Abstract
The proteolytic processing of amyloid precursor protein (APP) to generate the neurotoxic Aβ peptide is central to the pathogenesis of Alzheimer disease (AD). The endocytic system mediates the processing of APP by controlling its access to secretases that cleave APP. A key mediator of APP localization is SorL1 – a membrane protein that has been genetically linked to AD. The retromer complex is a conserved protein complex required for endosome-to-Golgi retrieval of a number of physiologically important membrane proteins including SorL1. Based on the prior suggestion that endocytosis and retromer sorting pathways might be involved, we hypothesized that variants in other genes in this pathway might also modulate AD risk. Genetic association of AD with 451 polymorphisms in 15 genes encoding retromer or retromer-associated proteins was tested in a Caucasian sample of 8,309 AD cases and 7,366 cognitively normal elders using individual SNP and gene-based tests. We obtained significant evidence of association with KIAA1033 (Paris p = 0.025), SNX1 (Paris p =0.035), SNX3 (p = 0.0057) and RAB7A (Paris p = 0.018). Ten KIAA1033 SNPs were also significantly associated with AD in a group of African Americans (513 AD cases, 504 controls). Findings with four significant SNX3 SNPs in the discovery sample were replicated in a community-based sample of Israeli-Arabs (124 AD cases, 142 controls). We show that Snx3 and Rab7A proteins interact with the cargo-selective retromer complex through independent mechanisms to regulate the membrane association of retromer and thereby are key mediators of retromer function. These data implicate additional AD risk genes in the retromer pathway and formally demonstrate a direct link between the activity of the retromer complex and the pathogenesis of AD.
1. Introduction
The localization of membrane proteins to discrete and specific compartments within eukaryotic cells is governed by a complex interplay of protein-protein interactions in which a sorting motif(s) in the cytoplasmic tail of a membrane protein is recognized by membrane-associated ‘coat’ proteins to direct the respective membrane proteins into a tubule or vesicle for transport to another compartment. A failure in the fidelity of sorting processes can lead to a range of pathologies. Sometimes the failure occurs when a sorting motif is mutated – a notable example being the mutation of the NPXY motif identified as causal in familial hypercholesterolemia by Brown and Goldstein [3]. Alternatively the molecular machinery that recognizes sorting motifs is at fault, for example, patients with deficient AP-3 function in Hermansky-Pudlack syndrome [10].
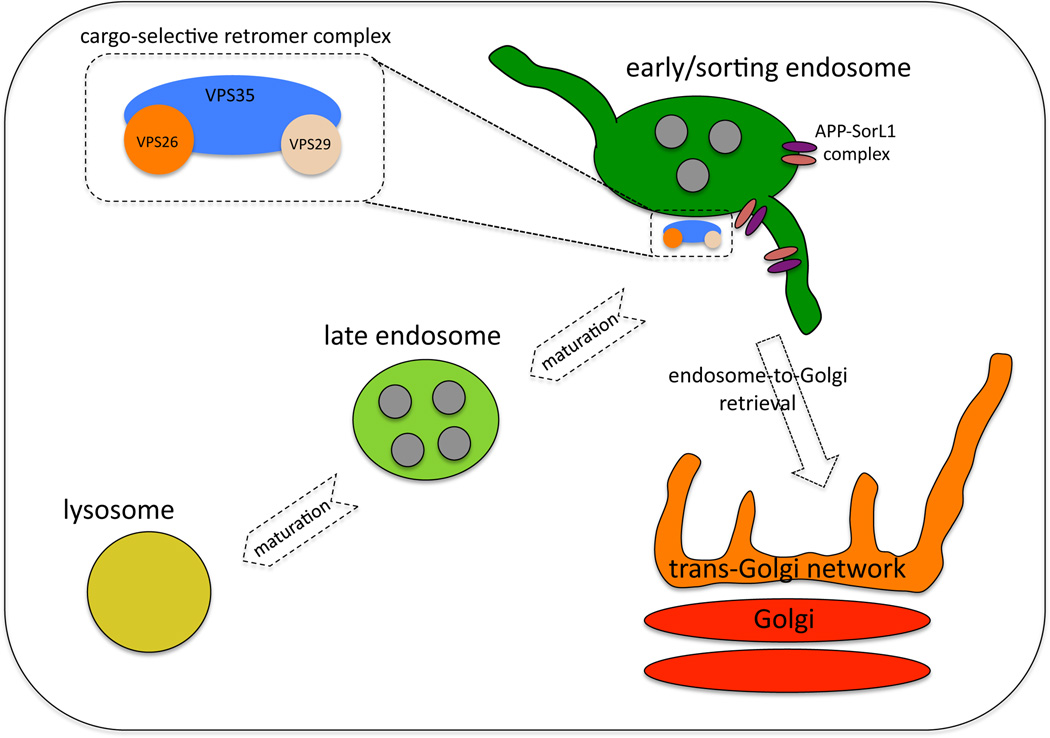
There has been a growing appreciation recently of the importance of correct protein sorting in regulating the processing of amyloid precursor protein (APP) and therefore the proteins that function in mediating localization to the post-Golgi endocytic system have been of great interest to studies of the underlying causes of late-onset Alzheimer disease (AD). Recently the retromer complex, an endosomally-localized protein complex, has been implicated in regulating APP processing (Figure 1) [8,61].
Figure 1.
Schematic diagram of the endocytic pathway and the role of retromer in sorting APP and SorL1. The SorL1 protein associates with APP. The cargo-selective retromer complex interacts with SorL1 to direct the APP-SorL1 complex into an endosome-to-Golgi retrieval pathway. Aberrant APP localization to late endosomal compartments increases processing to the neurotoxic Aβ peptide.
The retromer complex is a conserved endosome-associated protein complex that was first identified in yeast as essential for the endosome-to-Golgi retrieval of the CPY-sorting receptor, Vps10p. The studies first conducted in yeast revealed that retromer comprises five proteins, (encoded by vacuole protein sorting – VPS - genes) that are arranged into two functionally distinct subcomplexes; a cargo-selective trimer of Vps35p, Vps29p and Vps26p and a structural complex proposed to drive vesicle or tubule formation made of a dimer of the yeast sorting nexin proteins, Vps5p and Vps17p [53]. The retromer complex is conserved across all eukaryotes underscoring its vital role in mediating endosomal protein sorting [24].
Since retromer was first identified in yeast, studies in a variety of systems have identified cargo proteins that require retromer for their localization, and accessory proteins that function with retromer in endosomal protein sorting. For example, the small GTPase Rab7A associates with the cargo-selective retromer complex to mediate it’s localization to endosomes [47]. Other retromer-associated proteins include TBC1D5, a rab GTPase activating protein, Eps15-homology domain containing protein-1 (EHD1) and the WASH complex [15,17,52].
Membrane proteins that depend on retromer for their proper localization, and therefore are considered to be retromer cargo proteins, now include; the cation-independent mannose 6-phosphate receptor (CIMPR), Wntless, a protein required for the secretion of the Wnt morphogen, DMT-1, a divalent cation transporter and the Vps10-family members sortilin and SorL1 (also known as SorLA) [4,9,13,41,51,52,62]. Since SorL1 has been shown to associate with APP and regulate the processing of APP [27], it has been of interest to studies directed at understanding the pathology of AD. The pathophysiological importance of the physical association of SorL1 and its four type I membrane homologs (sortilin, sorCS1, sorCS2 and sorCS3) with APP has been elevated by the identification of AD-linked single nucleotide polymorphisms (SNPs) in the genes encoded by these loci [29,43–46].
Retromer is required to mediate the localization of SorL1 and loss of retromer results in increased production of the pro-aggregatory neurotoxic Aβ peptide. Indeed a reduction in the levels of retromer has been detected in neuronal tissue of AD-patients suggesting a causal link between retromer activity and AD [26,38,41,57] and carriers of some AD-associated SORL1 haplotypes have lower transcription of SORL1 [46]. Therefore genes encoding retromer subunits or retromer-associated proteins and genes that function in mediating endosomal protein sorting are prime candidates for analyses directed towards identifying AD-linked SNPs.
In this study, we analyzed the association of AD with SNPs in genes that have putative retromer function or interact with the retromer-complex to mediate endosomal protein sorting. Four out of 15 retromer or retromer-associated genes (SNX3, RAB7A, KIAA1033 and SNX1) showed significant evidence of association with AD in SNP-based and/or gene-based analyses. Intriguingly, loss of Snx3 or Rab7A function results in a reduction in endosome-associated retromer although the data presented herein suggests that Snx3 and Rab7A regulate retromer membrane association through distinct mechanisms.
2. Methods
2.1 Datasets
Summarized information from tests of genetic association of AD with SNPs located in the candidate gene regions was culled from a recent large genome-wide association study (GWAS) conducted by the Alzheimer Disease Genetics Consortium (ADGC) [39]. Naj et al. computed results for SNPs throughout the genome in their discovery sample composed of 8,309 AD cases and 7,366 cognitively normal elders from ten independent Caucasian data sets (Table 2). Details of the quality control and statistical analysis including genotype imputation procedures and genetic models has been published elsewhere [39]. GWAS datasets containing African Americans (513 AD cases, 504 controls) [35] and Israeli-Arabs (124 AD cases, 142 controls) from a consanguineous community [55] were used to replicate top-ranked findings in the Caucasian datasets. Although these samples are modest are modest in size, evidence of replication in populations which are genetically distinct from the discovery sample would make the findings more robust and generalizable [35, 55].
Table 2.
Characteristics of the GWAS datasets
| Cases | Controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases (N) |
Female (N, %) |
Age at onset (mean ± SD) |
Age at exam (mean ± SD) |
APOE ε4 (allele %) |
Controls (N) |
Female (N, %) |
Age at exam (mean ± SD) |
APOE ε4 (allele %) |
|
| ACT | 566 | 357(63%) | 83.90 (4.8) | 84.72 (4.9) | 0.26 | 1696 | 947 (56%) | 81.08 (6.0) | 0.11 |
| ADC1 | 1566 | 850 (54%) | 72.47 (7.1) | 81.61 (7.0) | 0.42 | 515 | 305 (59%) | 75.00 (8.0) | 0.16 |
| ADC2 | 738 | 377 (51%) | 73.19 (7.1) | 80.06 (7.2) | 0.39 | 160 | 110 (69%) | 75.68 (7.9) | 0.16 |
| ADNI | 268 | 113 (42%) | 75.30 (7.2) | 77.96 (6.5) | 0.42 | 173 | 70 (40%) | 78.6 (5.5) | 0.14 |
| GenADA | 669 | 380 (57%) | 74.59 (6.2) | 80.36 (6.2) | 0.38 | 713 | 456 (64%) | 74.21 (7.0) | 0.13 |
| UM/VU/MSSM | 1186 | 764 (64%) | 74.06 (7.8) | 77.48 (6.9) | 0.36 | 1135 | 696 (61%) | 74.00 (8.3) | 0.12 |
| MIRAGE | 509 | 324 (64%) | 71.16 (6.5) | 75.97 (6.6) | 0.36 | 753 | 440 (58%) | 72.04 (7.2) | 0.23 |
| NIA-LOAD | 1811 | 1176 (65%) | 73.57 (6.7) | 82.49 (7.1) | 0.46 | 1575 | 947 (60%) | 73.99 (8.5) | 0.20 |
| OHSU | 132 | 81 (61%) | 86.10 (5.5) | 90.40 (5.2) | 0.23 | 153 | 84 (55%) | 83.86 (7.6) | 0.08 |
| TGEN2 | 864 | 633 (73%) | 74.91 (7.2) | 82.00 (7.6) | 0.40 | 493 | 186 (38%) | 80.19 (8.7) | 0.11 |
| TOTAL | 8309 | 5055 (61%) | -- | -- | -- | 7366 | 4241 (58%) | -- | -- |
2.2 Gene selection
We tested the genetic association of AD with eight genes encoding the subunits of the mammalian retromer-complex [16,66] and with seven other retromer-associated genes (Table 1). The GTPase Rab7A mediates the localization of the retromer to the endosome [47]. FAM21C, KIAA1033, KIAA0196 form subunits of the human WASH-complex and are involved in retromer-dependent endosomal protein sorting [17]. The Eps15 homology domain-containing protein-1 (EHD1) interacts with the retromer to facilitate endosome-to-Golgi retrieval [15]. FK506 binding protein 15(FKBP15) has shown evidence of being involved in regulation of early endocytic transport [64]. The sorting nexin 3 (Snx3) has been shown to interact with the cargo-selective subcomplex of the retromer to sort Wntless (Wnt-binding protein) [19].
Table 1.
Retromer-complex and retromer-associated genes tested for genetic association with AD
| RETROMER COMPLEX |
RETROMER ASSOCIATED |
|---|---|
| SNX1 | EHD1 |
| SNX2 | FAM21C |
| VPS26A | FKBP15 |
| VPS26B | KIAA0196 |
| VPS29 | KIAA1033 |
| VPS35 | RAB7A |
| SNX5 | SNX3 |
| SNX6 |
2.3 Study heterogeneity and gene-based multiple testing correction
Heterogeneity of odds ratios for each SNP among datasets was assessed using the Cochran's Q and I2 statistics [20]. We corrected for testing multiple SNPs in a gene after accounting for correlation between SNP genotypes due to linkage disequilibrium. Each gene tested was treated as an independent hypothesis and the effective number of tests per gene was obtained by a method described by Li and Ji [32]. The Versatile Gene-based Association Study (VEGAS) approach [34] was used to summarize the strength of association of a gene with AD based on the number of SNPs tested in the gene and size of the gene. This method computes a gene-based test statistic based on the SNP p-values within the gene, and then uses simulation to calculate an empirical gene-based p-value taking into account all genes in the genome.
2.4 Antibodies, biochemicals and other reagents
Unless otherwise stated, general biochemical reagents were purchased from Sigma-Aldrich (Poole, Dorset, UK). Polyclonal antibodies against Vps26, Vps35, GFP and Snx1 have been described previously [51,52]. Polyclonal anti-actin, anti-Snx3 and monoclonal anti-Rab7a were purchased from Sigma-Aldrich (Poole, Dorset, UK). Monoclonal antibodies against Snx1 and EEA1 were purchased from BD Bioscience (Oxford, UK). The monoclonal anti-GFP was purchased from Invitrogen, (Paisley, Scotland, UK). The monoclonal anti-CD8 antibody was produced from a hydridoma cell line as described previously [50]. Fluorescently labeled secondary antibodies for immunofluorescence were purchased from Molecular Probes (Paisley, Scotland, UK). Small interfering (si) RNA for RNAi experiments was purchased from Dharmacon (Lafayette, CO, USA). 125I-protein A for western blotting was purchased from Perkin Elmer (Waltham, MA, USA). Effectene used in transfections was purchased from Qiagen (Crawley, Sussex, UK).
2.5 Generation of GFP-construct and cell lines
GFP-tagged Snx3 was produced by amplifying an EST encoding human Snx3 incorporating BamHI and SalI sites at the 5’ and 3’ ends respectively. The PCR product was cloned initially into pCR Blunt (Invitrogen – Paisley, Scotland, UK) sequenced and then subcloned into pEGFP C1 at the BglII and SalI sites. The GFP-Snx3 construct was then subcloned into pIRESneo2 following excision with NheI-BamHI and cloned into NheI-BamHI digested vector. Similar methodology was applied to generate the GFP-Syntaxin 6 cell line. The GFP-RhoB construct was generously provided by Professor H. Mellor (University of Bristol, UK) and was cloned into pIRESneo2 similarly to GFP-Snx3. The other GFP-tagged cell lines have been reported previously in [17,52].
2.6 Yeast two-hybrid analysis
The yeast two hybrid system (and most of the constructs) have been described previously [17]. The pGAD424-Snx3 construct was generated by subcloning Snx3 from pCR Blunt into a modified pGAD424 in which the BamHI and SalI sites in the multi-cloning site of pGAD424 were in the same reading frame as pEGFP C1.
2.7 Native immunoprecipitations
The immunoprecipitation of GFP-tagged proteins from HeLa cell lysates was performed as described previously [52] using the following lysis buffer; 100 mM Mes-NaOH, pH 6.5, 200 µM sodium orthovanadate; 0.5 mM EGTA, 1% digitonin and protease inhibitors using a polyclonal anti-GFP antisera described previously [52]. The CD8-CIMPR and CD8-SorL1 expressing cell lines have been reported elsewhere [14,50]. The immunoprecipitation of the CD8-reporter proteins was performed as described previously [14,50]. SDS-PAGE and western blotting was performed as described previously [51].
2.8 RNAi-mediated knockdowns
The silencing of Snx3, Rab7A etc expression was achieved by transfection of siRNA into the HeLa cells used as described previously [51]. For large scale siRNA knockdown experiments (e.g. in the CD8-reporter expressing cell lines) a single transfection of 500 pmol of siRNA was performed for cells grown on 140 mm tissue culture dishes. 48 hours post-transfection, each dish was trypsinised and the cells were seeded into two new 140 mm dishes. The cells were used for the respective experiment 24 hours later.
2.9 Automated microscopy
Following siRNA-mediated knockdown of target genes cells were seeded on 96-well plates and allowed to attach overnight. Cells were then fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton X-100 for 10 minutes and blocked with 3% BSA in PBS at 4°C overnight. Next, cells were labelled with anti-VPS26 and anti-Snx1 or anti-EEA1 antibodies for 1 hr at room temperature followed by Alexa Fluor 488-labeled secondary antibody. Finally, cells were also treated with a Whole Cell Stain Blue (Dharmacon, Lafayette, CO, USA) for 1 hr at room temperature before imaging on a Cellomics Arrayscan automated microscope, using a 2-channel Spot Detector protocol. The whole cell stain was used for focusing and for the definition of the cell outlines; in the 2nd channel endosomes were counted with the Spot Detector Algorithm using an identical intensity threshold for all conditions. To measure the overlap coefficient for the CD8-reporters and VPS26, cells treated with siRNA were seeded into four wells of a 24-well plate and allowed to adhere overnight. Following fixation and antibody staining using polyclonal anti-VPS26 and monoclonal anti-CD8, the cells were further stained with Hoecsht to visualize the nucleus. Using the nucleus for focusing, the cells were imaged on a Cellomics Arrayscan automated microscope and the intensity of the VPS26 staining that was coincident with the CD8-reporter protein was measured using the Colocalization Algorithm.
3. Results
3.1 AD is Associated With KIAA1033, RAB7A, SNX1, and SNX3
Nominally significant evidence of AD association was observed with at least one SNP in seven of the 15 retromer-related genes (Table 3 and Supplementary Table 1). In four of these seven genes (KIAA1033, RAB7A, SNX1 and SNX3), approximately 50% or more of the SNPs were significantly associated with AD at p=0.05. Two SNPs in SNX3 were significant after multiple testing correction (best p=0.0056 for rs12524840) and two SNPs in RAB7A nearly met the gene-based significance threshold (rs9831813, p=0.0058 rs7631994, p=0.0059). Analysis of the individual SNP results using the VEGAS approach revealed that AD was significantly associated with RAB7A, KIAA1033 and SNX1 at the gene level (Table 3). This gene-based analysis also showed suggestive evidence of association with SNX3 (p=0.06). This result may have been tempered by heterogeneity of odds ratios among studies for five SNX3 SNPs (p<0.05).
Table 3.
SNP-based and gene-based association test results
| GENE | #SNPs | #SNPs SIGNIFICANT AT 0.05 |
#SNPs SIGNIFICANT AFTER MULT TESTING |
BEST P-VALUE |
MULTIPLE TESTING THRESHOLD |
PARIS P-VALUE |
|---|---|---|---|---|---|---|
| EHD1 | 18 | 3 | 0 | 0.033 | 0.010 | 0.13 |
| FAM21C | 2 | 0 | 0 | 0.31 | 0.025 | 0.33 |
| FKBP15 | 95 | 0 | 0 | 0.11 | 0.0033 | 0.44 |
| KIAA0196 | 40 | 3 | 0 | 0.044 | 0.0056 | 0.18 |
| KIAA1033 | 56 | 27 | 0 | 0.0080 | 0.0045 | 0.025 |
| RAB7A | 32 | 17 | 0 | 0.0058 | 0.0050 | 0.018 |
| SNX1 | 17 | 16 | 0 | 0.035 | 0.017 | 0.035 |
| SNX2 | 43 | 0 | 0 | 0.15 | 0.0056 | 0.56 |
| SNX3 | 28 | 13 | 2 | 0.0057 | 0.0071 | 0.059 |
| SNX5 | 40 | 0 | 0 | 0.18 | 0.0071 | 0.32 |
| SNX6 | 24 | 0 | 0 | 0.14 | 0.0071 | 0.44 |
| VPS26A | 28 | 0 | 0 | 0.066 | 0.0062 | 0.21 |
| VPS26B | 12 | 1 | 0 | 0.034 | 0.0062 | 0.25 |
| VPS29 | 3 | 0 | 0 | 0.26 | 0.017 | 0.59 |
| VPS35 | 4 | 0 | 0 | 0.82 | 0.025 | 0.90 |
Examination of results for SNPs in these four genes in the replication datasets revealed no evidence of association in RAB7A or SNX1. However, in the Israeli-Arab dataset nearly significant results (p≤0.08) were obtained with four SNX3 SNPs all of which were nominally significant (p≤0.03) with the same pattern of effect in the Caucasians (Supplementary Table 2). Ten SNPs distributed across the proximal half of KIAA1033 were significantly associated with AD in AAs (Supplementary Table 2). Remarkably, these SNPs intercalate the 28 SNPs significantly associated with AD in Caucasians. Considering results from both single SNP and gene-based association analyses, SNX3 and RAB7A provided the most compelling evidence of association with AD. Consequently, these genes were studied further for genetic interactions with SORL1 and each other and for effects on retromer function.
All possible two-way interactions of SNPs from SNX3 (28 SNPs), RAB7A (32 SNPs) and SORL1 (152 SNPs) were tested in models including terms for the SNPs and their interaction. The most significant evidence of interaction was found involving rs2276346 in SORL1 and 14 RAB7A SNPs (0.0019 ≤ p ≤ 0.0037), however these results did not survive multiple testing correction (threshold p-value for 280 independent tests = 0.00018). Nominally significant interactions were also observed with SORL1 SNP rs11512475 and five SNX3 SNPs (0.0064 ≤ p ≤ 0.0080) which also did not remain significant after multiple test correction (threshold p-value for 196 independent tests = 0.00026). None of the interactions in models including RAB7A and SNX3 SNPs were significant at p < 0.01.
3.2 Interaction of Snx3 and Rab7A with retromer
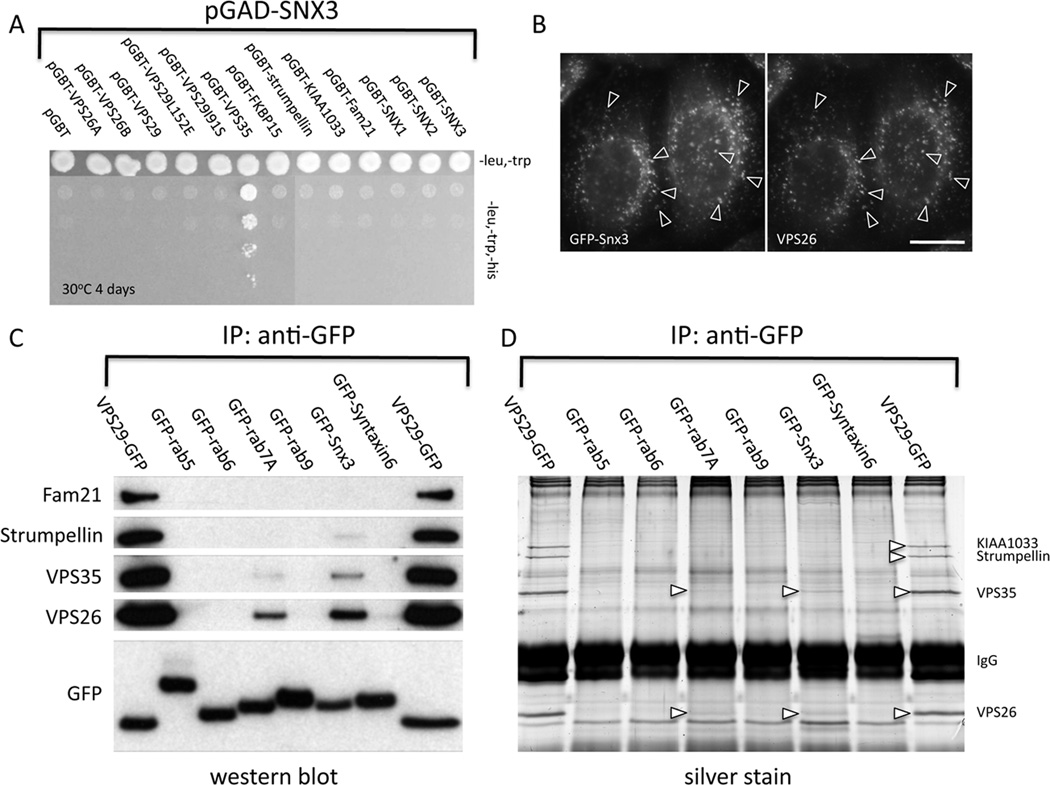
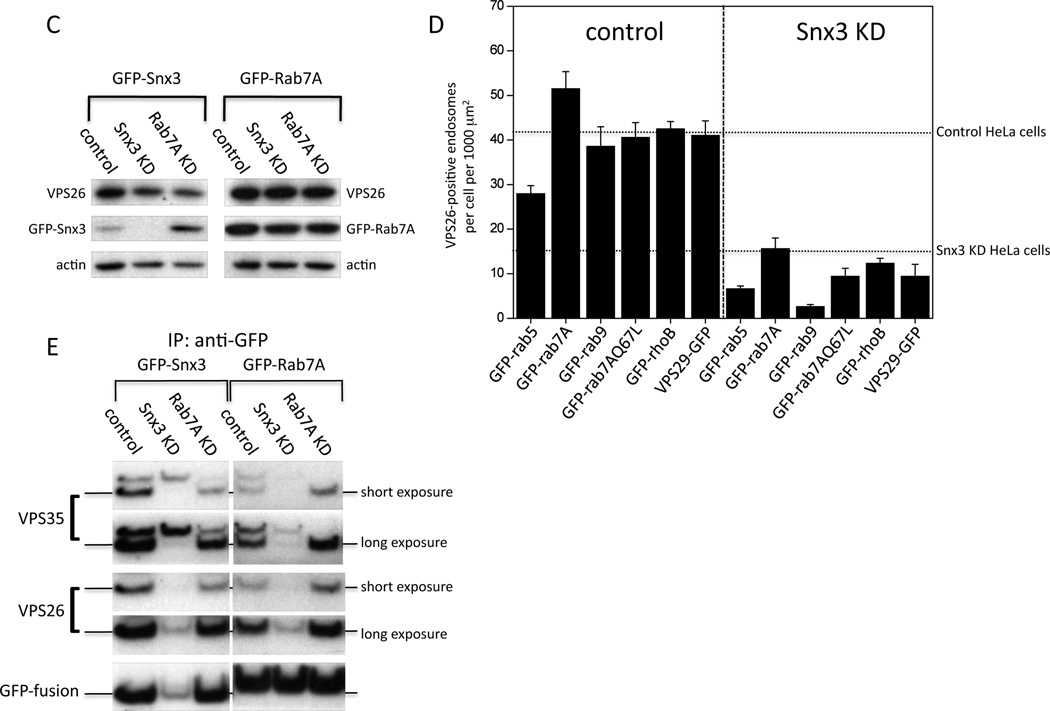
We investigated the interaction between mammalian retromer and Snx3 initially by using the yeast two-hybrid (Y2H) system. The Snx3 gene was expressed in the yeast HF7c strain from the pGAD424 (‘prey’) vector and thus tested for an interaction with retromer components and retromer-associated proteins (Figure 2A). Growth of the yeast on media lacking histidine is indicative of an interaction and was only observed for the combination of Snx3 and Vps35.
Figure 2.
Snx3 interacts with the cargo-selective retromer complex through VPS35. A. Yeast two-hybrid analysis of Snx3 interactions with retromer and retromer-associated proteins. The yeast HF7c strain was transformed with the pGAD-Snx3 plasmid along with pGBT9 plasmids expressing the various retromer proteins. Transformants were spotted out onto –leu,-trp media or onto –leu,-trp,-his media as a dilution series. Only yeast expressing both Snx3 and VPS35 grow on the triple dropout media indicating an interaction between Snx3 and VPS35. B. HeLa cells stably transfected with GFP-Snx3 were fixed and labeled with anti-GFP and anti-VPS26. The GFP-Snx3 protein colocalizes with VPS26 (indicated with arrow-heads) Bar = 20µm. C. HeLa cells stably expressing various GFP-tagged proteins were lysed and treated with anti-GFP to IP the respective proteins. GFP-Snx3 co-IPs the VPS26 and VPS35 proteins as does GFP-Rab7A (as reported previously). The VPS29-GFP construct is a positive control in this experiment and co-IPs significantly more retromer proteins. D. Identical samples to C were subjected to SDS-PAGE and silver staining. The VPS35 and VPS26 bands are visible in the GFP-Rab7A and GFP-Snx3 lanes (indicated with arrow-heads).
We next generated a HeLa cell line stably transfected with a GFP-Snx3 construct. The GFP-Snx3 fusion colocalizes with Vps26 on punctate endosomal structures (Figure 2B). The Y2H interaction of Snx3 and Vps35, and the colocalization of GFP-Snx3 with Vps26 suggest an in-vivo interaction is likely. Therefore we performed native immunoprecipitations (IPs) on a panel of HeLa cell lines stably expressing various endosomally localized GFP-tagged proteins. We observed that GFP-Snx3 co-IPs the Vps26 and Vps35 proteins consistent with an association with retromer (Figure 2C). In line with our previously published data [48], GFP-Rab7A co-IPs retromer as does Vps29-GFP that serves as a positive control for these experiments. Other GFP-fusion proteins (e.g. Rab5, Rab9) do not co-IP retromer demonstrating the specificity of the assay. In addition to analyzing the native IPs by western blotting we also subjected the samples to SDS-PAGE and silver staining (see figure 2D). The bands corresponding to Vps26 and Vps35, although faint, are visible in the GFP-Snx3 and GFP-Rab7A lanes.
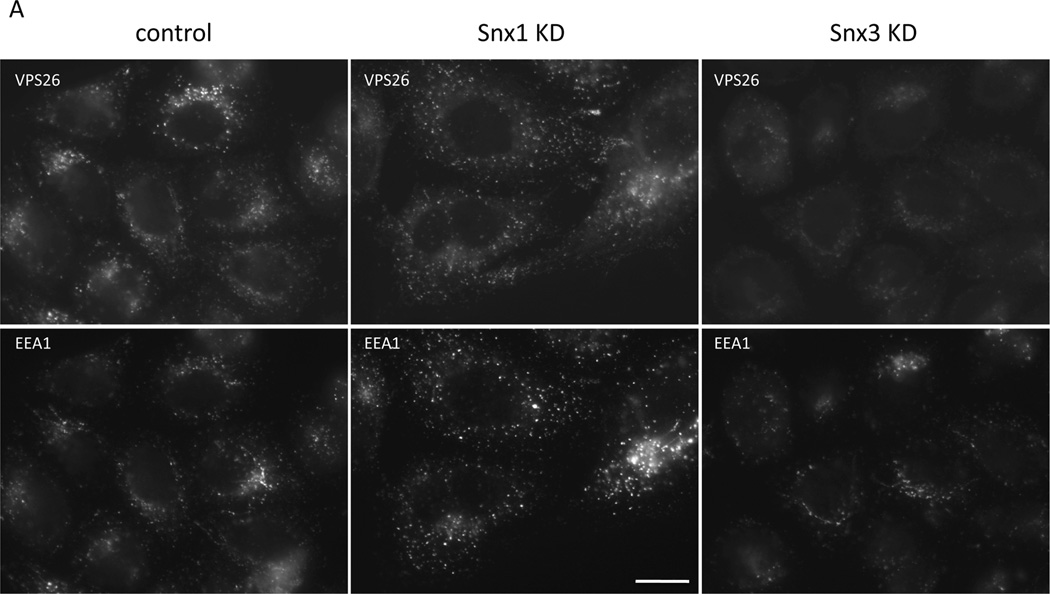
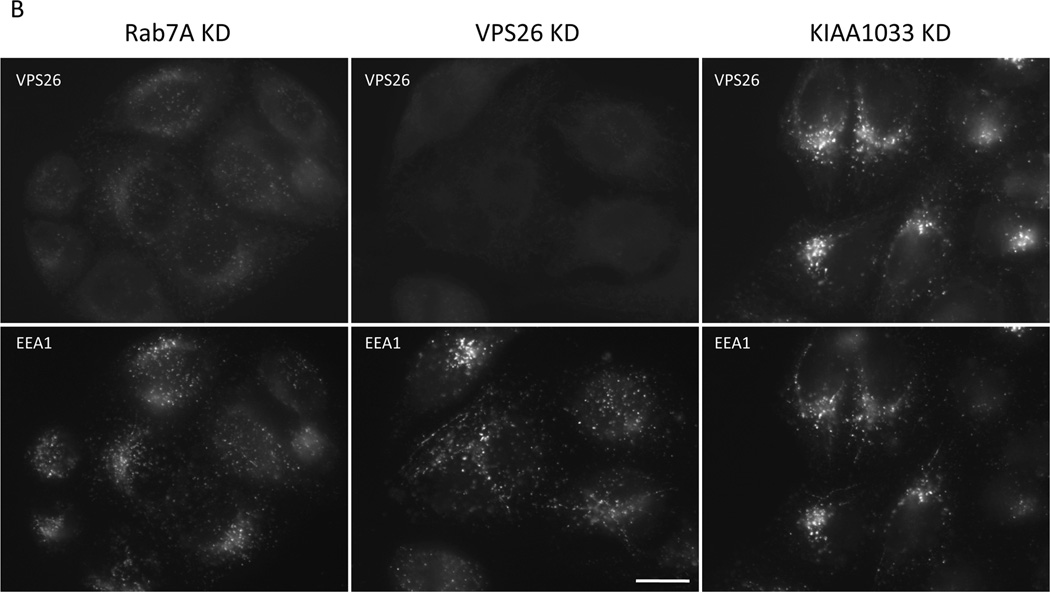
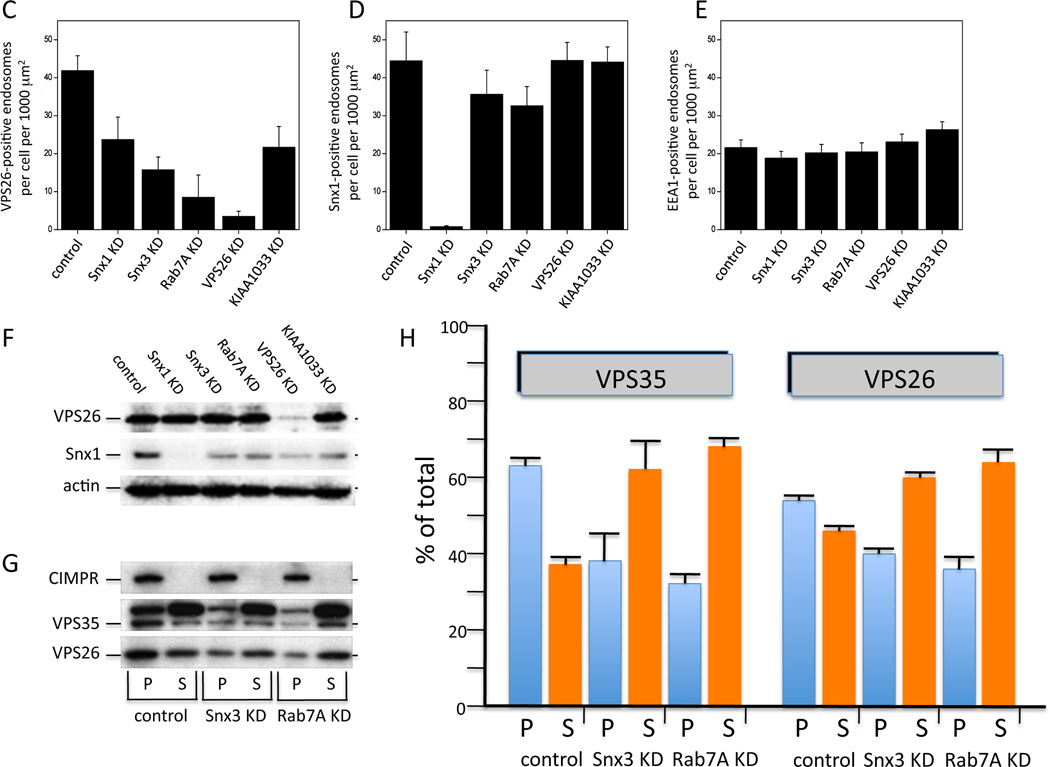
We next investigated the functional significance of the Snx3-retromer interaction by performing siRNA-mediated knockdowns of Snx3 and other retromer-associated proteins. Figure 3A and 3B shows cells that have been treated with siRNA to knockdown (KD) Snx1, Snx3, Rab7A, Vps26 and KIAA1033 and then fixed and labeled with antibodies to stain Vps26 and the endosomal protein EEA1. The Snx3 and Rab7A KDs both reduced the intensity of the Vps26 staining but did not markedly affect the EEA1 staining indicating that endosomal structures persist after Snx3 or Rab7A KD. To quantify the effect of the KDs on the endosomal localization of retromer, the cells were imaged using an automated microscope to determine the number of Vps26-positive endosomes. This experiment revealed that the loss of Rab7A and Snx3 both result in a pronounced reduction in the number of Vps26-positive endosomes (Figure 3C). The effect of the loss of Snx3 or Rab7A on Snx1-positive endosomes was much less (Figure 3D) and no effect on the number of EEA1-positive endosomes was observed for any knockdown performed (Figure 3E).
Figure 3.
Snx3 is required for the membrane association of the cargo-selective retromer complex. A & B. HeLa cells were treated with siRNA to abolish the expression of Snx1, Snx3, Rab7A, VPS26 or the WASH complex protein, KIAA1033. After fixation the cells were labeled with antibodies to VPS26 and EEA1. Loss of Snx3 expression results in a reduction of the VPS26 labeling comparable to loss of Rab7A. Bar = 20µm. C, D & E. Cells were treated with siRNA as in A and B. The number of VPS26-positive (C) endosomes was determined using a Cellomics ArrayScan Quantitative microscope along with the number of Snx1-positive (D) or EEA1-postive (E) endosomes. Loss of Snx3 or Rab7A results in a marked reduction in the number of VPS26-positive endosomes whilst little effect was observed for Snx1 or EEA1. F. Samples from the cells treated with the various siRNA were western blotted for VPS26 and Snx1. The reduction in the VPS26 signal observed in A, B and C is not due to the loss of the protein. G. HeLa cells were treated with siRNA to abolish Snx3 or Rab7A expression. The cells were then fractionated to separate membranes and membrane-bound proteins (P) from soluble cytosolic proteins (S). The samples were analyzed by SDS-PAGE and western blotting. The VPS26 and VPS35 proteins redistribute into the soluble (cytosolic) fraction after loss of Snx3 or Rab7A expression. H. The experiment performed in G was repeated a further two times and the data from the three experiments was averaged and is shown as a graph.
The reduction in the Vps26 staining observed after KD of Snx3 (or Rab7A) could be due to loss of membrane association or loss of the protein – the immunofluorescence based assays cannot distinguish these two possibilities. Therefore lysates from the various KDs were analyzed by western blotting. As shown in Figure 3F, there was no loss of the Vps26 protein following the Snx3 (or Rab7A) KD although Vps26 KD does result in loss of the Vps26 protein. The efficacy of the siRNA targeting Snx3 or Rab7A was confirmed by western blotting lysates of cells stably transfected with GFP-Snx3 or GFP-Rab7A that had been treated with siRNA to silence Snx3 or Rab7A and is shown in Supplemental Figure S1.
Thus we conclude that the reduction in Vps26 labeling observed in Figure 3A–3C is not caused by loss of the protein and is likely to be due to loss of membrane association. To test this hypothesis we performed a simple fractionation assay to separate membrane-associated (pelletable) and cytosolic (soluble) retromer. In Figure 3G, the pellet (P) and soluble (S) fractions from control, Snx3 and Rab7A KD cells have been analyzed by SDS-PAGE and western blotting. The Snx3 and Rab7A knockdowns both result in a shift of Vps26 and Vps35 from the pellet fraction to the soluble fraction. Data from three blots was quantified and is shown in Figure 3H. Loss of either Snx3 or Rab7A resulted in a pronounced change in the membrane association of retromer with both Vps26 and Vps35 becoming predominately soluble consistent with Snx3 and Rab7A regulating the membrane association of retromer. Simultaneous KD of both Snx3 and Rab7A did not cause further loss of retromer membrane association (data not shown) suggesting that there are additional factors responsible for a basal level of membrane association of the cargo-selective retromer complex.
If loss of Snx3 results in reduced membrane association of the cargo-selective retromer complex, does increased Snx3 promote retromer membrane association? To address this question, cells expressing Vps29-GFP or GFP-Vps35 where the elevated expression results in increased cytoplasmic localization were transfected with red fluorescent protein-tagged Snx3. We observed that in cells transiently transfected with RFP-Snx3 the localization of Vps29-GFP and GFP-Vps35 appears more punctate and endosomal (Supplementary Figure 2).
The data presented so far shows that Snx3 is required for the membrane association of retromer – similar to Rab7A. Therefore we wondered if Rab7A and Snx3 interact with each other either directly or through a third-party protein. Thus large scale anti-GFP native IPs were performed on cells stably expressing GFP-Snx3, GFP-Rab9 (a negative control), GFP-Rab7A and Vps29-GFP (a positive control). The native IPs were analyzed by mass spectrometry (see Supplementary Table 3). Other than retromer proteins, we did not identify any proteins that co-IPed with both GFP-Snx3 and GFP-Rab7 and therefore Snx3 and Rab7A are unlikely to interact with each other directly or indirectly unless via retromer.
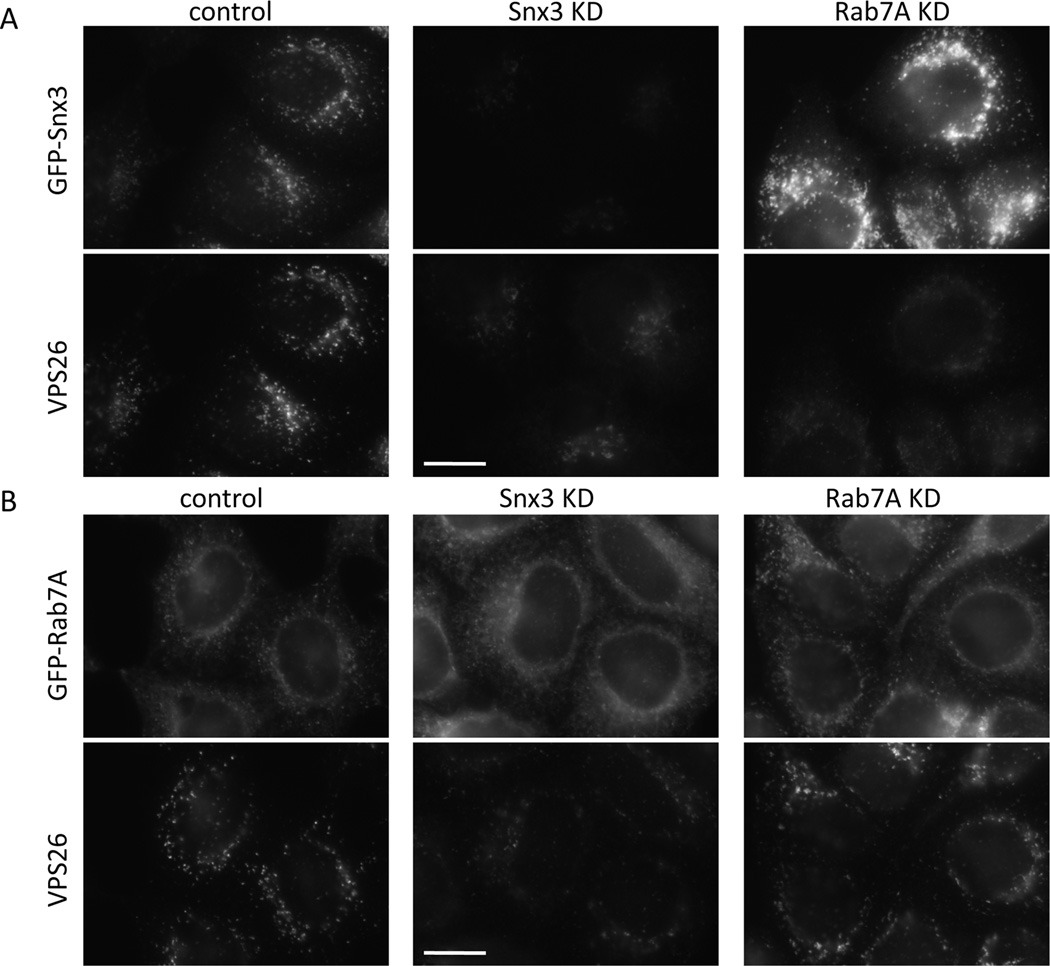
3.3 Snx3 and Rab7A act independently to regulate retromer membrane association
We next investigated whether Snx3 and Rab7 act together or independently to regulate retromer association. Initially, cells stably expressing GFP-Snx3 were treated with siRNA to KD either Snx3 or Rab7A. In Figure 4A, the Snx3 KD abolished expression of GFP-Snx3 and leads to a reduction in the Vps26 staining. Loss of Rab7A expression also caused a reduction in Vps26 staining but there was an apparent increase in the brightness of the GFP-Snx3 labeling in Rab7A knockdown cells. When the same KDs were performed in cells stably expressing GFP-Rab7A we noticed that loss of Snx3 resulted in reduced punctate GFP-Rab7A labeling along with diffuse Vps26 labeling. The Rab7A KD did not affect expression of the GFP-Rab7A as this construct utilizes murine Rab7A that is resistant to the siRNA that silences endogenous Rab7A expression in the HeLa cells and hence Vps26 labeling appears normal.
Figure 4.
Loss of Snx3 and Rab7A both affect retromer-association but through different mechanisms. A. HeLa cells stably expressing GFP-Snx3 were treated with siRNA to abolish Snx3 or Rab7A expression. Loss of Snx3 or Rab7A result in a similar reduction in VPS26 labeling but loss of Rab7 appears to increase the intensity of the GFP-Snx3 labeling. Bar = 20µm. B. HeLa cells stably expressing GFP-Rab7A were treated as in A. In this experiment, loss of Snx3 expression reduces the punctate labeling of the GFP-Rab7A and also results in a loss of VPS26 labeling. The GFP-Rab7A construct is of murine origin and is therefore resistant to the siRNA targeting human Rab7A and hence VPS26 labeling is similar to control cells. Bar = 20µm. C. Lysates from cells shown in A and B were subjected to SDS-PAGE and analyzed by western blotting. The Rab7A knockdown increases the GFP-Snx3 levels consistent with the increased intensity of GFP-Snx3 signal observed in A. D. Cells stably expressing various GFP-tagged endosomal proteins were treated with siRNA to abolish Snx3 expression. VPS26-positive endosomes were then quantified using the Cellomics ArrayScan microscope. In all cases, loss of Snx3 expression results in a marked reduction in the number of VPS26-positive endosomes indicating that increased expression of Rab7A or the constitutively active Rab7A Q67L mutant is unable to rescue the loss of Snx3. For comparison, the number of VPS26-positive endosomes determined for control and Snx3 KD cells in Figure 2C is shown by the dotted lines. E. Cells stably expressing GFP-Snx3 or GFP-Rab7A were treated with siRNA to KD Snx3 or Rab7A expression. Lysates were immunoprecipitated using anti-GFP and following SDS-PAGE, VPS35 and VPS26 were detected by western blotting. The KD of Snx3 abolishes the association of retromer with GFP-Rab7A, loss of Rab7A expression reduces but does not completely abolish the interaction between GFP-Snx3 and retromer.
Lysates from the KDs shown in Figure 4A and 4B were analyzed by SDS-PAGE and western blotting. We observed that the Rab7A KD in the GFP-Snx3 cells did result in an increase of GFP-Snx3 levels (Figure 4C) that is consistent with the increased fluorescence (Figure 4A). To test whether loss of Snx3 expression could be compensated by increased expression of Rab7A, Snx3 KDs were performed on a panel of cells expressing GFP-fusion proteins that localize to endosomes including the Rab7A-Q67L mutant that is constitutively active. The number of Vps26-positive endosomes was determined using an automated microscope (Figure 4D). In control cells there were ~40 Vps26 endosomes per 1000 µm2 (similar to untransfected HeLa cells) although increased Rab5 or Rab7 expression can respectively decrease or increase the number of Vps26 positive endosomes. In the Snx3 knockdown cells, the number of Vps26 positive endosomes was reduced to ~10 per 1000 µm2 although in cells expressing GFP-Rab7A it is ~15 per 1000 µm2, this number is no greater than was observed for untransfected HeLa cells. We conclude therefore that elevated expression of Rab7A (even constitutively active Rab7A-Q67L) cannot compensate for loss of Snx3. Furthermore, when native immunoprecipitation of GFP-Snx3 or GFP-Rab7A was performed on cells treated with siRNA to abolish Snx3 or Rab7A expression, we observed that knockdown of Snx3 abolishes the interaction of retromer with the GFP-Rab7A protein whereas some association of retromer with GFP-Snx3 was retained after loss of Rab7A expression (Figure 4E).
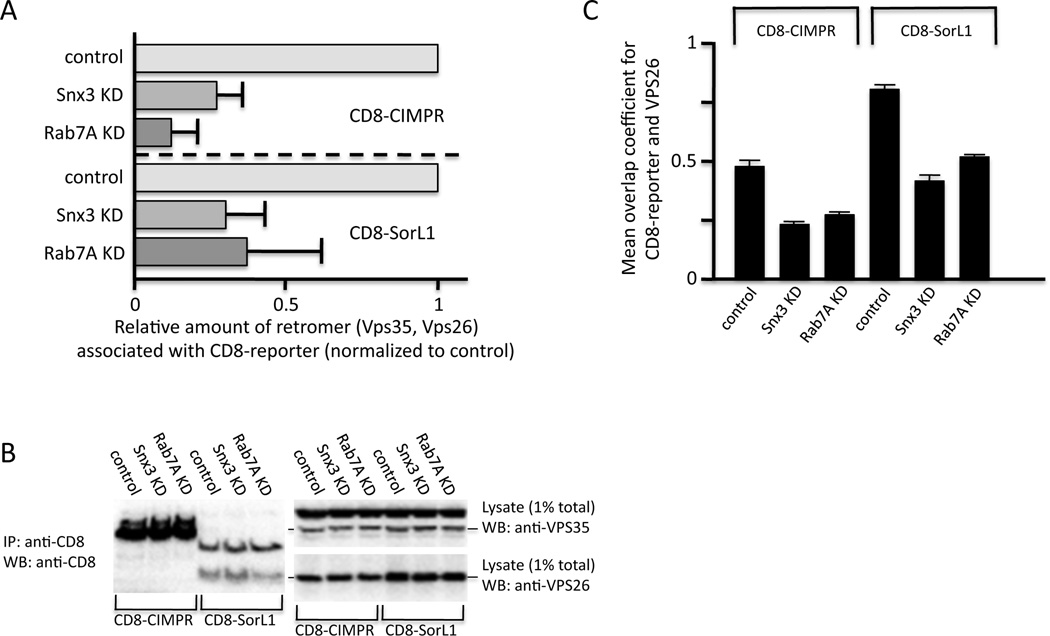
The inability of the cargo-selective retromer complex to associate with endosomal membranes following siRNA KD of Snx3 or Rab7A would be predicted to significantly impact the association of the cargo-selective complex with cargo proteins such as the CIMPR or SorL1. We tested this hypothesis by immunoprecipitating CD8-reporter proteins that contain the cytoplasmic tails of the CIMPR or SorL1 after siRNA KD of Snx3 or Rab7A. Figure 5A shows that loss of Snx3 or Rab7A greatly reduces the amount of retromer proteins (Vps35 and Vps26) that co-IP with the respective CD8-reporter protein. The KD of Snx3 or Rab7A does not however, alter the amount of CD8-reporter protein that IPs or levels of Vps35 or Vps26 proteins (Figure 5B) Additionally in Figure 5C, we also measured the intensity of the immunofluorescence signal of Vps26 protein that was coincident with the respective CD8-reporter. The KD of Snx3 or Rab7A markedly reduces the intensity of the Vps26 staining consistent with the loss of membrane association shown in Figure 3. This experiment supports the hypothesis that Snx3 and Rab7A mediate the association of retromer with cargo proteins such as SorL1.
Figure 5.
Snx3 and Rab7A are required for retromer association with cargo proteins. A. Cells stably expressing CD8-CIMPR or CD8-SorL1 were treated with siRNA to knock down Snx3 or Rab7A expression. Following lysis, the CD8-reporter proteins were immunoprecipitated with anti-CD8 and subjected to SDS-PAGE. Vps35 and Vps26 were detected by western blotting. Signals from three independent blots were quantified and normalized to the control, errors bars are mean deviation. B. The level of CD8-reporter protein immunoprecipitated is not affected by the Snx3 or Rab7A KDs and neither are levels of Vps35 or Vps26 in the lysates. C. The intensity of the Vps26 immunofluorescence signal that is coincident with the respective CD8-reporter protein was measured using an automated microscope. 250 cells per well and at least three wells for each condition were imaged. Error bars are standard deviation. These data show that the KD of Snx3 or Rab7A markedly reduces the association of retromer with cargo proteins.
4. Discussion
We identified significant association of AD with specific SNPs in or at the gene level for four of the 15 retromer and retromer-associated genes tested in a large group of Caucasian AD cases and controls. RAB7A, KIAA1033 and SNX1 attained gene-level significance and SNX3 contained SNPs significant after correction for multiple testing. Significant findings for several SNX3 SNPs were replicated in a sample of Israel-Arabs. We also obtained evidence of replication for KIAA1033 in African Americans, although with a completely different set of SNPs. These non-overlapping findings may be due to distinct population linkage disequilibrium patterns among SNPs in the KIAA1033 gene region reflecting association with the same or different causal variants.
No retromer or retromer-related genes including SORL1 contained a signal approaching genome-wide significance in the largest GWAS published to date [38]. However, failure to meet this threshold, due to small effect size or allelic heterogeneity, does not mean that a gene or variant is not associated as demonstrated by genetic and functional evidence for SORL1 [28–30,41,42,44–46,57,58]. Our use of GWAS data to address a specific hypothesis with far fewer tests requires much lower thresholds for significance.
Association of Rab7A protein with retromer has been observed in enteric amoeba, human tissue culture cells and yeast [5,40,47,52]. The Snx3 protein is a member of the sorting nexin family and contains a PX domain that binds to phosphotidylinositol 3-phosphate (PtdIns3-P). The yeast homolog of Snx3 is called Grd19p and interacts with the retromer complex to mediate endosomal protein sorting of specific cargo proteins [59,60]. The interaction of retromer with either Rab7A or Snx3 therefore appears to be conserved across species and through evolution suggesting that these interactions are of profound functional importance. Indeed, while this manuscript was in preparation, Harterink et al. [19] reported that Snx3 functions in retromer-mediated sorting of Wntless, a membrane protein required for Wnt secretion, although that study overlooked the evidence that Snx3 also affects Rab7A localization [36] thereby complicating the interpretation of the role of Snx3 in regulating retromer membrane association.
It is intriguing that both SNX3 and RAB7A contained AD-linked SNPs as herein we show that loss of either Snx3 or Rab7A expression leads to very similar phenotypes, specifically a reduction in the membrane association of the cargo-selective retromer complex. We have also previously reported that both Snx3 and Rab7A knockdowns result in a defect in endosome-to-Golgi retrieval [17,52]. Interestingly, data presented herein and in a previous study [7] indicate that loss of Snx3 expression also alters the localization of Rab7A hinting that the mechanism by which Snx3 affects the membrane association of retromer could be through altered Rab7A localization and necessitating a more detailed examination of the relationship between Snx3 and Rab7A.
We found no evidence for a direct association of Snx3 with Rab7A, therefore it is unclear why the loss of Snx3 should affect Rab7A localization. Attempts to compensate for the loss of Snx3 by increasing the expression of Rab7A, including the constitutively active Q67L mutant, were also unsuccessful. Given that loss of Rab7A results in a quantitative increase in the level of GFP-Snx3 but no rescue of the Vps26 localization we conclude that Rab7A and Snx3 regulate the membrane association of the cargo-selective retromer complex through distinct mechanisms. This conclusion is consistent with lack of evidence for genetic interaction of RAB7A with SNX3 on risk of AD.
Studies undertaken in yeast and nematode have implicated Snx3 in directing specific cargo proteins into a retromer-mediated retrieval pathway [24,29]. It is possible therefore that Snx3 aids in stabilizing the membrane association of the cargo-selective retromer complex by concentrating membrane proteins into discrete patches thereby elevating the local concentration of cytoplasmic tails for retromer to interact with. Additionally the Y2H data we present here shows that Snx3 can interact directly with Vps35 and therefore this interaction may also contribute to facilitating the stable association of retromer with endosomal membranes thereby promoting the association with cargo proteins such as CIMPR and SorL1 (see Figure 6). It is noteworthy however that, unlike Rab7A, Snx3 is not conserved across all eukaryotes being absent in plants. Therefore, while Snx3 is clearly important for retromer function in mammalian cells, it is dispensable in some other eukaryotes.
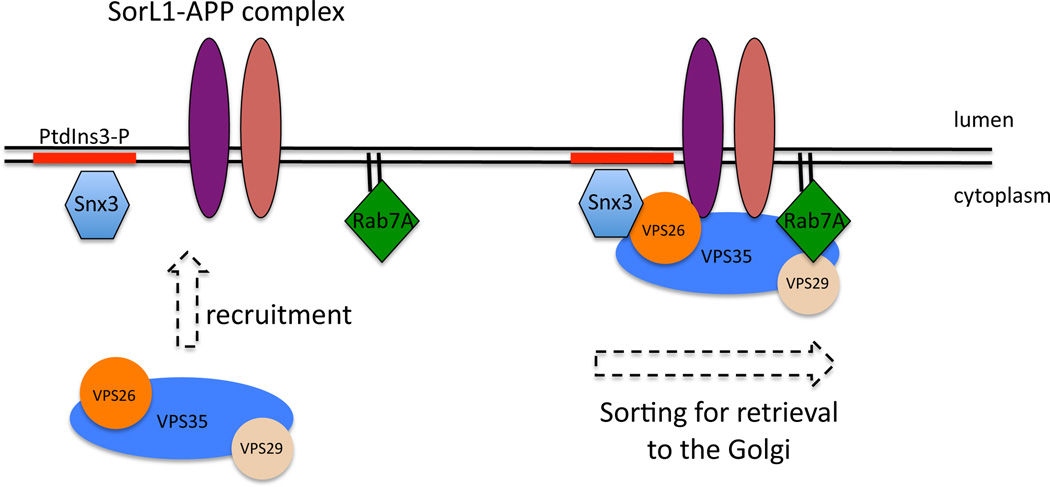
Figure 6.
Schematic diagram of the role of Snx3 and Rab7A in promoting the recruitment of the cargo-selective retromer complex to the endosomal membrane thereby facilitating the association of retromer with cargo proteins such as SorL1. This mechanism may therefore contribute to the proper trafficking of APP and regulate access of APP to the β-secretase.
Trafficking of the amyloid precursor protein (APP) from the cell surface via the endocytic pathways plays a key role in the generation of amyloid β-peptide (Aβ), the aberrant accumulation of which is postulated to be central to the pathogenesis of AD [36]. Thus, pathways modulating APP-sorting through the membrane or altering APP cleavage by the secretases are key to understating the pathophsyiology of AD. Multiple studies have shown that retromer deficiency elevates levels of endogenous Aβ in the brain and causes hippocampal dysfunction and neurodegeneration [38,56]. SorL1 is involved in trafficking of APP from the cell surface to the Golgi-complex and reduced expression of SORL1 leads to elevated Aβ levels and an increased risk of AD [1,2,12,42,49,58]. Several candidate-gene and targeted studies across different ethnic groups have shown the strong association of SORL1 with AD [6,22,23,27,28,30,33,46,63]. Recently, the other members of the Vps10p-domain sorting receptor family (SORCS1, SORCS2, SORCS3 and SORT1) have been demonstrated to be genetically and functionally associated with AD [26,43,45]. Because loss of Snx3 or Rab7A expression results in displacement of the cargo-selective retromer complex from the endosome membrane leading to a marked reduction in the ability of retromer to recognize cargo proteins (see Figure 6), the most likely explanation for the association of Snx3 and Rab7A with AD is through defective protein sorting of retromer cargoes such as SorL1. Connections of SORL1 with RAB7A and SNX3 are suggested by our genetic interaction analyses. Precise and more significant statistical evidence for interactions among these and other retromer-associated genes will likely emerge from studies of additional large datasets assembled by several international AD genetics consortia. A direct influence of the novel AD-associated genes reported herein (RAB7A, SNX1, SNX3, and KIAA1033) on APP processing or Aβ has not been reported, and thus future studies should explore these relationships. However, it has been reported from multiple studies that loss of retromer function or loss of retromer-association with SorL1 result in increased processing of APP to Aβ [14,26,37,45,55] and, therefore, it is expected that APP processing will be altered in Snx3 or Rab7A knockdowns.
Although none of the retromer or retromer-associated genes have yet emerged as strong candidates in agnostic approaches such as GWAS [25,39], perhaps due to their modest effect on genetic risk [43,45,46] or intragenic heterogeneity [45,46], large-scale GWAS have identified genome-wide significant association of AD with nine novel loci [17,21,25,54], three of which --- PICALM, BIN1 and CD2AP --- lend additional support to perturbation of membrane trafficking as an important mechanism in AD pathogenesis. Genetic studies in budding yeast indicate an essential role of the Bin1 homolog in endocytosis, however, this role appears to be non-essential for homologs in fission yeast, fruit flies, and mice [31]. CD2AP is involved in membrane trafficking occurring during receptor endocytosis and cytokinesis [37]. Although the role of PICALM in AD pathogenesis is poorly understood, it is believed to be involved in APP metabolism via the endocytic pathway [48].
A hypothesis-driven pathway approach allowed us to apply gene association methods with a much lower statistical significance threshold than required for a GWAS. Evidence for association of AD with the retromer-associated genes RAB7A and SNX3, as well as interactions of these genes with SORL1 on AD risk, present a compelling argument for further studies of protein-trafficking and APP sorting pathways in AD etiology. However, lack of association of the retromer-complex genes VPS35, VPS26 and VPS29 underscores the limitation of even hypothesis-driven genetic studies that typically evaluate common variants because they are known and, at best, are powered to detect association with them. The advent of high-throughput gene sequencing technologies has demonstrated that much of the genetic risk for common diseases is likely due to rare variants [11]. Recent studies have shown that rare mutations in VPS35 cause Parkinson disease [65,67]. It is also possible that these vacuolar protein sorting genes exert an effect on AD risk through interaction with other genes. This possibility can be studied statistically and by functional studies in vitro.
Supplementary Material
Supplemental Figure 1. Cells stably expressing GFP-Snx3 or GFP-Rab7A were treated with siRNA to silence Snx3 or Rab7A expression. Lysates were subjected to SDS-PAGE and then analysed by western blotting with antibodies against Snx3, Rab7A, or GFP (panels A–C, respectively). The band visible at 97 kD is present in all three blots and is likely to be a nonspecific protein that is recognized by the iodinated protein-A. D. The gel was stained with coomassie blue to show protein loading. The western blots confirm that the siRNA efficiently targets both endogenous and GFP-Snx3, and endogenous Rab7A. The GFP-tagged Rab7A construct utilizes the murine cDNA and is resistant to the siRNA targeting endogenous Rab7A.
Supplemental Figure 2. Cells stably expressing GFP-VPS35 or VPS29-GFP were transiently transfected with RFP-Snx3, fixed and then labeled with antibodies against GFP or RFP. The increased expression of Snx3 by the transient transfection of RFP-Snx3 is able to promote the membrane association of GFP-VPS35 or VPS29-GFP where a proportion of those constructs are cytosolic. Bar = 20 µm.
Acknowledgments
This work was supported in part by NIH grants R01 AG025259, R01-AG33193 and U01-AG032984, and grants from the Wellcome Trust, Medical Research Council and Canadian Institutes of Health Research. We thank the Alzheimer Disease Genetics Consortium for proving access to GWAS datasets and are grateful to Ms. Jacqueline Buros for data management assistance and Dr. Robin Antrobus for mass spectrometric analysis of anti-GFP immunoprecipitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement
The authors declare that they have no conflicts of interest.
References
- 1.Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CA, Breiderhoff T, Jansen P, Wu X, Bales KR, Cappai R, Masters CL, Gliemann J, Mufson EJ, Hyman BT, Paul SM, Nykjaer A, Willnow TE. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102(38):13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen OM, Schmidt V, Spoelgen R, Gliemann J, Behlke J, Galatis D, McKinstry WJ, Parker MW, Masters CL, Hyman BT, Cappai R, Willnow TE. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry. 2006;45(8):2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- 3.Anderson RG, Goldstein JL, Brown MS. A mutation that impairs the ability of lipoprotein receptors to localise in coated pits on the cell surface of human fibroblasts. Nature. 1977;270(5639):695–699. doi: 10.1038/270695a0. [DOI] [PubMed] [Google Scholar]
- 4.Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165(1):123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balderhaar HJ, Arlt H, Ostrowicz C, Brocker C, Sundermann F, Brandt R, Babst M, Ungermann C. The Rab GTPase Ypt7 is linked to retromer-mediated receptor recycling and fusion at the yeast late endosome. J Cell Sci. 123(Pt 23):4085–4094. doi: 10.1242/jcs.071977. [DOI] [PubMed] [Google Scholar]
- 6.Bettens K, Brouwers N, Engelborghs S, De Deyn PP, Van Broeckhoven C, Sleegers K. SORL1 is genetically associated with increased risk for late-onset Alzheimer disease in the Belgian population. Hum Mutat. 2008;29(5):769–770. doi: 10.1002/humu.20725. [DOI] [PubMed] [Google Scholar]
- 7.Braun V, Wong A, Landekic M, Hong WJ, Grinstein S, Brumell JH. Sorting nexin 3 (SNX3) is a component of a tubular endosomal network induced by Salmonella and involved in maturation of the Salmonella-containing vacuole. Cell Microbiol. 12(9):1352–1367. doi: 10.1111/j.1462-5822.2010.01476.x. [DOI] [PubMed] [Google Scholar]
- 8.Burd CG. Physiology and pathology of endosome-to-Golgi retrograde sorting. Traffic. 12(8):948–955. doi: 10.1111/j.1600-0854.2011.01188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canuel M, Lefrancois S, Zeng J, Morales CR. AP-1 and retromer play opposite roles in the trafficking of sortilin between the Golgi apparatus and the lysosomes. Biochem Biophys Res Commun. 2008;366(3):724–730. doi: 10.1016/j.bbrc.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Dell'Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell. 1999;3(1):11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- 11.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 8(1) doi: 10.1371/journal.pbio.1000294. e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodson SE, Andersen OM, Karmali V, Fritz JJ, Cheng D, Peng J, Levey AI, Willnow TE, Lah JJ. Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer's disease. J Neurosci. 2008;28(48):12877–12886. doi: 10.1523/JNEUROSCI.4582-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton S. Retromer retrieves wntless. Dev Cell. 2008;14(1):4–6. doi: 10.1016/j.devcel.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Fjorback AW, Seaman M, Gustafsen C, Mehmedbasic A, Gokool S, Wu C, Militz D, Schmidt V, Madsen P, Nyengaard JR, Willnow TE, Christensen EI, Mobley WB, Nykjær A, Andersen OM. Retromer Binds the FANSHY Sorting Motif in SorLA to Regulate Amyloid Precursor Protein Sorting and Processing. J Neuroscience. 2012;32(4):1467–1480. doi: 10.1523/JNEUROSCI.2272-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gokool S, Tattersall D, Seaman MN. EHD1 interacts with retromer to stabilize SNX1 tubules and facilitate endosome-to-Golgi retrieval. Traffic. 2007;8(12):1873–1886. doi: 10.1111/j.1600-0854.2007.00652.x. [DOI] [PubMed] [Google Scholar]
- 16.Haft CR, de la Luz Sierra M, Bafford R, Lesniak MA, Barr VA, Taylor SI. Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes. Mol Biol Cell. 2000;11(12):4105–4116. doi: 10.1091/mbc.11.12.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbour ME, Breusegem SY, Antrobus R, Freeman C, Reid E, Seaman MN. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J Cell Sci. 123(Pt 21):3703–3717. doi: 10.1242/jcs.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harterink M, Port F, Lorenowicz MJ, McGough IJ, Silhankova M, Betist MC, van Weering JR, van Heesbeen RG, Middelkoop TC, Basler K, Cullen PJ, Korswagen HC. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 13(8):914–923. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 21.Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, Buxbaum JD, Ertekin-Taner N, Fallin MD, Friedland R, Inzelberg R, Kramer P, Rogaeva E, St George-Hyslop P, Cantwell LB, Dombroski BA, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Lunetta KL, Martin ER, Montine TJ, Goate AM, Blacker D, Tsuang DW, Beekly D, Cupples LA, Hakonarson H, Kukull W, Foroud TM, Haines J, Mayeux R, Farrer LA, Pericak-Vance MA, Schellenberg GD. Meta-analysis confirms CR1, CLU, PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 67(12):1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura R, Yamamoto M, Morihara T, Akatsu H, Kudo T, Kamino K, Takeda M. SORL1 is genetically associated with Alzheimer disease in a Japanese population. Neurosci Lett. 2009;461(2):177–180. doi: 10.1016/j.neulet.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Kolsch H, Jessen F, Wiltfang J, Lewczuk P, Dichgans M, Teipel SJ, Kornhuber J, Frolich L, Heuser I, Peters O, Wiese B, Kaduszkiewicz H, van den Bussche H, Hull M, Kurz A, Ruther E, Henn FA, Maier W. Association of SORL1 gene variants with Alzheimer's disease. Brain Res. 2009;1264:1–6. doi: 10.1016/j.brainres.2009.01.044. [DOI] [PubMed] [Google Scholar]
- 24.Koumandou VL, Klute MJ, Herman EK, Nunez-Miguel R, Dacks JB, Field MC. Evolutionary reconstruction of the retromer complex and its function in Trypanosoma brucei. J Cell Sci. 124(Pt 9):1496–1509. doi: 10.1242/jcs.081596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 26.Lane RF, Raines SM, Steele JW, Ehrlich ME, Lah JA, Small SA, Tanzi RE, Attie AD, Gandy S. Diabetes-associated SorCS1 regulates Alzheimer's amyloid-beta metabolism: evidence for involvement of SorL1 and the retromer complex. J Neurosci. 2010;30(39):13110–13115. doi: 10.1523/JNEUROSCI.3872-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JH, Barral S, Reitz C. The neuronal sortilin-related receptor gene SORL1 and late-onset Alzheimer's disease. Curr Neurol Neurosci Rep. 2008;8(5):384–391. doi: 10.1007/s11910-008-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, Cheng R, Honig LS, Vonsattel JP, Clark L, Mayeux R. Association between genetic variants in SORL1 and autopsy-confirmed Alzheimer disease. Neurology. 2008;70(11):887–889. doi: 10.1212/01.wnl.0000280581.39755.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JH, Cheng R, Schupf N, Manly J, Lantigua R, Stern Y, Rogaeva E, Wakutani Y, Farrer L, St George-Hyslop P, Mayeux R. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64(4):501–506. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, Shibata N, Cheng R, Mayeux R. Possible association between SORL1 and Alzheimer disease? Reanalysing the data of Shibata et al. Dement Geriatr Cogn Disord. 2008;26(5):482. doi: 10.1159/000167792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leprince C, Romero F, Cussac D, Vayssiere B, Berger R, Tavitian A, Camonis JH. A new member of the amphiphysin family connecting endocytosis and signal transduction pathways. J Biol Chem. 1997;272(24):15101–15105. doi: 10.1074/jbc.272.24.15101. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Rowland C, Catanese J, Morris J, Lovestone S, O'Donovan MC, Goate A, Owen M, Williams J, Grupe A. SORL1 variants and risk of late-onset Alzheimer's disease. Neurobiol Dis. 2008;29(2):293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 87(1):139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Logue MW, Schu M, Vardarajan BN, Buros J, Green RC, Go RCP, Griffith P, Akomolafe A, Obisesan TO, Shatz R, Borenstein A, Cupples LA, Lunetta KL, Fallin MD, Baldwin CT, Farrer LA. Genetic Variants at Multiple Loci Influence Alzheimer Disease Risk in African Americans. Arch Neurol. 2011 doi: 10.1001/archneurol.2011.646. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattson MP. Pathways towards and away from Alzheimer's disease. Nature. 2004;430(7000):631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monzo P, Gauthier NC, Keslair F, Loubat A, Field CM, Le Marchand-Brustel Y, Cormont M. Clues to CD2-associated protein involvement in cytokinesis. Mol Biol Cell. 2005;16(6):2891–2902. doi: 10.1091/mbc.E04-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muhammad A, Flores I, Zhang H, Yu R, Staniszewski A, Planel E, Herman M, Ho L, Kreber R, Honig LS, Ganetzky B, Duff K, Arancio O, Small SA. Retromer deficiency observed in Alzheimer's disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc Natl Acad Sci U S A. 2008;105(20):7327–7332. doi: 10.1073/pnas.0802545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakada-Tsukui K, Saito-Nakano Y, Ali V, Nozaki T. A retromerlike complex is a novel Rab7 effector that is involved in the transport of the virulence factor cysteine protease in the enteric protozoan parasite Entamoeba histolytica. Mol Biol Cell. 2005;16(11):5294–5303. doi: 10.1091/mbc.E05-04-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen MS, Gustafsen C, Madsen P, Nyengaard JR, Hermey G, Bakke O, Mari M, Schu P, Pohlmann R, Dennes A, Petersen CM. Sorting by the cytoplasmic domain of the amyloid precursor protein binding receptor SorLA. Mol Cell Biol. 2007;27(19):6842–6851. doi: 10.1128/MCB.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, Lah JJ. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26(5):1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reitz C. Genetic variants in SORCS2, SORCS3 and SORT1 affect amyloid processing and the risk of Alzheimer's disease. Submitted. [Google Scholar]
- 44.Reitz C, Cheng R, Rogaeva E, Lee JH, Tokuhiro S, Zou F, Bettens K, Sleegers K, Tan K, Kimura R, Shibata N, Arai H, Kamboh MI, Prince JA, Maier W, Riemenschneider M, Owen M, Harold D, Hollingworth P, Cellini E, Sorbi S, Nacmias B, Takeda M, Pericak-Vance MA, Haines JL, Younkin S, Williams J, van Broeckhoven C, Farrer LA, St George-Hyslop PH, Mayeux R. Meta-analysis of the association between variants in SORL1 and Alzheimer disease. Arch Neurol. 68(1):99–106. doi: 10.1001/archneurol.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reitz C, Tokuhiro S, Clark LN, Conrad C, Vonsattel JP, Hazrati LN, Palotas A, Lantigua R, Medrano M, I ZJ-V, Vardarajan B, Simkin I, Haines JL, Pericak-Vance MA, Farrer LA, Lee JH, Rogaeva E, George-Hyslop PS, Mayeux R. SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer's disease risk. Ann Neurol. 69(1):47–64. doi: 10.1002/ana.22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39(2):168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJ, Raposo G, van der Sluijs P, Bonifacino JS. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol. 2008;183(3):513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudinskiy N, Grishchuk Y, Vaslin A, Puyal J, Delacourte A, Hirling H, Clarke PG, Luthi-Carter R. Calpain hydrolysis of alpha- and beta2-adaptins decreases clathrin-dependent endocytosis and may promote neurodegeneration. J Biol Chem. 2009;284(18):12447–12458. doi: 10.1074/jbc.M804740200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scherzer CR, Offe K, Gearing M, Rees HD, Fang G, Heilman CJ, Schaller C, Bujo H, Levey AI, Lah JJ. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61(8):1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- 50.Seaman MNJ. Identification of a novel conserved sorting motif necessary for retromer-mediated endosome-to-Golgi retrieval. J Cell Sci. 2007;120:2378–2389. doi: 10.1242/jcs.009654. [DOI] [PubMed] [Google Scholar]
- 51.Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165(1):111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seaman MN, Harbour ME, Tattersall D, Read E, Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci. 2009;122(Pt 14):2371–2382. doi: 10.1242/jcs.048686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142(3):665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT, Jr, Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM. Genome-wide analysis of genetic loci associated with Alzheimer disease. Jama. 303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherva R, Baldwin CT, Inzelberg R, Vardarajan B, Cupples LA, Lunetta K, Bowirrat A, Naj A, Pericak-Vance M, Friedland RP, Farrer LA. Identification of novel candidate genes for Alzheimer's disease by autozygosity mapping using genome wide SNP data. J Alzheimers Dis. 23(2):349–359. doi: 10.3233/JAD-2010-100714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Small SA. Retromer sorting: a pathogenic pathway in late-onset Alzheimer disease. Arch Neurol. 2008;65(3):323–328. doi: 10.1001/archneurol.2007.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Small SA, Kent K, Pierce A, Leung C, Kang MS, Okada H, Honig L, Vonsattel JP, Kim TW. Model-guided microarray implicates the retromer complex in Alzheimer's disease. Ann Neurol. 2005;58(6):909–919. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- 58.Spoelgen R, von Arnim CA, Thomas AV, Peltan ID, Koker M, Deng A, Irizarry MC, Andersen OM, Willnow TE, Hyman BT. Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and beta-secretase beta-site APP-cleaving enzyme. J Neurosci. 2006;26(2):418–428. doi: 10.1523/JNEUROSCI.3882-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strochlic TI, Schmiedekamp BC, Lee J, Katzmann DJ, Burd CG. Opposing activities of the Snx3-retromer complex and ESCRT proteins mediate regulated cargo sorting at a common endosome. Mol Biol Cell. 2008;19(11):4694–4706. doi: 10.1091/mbc.E08-03-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177(1):115–125. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sullivan CP, Jay AG, Stack EC, Pakaluk M, Wadlinger E, Fine RE, Wells JM, Morin PJ. Retromer disruption promotes amyloidogenic APP processing. Neurobiol Dis. 2011;43(2):338–345. doi: 10.1016/j.nbd.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tabuchi M, Yanatori I, Kawai Y, Kishi F. Retromer-mediated direct sorting is required for proper endosomal recycling of the mammalian iron transporter DMT1. J Cell Sci. 123(Pt 5):756–766. doi: 10.1242/jcs.060574. [DOI] [PubMed] [Google Scholar]
- 63.Tan EK, Lee J, Chen CP, Teo YY, Zhao Y, Lee WL. SORL1 haplotypes modulate risk of Alzheimer's disease in Chinese. Neurobiol Aging. 2009;30(7):1048–1051. doi: 10.1016/j.neurobiolaging.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 64.Viklund IM, Aspenstrom P, Meas-Yedid V, Zhang B, Kopec J, Agren D, Schneider G, D'Amato M, Olivo-Marin JC, Sansonetti P, Van Nhieu GT, Pettersson S. WAFL, a new protein involved in regulation of early endocytic transport at the intersection of actin and microtubule dynamics. Exp Cell Res. 2009;315(6):1040–1052. doi: 10.1016/j.yexcr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 65.Vilarino-Guell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, Behrouz B, Melrose HL, Hentati E, Puschmann A, Evans DM, Conibear E, Wasserman WW, Aasly JO, Burkhard PR, Djaldetti R, Ghika J, Hentati F, Krygowska-Wajs A, Lynch T, Melamed E, Rajput A, Rajput AH, Solida A, Wu RM, Uitti RJ, Wszolek ZK, Vingerhoets F, Farrer MJ. VPS35 mutations in Parkinson disease. Am J Hum Genet. 89(1):162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wassmer T, Attar N, Bujny MV, Oakley J, Traer CJ, Cullen PJ. A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer. J Cell Sci. 2007;120(Pt 1):45–54. doi: 10.1242/jcs.03302. [DOI] [PubMed] [Google Scholar]
- 67.Zimprich A, Benet-Pages A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, Rossle SC, Klopp N, Wolf E, Seppi K, Pirker W, Presslauer S, Mollenhauer B, Katzenschlager R, Foki T, Hotzy C, Reinthaler E, Harutyunyan A, Kralovics R, Peters A, Zimprich F, Brucke T, Poewe W, Auff E, Trenkwalder C, Rost B, Ransmayr G, Winkelmann J, Meitinger T, Strom TM. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 89(1):168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Cells stably expressing GFP-Snx3 or GFP-Rab7A were treated with siRNA to silence Snx3 or Rab7A expression. Lysates were subjected to SDS-PAGE and then analysed by western blotting with antibodies against Snx3, Rab7A, or GFP (panels A–C, respectively). The band visible at 97 kD is present in all three blots and is likely to be a nonspecific protein that is recognized by the iodinated protein-A. D. The gel was stained with coomassie blue to show protein loading. The western blots confirm that the siRNA efficiently targets both endogenous and GFP-Snx3, and endogenous Rab7A. The GFP-tagged Rab7A construct utilizes the murine cDNA and is resistant to the siRNA targeting endogenous Rab7A.
Supplemental Figure 2. Cells stably expressing GFP-VPS35 or VPS29-GFP were transiently transfected with RFP-Snx3, fixed and then labeled with antibodies against GFP or RFP. The increased expression of Snx3 by the transient transfection of RFP-Snx3 is able to promote the membrane association of GFP-VPS35 or VPS29-GFP where a proportion of those constructs are cytosolic. Bar = 20 µm.