Summary
Hfq is an RNA-binding protein that functions in post-transcriptional gene regulation by mediating interactions between mRNAs and small regulatory RNAs (sRNAs). Two proteins encoded by BAB1_1794 and BAB2_0612 are highly overproduced in a Brucella abortus hfq mutant compared to the parental strain, and recently, expression of orthologs of these proteins in Agrobacterium tumefaciens was shown to be regulated by two sRNAs, called AbcR1 and AbcR2. Orthologous sRNAs (likewise designated AbcR1 and AbcR2) have been identified in B. abortus 2308. In Brucella, abcR1 and abcR2 single mutants are not defective in their ability to survive in cultured murine macrophages, but an abcR1 abcR2 Sdouble mutant exhibits significant attenuation in macrophages. Additionally, the abcR1 abcR2 double mutant displays significant attenuation in a mouse model of chronic Brucella infection. Quantitative proteomics and microarray analyses revealed that the AbcR sRNAs predominantly regulate genes predicted to be involved in amino acid and polyamine transport and metabolism, and Northern blot analyses indicate that the AbcR sRNAs accelerate the degradation of the target mRNAs. In an E. coli two-plasmid reporter system, over-expression of either AbcR1 or AbcR2 was sufficient for regulation of target mRNAs, indicating that the AbcR sRNAs from Brucella abortus 2308 perform redundant regulatory functions.
Introduction
Small regulatory RNAs (sRNAs) play critical roles in bacterial gene regulation (Gottesman, 2005; Storz et al., 2011). These small molecules are typically 50–500 nucleotides in length and are often encoded in the intergenic regions between protein-encoding genes (Altuvia, 2007). Additionally, sRNAs are usually encoded in a different location from the genes they regulate; however, it is becoming increasingly clear that bacteria also encode an abundance of cis-acting sRNAs (Liu and Camilli, 2010). In terms of gene regulation, sRNAs can affect gene expression both positively and negatively. sRNAs can bind to the 5′-untranslated region (UTR) of target mRNAs, and these interactions alleviate secondary structure in the ribosome-binding site (RBS) region of the mRNA and allow translation to proceed. Additionally, a sRNA can bind to the 3′ end of the mRNA, stabilizing the transcript and allowing for increased gene expression (Fröhlich and Vogel, 2009). sRNAs can also exert inhibitory effects on gene expression by binding to the 5′-UTR of mRNAs, which results in occlusion of the RBS, leading to the inhibition of translation. Alternatively, binding of a sRNA to an mRNA 5′-UTR can also result in the decreased stability of a target mRNA, leading to degradation of the transcript (Aiba, 2007).
The RNA chaperone Hfq is usually required to facilitate sRNA-mRNA interactions, because these interactions generally occur through short stretches of imperfect base pairing (Moller et al., 2002; Sauter et al., 2003). Because Hfq is a central component of sRNA-mediated gene regulation, mutation of the hfq locus often leads to pleiotropic phenotypes and significant attenuation of virulence (Sonnleitner et al., 2003; Ding et al., 2004; Sittka et al., 2007; Meibom et al., 2009; Fantappiè et al., 2009). Indeed, an hfq mutant strain derived from the Gram-negative α-proteobacterium Brucella abortus 2308 displays extreme attenuation in mice and increased sensitivity to oxidative stress, decreased survival in low pH, and increased sensitivity to starvation conditions compared to the parental strain (Robertson and Roop, 1999). In recent years, our laboratory has identified several genes that are disregulated in a B. abortus hfq mutant, including those encoding the periplasmic superoxide dismutase, SodC (Gee et al., 2005), the acid resistance protein, HdeA (Valderas et al., 2005), the LuxR-type transcriptional regulator, BabR, and the VirB type IV secretion machinery (Caswell et al., 2012). While these findings have added to our understanding of the role of Hfq in Brucella gene regulation, the sRNAs involved in these regulatory events, as well as the genes directly regulated by those sRNAs, remain unknown.
As mentioned above, the Brucella species are members of the α-proteobacteria, specifically the α2 subclass of proteobacteria (Moreno et al., 1990). This group is comprised of several bacteria whose livelihoods depend on close associations with eukaryotic cells, particularly plant or animal cells. Members of this group include plant symbionts (Rhizobium sp., Sinorhizobium sp., and Bradyrhizobium sp.), plant pathogens (Agrobacterium sp.), and animal pathogens (Bartonella sp. and Brucella sp.), but despite the diversity of their hosts, these bacteria have retained similar genes and common strategies for surviving in their unique niches (Batut et al., 2004). Not surprisingly, the importance of Hfq for the proper association with the host is one common feature shared by some of these bacteria, including Sinorhizobium meliloti (Barra-Bily et al., 2010; Torres-Quesada et al., 2010) and Brucella abortus (Robertson and Roop, 1999), and it is becoming more apparent that the α2 proteobacteria also encode orthologous sRNAs (Del Val et al., 2007; Ulvé et al., 2007; Valverde et al., 2008; Schlüter et al., 2010; Vercruysse et al., 2010; Wilms et al., 2011; Del Val et al., 2012; Madhugiri et al., 2012; Wilms et al., 2012); however, the biological function(s) of these closely related sRNAs is currently not well defined.
In the present study, we have identified two sRNAs in Brucella abortus 2308 that are orthologs of the SmrC15/SmrC16 sRNAs of Sinorhizobium meliloti (Del Val et al., 2007), the ReC58/ReC59 sRNAs of Rhizobium etli (Vercruysse et al., 2010), and the AbcR1/AbcR2 sRNAs of Agrobacterium tumefaciens (Wilms et al., 2011). By defining the role of these sRNAs in virulence and characterizing genes regulated by the AbcR sRNAs in Brucella, this study sheds light on the shared functions of these sRNAs and their contribution to the biology of the α2 proteobacteria as a whole.
Results
Identification of two proteins highly over-expressed in a B. abortus hfq mutant
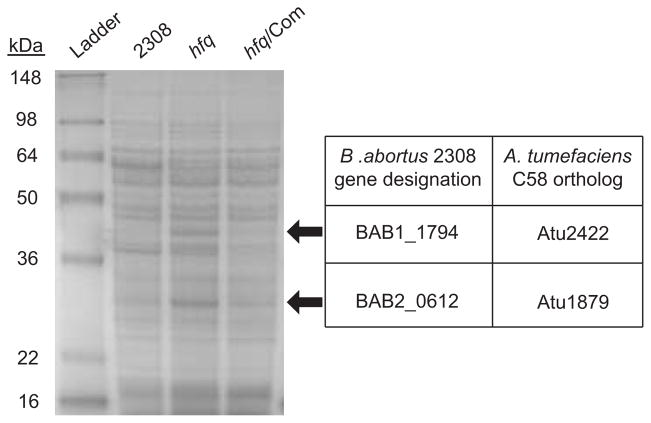
Previous studies in our laboratory have sought to identify Hfq-regulated gene products, and determine how these proteins contribute to the pleiotropic phenotype of a B. abortus hfq mutant. During the course of these studies, SDS-PAGE analysis was performed using whole protein lysates from the wild-type B. abortus strain 2308 and the isogenic hfq mutant strain, B8 (Caswell et al., 2012) (Fig. 1) grown in a rich medium (brucella broth) to early stationary phase. It was noted that two proteins, one approximately 30 kDa and another approximately 40 kDa, were highly over-produced in the hfq mutant strain compared to the wild-type strain (Fig. 1). Additionally, complementation of hfq expression in the mutant strain restored the level of these proteins to that of the wild-type strain. In order to identify these over-produced proteins, the protein bands were excised from the gel, and electrospray ionization tandem mass spectrometric analysis of a tryptic digest identified the ~30 kDa protein as the product of the gene designated BAB2_0612 in the B. abortus 2308 genome sequence. The ~40 kDa protein was determined to be encoded by the gene designated BAB1_1794 in the B. abortus 2308 genome. Both BAB2_0612 and BAB1_1794 encode proteins that are putative periplasmic-binding proteins, but currently it is not known what these proteins bind or transport in B. abortus 2308.
Figure 1. Over-production of two putative periplasmic-binding proteins in a B. abortus hfq mutant.
Whole protein lysates from the parental B. abortus strain 2308, the hfq mutant strain, and the hfq complemented strain growth in brucella both to early stationary phase were separated on 15% SDS-PAGE, and the gel was stained with RAPIDstain™. Two proteins (indicated by black arrows) were over-produced in the hfq mutant compared to the parental and complemented strains, and these two proteins were excised from the gel and subjected to electrospray ionization tandem mass spectrometric analysis. The ~30 kDa protein was identified as the B. abortus 2308 protein BAB2_0612, which is an ortholog of the Agrobacterium tumefaciens protein Atu1879. The ~40 kDa protein was identified as the B. abortus 2308 protein BAB1_1794, which is an ortholog of the A. tumefaciens protein Atu2422.
Interestingly, the closely related α-proteobactrium, Agrobacterium tumefaciens, encodes proteins highly similar to BAB1_1794 and BAB2_0612. These orthologs are designated Atu2422 and Atu1879, respectively, in A. tumefaciens C58. BAB1_1794 and Atu2422 are 88% similar (73% identical) at the amino acid level, while BAB2_0612 and Atu1879 are 70% similar (52% identical). Of direct relevance to this work, it was reported that production of Atu2422 and Atu1879 in A. tumefaciens is regulated post-transcriptionally by two small regulatory RNAs (sRNAs) called AbcR1 and AbcR2 (Wilms et al., 2011).
Identification of two small RNAs in B. abortus that are orthologous to the AbcR sRNAs of Agrobacterium tumefaciens
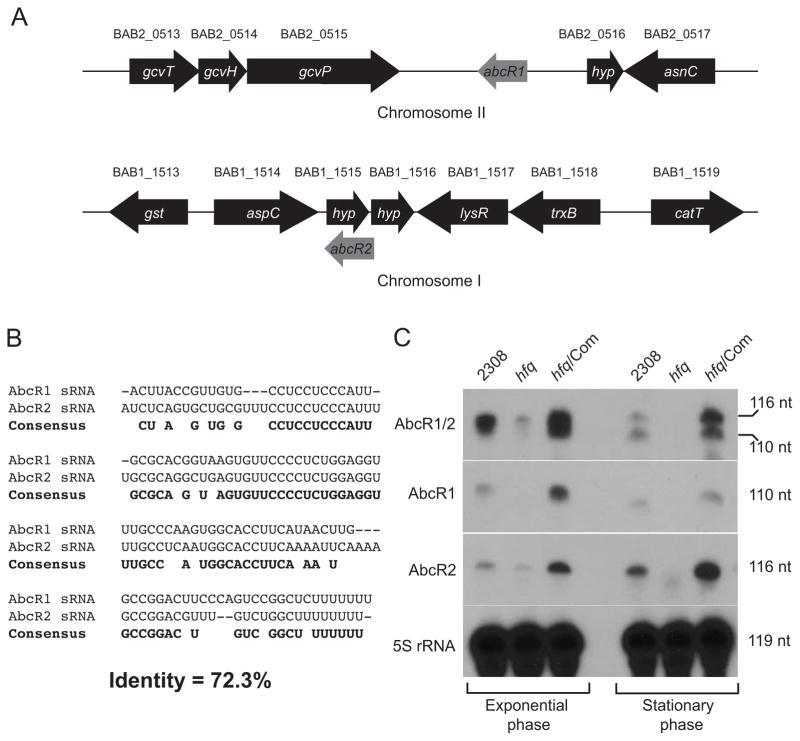
Because Hfq is known to bridge interactions between mRNAs and sRNAs, we hypothesized that similar AbcR sRNAs were present and functioning in B. abortus to affect production of BAB1_1794 and BAB2_0612. In fact, the study describing the A. tumefaciens AbcR sRNAs predicts that Brucella sp. encode AbcR1, but the presence of AbcR2 in Brucella was not predicted (Wilms et al., 2011). The predicted AbcR1 locus is found in the intergenic region between gcvP of the glycine cleavage system (BAB2_0515) and a hypothetical protein-encoding gene (BAB2_0516) on chromosome 2 of B. abortus 2308 (Fig. 2A). Using computational searches, a potential region encoding AbcR2 was identified on chromosome 1 in the region of BAB1_1515, which potentially encodes a small, hypothetical protein.
Figure 2. Identification of two small RNAs (sRNAs) in Brucella abortus 2308.
A. Schematic of the loci encoding the AbcR sRNAs.
Due to the high degree of similarity to the A. tumefaciens AbcR sRNAs, the two putative sRNAs in B. abortus 2308 were similarly named AbcR1 and AbcR2. Bioinformatic analyses predicted the B. abortus AbcR1 sRNA to be encoded on chromosome II in the intergenic region between gcvP (BAB2_0515) and a small hypothetical protein-encoding gene (BAB2_0516). The B. abortus AbcR2 sRNA was predicted to be encoded on chromosome I near a small hypothetical protein-encoding gene (BAB1_1515)
B. Alignment of the B. abortus 2308 AbcR sRNAs
The nucleotide sequence of AbcR1 and AbcR2 were aligned using the AlignX® software from Vector NTI® (Invitrogen). Identical nucleotides between the two sRNAs are shown as the consensus sequence in bold font
C.Northern blot detection of AbcR1 and AbcR2
Total RNA isolated from exponential or stationary phase cultures of B. abortus 2308, the hfq mutant strain, and the complemented hfq mutant strain was separated on denaturing polyacrylamide gels. Following transfer to a membrane, a single probe designed to detect both AbcR1 and AbcR2, or probes designed to detect either AbcR1 or AbcR2 were used in the Northern blot analyses. Detection of 5S rRNA was also performed as a loading control. The sizes (in nucleotides) of the detected sRNAs are indicated to the right of the panels.
Alignment of the potential AbcR1 and AbcR2 sRNAs of Brucella revealed a high level of nucleotide identity (Fig. 2B). Using this alignment, oligonucleotide probes were designed, and Northern blot analyses were performed to test if these potential sRNAs were expressed in B. abortus 2308 (Fig. 2C). A common AbcR1/AbcR2 probe detected two sRNAs in B. abortus 2308, while specific AbcR1 or AbcR2 probes detected individual sRNAs in the Northern analyses. Interestingly, both AbcR1 and AbcR2 were almost completely absent in the B. abortus hfq mutant strain, but expression of the sRNAs was restored by in trans complementation of hfq expression in the mutant strain. Additionally, recombinant B. abortus Hfq protein bound to both AbcR1 and AbcR2 in electrophoretic mobility shift assays (EMSAs) (Fig. S4). Altogether, these experiments determined that B. abortus 2308 expresses the AbcR sRNAs, and it appears that Hfq plays a role in the stability of these sRNAs.
In order to identify the transcriptional start sites of the AbcR sRNAs, 5′-RACE was performed, and it was determined that AbcR1 is 110 nucleotides in length, while AbcR2 is 116 nucleotides in length. Interestingly, the AbcRs of A. tumefaciens are encoded in tandem in a single intergenic region, while AbcR1 and AbcR2 in B. abortus are found on separate chromosomes. Due to the high level of similarity between the AbcR sRNAs of A. tumefaciens and B. abortus (Fig. S1), we have adopted the AbcR nomenclature for these sRNAs in B. abortus 2308. Additionally, the Brucella AbcR sRNAs are not limited to B. abortus 2308, as they were also detected in RNA isolated from B. melitensis 16M and B. suis 1330 (Fig. S2), and interestingly, similar genetic organizations of the loci (e.g., two hypothetical genes within the region encoding AbcR2) are conserved among the different Brucella spp. Similar to the AbcR sRNAs from Agrobacterium, the Brucella AbcR sRNAs are predicted to form secondary structures containing three stem loops (Fig. S3). Overall, these experiments have identified two sRNAs in Brucella abortus that are highly similar in sequence and structure to two sRNAs of Agrobacterium tumefaciens, and the next goal was to determine the function of these sRNAs in the biology and virulence of B. abortus 2308.
The AbcR sRNAs are required for wild-type virulence of B. abortus 2308
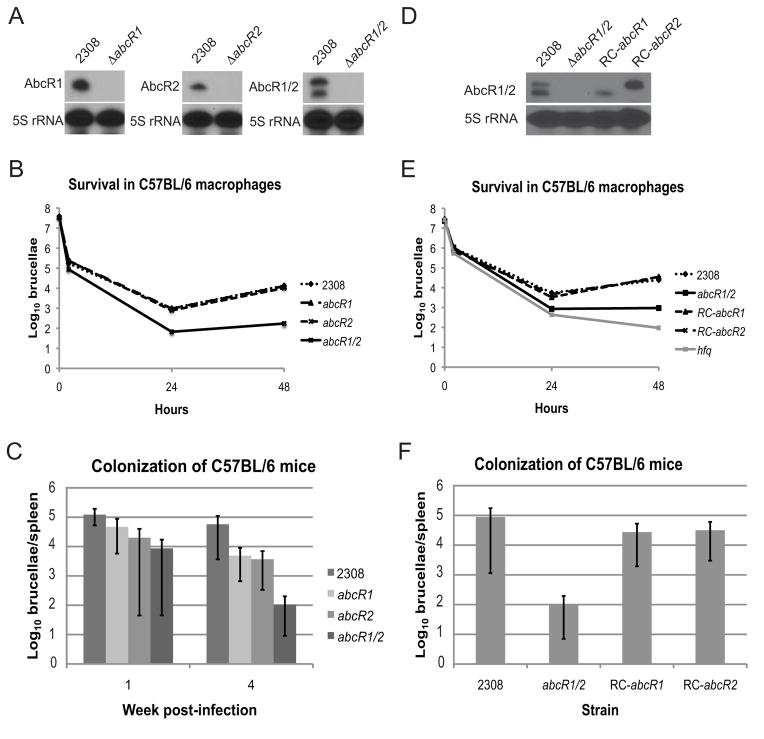
To begin to define the function(s) of the AbcR sRNAs in B. abortus 2308, isogenic abcR1 and abcR2 mutants, as well as an abcR1 abcR2 double mutant, were constructed using an unmarked gene excision strategy. Northern blot analysis was performed to ensure that the mutant strains did not express the corresponding sRNAs (Fig. 3A). Importantly, deletion of the abcR loci did not alter the in vitro growth of Brucella strains in nutrient-replete (brucella broth) or nutrient-limiting (Gerhardt’s minimal media [Gerhardt, 1958]) media. The abcR mutant strains, along with the parental strain 2308, were tested for their virulence in primary murine macrophages (Fig. 3B). The abcR1 and abcR2 isogenic mutant strains exhibited no difference from the parental strain 2308 in their ability to survive and replicate in macrophages; however, the abcR1 abcR2 double mutant showed significantly decreased levels of survival at both 24 and 48 hours post-infection compared to the parental strain 2308.
Figure 3. AbcR1 and AbcR2 are required for wild-type virulence of Brucella abortus 2308.
A. Absence of sRNAs in mutant strains
Isogenic abcR1 and abcR2 mutants, as well as a double abcR1 abcR2 mutant, were constructed using an unmarked gene excision strategy. Following mutagenesis, Northern blot analyses were performed to ensure that the mutants do not express the corresponding sRNAs
B. Virulence of the sRNA mutants in primary murine macrophaes
Primary peritoneal macrophages were isolated from C57BL/6 mice, and the macrophages were infected with B. abortus 2308, the abcR1 mutant strain, the abcR2 mutant strain, or the abcR1 abcR2 double mutant strain. Extracellular bacteria were killed by treatment with gentamicin. At the indicated time points, the macrophages were lysed, and serial dilutions of the lysates were plated on blood agar to enumerate Brucella colony-forming units. The data are represented as the mean of three independent wells for each indicated Brucella strain, and the graph shown is from a single representative experiment.
C. Spleen colonization of mice infected with the sRNA mutants.
6–8 week-old female C57BL/6 mice were infected intraperitoneally with B. abortus 2308, the abcR1 mutant strain, the abcR2 mutant strain, or the abcR1 abcR2 double mutant strain. 5 mice were used for each Brucella strain for each time point. At the indicated time points, the mice were sacrificed, and the spleens were removed. Spleen homogenates were serially diluted and plated on blood agar to enumerate Brucella colony-forming units. The data are depicted as the mean and standard deviation for each Brucella strain from a single experiment, and the (*) denotes a statistically significant difference between the parental strain 2308 and the abcR1/2 double mutant strain (T-test; p<0.05)
D. Reconstruction of abcR loci restores production of the AbcR sRNAs
The abcR loci were individually reconstructed in the B. abortus abcR1 abcR2 double mutant strain, and Northern blot analysis was used to show the restoration of abcR1 (RC-abcR1) or abcR2 (RC-abcR2) expression in the reconstructed strains.
E. Virulence of the reconstructed AbcR sRNA strains in primary murine macrophages. These experiments were performed as described in part B, but here, the strains tested included: the parental strain B. abortus 2308, the abcR1 abcR2 double mutant (abcR1/2), the abcR1 reconstructed strain (RC-abcR1), the abcR2 reconstructed strain (RC-abcR2), and the B. abortus hfq mutant strain. The data are represented as the mean of three independent wells for each indicated Brucella strain, and the graph shown is from a single representative experiment.
F. Spleen colonization of mice infected with the reconstructed AbcR sRNA strains.
These experiments were performed as described in part C, but here, spleen colonization was assessed after 4 weeks post-infection. The data are depicted as the mean and standard deviation for each Brucella strain from a single experiment, and the (*) denotes a statistically significant difference between the parental strain 2308 and the abcR1/2 double mutant strain (T-test; p<0.05).
The abcR mutant strains were next tested in a mouse model of Brucella infection (Fig. 3C). C57BL/6 mice were infected intraperitoneally with the Brucella strains, and spleen colonization by the brucellae was assessed after 1 and 4 weeks post-infection. Similar to the macrophage experiments, the isogenic abcR1 and abcR2 mutant strains colonized the spleens of the infected mice at levels similar to those of the parental strain 2308 at 1 and 4 weeks post-infection. At 1 week post-infection, the abcR1 abcR2 double mutant stain displayed an approximately 1-log defect in spleen colonization compared to 2308, although this decrease was not significant. However, at 4 weeks post-infection, the abcR1 abcR2 double mutant strain was significantly attenuated compared to the other strains tested.
To confirm a link between the AbcR sRNAs and the virulence of B. abortus 2308, the abcR loci were individually reconstructed in the abcR1 abcR2 double mutant strain, creating two strains that express either abcR1 or abcR2 (Fig. 3D). The reconstructed strains were tested for their ability to survive and replicate in primary murine macrophages (Fig. 3E). As shown previously in Fig. 3B, the abcR1 abcR2 double mutant strain was attenuated at 24 and 48 hours post-infection compared to the parental strain 2308 in these experiments, and importantly, reconstruction of either the abcR1 or abcR2 locus was sufficient to restore wild-type virulence to the double mutant strain. The B. abortus hfq mutant strain, B8, constructed previously (Caswell et al., 2012) was also included in these assays, and the hfq mutant strain was more significantly attenuated than the abcR1 abcR2 double mutant, indicating that other sRNAs, in addition to the AbcR sRNAs, are required for the full virulence of B. abortus 2308.
In addition to the macrophage experiments, the reconstructed abcR strains were tested for their capacity to colonize the spleens of experimental infected mice, and following 4 weeks of infection, spleen colonization by brucellae was assessed (Fig. 3F). As in the previous experiment (Fig. 3C), the abcR1 abcR2 double mutant exhibited a significant reduction in spleen colonization compared to the parental strain 2308. Importantly, both the abcR1- and abcR2-reconstructed strains displayed levels of spleen colonization similar to those of the parental strain 2308, confirming that production of only one AbcR sRNA is adequate for full virulence of B. abortus. Altogether, from the macrophage and animal experiments, it was concluded that the AbcR sRNAs are required for wild-type virulence of B. abortus 2308, but additionally, AbcR1 and AbcR2 appear to serve redundant functions, as mutation of both abcR1 and abcR2 is required for attenuation.
Identification of genes regulated by the AbcR sRNAs in B. abortus 2308
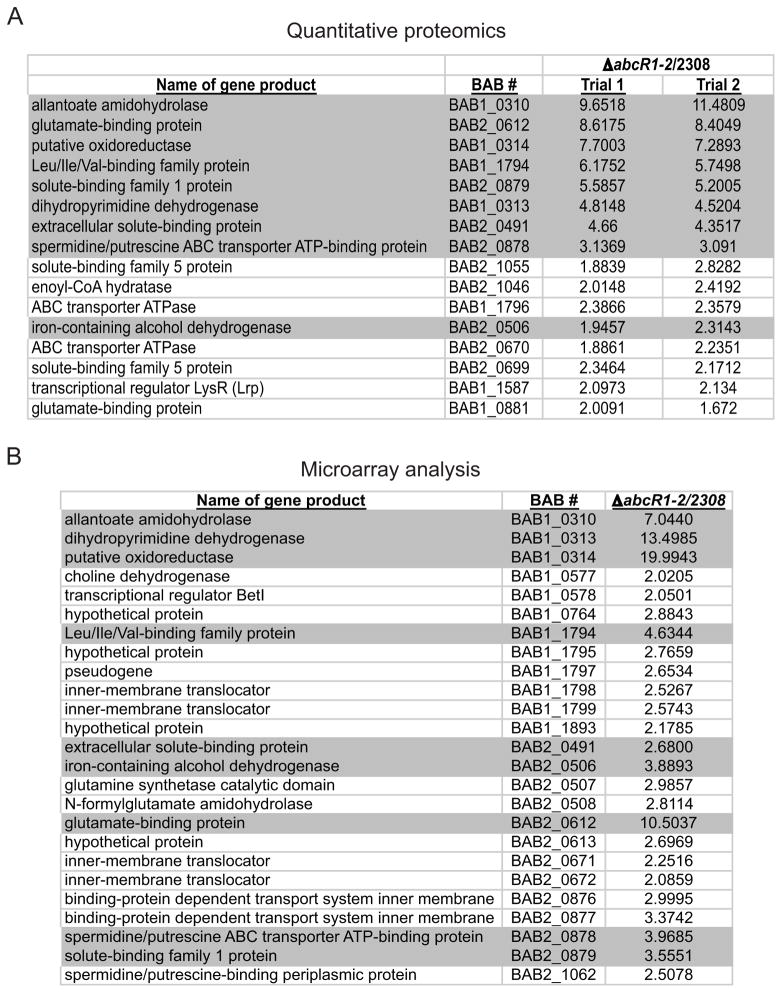
We next sought to identify the targets of AbcR regulation in B. abortus 2308, and for this, both quantitative proteomic (iTRAQ) and microarray analyses were employed (Fig. 4). Because AbcR1 and AbcR2 are potentially serving redundant functions in the bacterium (Fig. 3), the abcR1 abcR2 double mutant was used in these studies. Quantitative proteomics detected more than 1100 proteins from B. abortus 2308, covering approximately 1/3 of the total predicted proteins of this strain (Fig. S6). Of the >1100 detected, only 16 proteins were determined to be more than 2-fold overproduced in the abcR1 abcR2 double mutant compared to 2308. These experiments were performed in duplicate, and a strict ≥2-fold difference in at least one experiment was used as a parameter to determine if a protein was considered significantly different between the parental strain and the abcR1 abcR2 double mutant strain (Fig. 4A). Importantly, the proteins originally identified as being over-produced in the hfq mutant strain (Fig. 1), BAB1_1794 and BAB2_0612, were detected by iTRAQ and shown to be >5.5- and >8-fold increased in the abcR1 abcR2 double mutant, respectively. In general, quantitative proteomics revealed that the levels of several proteins related to ABC transport systems, putatively of amino acids and polyamines, are affected by the AbcR sRNAs in B. abortus 2308 (Fig. 4A). It is noteworthy to point out that only increases in proteins levels were observed in the abcR1 abcR2 knockout strain compared to 2308, and this was the first indication that these sRNAs are inhibitory for the ex pression of their targets.
Figure 4. Identification of the targets of regulation by the Brucella abortus 2308 AbcR sRNAs.
A. List of proteins differentially produced in the abcR1 abcR2 double mutant compared to the parental strain 2308
Quantitative proteomics analysis (iTRAQ mass spectrometry) was performed on whole protein lysates from Burcella strains grown in brucella broth to early stationary phase, and several proteins were found to be more than 2-fold increased in the abcR1 abcR2 double mutant compared to strain 2308. The analysis was performed on proteins isolated from two independent sets of cultures, and those proteins exhibiting a >2-fold increase in at least one experiment are shown. The entire data set from the proteomics experiments can be found in Fig. S6
B. List of genes differentially expressed in the abcR1 abcR2 double mutant compared to the parental strain 2308
Microarray analysis was performed using total cellular RNA from Brucella strains grown in brucella broth to early stationary phase, and those genes shown to be more than 2-fold increased in the abcR1 abcR2 double mutant compared to strain 2308 are shown in the list. The AbcR-regulated genes identified in the microarray whose protein products were also determined to be different in iTRAQ analysis are highlighted in grey. The GenBank accession number for the microarray data is GSE38017.
Microarray analysis was also performed comparing total RNA from B. abortus 2308 and the abcR1 abcR2 double mutant (Fig. 4B). These experiments revealed 25 transcripts that were significantly increased in the abcR1 abcR2 double mutant compared to strain 2308 (Fig. 4B). Similar to the proteomics data (Fig. 4A), the microarray determined that several genes predicted to encode ABC transport systems showed increased expression in the abcR double mutant. While the molecules transported by these systems are not currently known, many of them are annotated as amino acid and polyamine transporters.
It is also noteworthy that the proteomic and microarray data closely parallel each other: both protein and transcript levels of individual genes are increased. This indicated that the AbcR sRNAs might be directly affecting the mRNA levels of the target genes in order to facilitate gene expression. However, there are examples of differences that were detected by microarray analysis but not by proteomic analysis, and vice versa. For example, expression of the genes BAB2_0507 and BAB2_0508 was shown by microarray analysis to be more than 2-fold increased in the abcR double mutant strain, but the corresponding proteins were not detected by iTRAQ proteomic analysis. While the microarrays provide full coverage of the genes of B. abortus 2308, the iTRAQ analysis does not necessarily detect every protein produced by the bacterium. Therefore, it is possible that some discrepancies between the microarray and proteomic analyses result from proteins simply not being detected via iTRAQ.
The AbcR sRNAs promote degradation of target mRNAs in B.abortus 2308
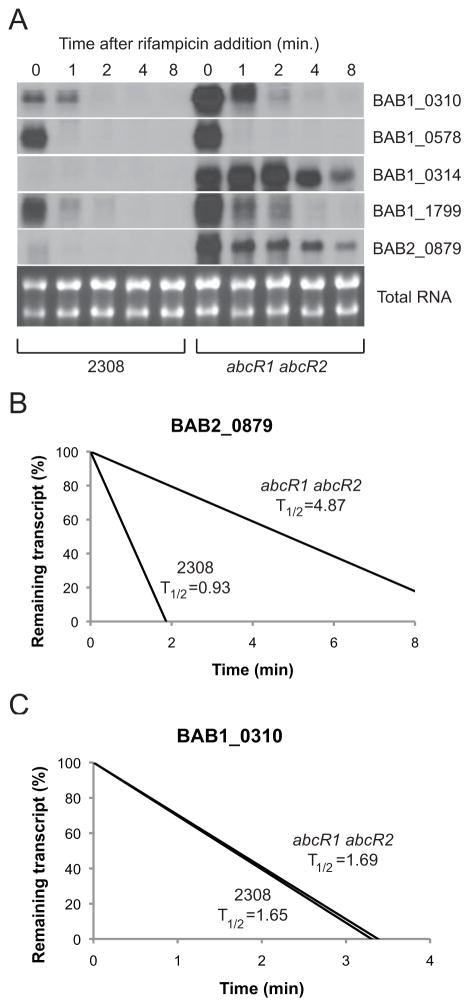
Because both protein and mRNA levels for specific genes are elevated in the abcR1 abcR2 double mutant, it was hypothesized that these sRNAs are affecting the stability of the target mRNAs. In support of this hypothesis, the AbcR sRNAs were shown to destabilize specific target mRNAs in Agrobacterium tumefaciens (Wilms et al., 2011). To gain insight into the ability of the AbcR sRNAs to affect target mRNA stability, rifampicin block experiments followed by Northern blot analyses were performed using the parental strain 2308 and the abcR1 abcR2 double mutant strain (Fig. 5). Several of the mRNA transcripts tested, including those corresponding to BAB1_0314, BAB1_1799, and BAB2_0879, appeared to be stabilized in the abcR1 abcR2 double mutant compared to the parental strain 2308, while other mRNA transcripts (BAB1_0310 and BAB1_0578) did not appear to be significantly stabilized in the abcR1 abcR2 mutant (Fig. 5A).
Figure 5. The AbcR sRNAs target mRNAs for degradation.
A. Northern blot analysis of Brucella mRNA transcripts from cultures treated with rifampicin
Cultures of B. abortus 2308 or the abcR1 abcR2 double mutant were treated with rifampicin, and RNA was collected at the indicated time points. Northern blot analysis was performed for specific mRNA transcripts (indicated to the right of each panel), and the total RNA is shown as a loading control in the bottom panel
B and C. Determination of the half-lives of Brucella mRNA transcripts
Using ImageJ software (http://rsbweb.nih.gov/ij/), the amount of signal for each transcript at each time point was quantified, and these values were used to calculate the half-life of a specific mRNA transcript in B. abortus 2308 or the abcR1 abcR2 double mutant. Part B depicts the half-life (in min.) of the BAB2_0879 transcript in these two strains, while part C shows the half-life of the BAB1_0310 transcript.
In order to determine the half-lives of the transcripts in the different strains, the Northern blot signals were quantified using ImageJ software (http://rsbweb.nih.gov/ij/). The half-life of the BAB2_0879 mRNA was approximately 0.93 min. in B. abortus 2308, while in the abcR1 abcR2 double mutant, the half-life was ~4.87 min. (Fig. 5B). Similarly, the half-lives of the BAB1_0314 and BAB1_1799 mRNA transcripts were significantly increased in the abcR1 abcR2 mutant compared to strain 2308 (data not shown). On the contrary, the half-life of the BAB1_0310 mRNA was about the same in both the parental strain 2308 and the abcR1 abcR2 mutant. It is important to note that while the half-life of the BAB1_0310 mRNA was the same in both strains, there is clearly more transcript present in the abcR1 abcR2 double mutant compared 2308 (Fig. 5A), and this supports the microarray data (Fig. 4B) indicating a significant increase in BAB1_0310 mRNA in the abcR1 abcR2 mutant. The increase in specific mRNA levels that are not accompanied by increased half-lives, such as the case of BAB1_0310, may be the result of indirect regulation by the AbcR sRNAs. In this situation, the AbcRs may control the expression of a transcriptional regulator, such as Lrp (see Discussion), which may, in turn, control the expression of downstream genes. Because the AbcR sRNAs are not directly controlling the stability of these messages, the mRNA half-lives would remain the same in both the wild-type strain and the abcR double mutant strain; however, the overall abundance of these mRNAs would be disparate between the two strains due to difference in the levels of the transcriptional regulator. Nonetheless, these data show that the AbcR sRNAs promote the degradation of certain mRNA transcripts in B. abortus 2308.
AbcR1 and AbcR2 perform redundant functions in B. abortus 2308
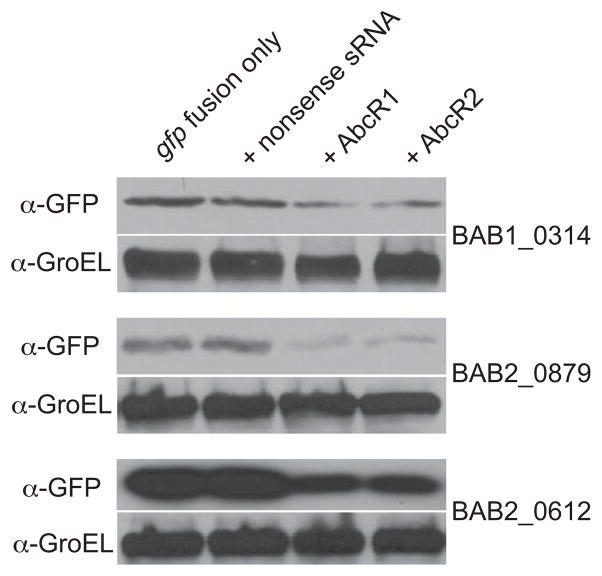
Based on the virulence studies showing that only the abcR1 abcR2 double mutant strain, but not the single abcR mutant strains, are attenuated in macrophages and mice (Fig. 3), it was hypothesized that AbcR1 and AbcR2 perform redundant regulatory functions in B. abortus 2308. In order to test this hypothesis, an in vivo E. coli reporter system for assessing regulation by sRNAs was employed (Urban and Vogel, 2007). For this, the 5′-untranslated region and approximately 10 codons of a gene of interest was cloned in-frame with gfp into a low-copy plasmid. Likewise, the genes encoding AbcR1, AbcR2, or a nonsense sRNA (the AbcR2 sequence in reverse) were cloned into a high-copy number plasmid. The gfp fusion plasmid alone, or a combination of both the gfp fusion plasmid and one of the sRNA-encoding plasmids, was transformed into E. coli, and the amount of GFP in the cells was assessed by immunoblotting (Fig. 6). It is important to note that while the sequences of the gfp constructs were confirmed by DNA sequence analysis, there was no visual GFP fluorescence detectable when the strains were grown on agar plates. However, immunoblot analysis allowed for the detection of GFP in the strains harboring the various plasmid combinations. For the BAB1_0314-gfp fusion, the levels of GFP were similar between the “fusion-only” strain and the strain carrying the gfp fusion and the nonsense sRNA, but when either AbcR1 or AbcR2 was co-expressed with the BAB1_0314-gfp fusion, there was reduction in the amount of GFP detected. Similarly, the levels of GFP did not differ between a strain carrying a BAB2_0879-gfp or a BAB2_0612-gfp fusion only and a strain carrying the gfp fusions and the nonsense sRNA. However, co-expression of either AbcR1 or AbcR2 with the BAB2_0879-gfp or the BAB2_0612-gfp fusions resulted in a significant decrease in the amount of GFP in the cells. Overall, these data confirm that, at least in the cases of BAB1_0314, BAB2_0879, and BAB2_0612, AbcR1 and AbcR2 perform redundant regulatory functions in B. abortus 2308.
Figure 6. AbcR1 and AbcR2 perform redundant regulatory functions.
The 5′-untranslated regions and the first 10 codons of BAB1_0314, BAB2_0879, or BAB2_0612 were cloned in-framed with gfp into a low-copy plasmid, and abcR1, abcR2, or a DNA fragment encoding a nonsense sRNA (the abcR2 gene in reverse) were cloned into a high-copy plasmid (see Experimental procedures). E. coli strains carrying only a gfp fusion construct, or carrying both a gfp fusion construct and a sRNA-encoding construct, were grown in LB broth, and immunoblot analyses were performed on total protein lysates to detect levels of GFP or GroEL.
Discussion
The Brucella spp. naturally infect a variety of animals, including cattle, goats and sheep, and swine (Pappas et al., 2005), and in these hosts, the brucellae infect cells of the reticuloendothelial system, particularly macrophages, during the course of a chronic infection (Celli and Gorvel, 2004). During infection of and residence within these cells, the brucellae encounter a variety of stress conditions, including low pH, oxidative stresses, and nutrient deprivation (Roop et al., 2009). In light of these harsh conditions, it is not surprising that Hfq and sRNAs play an important role during the intramacrophagic life of the brucellae. The expression of sRNAs is many times driven by specific stimuli that, in turn, allow for rapid alterations in gene expression to cope with the stimulus/stress (Wassarman, 2002; Repoila et al., 2003). In the present study, the Brucella sRNAs, AbcR1 and AbcR2, were shown to be important for survival in macrophages and virulence in a mouse model of infection (Fig. 3). sRNAs have been linked to the proper expression of virulence factors in a variety of pathogenic bacteria (Papenfort and Vogel, 2010), and several recent studies show that sRNAs are directly linked to the virulence of organisms such as Listeria (Mraheil et al., 2011), Salmonella (Gong et al., 2011), Vibrio (Song et al., 2008), and Yersinia (Koo et al., 2011). In addition to the well-described role of sRNAs in stress adaptation, these reports, along with the present characterization of the Brucella AbcR sRNAs, underscore the important role of sRNAs in bacterial pathogenesis.
Interestingly, deletion of a single abcR locus did not render Brucella more vulnerable to killing in macrophages and mice, but rather, deletion of both abcR loci was required to observe a significant phenotype (Fig. 3). This offered the first evidence that the AbcR sRNAs perform redundant and compensatory functions, a hypothesis that was later tested and confirmed using an in vivo reporter system (Fig. 6). It is not uncommon for bacterial genomes to contain redundant sRNAs, as exemplified by the 4 Qrr sRNAs of Vibrio cholerae that are involved in the regulation of quorum sensing (Lenz et al., 2004). In this system, any one Qrr sRNA is sufficient for the intact regulatory circuit, and mutation of all four qrr loci is required to disrupt quorum sensing. A similar situation is observed for the Brucella AbcR sRNAs, and it is possible that redundancy of this kind may be an evolutionary adaption ensuring the proper expression of essential genes. In the case of Brucella, and perhaps the other α-proteobacteria, retaining multiple copies of the AbcR-type sRNAs would ensure the proper expression of transporters required for the pathogenic/symbiotic lifestyle.
Presently, nothing is known about the expression of the AbcR sRNAs during residence of the brucellae within host macrophages. In vitro, deletion of the AbcRs in Brucella results in increased mRNA and protein levels of several different transporters and enzymes (Fig. 4), and the constitutive expression of these genes results in significant attenuation in macrophages and mice (Fig. 3). It is conceivable that the AbcR sRNAs function to modulate the expression of their target genes in response to environmental signals, allowing for temporal activation of these target systems when they are needed, and likewise, repressing expression of the genes when they are no longer required. In support of this proposition, some Brucella AbcR targets are differentially expressed during intracellular growth. The brucellae exhibit differential levels of the protein encoded by BAB1_1794 and the transcripts of BAB2_0878 and BAB1_0577 when the bacteria are growing within macrophages (Lamontagne et al., 2009) and HeLa cells (Rossetti et al., 2011), respectively, compared to brucellae grown in vitro. However, for this type of temporal regulatory mechanism to function between the AbcRs and their target genes, expression of the genes encoding abcR1 and abcR2 must be regulated in some fashion, and our laboratory is currently assessing how different environmentally relevant signals (e.g., low pH, oxidative stress, and nutrient deprivation), as well as various transcriptional regulatory proteins in Brucella, influence abcR expression. Defining the circumstances and mechanisms that modulate AbcR levels intracellularly will provide valuable information about the biological function of these sRNAs, and their target genes, in Brucella.
Wilms and colleagues have suggested that as a group, the AbcR-type sRNAs may be serving a common function in regulating ABC transport systems in the α-proteobacteria (Wilms et al., 2011), and our data provide additional support for this proposal (Fig. 4). We show that in Brucella several components of ABC transport systems are disregulated in the abcR1 abcR2 double mutant, and these systems are putative amino acid and polyamine transporters. In Agrobacterium tumefaciens, AbcR1 controls the expression of the periplasmic-binding protein Atu2422, which is known to be involved in the transport of γ-aminobutyric acid (GABA) (Wilms et al., 2011; Haudecoeur et al., 2009). The detection and transport of GABA by Agrobacterium is important for proper quorum sensing in the bacterium and the subsequent infection of the plant (Chevrot et al., 2006). The Atu2422 ortholog in Brucella, BAB1_1794, is regulated by the AbcR sRNAs; however, there is currently no documented role for GABA transport in Brucella sp. Nonetheless, GABA has recently been detected in human macrophages (Stuckey et al., 2005) and has been shown to play a role in the inhibition of autoimmune inflammation (Bhat et al., 2010). This raises some interesting questions regarding a potential link between Brucella and GABA: Does GABA play a previously unrecognized role in the biology of Brucella within host macrophages? If so, does GABA detection and transport affect quorum sensing in Brucella in a manner similar to Agrobacterium?
GcvB is a sRNA found in many Gram-negative bacteria, including E. coli and Salmonella, and GcvB controls the expression of many genes involved in amino acid transport and metabolism (Urbanowski et al., 2000; Papenfort and Vogel, 2007; Sharma et al., 2011). While there is no apparent sequence homology between GcvB and the AbcR sRNAs, there appears to be “functional homology” between them, as the regulons of these sRNAs are strikingly similar in that both GcvB and the AbcRs regulate expression of amino acid transport and metabolism (Sharma et al., 2011 and Fig. 4). The leucine-responsive regulatory protein, Lrp, is one of the commonly regulated proteins, as Lrp levels are modulated in Salmonella by GcvB (Sharma et al., 2011) and in Brucella by the AbcR sRNAs (Fig. 4A, designated BAB1_1587). In E. coli, Lrp is considered a global regulator that controls the expression of many genes involved in the transport and metabolism of amino acids (Newman and Lin, 1994), but in Brucella, it is not known what genes are controlled by Lrp. It is possible that AbcR-mediated fluctuations in Lrp levels are responsible for some of the differential expression of genes observed in the microarray analyses that do not appear to be directly controlled by the AbcR sRNAs (Fig. 4 and 5A), such as BAB1_0310, and we are currently exploring this possibility. Nonetheless, the similarities between GcvB- and AbcR-mediated regulation of amino acid transport/metabolism highlight the importance of amino acids in the biology of bacteria as diverse as Salmonella and Brucella, and moreover, these bacteria have conserved sRNAs as a means of modulating these systems.
Finally, the mechanism by which the AbcR sRNAs directly regulate gene expression involves degradation of target mRNAs (Fig. 5). Currently though, it is not clear how the AbcRs are promoting mRNA degradation. In other bacteria, sRNA can target mRNA transcripts to RNase E for cleavage and degradation (Massé et al., 2003; Ikeda et al., 2011; Prévost et al., 2011). It is possible that a similar mechanism is in place in Brucella. In fact, the gene BAB1_0930 in the B. abortus 2308 genome is annotated as “ribonuclease E and G” and exhibits 45% identity and 64% similarity at the amino acid level to RNase E of E. coli K12. Thus, AbcR-mediated degradation of mRNAs may involve targeting to a specific ribonuclease, but additional experiments are needed to confirm this proposition. Also in regard to direct AbcR-mediated regulation of genes, it should be noted that the Brucella AbcR sRNAs contain a sequence motif (CUCCCA) that is identical to the sequence found in AbcR1 of Agrobacterium tumefaciens that is known to bind to target mRNAs in A. tumefaciens (Wilms et al., 2011). In B. abortus 2308, this motif is present in both AbcR sRNAs, and several of the known Brucella AbcR mRNA targets contain a complete, or very similar, complementary sequence in their 5′-untranslated regions (Fig. S5). However, the authenticity of this sequence as a regulatory motif in the Brucella AbcR sRNAs has not been experimentally validated, and future work employing site-directed mutagenesis will be helpful in defining and/or uncovering the true consensus regulatory sequence.
Experimental procedures
Bacterial strains and growth conditions
Brucella abortus 2308 and derivative strains were routinely grown on Schaedler agar (BD, Franklin Lakes, NJ USA) containing 5% defibrinated bovine blood (Quad Five, Ryegate, MT, USA) (SBA) or in brucella broth (BD). Escherichia coli strains were grown routinely on tryptic soy agar (BD) or in Luria-Bertani broth. When appropriate, growth media were supplemented with ampicillin (100 μg ml−1), kanamycin (45 μg ml−1), or chloramphenicol (30 μg ml−1; E. coli strains only). All cultures of and experiments with Brucella strains were carried out in a Biosafety Level-3 (BSL-3) laboratory.
Construction and genetic complementation of B. abortus abcR1, abcR2, and abcR1 abcR2 mutants
The abcR1 and abcR2 loci were mutated using an unmarked gene excision strategy described previously (Caswell et al., 2012). An approximately 1-kb fragment of the abcR1 upstream region was amplified by PCR using the primers ΔabcR1-Up-For and Δ abcR1-Up-Rev (Table S1), genomic DNA from B. abortus 2308 as a template, and Platinum® Pfx DNA Polymerase (Invitrogen, Carlsbad, California, USA). Similarly, the abcR2 upstream region was amplified by PCR using the same parameters with the primers ΔabcR2-Up-For and ΔabcR2-Up-Rev (Table S1). Approximately 1 kb ownstream of the abcR1 or abcR2 genes was amplified with the primers ΔabcR1-Dn-For and ΔabcR1-Dn-Rev or ΔabcR2-Dn-For and ΔabcR2-Dn-Rev, respectively (Table S1). The upstream fragments were digested with BamHI, while the downstream fragments were digested with PstI, and both fragments were treated with polynucleotide kinase. The appropriate DNA fragments (either abcR1-Up and abcR1-Dn, or abcR2-Up and abcR2-Dn) were combined in a single ligation mix with BamHI/PstI-digested pNPTS138 (Spratt et al., 1986), which contains a kanamycin resistance marker and sacB gene for counter-selection with sucrose. The resulting plasmids (pC3022 [ΔabcR1] or pC3023 [ΔabcR2]) (Table S2) were introduced individually into B. abortus 2308, and merodiploid clones were obtained by selection on SBA+kanamycin. A single kanamycin-resistant clone for each construct was grown overnight in brucella broth, and then plated onto SBA containing 10% sucrose. Genomic DNA from sucrose-resistant, kanamycin-sensitive colonies was isolated and screened by PCR for loss of the abcR1 or abcR2 genes. The isogenic abcR1 mutant derived from B. abortus 2308 was named CC094, and the isogenic abcR2 mutant derived from B. abortus 2308 was named CC095.
To generate an abcR1 abcR2 double mutant, the plasmid pC3023 was introduced into strain CC094, and selection and screening was performed as described above. The abcR1 abcR2 double mutant derived from B. abortus 2308 was named CC097. Total cellular RNA from all mutant strains was assessed by Northern blot analysis to ensure that the AbcR1 and AbcR2 sRNAs were not produced by these strains (Figure 3A).
Reconstruction of the abcR loci was achieved using the sacB counter-selection strategy described above, but with modification. Here, the wild-type abcR1 locus was amplified using primers ΔabcR1-Up-For and ΔabcR1-Dn-Rev, genomic DNA from B. abortus 2308 as a template, and Platinum® Pfx DNA Polymerase (Invitrogen, Carlsbad, California, USA), while the wild-type abcR2 locus was amplified using similar conditions and the primers ΔabcR2-Up-For and ΔabcR2-Dn-Rev. These DNA fragments were treated with BamHI and PstI, and ligated into BamHI/PstI-digested pNPTS138. The resulting plasmids (pC3032 [RC-abcR1] or pC3033 [RC-abcR2]) were introduced individually into B. abortus CC097 (abcR1 abcR2 double mutant), and selection and screening were carried out as described above. The strain carrying the reconstructed abcR1 locus was named CC119, and the strain carrying the reconstructed abcR2 locus was named CC120.
Northern blot analysis
For small RNAs
RNA was isolated from Brucella cultures as described previously (Caswell et al., 2012). 10μg of RNA was separated on a denaturing 10% polyacrylamide gel containing 7M urea and 1X TBE (89 mM Tris-base, 89 mM boric acid, and 2 mM EDTA). A low molecular weight DNA ladder (New England BioLabs, Ipswich, MA, USA) was labeled with [γ-32P]ATP and polynucleotide kinase, and this radiolabeled ladder was also separated on the polyacrylamide gel. Following electrophoresis in 1X TBE buffer, the ladder and RNA samples were transferred to an Amersham Hybond™-N+ membrane (GE Healthcare, Piscataway, NJ, USA) in 1X TBE buffer. The samples were UV-crosslinked to the membrane, and the membrane was pre-hybridized in ULTRAhyb®-Oligo Buffer (Ambion, Austin, TX, USA) for 2 hours at ~45°C on a rocking shaker. The oligonucleotide probes (AbcR1-Northern, AbcR2-Northern, or AbcR-Northern [for detection of both AbcR1 and AbcR2]) were end-labeled with [γ-32P]ATP and polynucleotide kinase. The radiolabeled probes were incubated with the pre-hybridized membranes at ~45°C on a rocking shaker overnight (~12 hours). The membranes were then washed 3 times for 30 min each with 2X SSC (300 mM sodium chloride and 30 mM sodium citrate), 1X SSC, and 0.5X SSC, respectively, at ~45°C on a rocking shaker. All SSC wash buffers contained 0.1% sodium dodecyl sulfate (SDS). The membranes were then exposed to X-ray film and visualized by autoradiography.
For messenger RNAs
Brucella strains were grown in brucella broth to late exponential phase, and transcription was terminated by the addition of rifampicin (final concentration of 250 μg ml−1). Aliquots of the cultures were killed by mixing with an equal volume of 1:1 ethanol:acetone at time points following rifampicin addition (Time 0 [immediately after the addition of rifampicin], 1, 2, 4, and 8 min. after the addition of rifampicin). The cells were collected by centrifugation (18,000 X g for 5 min at room temperature), and RNA was isolated as described previously (Caswell et al., 2012). 10 μg of total RNA was separated by electrophoresis in a 1.2% agarose formaldehyde gel, and following electrophoresis, the RNA was transferred to an Amersham Hybond™-N+ membrane (GE Healthcare, Piscataway, NJ, USA) by capillary action. The samples were UV-crosslinked to the membrane, and the membrane was washed several times with 5X SSC containing 0.1% SDS at room temperature to remove any remaining formaldehyde. Membranes were then pre-hybridized in ULTRAhyb® Ultrasensitive Hybridization Buffer (Ambion) for 2–3 hours at ~68°C in a hybridization oven with constant rotation. Probes for mRNA detection were generated by in vitro transcription using the T7 RNA polymerase promoter and the MAXIscript® T7 Kit (Ambion). For this, a DNA fragment corresponding to the coding region of the gene of interest was amplified by PCR using Taq polymerase, genomic B. abortus 2308 DNA as a template, and specific oligonucleotide primers, where the T7 promoter (TATAATACGACTCACTATAGGG) was added to the 5′ end of the forward primer (Table S1). The probe was generated using this PCR-generated DNA fragment as a template for transcription and uniform incorporation of [α-32P]UTP according to the manufacturer’s instructions (Ambion), and the radiolabeled probe was then incubated with the membrane for ~12 hours at ~68°C. Membranes were then washed with 4 times for 30 min each with 2X SSC, 1X SSC, 0.5X SSC, and 0.25X SSC respectively, at 68°C. The membranes were then exposed to X-ray film and visualized by autoradiography. In order to determine the half-life of the specific mRNAs, the amount of transcript present at each time point was quantified using ImageJ software (http://rsbweb.nih.gov/ij/), and the amount of transcript present at each time point after rifampicin treatment was compared to the amount of transcript present at the addition of rifampicin (i.e., time 0).
5′-rapid amplification of cDNA ends (5′-RACE)
RNA was isolated from Brucella cultures as described previously (Caswell et al., 2012), and contaminating genomic DNA was removed by treatment with RNase-free DNase I (Ambion). 5′-rapid amplification of cDNA ends (RACE) was carried out using the FirstChoice® RLM-RACE kit (Ambion) according to the manufacturer’s instructions. The abcR1- or abcR2-specific primer AbcR1-5′-RACE or AbcR2-5′-RACE, respectively, was used in the PCR steps of the 5′-RACE protocol where Taq polymerase was also employed. The 5′-RACE products were gel purified, and cloned into pGEM®-T Easy (Promega, Madison, WI, USA). Plasmid DNA was purified from potential clones, and DNA sequencing was performed to identify the transcriptional start site of AbcR1 and AbcR2.
Proteomics
Electrospray ionization tandem mass spectrometric analysis
Proteins were isolated from cultures of B. abortus 2308 and its derivatives grown to early stationary phase in brucella broth (rich medium). Total protein lysates were collected as previously described (Caswell et al., 2012), and 5 μg of protein was separated by 12% SDS-PAGE. The gel was then stained with RAPIDstrain™ (G-Biosciences, Maryland Heights, MO) at room temperature. The protein bands of interest as indicated in Figure 1 by arrows were excised and the gel slices were dried in a speed vacuum at 37°C. Gel bands were de-stained for 1 hour at room temperature using 100 μL of 50% acetonitrile (ACN), 50 mM NH4HCO3/50% ACN and 10 mM NH4HCO3/50% ACN. The gel bands were dried and incubated with trypsin (Promega, Madison, WI, USA) in 10 mM NH4HCO3 overnight at 37°C. Resulting peptides were extracted from trypsinized gels by washing gel pieces for 2 hours with 0.1% trifluoroacetic acid (TFA) and 60 % ACN and were purified using C18 Zip-Tip™ (Millipore, Billerica, MA) (Wiederin et al., 2009).
Peptides were fractionated on a microcapillary RP-C18 column (NewObjectives, Woburn, MA, USA) followed by fragmentation using the nano-ESI–LC–MS/MS system (Eksigent LC and LTQ Orbitrap from Thermo Scientific, San Jose, CA, USA). For protein identification, spectra obtained from mass spectrometric analyses were analyzed using the Sequest algorithm: Proteome Discoverer 1.1 software. The following parameters were used: Database: Bacterial (nr.fasta) ftp.ncbi.nih.gov/blast/db/FASTA; Max precursor Mass: 2000; Min precursor Mass: 300; Precursor Mass tolerance: 10 ppm; Fragment Mass tolerance: 0.8 Da; Average Precursor Mass: False; Fragmentation Mass: True; Dynamic modification: Oxidation (M); Static Modification: Carboxymethyl (C).
Isobaric tag for relative and absolute quantitation (iTRAQ) proteomic analysis
iTRAQ was performed as described previously (Eyford et al., 2011). Briefly, B. abortus 2308 and B. abortus CC097 (ΔabcR1 abcR2) were grown to early stationary phase in brucella broth, and the cells were collected by centrifugation (15,000 X g for 10 min at 4°C) and suspended in ~700 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH7.5). The cells were then boiled for 1 hour, followed by 20 cycles (setting 4; 20 seconds per cycle) of disintegration in Lysing Matrix B (MP Biomedicals, Solon, OH) using a BIO101 FastPrep FP120 cell disintegrator (Thermo Savant [Thermo Fisher Scientific Inc.], Waltham, MA). Approximately 100 μg of protein from each sample was digested with sequencing grade modified porcine trypsin (Promega), and the tryptic peptides were labeled with isobaric tags according to the guidelines in the iTRAQ® Reagents Multiplex Kit (AB SCIEX, Foster City, CA, USA). The labeled peptides were then purified using strong cation exchange (SCX) high performance liquid chromatography (HPLC) before being analyzed via liquid chromatography and tandem mass spectrometry (LC-MS/MS). ProteinPilot V2.0.1 software (AB SCIEX, Foster City, CA, USA) was used to search the annotated Brucella melitensis biovar abortus 2308 genome sequence, and differences in the amounts of identical peptides between the parental strain 2308 and the hfq mutant were determined using the software. The data are shown as the ratio of CC097:2308; therefore, a value >1 indicates that a given protein is more abundant in the abcR1 abcR2 double mutant compared to the parental strain 2308.
Microarray analysis
RNA was isolated from Brucella cultures grown to early stationary phase in brucella broth (Caswell et al., 2012), and contaminating genomic DNA was removed by treatment with RNase-free DNase I (Ambion). Ten micrograms of each RNA sample were reverse transcribed, fragmented and 3′ biotinylated as previously reported (Beenken et al., 2004). The labeled cDNA (1.5 μg) was hybridized to custom-made Brucella abortus GeneChips (PMD2308a520698F) according to the manufacturer’s recommendations for antisense prokaryotic arrays (Affymetrix, Inc., Santa Clara, CA, USA). Signal intensities were normalized to the median signal intensity value for each GeneChip, averaged and analyzed with GeneSpring Software X. RNA species exhibiting ≥ 2-fold change in expression, as determined by Affymetrix algorithms to be statistically differentially expressed (T-test; p ≤ 0.05), between the parental strain 2308 and the abcR1 acbR2 double mutant strain were reported. The microarrays used in this study were developed based on B. melitensis biovar abortus 2308 and all B. abortus GenBank entries that were available at the time of design. In total, 3019 predicted Brucella open reading frames and 1892 intergenic regions greater than 50 bp in length are represented on PMD2308a520698F. The GenBank accession number for the microarray data is GSE38017.
Two-plasmid system for assessing target regulation by AbcR1/2
The E. coli-based system for studying gene regulation by sRNAs developed by Urban and Vogel was used here to assess the regulation of target mRNAs by Brucella AbcR1 and AbcR2 (Urban and Vogel, 2007). The genes encoding AbcR1, AbcR2, or a nonsense sRNA were amplified by PCR using genomic DNA from B. abortus 2308 as a template, Platinum® Pfx DNA Polymerase (Invitrogen), and the primer sets AbcR1-express-For/Rev, AbcR2-express-For/Rev, or nonsense sRNA-express-For/Rev, respectively. The amplified DNA fragments were digested with XbaI and treated with polynucleotide kinase, and the digested/treated fragments were then cloned into a derivative of pZE12 as described previously (Urban and Vogel, 2007). To construct the gfp fusion constructs, the 5′-untranslated region (UTR) to the first 10 codons of a target Brucella gene was amplified by PCR using genomic DNA from B. abortus 2308 as a template, Platinum® Pfx DNA Polymerase (Invitrogen), and specific primers (Table S1). The target genes tested were BAB1_0314, BAB2_0879 and BAB2_0612. The PCR-amplified DNA fragments were digested with NsiI and NheI, and ligated into pXG10 (Urban and Vogel, 2007). The authenticity of all constructs was confirmed by DNA sequence analysis. E. coli TOP10 cells (Invitrogen) were transformed with either a target gfp fusion plasmid alone, or with a combination of a target gfp fusion plasmid and a sRNA expression plasmid. The levels of GFP in the E. coli strains were assessed by immunoblot analysis using the methods described previously (Caswell et al., 2012). Here, GFP was detected using anti-GFP anibodies (Roche), and detection of GroEL as a loading control was performed with anti-GroEL antibodies (Sigma).
Experimental infection of murine macrophages and mice
Experiments to test the virulence of Brucella strains in primary, peritoneal murine macrophages were carried out as described previously (Gee et al., 2005). Briefly, macrophages were isolated from mice and seeded in 96-well plates in Dulbecco’s modified Eagle’s medium with 5% fetal bovive serum, and the following day, the macrophages were infected with opsonized brucellae at an MOI of 50:1. After 2 hours of infection, extracellular bacteria were killed by treatment with gentamicin (50 μg ml−1). For the 2-hour time point, the macrophages were then lysed with 0.1% deoxycholate in PBS, and serial dilutions were plated on Schadler blood agar (SBA). For the 24- and 48-hour time points, the cells were washed with PBS following gentamicin treatment, and fresh cell culture medium containing gentamicin (20 μg ml−1) was added to the monolayer. At the indicated time point, the macrophages were lysed, and serial dilutions were plated on SBA. Triplicate wells were used for each Brucella strain tested.
The infection and colonization of mice by Brucella strains was performed as described previously (Gee et al., 2005). 6–8 week-old female C57BL/6 mice (5 per Brucella strain per time point) were infected intraperitoneally with ~5 × 104 CFU of each Brucella strain in sterile PBS. The mice were sacrificed at 1 and 4 weeks post-infection, and serial dilutions of spleen homogenates were plated on SBA.
Supplementary Material
Fig. S1. Alignment of the AbcR sRNAs from Agrobacterium tumefaciens C58 and Brucella abortus 2308
The nucleotide sequence of AbcR1 and AbcR2 from Brucella abortus 2308 and Agrobacterium tumefaciens C58 were aligned using the AlignX® software from Vector NTI® (Invitrogen). Identical nucleotides between the two sRNAs are shown as the consensus sequence in bold font
A. Alignment of AbcR1 sRNAs
B. Alignment of AbcR2 sRNAs
Fig. S2. Northern blot analysis of AbcR sRNAs from different Brucella species
Total RNA isolated from cultures of B. abortus 2308, B. suis 1330, and B. melitensis 16M was separated on denaturing polyacrylamide gels. Following transfer to a membrane, a single probe designed to detect either AbcR1 or AbcR2 were used in the northern blot analyses. Detection of 5S rRNA was also performed as a loading control.
Fig. S3. Folding predictions of the AbcR1 and AbcR2 secondary structures
Secondary structure predictions of the AbcR sRNAs from Brucella abortus 2308 were determined using mfold (http://mfold.rna.albany.edu/?q=mfold).
Fig. S4. Hfq binding to AbcR1 and AbcR2
Electrophoretic mobility shift assays (EMSAs) were employed to test the binding between of Hfq to the AbcR sRNAs, and methods described previously were used for these experiments (Caswell et al., 2012). The AbcR1 and AbcR2 sRNAs were transcribed in vitro and radiolabeled by [α-32P]UTP incorporation, and EMSAs were performed using the labeled in vitro transcribed sRNAs and purified recombinant Brucella abortus Hfq. Increasing concentrations of rHfq were incubated with the labeled RNA probe, and the binding reactions were resolved in 5% native polyacrylamide gels and visualized by autoradiography.
Fig. S5. Schematic of putative interacting motif between the AbcR sRNAs and target mRNAs in B. bortus 2308.
The numbers in parentheses depict the nucleotide position of the motif within the AbcR sRNAs, and similarly, the distance from the putative binding motif to the start codon is shown for the mRNAs.
Fig. S6. Proteomic analysis of B. abortus 2308 and the B. abortus abcR1 abcR2 double mutant strains
iTRAQ analysis was performed as described in the Experimental procedures. The value for a given protein represents the fold difference in the level of that protein between the two indicated strains. For example, values in the “abcR/WT-1” column depict protein levels in the abcR1 abcR2 double mutant strain compared to protein levels in the parental strain 2308.
Acknowledgments
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases to R.M.R. (AI48499). The authors would also like to thank Edson Rocha for assistance with experiments involving rifampicin treatment and subsequent Northern blot analysis. The iTRAQ proteomics work was supported by Genome Canada and Genome BC platform funding grants.
References
- Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Altuvia S. Identification of bacterial small non-coding RNAs: experimental approaches. Curr Opin Microbiol. 2007;10:257–261. doi: 10.1016/j.mib.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Barra-Bily L, Pandey SP, Trautwetter A, Blanco C, Walker GC. The Sinorhizobium meliloti RNA chaperone Hfq mediates symbiosis of S. meliloti and alfalfa. J Bacteriol. 2010;192:1710–1718. doi: 10.1128/JB.01427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut J, Andersson SG, O’Callaghan D. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat Rev Microbiol. 2004;2:933–945. doi: 10.1038/nrmicro1044. [DOI] [PubMed] [Google Scholar]
- Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat R, Axtell R, Mitra A, Miranda M, Lock C, Tsien RW, Steinman L. Inhibitory role for GABA in autoimmune inflammation. Proc Natl Acad Sci USA. 2010;107:2580–2585. doi: 10.1073/pnas.0915139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell CC, Gaines JM, Roop RM., II The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator, BabR, in Brucella abortus 2308. J Bacteriol. 2012;194:3–14. doi: 10.1128/JB.05623-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Gorvel JP. Organelle robbery: Brucella interactions with the endoplasmic reticulum. Curr Opin Microbiol. 2004;7:93–97. doi: 10.1016/j.mib.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Chevrot R, Rosen R, Haudecoeur E, Cirou A, Shelp BJ, Ron E, Faure D. GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2006;103:7460–7464. doi: 10.1073/pnas.0600313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Val C, Rivas E, Torres-Quesada O, Toro N, Jiménez-Zurdo JI. Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol Microbiol. 2007;66:1080–1091. doi: 10.1111/j.1365-2958.2007.05978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Val C, Romero-Zaliz R, Torres-Quesada O, Peregrina A, Toro N, Jiménez-Zurdo JI. A survey of sRNA families in α-proteobacteria. RNA Biol. 2012;9 doi: 10.4161/rna.18643. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- Eyford BA, Sakurai T, Smith D, Loveless B, Hertz-Fowler C, Donelson JE, Inoue N, Pearson TW. Differential protein expression throughout the life cycle of Trypanosoma congolense, a major parasite of cattle in Africa. Mol Biochem Parasitol. 2011;177:116–125. doi: 10.1016/j.molbiopara.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantappiè L, Metruccio MM, Seib KL, Oriente F, Cartocci E, Ferlicca F, Giuliani MM, Scarlato V, Delany I. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect Immun. 2009;77:1842–1853. doi: 10.1128/IAI.01216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Gee JM, Valderas MW, Kovach ME, Grippe VK, Robertson GT, Ng WL, Richardson JM, Winkler ME, Roop RM., II The Brucella abortus Cu/Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect Immun. 2005;73:2873–2880. doi: 10.1128/IAI.73.5.2873-2880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt P. The nutrition of brucellae. Bacteriol Rev. 1958;22:81–98. doi: 10.1128/br.22.2.81-98.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Vu GP, Bai Y, Chan E, Wu R, Yang E, Liu F, Lu S. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog. 2011;7:e1002120. doi: 10.1371/journal.ppat.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Haudecoeur E, Planamente S, Cirou A, Tannières M, Shelp BJ, Moréra S, Faure D. Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2009;106:14587–14592. doi: 10.1073/pnas.0808005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol. 2011;79:419–432. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol. 2011;79:419–432. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- Koo JT, Alleyne TM, Schiano CA, Jafari N, Lathem WW. Global discovery of small RNAs in Yersinia pseudotuberculosis identifies Yersinia-specific small, noncoding RNAs required for virulence. Proc Natl Acad Sci USA. 2011;108:E709– 717. doi: 10.1073/pnas.1101655108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakov YK, Guigue-Talet PG, Ramuz MR, O’Callaghan D. Response of Brucella suis 1330 and B. canis RM6/66 to growth at acid pH and induction of an adaptive acid tolerance response. Res Microbiol. 1997;148:145–151. doi: 10.1016/S0923-2508(97)87645-0. [DOI] [PubMed] [Google Scholar]
- Lamontagne J, Forest A, Marazzo E, Denis F, Butler H, Michaud JF, Boucher L, Pedro I, Villeneuve A, Sitnikov D, Trudel K, Nassif N, Boudjelti D, Tomaki F, Chaves-Olarte E, Guzmán-Verri C, Brunet S, Côté-Martin A, Hunter J, Moreno E, Paramithiotis E. Intracellular adaptation of Brucella abortus. J Proteome Res. 2009;8:1594–1609. doi: 10.1021/pr800978p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Liu JM, Camilli A. A broadening world of bacterial small RNAs. Curr Opin Microbiol. 2010;13:18–23. doi: 10.1016/j.mib.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhugiri R, Pessi G, Voss B, Hahn J, Sharma CM, Reinhardt R, Vogel J, Hess WR, Fischer HM, Evguenieva-Hackenberg E. Small RNAs of the Bradyrhizobium/Rhodopseudomonas lineage and their analysis. RNA Biol. 2012;9 doi: 10.4161/rna.9.1.18008. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom KL, Forslund AL, Kuoppa K, Alkhuder K, Dubail I, Dupuis M, Forsberg A, Charbit A. Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect Immun. 2009;77:1866–1880. doi: 10.1128/IAI.01496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller T, Franch T, Hojrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. Hfq: A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J Bacteriol. 1990;172:3569–3576. doi: 10.1128/jb.172.7.3569-3576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 2011;39:4235–4248. doi: 10.1093/nar/gkr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman EB, Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu Rev Microbiol. 1994;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res Microbiol. 2007;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe. 2010;8:116–127. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Pappas G, Akritidis N, Bosilkovski M, Tsianos E. Brucellosis. N Engl J Med. 2005;352:2325–2336. doi: 10.1056/NEJMra050570. [DOI] [PubMed] [Google Scholar]
- Prévost K, Desnoyers G, Jacques JF, Lavoie F, Massé E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25:385–396. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repoila F, Majdalani N, Gottesman S. Small non-coding RNAs, coordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- Robertson GT, Roop RM., II The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol Microbiol. 1999;34:690–700. doi: 10.1046/j.1365-2958.1999.01629.x. [DOI] [PubMed] [Google Scholar]
- Roop RM, II, Gaines JM, Anderson ES, Caswell CC, Martin DW. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol. 2009;198:221–238. doi: 10.1007/s00430-009-0123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetti CA, Galindo CL, Garner HR, Adams LG. Transcriptional profile of the intracellular pathogen Brucella melitensis following HeLa cells infection. Microb Pathog. 2011;51:338–344. doi: 10.1016/j.micpath.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acid Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter JP, Reinkensmeier J, Daschkey S, Evguenieva-Hackenberg E, Janssen S, Jänicke S, Becker JD, Giegerich R, Becker A. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti. BMC Genomics. 2010;11:245. doi: 10.1186/1471-2164-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol. 2011;81:1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, Wai SN. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jäger KE, Bläsi U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- Spratt BG, Hedge PJ, te Heesen S, Edelman A, Broome-Smith JK. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey DJ, Anthony DC, Lowe JP, Miller J, Palm WM, Styles P, Perry VH, Blamire AM, Sibson NR. Detection of the inhibitory neurotransmitter GABA in macrophages by magnetic resonance spectroscopy. J Leukoc Biol. 2005;78:393–400. doi: 10.1189/jlb.1203604. [DOI] [PubMed] [Google Scholar]
- Torres-Quesada O, Oruezabal RI, Peregrina A, Jofré E, Lloret J, Rivilla R, Toro N, Jiménez-Zurdo JI. The Sinorhizobium meliloti RNA chaperone Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa. BMC Microbiol. 2010;10:71. doi: 10.1186/1471-2180-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvé VM, Sevin EW, Chéron A, Barloy-Hubler F. Identification of chromosomal alpha-proteobacterial small RNAs by comparative genome analysis and detection in Sinorhizobium meliloti strain 1021. BMC Genomics. 2007;8:467. doi: 10.1186/1471-2164-8-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Valderas MW, Alcantara RB, Baumgartner JE, Bellaire BH, Robertson GT, Ng WL, Richardson JM, Winkler ME, Roop RM., II Role of HdeA in acid resistance and virulence in Brucella abortus 2308. Vet Microbiol. 2005;107:307–312. doi: 10.1016/j.vetmic.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Valverde C, Livny J, Schlüter JP, Reinkensmeier J, Becker A, Parisi G. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genomics. 2008;9:416. doi: 10.1186/1471-2164-9-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse M, Fauvart M, Cloots L, Engelen K, Thijs IM, Marchal K, Michiels J. Genome-wide detection of predicted non-coding RNAs in Rhizobium etli expressed during free-living and host-associated growth using a high-resolution tiling array. BMC Genomics. 2010;11:53. doi: 10.1186/1471-2164-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell. 2002;109:141–144. doi: 10.1016/s0092-8674(02)00717-1. [DOI] [PubMed] [Google Scholar]
- Wiederin J, Duan F, Rozek W, Ciborowski P. Biomarkers of HIV-1 associated neurocognitive disorders: Proteomic investigation of sera. Proteome Sci. 2009;7:8. doi: 10.1186/1477-5956-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilms I, Overlöper A, Nowrousian M, Sharma CM, Narberhaus F. Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens. RNA Biol. 2012;9 doi: 10.4161/rna.17212. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilms I, Voss B, Hess WR, Leichert LI, Narberhaus F. Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol Microbiol. 2011;80:492–506. doi: 10.1111/j.1365-2958.2011.07589.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Alignment of the AbcR sRNAs from Agrobacterium tumefaciens C58 and Brucella abortus 2308
The nucleotide sequence of AbcR1 and AbcR2 from Brucella abortus 2308 and Agrobacterium tumefaciens C58 were aligned using the AlignX® software from Vector NTI® (Invitrogen). Identical nucleotides between the two sRNAs are shown as the consensus sequence in bold font
A. Alignment of AbcR1 sRNAs
B. Alignment of AbcR2 sRNAs
Fig. S2. Northern blot analysis of AbcR sRNAs from different Brucella species
Total RNA isolated from cultures of B. abortus 2308, B. suis 1330, and B. melitensis 16M was separated on denaturing polyacrylamide gels. Following transfer to a membrane, a single probe designed to detect either AbcR1 or AbcR2 were used in the northern blot analyses. Detection of 5S rRNA was also performed as a loading control.
Fig. S3. Folding predictions of the AbcR1 and AbcR2 secondary structures
Secondary structure predictions of the AbcR sRNAs from Brucella abortus 2308 were determined using mfold (http://mfold.rna.albany.edu/?q=mfold).
Fig. S4. Hfq binding to AbcR1 and AbcR2
Electrophoretic mobility shift assays (EMSAs) were employed to test the binding between of Hfq to the AbcR sRNAs, and methods described previously were used for these experiments (Caswell et al., 2012). The AbcR1 and AbcR2 sRNAs were transcribed in vitro and radiolabeled by [α-32P]UTP incorporation, and EMSAs were performed using the labeled in vitro transcribed sRNAs and purified recombinant Brucella abortus Hfq. Increasing concentrations of rHfq were incubated with the labeled RNA probe, and the binding reactions were resolved in 5% native polyacrylamide gels and visualized by autoradiography.
Fig. S5. Schematic of putative interacting motif between the AbcR sRNAs and target mRNAs in B. bortus 2308.
The numbers in parentheses depict the nucleotide position of the motif within the AbcR sRNAs, and similarly, the distance from the putative binding motif to the start codon is shown for the mRNAs.
Fig. S6. Proteomic analysis of B. abortus 2308 and the B. abortus abcR1 abcR2 double mutant strains
iTRAQ analysis was performed as described in the Experimental procedures. The value for a given protein represents the fold difference in the level of that protein between the two indicated strains. For example, values in the “abcR/WT-1” column depict protein levels in the abcR1 abcR2 double mutant strain compared to protein levels in the parental strain 2308.