scholar]
- 50.Li L, et al. PACT is a negative regulator of p53 and essential for cell growth and embryonic development. Proc Natl Acad Sci U S A. 2007;104(19):7951–6. doi: 10.1073/pnas.0701916104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grüter P, et al. TAP, the Human Homolog of Mex67p, Mediates CTE-Dependent RNA Export from the Nucleus. Molecular Cell. 1998;1(5):649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 52.Pushkin A, et al. Cloning, tissue distribution, genomic organization, and functional characterization of NBC3, a new member of the sodium bicarbonate cotransporter family. J Biol Chem. 1999;274(23):16569–75. doi: 10.1074/jbc.274.23.16569. [DOI] [PubMed] [Google Scholar]
- 53.Park M, et al. The cystic fibrosis transmembrane conductance regulator interacts with and regulates the activity of the HCO3- salvage transporter human Na+-HCO3- cotransport isoform 3. J Biol Chem. 2002;277(52):50503–9. doi: 10.1074/jbc.M201862200. [DOI] [PubMed] [Google Scholar]
- 54.Mihalik SJ, et al. Participation of two members of the very long-chain acyl-CoA synthetase family in bile acid synthesis and recycling. J Biol Chem. 2002;277(27):24771–9. doi: 10.1074/jbc.M203295200. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, et al. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118(1):57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 56.Diekwisch TG, et al. Cloning, gene expression, and characterization of CP27, a novel gene in mouse embryogenesis. Gene. 1999;235(1–2):19–30. doi: 10.1016/s0378-1119(99)00220-6. [DOI] [PubMed] [Google Scholar]
- 57.The_Wellcome_Trust_Case_Control_Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson AD, O’Donnell CJ. An open access database of genome-wide association results. BMC Med Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ehrig K, et al. Merosin, a tissue-specific basement membrane protein, is a laminin-like protein. Proc Natl Acad Sci U S A. 1990;87(9):3264–8. doi: 10.1073/pnas.87.9.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 61.Raychaudhuri S, et al. HYPK, a Huntingtin interacting protein, reduces aggregates and apoptosis induced by N-terminal Huntingtin with 40 glutamines in Neuro2a cells and exhibits chaperone-like activity. Hum Mol Genet. 2008;17(2):240–55. doi: 10.1093/hmg/ddm301. [DOI] [PubMed] [Google Scholar]
- 62.Koc EC, et al. Identification of four proteins from the small subunit of the mammalian mitochondrial ribosome using a proteomics approach. Protein Sci. 2001;10(3):471–81. doi: 10.1110/ps.35301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koc EC, et al. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem. 2001;276(47):43958–69. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 64.Chen C, et al. GEC1 interacts with the kappa opioid receptor and enhances expression of the receptor. J Biol Chem. 2006;281(12):7983–93. doi: 10.1074/jbc.M509805200. [DOI] [PubMed] [Google Scholar]
- 65.Bixel MG, et al. A CD99-related antigen on endothelial cells mediates neutrophil but not lymphocyte extravasation in vivo. Blood. 2007;109(12):5327–36. doi: 10.1182/blood-2006-08-043109. [DOI] [PubMed] [Google Scholar]
- 66.Shiga K, Yamamoto H, Okamoto H. Isolation and characterization of the human homologue of rig and its pseudogenes: the functional gene has features characteristic of housekeeping genes. Proc Natl Acad Sci U S A. 1990;87(9):3594–8. doi: 10.1073/pnas.87.9.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horii M, et al. CHMP7, a novel ESCRT-III-related protein, associates with CHMP4b and functions in the endosomal sorting pathway. Biochem J. 2006;400(1):23–32. doi: 10.1042/BJ20060897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loeber G, Maurer-Fogy I, Schwendenwein R. Purification, cDNA cloning and heterologous expression of the human mitochondrial NADP(+)-dependent malic enzyme. Biochem J. 1994;304(Pt 3):687–92. doi: 10.1042/bj3040687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.MacDonald MJ, et al. Decreased levels of metabolic enzymes in pancreatic islets of patients with type 2 diabetes. Diabetologia. 2009;52(6):1087–91. doi: 10.1007/s00125-009-1319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duwel M, Ungewickell EJ. Clathrin-dependent association of CVAK104 with endosomes and the trans-Golgi network. Mol Biol Cell. 2006;17(10):4513–25. doi: 10.1091/mbc.E06-05-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Culi J, Springer TA, Mann RS. Boca-dependent maturation of beta-propeller/EGF modules in low-density lipoprotein receptor proteins. EMBO J. 2004;23(6):1372–80. doi: 10.1038/sj.emboj.7600132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lv BF, et al. Protein tyrosine phosphatase interacting protein 51 (PTPIP51) is a novel mitochondria protein with an N-terminal mitochondrial targeting sequence and induces apoptosis. Apoptosis. 2006;11(9):1489–501. doi: 10.1007/s10495-006-8882-9. [DOI] [PubMed] [Google Scholar]
- 73.Seong HA, Kim KT, Ha H. Enhancement of B-MYB transcriptional activity by ZPR9, a novel zinc finger protein. J Biol Chem. 2003;278(11):9655–62. doi: 10.1074/jbc.m207478200. [DOI] [PubMed] [Google Scholar]
- 74.Barrowman J, et al. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat Rev Mol Cell Biol. 2010;11(11):759–63. doi: 10.1038/nrm2999. [DOI] [PubMed] [Google Scholar]
- 75.Burstein E, et al. COMMD proteins, a novel family of structural and functional homologs of MURR1. J Biol Chem. 2005;280(23):22222–32. doi: 10.1074/jbc.M501928200. [DOI] [PubMed] [Google Scholar]
- 76.del Castillo FJ, et al. Consortin, a trans-Golgi network cargo receptor for the plasma membrane targeting and recycling of connexins. Hum Mol Genet. 2010;19(2):262–75. doi: 10.1093/hmg/ddp490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Winzell MS, Ahren B. G-protein-coupled receptors and islet function-implications for treatment of type 2 diabetes. Pharmacol Ther. 2007;116(3):437–48. doi: 10.1016/j.pharmthera.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 78.Tran PV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 2008;40(4):403–10. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.