Abstract
Lynch syndrome (hereditary nonpolyposis colon cancer) and adenomatous polyposis syndromes frequently have overlapping clinical features. Current approaches for molecular genetic testing are often stepwise, taking a best-candidate gene approach with testing of additional genes if initial results are negative. We report a comprehensive assay called ColoSeq that detects all classes of mutations in Lynch and polyposis syndrome genes using targeted capture and massively parallel next-generation sequencing on the Illumina HiSeq2000 instrument. In blinded specimens and colon cancer cell lines with defined mutations, ColoSeq correctly identified 28/28 (100%) pathogenic mutations in MLH1, MSH2, MSH6, PMS2, EPCAM, APC, and MUTYH, including single nucleotide variants (SNVs), small insertions and deletions, and large copy number variants. There was 100% reproducibility of mutation detection between independent runs. The assay correctly identified 222 of 224 heterozygous SNVs (99.4%) in HapMap samples, demonstrating high sensitivity of calling all variants across each captured gene. Average coverage was greater than 320 reads per base pair when the maximum of 96 index samples with barcodes were pooled. In a specificity study of 19 control patients without cancer from different ethnic backgrounds, we did not find any pathogenic mutations but detected two variants of uncertain significance. ColoSeq offers a powerful, cost-effective means of genetic testing for Lynch and polyposis syndromes that eliminates the need for stepwise testing and multiple follow-up clinical visits.
Defects in mismatch repair (MMR) are responsible for hereditary nonpolyposis colorectal cancer, also known as Lynch syndrome. Inherited loss of function mutations in MLH1, MSH2, MSH6, PMS2, and EPCAM result in a 25% to 75% lifetime risk of colon cancer and up to a 60% lifetime risk of endometrial cancer in women.1 The population prevalence of Lynch syndrome is estimated to be as high as 1 in 440,2 making it the most common inherited cancer predisposition syndrome. Germline mutations in APC and MUTYH result in adenomatous polyposis syndromes that are also associated with a very high lifetime risk of colorectal cancer. Mutations in APC cause familial adenomatous polyposis, Gardner's syndrome, Turcot's syndrome, and attenuated familial adenomatous polyposis, whereas MUTYH mutations are the cause of autosomal recessive MUTYH-associated polyposis syndrome.3 Even with clinical guidelines such as the Revised Bethesda and Amsterdam II criteria,4 it can be challenging to distinguish Lynch syndrome from adenomatous polyposis syndromes on the basis of clinical features, particularly with regard to attenuated familial adenomatous polyposis and Lynch syndrome, which present at a similar median age of cancer onset (∼45 to 60 years), and with similar numbers of colonic polyps.5,6 Clinicians are therefore frequently faced with the dilemma of which gene or genes to test first when ordering expensive genetic testing using standard Sanger sequencing.
A complex battery of tumor-based screening tests have been developed for Lynch syndrome, in part because of the high cost of conventional germline genetic testing. These tests include functional assessment of defective MMR genes either by demonstration of genomic microsatellite instability (MSI) or loss of MLH1, MSH2, MSH6, or PMS2 protein expression by immunohistochemistry (IHC), BRAF V600E mutational analysis, and MLH1 promoter hypermethylation analysis (screening algorithms are reviewed in Pritchard and Grady7). Loss of a specific MMR protein revealed by IHC can be helpful in suggesting which gene most likely harbors a germline mutation. MMR proteins are often lost as pairs (MLH1/PMS2 or MSH2/MSH6) because they function as heterodimers. However, mutations in PMS2 and MSH6 are more likely to result in isolated loss of the corresponding protein by IHC because they are minor partners in the heterodimer.8 No tumor-based screening tests are currently available for polyposis syndromes.
Massively parallel next-generation sequencing technology has dramatically increased throughput and reduced the cost per nucleotide sequenced compared with traditional Sanger methods, enabling cost-effective sequencing of multiple genes simultaneously in the clinical laboratory setting.9–13 Target enrichment is generally required to achieve adequate read depth for accurate identification of the spectrum of mutations, including large genomic rearrangements, small insertions and deletions (indels), and SNVs.14 We recently reported a proof-of-principle study demonstrating the accuracy and feasibility of solution-based targeted capture and next-generation sequencing for 21 genes that are associated with breast and ovarian cancer risk.12 Here we report the validation results of ColoSeq, a clinical diagnostic assay for hereditary colon cancer that detects single nucleotide, indel, and deletion/duplication mutations in MLH1, MSH2, MSH6, PMS2, EPCAM, APC, and MUTYH. The ColoSeq assay can be performed for approximately the same cost as performing sequencing and deletion/duplication analysis of a single gene by traditional methods, and with equivalent or better sensitivity and accuracy, eliminating the need for stepwise molecular genetic testing in patients with suspected Lynch or polyposis syndromes.
Materials and Methods
DNA Samples
We tested a total of 82 unique DNA samples, including 23 peripheral blood DNA samples from patients with known mutations in MLH1, MSH2, MSH6, PMS2, EPCAM, APC, or MUTYH; 31 peripheral blood DNA samples from patients with a clinical history suggestive of Lynch or polyposis syndrome; 19 peripheral blood DNA samples from patients without a known family history of cancer; 6 publicly available DNA samples from the HapMap project;15 and 3 DNA samples from colon cancer cell lines known to harbor mutations in MMR genes and/or APC. Tumor cell lines (LoVo, HCT116, LS174T) were obtained from William M. Grady (University of Washington). HapMap samples (NA18507, NA18558, NA07019, NA07348, NA10857, NA10851) were obtained from Coriell Cell Repositories (Camden, New Jersey). Clinical specimens were obtained in accordance with the Declaration of Helsinki and the study was approved by the Human Subjects Division of the University of Washington (protocol 34173) and the University of Chicago Institutional Review Board.
Library Construction, Gene Capture, and Massively Parallel Sequencing
Three micrograms of DNA was sonicated to a peak of 200 bp on a Covaris S2 instrument (Covaris, Woburn, MA) in 1× low TE (10 mmol/L Tris/0.1 mmol/L EDTA) for 6 minutes using frequency sweeping mode with duty cycle, 10%; intensity, 5; cycles per burst, 200 at a temperature 4° to 7°C. After sonication, DNA was purified with AMPure XP beads (Beckman Coulter, Brea CA) and subjected to three enzymatic steps: end repair, A-tailing, and ligation to Illumina paired-end adapters as described in the SureSelectXT Target Enrichment for Illumina multiplexed sequencing that is available for free download. All liquid handling steps were performed in 96-well plates on a Bravo liquid-handling instrument (Agilent Technologies, Santa Clara, CA). The adapter-ligated library was amplified by polymerase chain reaction (PCR) for five cycles with Illumina primers 1.0 and 2.0 and quantified by a DNA1000 chip on a Bioanalyzer 2100 instrument (Agilent Technologies). Individual paired-end libraries (500 ng) were hybridized to a custom design of complementary RNA (cRNA) biotinylated oligonucleotides targeting 31 genes in 30 genomic regions (see Supplemental Table S1 at http://jmd.amjpathol.org). The 120-mer oligonucleotide baits were designed in Agilent's eArray web portal with the following parameters: centered tiling, 3x bait overlap and a maximum overlap of 20 bp into repetitive regions. The custom design targets a total of 1.1 Mb of DNA including 209 kb in MLH1, MSH2, MSH6, PMS2, EPCAM, APC, and MUTYH (Table 1). The BED file of probe sequences is available on request. After 24 hours of hybridization at 65°C, the library-bait hybrids were purified by incubation with streptavidin-bound T1 Dynabeads (LifeTechnologies, Carlsbad CA) and washed with increasing stringency to remove nonspecific binding. After capture, each library was amplified by PCR directly on the Dynabeads for 13 cycles with primers containing a unique 6-bp index (Table 2). After PCR amplification, the libraries were quantified by a high-sensitivity chip on a Bioanalyzer 2100 instrument (Agilent Technologies). Equimolar concentrations of 96 libraries were pooled to a final concentration of 10 pM, denatured with 3N NaOH, and cluster amplified with a cBot instrument on a single lane of an Illumina v3 flow cell. Sequencing was performed with 2 × 101-bp paired-end reads and a 7-bp index read using SBS v3 chemistry on a HiSeq2000 (Illumina, Inc, San Diego, CA).
Table 1.
Genes Validated for ColoSeq
| Gene | Chromosome | Kb targeted⁎ | Genomic region targeted (Hg 19) |
|
|---|---|---|---|---|
| Start | End | |||
| MLH1 | 3 | 35 | 37029979 | 37097337 |
| MSH2 + EPCAM | 2 | 48 | 47595263 | 47715360 |
| MSH6 | 2 | 19 | 48005221 | 48039092 |
| PMS2 | 7 | 17 | 6007870 | 6053737 |
| APC | 5 | 77 | 112038202 | 112186936 |
| MUTYH | 1 | 14 | 45789914 | 45811142 |
A total of 30 genomic regions representing 31 genes related to hereditary cancer risk were targeted19, but here we focus on validation of these 7 genes.
Repetitive DNA elements were not targeted.
Table 2.
Postindex Barcodes
| ATCACG | AACAAA | AATAGG | ACTCTC |
| CGATGT | CACGAT | CCACGC | CGGAAT |
| TTAGGC | GATATA | GCTCCA | GTGGCC |
| TGACCA | TATAAT | TCGGCA | TGCTGG |
| ACAGTG | AACCCC | ACAAAC | ACTGAT |
| GCCAAT | CACTCA | CCCATG | CTAGCT |
| CAGATC | GATGCT | GGCACA | GTTTCG |
| ACTTGA | TCATTC | TCTACC | TGGCGC |
| GATCAG | AACTTG | ACATCT | AGAAGA |
| TAGCTT | CAGGCG | CCCCCT | CTATAC |
| GGCTAC | GCAAGG | GGCCTG | CGTACG |
| CTTGTA | ATAATT | ATCCTA | ATGAGC |
| AAACAT | AAGACT | ACCCAG | AGATAG |
| CAAAAG | CATGGC | CCGCAA | CTCAGA |
| GAAACC | GCACTT | GTAGAG | GAGTGG |
| TAATCG | TCCCGA | TGAATG | TTCGAA |
| AAAGCA | AAGCGA | ACCGGC | AGCATC |
| CAACTA | CATTTT | CCTTAG | CTGCTG |
| GAATAA | GCCGCG | GTCCGC | GGTAGC |
| TACAGC | TCGAAG | TGCCAT | TTCTCC |
| AAATGC | AAGGAC | ACGATA | AGCGCT |
| CACCGG | CCAACA | CGAGAA | CCGTCC |
| GACGGA | GCCTTA | GTGAAA | ATTCCT |
| AGGCCG | ATACGG | ATCTAT | AGGTTT |
Mutation Analysis
Sequence alignment and variant calling were performed against the reference human genome (UCSC hg19). Sequencing reads were aligned using MAQ,16 and SNVs and insertions and deletions were detected as previously described.12 Each variant was annotated with respect to gene location and predicted function in Human Genome Variation Society nomenclature. Deletions and duplications of exons were detected by depth of coverage analysis.17 All previously unidentified frameshift, nonsense, and splice site mutations predicted to be deleterious to protein function were confirmed by PCR amplification and Sanger sequencing. Exonic deletions and duplications were confirmed by multiplex ligation-dependent probe amplification or gap-PCR and direct sequencing.18
Results
ColoSeq Assay
The objective of our study was to evaluate the performance of targeted DNA capture and massively parallel sequencing for the detection of inherited mutations in colon cancer in the clinical laboratory setting. We designed oligonucleotides to target all exons, introns, and approximately 10 kb of 5′ and 3′ flanking genomic regions of the seven genes that are most commonly responsible for inherited risk of colon cancer (Table 1). Our capture panel also includes 24 other genes that rarely harbor mutations causing colon cancer, endometrial cancer, and other solid tumors,19 for a total of 31 captured genes and 1.1 Mb of captured DNA after removal of repetitive sequences. Here we focus on validation results of the seven genes listed in Table 1 that constitute the ColoSeq assay for Lynch and polyposis syndromes.
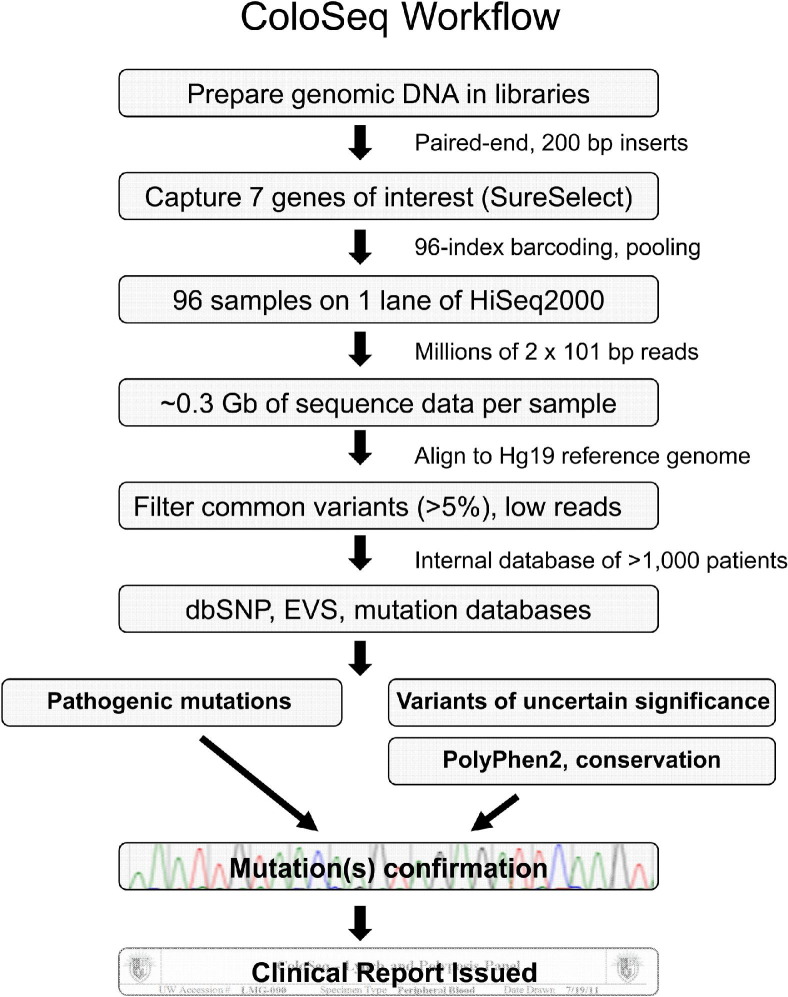
Workflow of the ColoSeq assay is summarized in Figure 1. We begin by fragmenting genomic DNA extracted from peripheral blood and preparing libraries with an average insert size of approximately 200 bp. ColoSeq genes are captured via solution hybridization with complementary oligonucleotide RNA (cRNA) baits (Agilent SureSelect). Each captured sample library is amplified by PCR to incorporate a unique index barcode, and up to 96 barcoded samples are pooled and run on a single lane of an Illumina HiSeq2000 instrument.
Figure 1.
ColoSeq workflow.
It is unusual to pool patient samples in the clinical setting because of the risks of sample misidentification and analytic interference. To control for specimen mix-up and for potential barcode-specific sequencing biases, each clinical sample is indexed with two distinct barcodes and run in duplicate. Barcodes were selected such that a single base change could not lead to one barcode being misidentified as another (Table 2). Less than 0.005% of sequencing reads were assigned to the incorrect patient because of barcode misidentification.
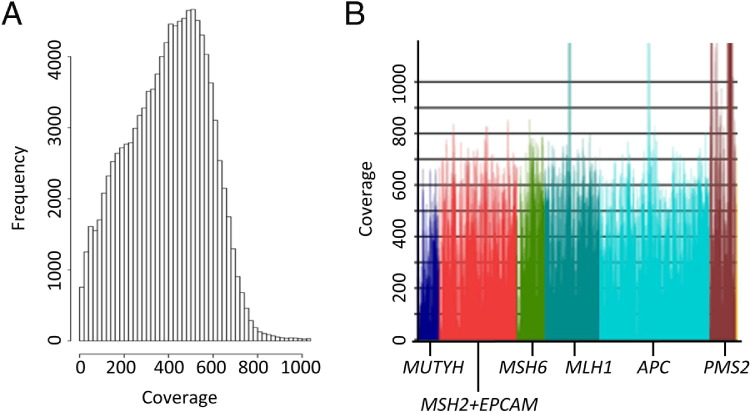
Paired-end 2 × 101-bp reads are generated at a median of 320-fold coverage per nucleotide across the entire targeted region (range of median coverage between samples 145-fold to 556-fold; Figure 2A). The median coverage specifically across the seven ColoSeq genes is 475-fold (Figure 2B). Raw sequence of 0.3 gigabases (Gb) (range 0.15 to 0.56 Gb) of high-quality (>Q30) sequence per sample is generated when the maximum 96 samples are pooled within a lane. On average, 62% of sequence reads are on target, mapping specifically to the captured regions. We align sequences to the reference genome (build Hg 19) and select variants for further analysis that meet the following criteria: i) variant is present in sequence reads from both strands (mean ± SD, 433 ± 97 variants/sample), ii) population frequency of the variant is less than 5% (44 ± 31 variants/sample remain), and iii) variant represents at least 5% of the sequence reads at a particular site (26 ± 21 variants/sample remain). We chose a threshold of 5% variant reads to ensure that variants in highly segmentally duplicated regions of genes would be detected. Frequencies of variants among individuals without colon cancer are based on an internal database of more than 1000 unique patients from different ethnic backgrounds who have had targeted capture and massively parallel sequencing performed for these genes.12,19 By using an internal frequency database, we can quickly filter out both common benign variants and potential recurrent artifactual variant calls, leaving an average of 20 noncoding and only 6 rare coding or splice junction variants per patient across the seven genes. These filtered variants are then compared with the exome variant server (NHLBI Exome Sequencing Project [ESP], Seattle, WA), dbSNP, and public mutation databases such as InSight20 (http://www.insight-group.org/mutations) and results reviewed by a laboratory director. Novel variants of uncertain significance (VUS) are analyzed by PolyPhen2,21 SIFT,22 and MutationTaster23 to predict potential deleterious effects and are assessed for evolutionary conservation. Pathogenic mutations and VUS are confirmed by Sanger sequencing or multiplex ligation-dependent probe amplification (for deletions/duplications). Missense variants that are not well characterized are reported as VUS when they meet the following criteria: i) population frequency <5%, ii) evolutionarily conserved, iii) predicted to be at least possibly damaging by in silico prediction tools, and iv) confirmed by Sanger sequencing.
Figure 2.
Depth of coverage. Depth of coverage from a representative patient sample run on a single lane of a HiSeq2000 instrument in a 96-plexed ColoSeq run. A: Distribution of coverage for each targeted base. Each bar represents the number of base pairs (in 1000s) at a particular depth of coverage. Of the targeted bases, 98.5% have a minimum of 50-fold coverage. B: Median coverage across each ColoSeq gene is shown (average coverage of 475-fold). Areas with depth of coverage >1000-fold reflect capture of highly homologous genomic regions.
Sensitivity
To assess the sensitivity of the ColoSeq assay to detect pathogenic mutations, we blindly tested peripheral blood DNA samples from 23 cancer patients and 3 colon cancer cell lines (LS174T, HCT116, LoVo) with previously defined germline mutations in MLH1, MSH2, MSH6, PMS2, EPCAM, MUTYH, or APC. Some cell lines harbored more than one mutation. ColoSeq correctly identified all mutations (23/23 patient mutations, 5/5 cell line mutations, 100% sensitivity) including nonsense, missense, frameshift, in-frame deletions, splice site, and large deletions and duplications (Table 3, cohorts: known and cell line).24–39
Table 3.
Mutations and Variants of Uncertain Significance Identified by ColoSeq
| Cohort | ID | Gene | Mutation/VUS† | Effect | Chr | Start (Hg19) | End (Hg19) | Total reads‡ | Variant reads‡ |
|---|---|---|---|---|---|---|---|---|---|
| Cell line | HCT116 | MLH1 | c.755C>A20 | S252⁎ | 3 | 37,056,000 | 244 | 244 | |
| Cell line | LoVo | APC | c.3340C>T24,25 | R1114⁎ | 5 | 112,174,631 | 417 | 194 | |
| Cell line | LoVo | APC | c.4289delC25 | M1431Cfs⁎42 | 5 | 112,175,580 | 112,175,581 | 174 | 97 |
| Cell line | LoVo | MSH2 | ∼51kb deletion26 | del exons 3-8 | 2 | ∼47,637,277 | ∼47,688,664 | NA | NA |
| Cell line | LS174T | MLH1 | c.350C>T27 | T117M | 3 | 37,045,935 | 325 | 323 | |
| Known | FO5 | MSH2 | ∼31kb deletion28 | del ex 9-16 | 2 | ∼47,688,689 | ∼47,719,344 | 322 | 160 |
| Known | 011 | MSH2 | c.513delG29 | K172Afs⁎2 | 2 | 47,637,378 | 47,637,379 | 64 | 31 |
| Known | 012 | MSH6 | c.3103C>T30 | R1035⁎ | 2 | 48,028,225 | 389 | 117 | |
| Known | 013 | MSH2 | c.1609A>T | K537⁎ | 2 | 47,693,895 | 151 | 73 | |
| Known | 014 | MLH1 | c.1668-1G>A28 | Splice | 3 | 37,083,758 | 180 | 91 | |
| Known | 017 | MLH1 | c.1517 T>C31 | V506A | 3 | 37,070,382 | 227 | 114 | |
| Known | 022 | APC | c.221-2A>G | Splice | 5 | 112,102,884 | 143 | 80 | |
| Known | 023 | MLH1 | c.1381A>T32 | K461⁎ | 3 | 37,067,470 | 139 | 75 | |
| Known | 025 | MSH2 | c.380A>G | N127S (VUS) | 2 | 47,637,246 | 64 | 35 | |
| Known | 031 | MSH6 | c.694C>T | Q232⁎ | 2 | 48,025,816 | 152 | 76 | |
| Known | 032 | MSH2 + EPCAM | ∼40kb deletion | del ex 1-2 + EPCAM | 2 | ∼47,595,376 | ∼47,635,837 | 322 | 172 |
| Known | 034 | MSH2 | c.2501_2507delCTAATTT | N835Lfs⁎4 | 2 | 47,707,877 | 47,707,884 | 129 | 47 |
| Known | 035 | PMS2 | c.1927C>T33 | Q643⁎ | 7 | 6026,469 | 97 | 48 | |
| Known | 037 | MLH1 | ∼18kb duplication | dup ex 6-12 | 3 | ∼37,050,170 | ∼37,067,982 | 351 | 509 |
| Known | 086 | MSH6 | c.3485_3487delCTG34 | A1162del | 2 | 48,032,092 | 48,032,095 | 134 | 58 |
| Known | 087 | MSH2 | ∼20kb deletion | del exon 1-6 | 2 | 47,629,721 | 47,649,842 | 274 | 120 |
| Known | 088 | MSH6 | c.3956_3959delAAGC34 | A1320Gfs⁎6 | 2 | 48,033,745 | 48,033,749 | 174 | 72 |
| Known | 090 | MSH6 | c.2731C>T34 | R911⁎ | 2 | 48,027,853 | 130 | 61 | |
| Known | 093 | MSH6 | c.2731C>T34 | R911⁎ | 2 | 48,027,853 | 74 | 41 | |
| Known | 094 | MSH6 | c.3939_3940ins1934 | Q1314Sfs⁎5 | 2 | 48,033,728 | 48,033,728 | 166 | 20 |
| Known | 096 | MLH1 | ∼3kb deletion35 | del exon 14-15 | 3 | ∼37,081,516 | ∼37,084,593 | 345 | 177 |
| Known | 147 | MUTYH | c.1187C>T36 | G396D§ | 1 | 45797228 | 147 | 68 | |
| Known | 148 | MUTYH | c.1187C>T36 | G396D§ | 1 | 45797228 | 102 | 32 | |
| Control | 047 | MLH1 | c.2161T>C | Y721H (VUS) | 3 | 37,092,034 | 220 | 137 | |
| Control | 063 | MLH1 | c.1151T>A31 | V384D (VUS) | 3 | 37,067,240 | 167 | 77 | |
| Unknown | 003 | APC | c.1959G>A37 | ?Splice (VUS) | 5 | 112,173,250 | 199 | 92 | |
| Unknown | 004 | MSH6 | c.2147_2150delCAGT | V717Afs⁎18 | 2 | 48,027,269 | 48,027,273 | 100 | 41 |
| Unknown | 006 | APC | c.426_427delAT38 | L143Afs⁎4 | 5 | 112,111,329 | 112,111,331 | 143 | 66 |
| Unknown | 018 | MUTYH | c.158-1G>A | Splice (VUS) | 1 | 45,799,276 | 61 | 33 | |
| Unknown | 036 | PMS2 | c.2192_2196delTAACT39 | L731Cfs⁎3 | 7 | 6018,306 | 6018,311 | 255 | 64 |
| Unknown | 089 | PMS2 | c.2192_2196delTAACT39 | L731Cfs⁎3 | 7 | 6018,306 | 6018,311 | 545 | 81 |
| Unknown | 040A | MSH6 | c.2588A>G | E863G (VUS) | 2 | 48,027,710 | 278 | 140 | |
| Unknown | 041 | APC | c.646C>T38 | R216⁎ | 5 | 112,128,143 | 179 | 83 | |
| Unknown | 080 | PMS2 | 3.3kb deletion | del exon 8 | 7 | 6033,256¶ | 6036,607¶ | 409 | 192 |
Chr, chromosome; NA, not applicable.
Note: Asterisk (*) refers to stop codon (per Human Genome Variation Society standard nomenclature).
Reference sequences: MLH1 NM_000249.3; MSH2 NM_000251.1; MSH6 NM_000179.2; PMS2 NM_000535.5; EPCAM NM_002354.2; APC NM_000038.5; MUTYH NM_001128425.1.
Numbers reflect perfect matches only. For CNVs, total reads refers to the normalized read depth and variant reads to the read depth in the sample.
This mutation is commonly referred to as G382D (NM_001048171.1:c.1145G>A).
Exact breakpoints were determined for the PMS2 exon 8 deletion, which we detected in patient 080.
To assess the analytic sensitivity of detecting all heterozygous single base pair variants across these genes, we evaluated six DNA samples that had been genotyped by the HapMap Project15 (http://www.hapmap.org, last accessed December 6, 2011). HapMap genotype data were available for a total of 1388 sites in aggregate across the six samples, including 226 heterozygous SNVs (38 ± 23 per sample), 144 homozygous variants (27 ± 15 per sample), and 1018 sites that matched the reference genome (wild type, 170 ± 23 per sample). We focused on defining the sensitivity of heterozygous variant detection because pathogenic mutations in Lynch and polyposis syndromes are almost always heterozygous, except in MUTYH. ColoSeq identified 222/226 heterozygous SNVs in the seven genes captured by the assay. Two of the four discrepant variants were found by Sanger sequencing to be errors in the HapMap annotation, for a sensitivity of 99.4% (222/224) (Table 4). The two HapMap variants that were missed by ColoSeq corresponded to a single intronic SNV (rs3771280) that was missed in two different samples. This SNV is in close proximity to two other SNVs (rs3771278, rs3771279), which were both either heterozygous or homozygous for the rarer allele in the two samples, resulting in the reads mapping to this region being discarded because of poor alignment with the reference genome. ColoSeq correctly identified all HapMap variants within exons and splice junctions (27/27; 100% sensitivity). Among homozygous SNVs, all were correctly identified except rs3771280.
Table 4.
Analytic Sensitivity, Specificity, and Accuracy
| Sensitivity | Specificity | Accuracy | |
|---|---|---|---|
| All variants (with introns) | 99.4% (222/224) | 99.4% (1012/1018) | 99.4% (1234/1242) |
| Exons only | 100% (27/27) | 100% (198/198) | 100% (225/225) |
Calculations are based on heterozygous variant detection in 6 HapMap samples.
We next evaluated the performance of ColoSeq in a cohort of 31 prospectively collected blood samples from patients with a clinical history suggestive of Lynch or polyposis syndrome, but without a previously determined mutation. In this cohort, we identified six patients with pathogenic mutations and three additional patients with VUS; the mutations were all confirmed using alternative methods (Table 3; cohort: unknown). Most of the patients in this cohort had previous clinical testing performed by outside laboratories for one or more of the ColoSeq genes, with negative results. Even though the group was enriched for patients who had negative test results for one or more of the ColoSeq genes, our mutation detection in 6 of 31 patients was similar to published rates among patients meeting Bethesda criteria,40 highlighting the utility of a comprehensive Lynch and polyposis panel.
Specificity and Accuracy
We determined analytic specificity by analyzing the 6 HapMap characterized samples at 1018 known nonvariant (reference sequence) sites in the seven ColoSeq genes. ColoSeq correctly identified 1012/1018 sites as nonvariant from the reference genome for an analytic specificity of 99.4% (Table 4). The six false-positive variants that we detected but that were not reported as present in HapMap samples were in MLH1 intron 11 (rs3774339, detected in one sample), and in segmentally duplicated exons 11 and 14 of PMS2 (rs1805321, detected in three samples; rs1059060, detected in two samples). Analytic specificity was 100% within single-copy exons and splice junctions (198/198). We calculated analytic accuracy as [number of true positives + number of true negatives]/[total number of HapMap genotyped sites]. Accuracy was 99.4% and 100% for all variants and exons only, respectively (Table 4).
To assess clinical specificity we selected 19 control patients without a personal or family history of colon cancer or other Lynch syndrome–associated cancers. Patients were selected to represent diverse ethnic backgrounds including African American, Pacific Islander, white, Indian, Cambodian, Vietnamese, Iranian, Hispanic, Alaskan Native, Chinese, Laotian, and Filipino (see Supplemental Table S2 at http://jmd.amjpathol.org). We did not find any pathogenic mutations in this cohort, for a specificity of 100% (19/19). However, we identified a VUS that met our criteria for clinical reporting in two control samples; one each from an African-American and a Vietnamese control subject (Table 3, cohort: control), suggesting that about 10% of nonwhite patients without Lynch or polyposis syndromes will have a VUS detected (2/17).
Deletion/Duplication Analysis
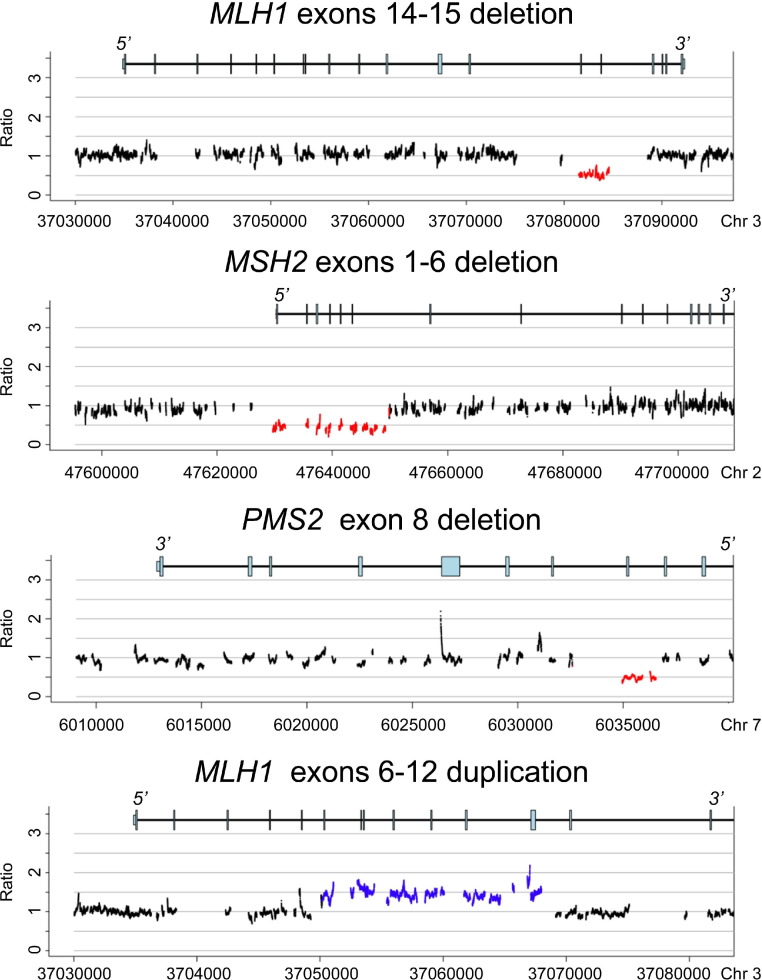
The very high fold coverage (mean, 320-fold) allowed accurate detection of large deletions and duplications using normalized depth of coverage and split-read analysis (Figure 3).12,17 Exact breakpoints could not be determined for most large deletions and duplications because breakpoints are commonly in Alu or other repetitive DNA elements that are not captured by our design. However, the assay demonstrated at least exon-level resolution for all large deletions and duplications in all cases, which was comparable or better than the resolution of traditional approaches to deletion/duplication analysis such as multiplex ligation-dependent probe amplification. ColoSeq correctly identified 6/6 known large deletions/duplications in blinded challenge specimens (Table 3, cohort: known). In addition, in DNA from a patient with microsatellite instability–positive endometrial cancer and isolated loss of PMS2 protein expression (by IHC) who had not had previous genetic testing, we detected a PMS2 exon 8 deletion. We confirmed the exact breakpoints of this mutation by gap-PCR and Sanger sequencing (Table 3, patient 080).
Figure 3.
Deletions and duplications detected by normalized depth of coverage. Three examples of genomic deletions (MLH1 exons 14-15, MSH2 exons 1-6, and PMS2 exon 8) and one example of a genomic duplication (MLH1 exons 6-12) detected by ColoSeq. Read depth is normalized across the 96 samples on a run. Significant deviations less than or greater than a normalized ratio of 1 across adjacent baited regions reflect genomic deletions (red) and duplications (blue). Genomic coordinates shown on the x axis are hg19.
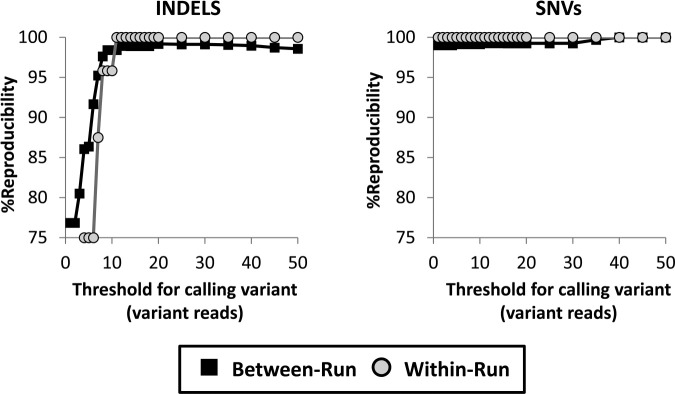
Reproducibility and Cutoff
To determine the between-run assay reproducibility, we ran 75 samples on at least two independent runs. We detected 39/39 pathogenic mutations and VUS (Table 3) in replicate runs, yielding 100% between-run reproducibility of mutation detection. We next assessed the between-run reproducibility for all variant calls in unique exons and splice junctions (exons only, mean = 39 ± 9 variants per sample), or for the entire captured region, including deeply intronic, nonunique, and repetitive DNA elements (with introns, mean = 451 ± 70 variants per sample). We separated SNVs and indel variants for this analysis. Not surprisingly, assay reproducibility increased as a function of the number of variant sequence reads, particularly for indel variants (Figure 4). We selected a minimum threshold of 15 variant sequencing reads to assess assay reproducibility based on the following two criteria: all pathogenic mutations and VUS were reliably detected with at least 15 variant reads, and the threshold of 15 variant reads substantially improved assay reproducibility without sacrificing sensitivity (all detected HapMap variants had at least 15 reads). We did not use receiver operating characteristic analysis to select the cutoff because the analytic specificity was >99% at all cutoff values (based on the 1018 HapMap known wild-type SNVs) (see Supplemental Figure S1 at http://jmd.amjpathol.org). For exons only, for which the performance of the assay is most critical, between-run reproducibility was 99.3% for SNVs and 98.9% for indels at a cutoff of 15 variant reads (Figure 4). Reproducibility improved to 100% for indels and SNVs at cutoff values higher than 40, which is representative of the majority of variant calls (66% of indels and 86% of SNVs had 40 or more variant reads). Reproducibility was lower with introns, at 93.9% for SNVs and 92.5% for indels at the cutoff of 15 reads (see Supplemental Figure S2 at http://jmd.amjpathol.org). Variant calls that did not replicate on repeated runs were most prevalent in introns of PMS2, which has a family of highly homologous pseudogenes on chromosome 7.41 Importantly, we were able to reproducibly detect all SNV, indel, and deletion mutations in exons of PMS2, even in regions of the gene that are segmentally duplicated (Table 3).
Figure 4.
Between-run and within-run reproducibility. Between-run and within-run reproducibility are shown as a function of minimum threshold for indel variants and calling single nucleotide (SNV). Variants were considered reproducible if they met or exceeded the specified cutoff value in the index sample and appeared in the replicate sample regardless of cutoff value. Reproducibility is shown for all variants in single-copy regions of exons and splice junctions. A threshold of 15 variant reads was chosen for the assay. Black squares, between-run; grey circles, within run.
Within-run reproducibility was determined by running 12 samples in duplicate on the same run. The within-run reproducibility for exons only was 100% for both SNVs and indels at the cutoff of 15 reads (Figure 4). Within-run reproducibility with introns was 95.0% for SNVs and 92.1% for indels at the cutoff of 15 variant reads, which was qualitatively similar to the between-run results (see Supplemental Figure S2 at http://jmd.amjpathol.org).
Discussion
Next-generation sequencing technology is rapidly changing the landscape of genetic testing. During a plenary session at the 2011 Association of Molecular Pathology annual meeting, Dr. Stephen Kingsmore suggested that molecular pathologists will look back to this time as a golden age in the field of molecular diagnostics because of the new possibilities afforded by massively parallel sequencing. Much of the excitement surrounding this technology is directed at the possibility of clinical exome and whole-genome sequencing as a one-stop, all-inclusive genetic test. Clinical laboratories such as Ambry Genetics and Baylor Whole Genome Laboratory are already offering exome sequencing.42,43 However, exome and genome sequencing currently have important limitations compared with targeted approaches with regard to both cost-effectiveness and analytic performance in the clinical setting. Exome sequencing often does not achieve adequate depth of coverage to detect structural rearrangements such as large deletions and duplications that are a significant component of the mutational spectrum for many inherited disorders. Considering that up to 15% of disease-causing mutations in the Lynch syndrome are due to large deletions,2 it is clinically relevant if this class of mutation is missed. An important advantage of the targeted approach we have taken with ColoSeq is that it can readily detect structural rearrangements through a very high depth of coverage.
Additional advantages of our targeted capture approach for clinical diagnostic testing include the following: i) For scalability, we were able to multiplex and pool 96 index barcoded samples onto one lane of an Illumina HiSeq flow cell, facilitating cost-effectiveness and allowing samples to be run in duplicate to control for sample mix-up and analytic variability. ii) Clinical validity was achieved by targeting and validating only genes that, when mutated, are well-established causes of hereditary colon cancer; it is therefore straightforward to incorporate the results of ColoSeq testing into clinical decision-making. iii) Limiting VUS, which can cause stress for both patients and providers, was achieved by our focus on a relatively limited number of genes and the existence of well-annotated hereditary colon cancer mutation databases such as InSight,20 and MMRUV44; we found a reportable VUS in only about 1 in 10 patients. iv) Clinical expertise is strengthened by focusing on just a handful of genes, making it reasonable for a molecular pathologist to gain familiarity with their mutational spectrum and to consider the data at each variant position individually before signing out a case.
At least one previous study has evaluated targeted capture and massively parallel sequencing for the detection of Lynch and polyposis syndromes.45 In that study, solid-phase NimbleGen 385K Custom Sequence Capture Arrays were used to examine 22 genes that included MLH1, MSH2, MSH6, and APC. One of four samples with a known Lynch syndrome indel mutations was missed using both Roche 454 (Roche, Branford, CT) and Illumina GAII technology, along with many other neutral variants that were detectable by Sanger sequencing. The reason for the relatively poor performance could be attributed to inefficient/off-target capture, limitations in sequencing homopolymer regions on the 454 platform, and difficulties calling indels on the Illumina GAII platform due to short (36 bp) single-end read length. Both gene capture and sequencing technology has matured in the past 2 years, allowing us to overcome these limitations through more efficient capture and much longer 101-bp paired-end reads.
Methods and guidelines to rigorously validate clinical diagnostic assays using next-generation sequencing technology are just beginning to emerge. The Association of Molecular Pathology is leading an effort to establish formal guidelines for the validation of tests using this technology. This includes a statement from the Association of Molecular Pathology in June 2011 that outlines aspects of method validation and ongoing quality control, including sensitivity and specificity, and recommends that precharacterized control samples such as HapMap DNAs be analyzed on each run (http://www.amp.org/documents/AMPCommentsNGS_June2011_Final.pdf, last accessed December 1, 2011). Additional groups such as the Centers for Disease Control–led Nex-StoCT working group (http://www.cdc.gov/osels/lspppo/Genetic_Testing_Quality_Practices/Nex-StoCT.html, last accessed December 1, 2011) and the College of American Pathologists are also working to establish guidelines. We have taken a multifaceted approach to validate the ColoSeq assay that includes measurements of both analytic and clinical sensitivity and specificity, between- and within-run reproducibility, and selection of cutoffs (analytic measurement range). In addition to using blinded challenge specimens with defined mutations at a single locus, we determined analytic sensitivity using HapMap samples that had between 50 and 100 defined variants each in the seven ColoSeq genes. Through this process, we discovered a false-negative intronic SNV (rs3771280) and, importantly, were alerted to the mechanism by which this SNV was missed.
We were particularly concerned with defining the VUS detection rate of the assay, given the large amount of DNA sequenced and the potential consequence that the assay would cause undue worry among patients and their families if a high number of VUS were reported. We were pleased to find a reportable VUS for only two nonwhite patients of 19 patients without Lynch or polyposis syndrome, even among patients of diverse ethnic backgrounds in which missense variants are not well characterized. Still, we will undoubtedly encounter VUS more frequently than in standard single-gene assays. Better in silico tools to predict the relative pathogenicity of VUS will be valuable, such as gene-specific algorithms46 and aggregate pathogenicity scores47 that can integrate established algorithms such as PolyPhen2,21 SIFT,22 Genomic Evolutionary Rate Profiling,48 and MutationTaster.23
We designed the seven-gene ColoSeq panel to be focused only on genes that have a well-established role in clinical decision making for patients with Lynch or polyposis syndromes. Mutations in genes such as MLH3, MSH3, PMS1, and EXO1 that have been reported to cause Lynch syndrome in rare cases2 are not included in the ColoSeq panel because of lack of knowledge regarding their clinical significance. Our design targets additional genes for rare hereditary cancer syndromes that may predispose to gastrointestinal cancers, including Cowden syndrome (PTEN), hereditary diffuse gastric cancer (CDH1), Peutz-Jeghers syndrome (STK11), and Li-Fraumeni syndrome (TP53), and we are optimistic about validating these genes in a future expanded version of the ColoSeq panel.
A limitation of the assay is the long turnaround time necessitated by the 9-day run time on the HiSeq2000 instrument. ColoSeq is amenable to transition to a smaller, more clinically oriented instrument, with faster turnaround time such as the MiSeq (Illumina) or Ion Torrent (Guilford, CT). However, an important drawback of these more rapid instruments is that only about 1 to 2 Gb of sequence data are generated per run, compared with 30 to 40 Gb for a single lane of a HiSeq2000. We estimate that only about three to five index barcoded samples could be pooled per run on the MiSeq using 100-bp or 150-bp paired-end reads to achieve the same depth of coverage as our current 96-plexed assay on the HiSeq2000. The assay workflow and turnaround time might also be improved through newer, more streamlined library preparation strategies such as transposase-based methods.49
In conclusion, we have developed and validated a comprehensive and cost-effective test for hereditary colon cancer syndromes that uses solution-based targeted capture and next-generation sequencing. The assay detected 100% of known mutations in challenge specimens, including all indel mutations and large deletions and duplications. We believe that ColoSeq will streamline and simplify genetic testing in patients with suspected hereditary colon cancer syndromes, sparing patients from needing to have multiple rounds of expensive genetic testing to establish a diagnosis.
Acknowledgments
We thank William Grady for providing colon cancer cell lines, Daniel Sabath for help reviewing the manuscript, Fuki Hisama, Wylie Burke, Wendy Raskind, Gideon Steinbach, Martha Horike-Pyne, Angela Jacobson, Heidi Holmes, and Britta Sjoding for help recruiting and obtaining consent from patients, and Deborah Barden, Rachel Slusher, Laura Akagi, Jane Ranchalis, and Patty Callahan for help preparing genomic DNA.
Footnotes
Supported by grants from the Department of Defense Ovarian Cancer Research ProgramOC093285 (T.W.) and NIHR01CA157744 (T.W. and M.-C.K.).
Supplemental material for this article can be found at http://jmd.amjpathol.org or at http://dx.doi.org/10.1016/j.jmoldx.2012.03.002.
Disclosure: R.J.B. serves on an expert panel for Myriad Genetics Laboratory.
Supplementary data
Receiver operating characteristic analysis. The sensitivity of the assay to detect HapMap-characterized single nucleotide polymorphisms is plotted versus 1-specificity at different minimum fold-coverage (number of reads) cutoffs. Even at the lowest reads cutoff, the specificity was >99%, which is why 1-specificity does not go to 0.
Assay reproducibility for introns, nonunique, and repetitive DNA. Between-run (black squares) and within-run (gray circles) reproducibility is shown as a function of minimum threshold for calling SNV and indel variants. Variants were considered reproducible if they met or exceeded the specified cutoff value in the index sample and appeared in the replicate sample regardless of cutoff value. All variants (including in deeply intronic, nonunique, and repetitive DNA) were included in this analysis. Nonreproducible variant calls were not in clinically important regions and were most prevalent in segmentally duplicated introns of PMS2.
References
- 1.Jenkins M.A., Hayashi S., O'Shea A.M., Burgart L.J., Smyrk T.C., Shimizu D., Waring P.M., Ruszkiewicz A.R., Pollett A.F., Redston M., Barker M.A., Baron J.A., Casey G.R., Dowty J.G., Giles G.G., Limburg P., Newcomb P., Young J.P., Walsh M.D., Thibodeau S.N., Lindor N.M., Lemarchand L., Gallinger S., Haile R.W., Potter J.D., Hopper J.L., Jass J.R. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology. 2007;133:48–56. doi: 10.1053/j.gastro.2007.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohlmann W., Gruber S.B. Copyright University of Washington; Seattle: 1993–2012. Lynch Syndrome: In GeneReviews [Internet]http://www.ncbi.nlm.nih.gov/books/NBK1211 Last revised August 11, 2011. [Google Scholar]
- 3.Jasperson K.W., Tuohy T.M., Neklason D.W., Burt R.W. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Gastroenterological Association medical position statement: hereditary colorectal cancer and genetic testing. Gastroenterology. 2001;121:195–197. doi: 10.1053/gast.2001.25580. [DOI] [PubMed] [Google Scholar]
- 5.Cao Y., Pieretti M., Marshall J., Khattar N.H., Chen B., Kam-Morgan L., Lynch H. Challenge in the differentiation between attenuated familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer: case report with review of the literature. Am J Gastroenterol. 2002;97:1822–1827. doi: 10.1111/j.1572-0241.2002.05850.x. [DOI] [PubMed] [Google Scholar]
- 6.Jasperson K.W., Blazer K.R., Lowstuter K., Weitzel J.N. Working through a diagnostic challenge: colonic polyposis: Amsterdam criteria, and a mismatch repair mutation. Fam Cancer. 2008;7:281–285. doi: 10.1007/s10689-007-9179-z. [DOI] [PubMed] [Google Scholar]
- 7.Pritchard C.C., Grady W.M. Colorectal cancer molecular biology moves into clinical practice. Gut. 2011;60:116–129. doi: 10.1136/gut.2009.206250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall G., Clarkson A., Shi A., Langford E., Leung H., Eckstein R.P., Gill A.J. Immunohistochemistry for PMS2 and MSH6 alone can replace a four antibody panel for mismatch repair deficiency screening in colorectal adenocarcinoma. Pathology. 2010;42:409–413. doi: 10.3109/00313025.2010.493871. [DOI] [PubMed] [Google Scholar]
- 9.Hu H., Wrogemann K., Kalscheuer V., Tzschach A., Richard H., Haas S.A., Menzel C., Bienek M., Froyen G., Raynaud M., Van Bokhoven H., Chelly J., Ropers H., Chen W. Mutation screening in 86 known X-linked mental retardation genes by droplet-based multiplex PCR and massive parallel sequencing. Hugo J. 2009;3:41–49. doi: 10.1007/s11568-010-9137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meder B., Haas J., Keller A., Heid C., Just S., Borries A., Boisguerin V., Scharfenberger-Schmeer M., Stahler P., Beier M., Weichenhan D., Strom T.M., Pfeufer A., Korn B., Katus H.A., Rottbauer W. Targeted next-generation sequencing for the molecular genetic diagnostics of cardiomyopathies. Circ Cardiovasc Genet. 2011;4:110–122. doi: 10.1161/CIRCGENETICS.110.958322. [DOI] [PubMed] [Google Scholar]
- 11.Voelkerding K.V., Dames S., Durtschi J.D. Next generation sequencing for clinical diagnostics-principles and application to targeted resequencing for hypertrophic cardiomyopathy: a paper from the 2009 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2010;12:539–551. doi: 10.2353/jmoldx.2010.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh T., Lee M.K., Casadei S., Thornton A.M., Stray S.M., Pennil C., Nord A.S., Mandell J.B., Swisher E.M., King M.C. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:12629–12633. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan J.E., Carr I.M., Sheridan E., Chu C.E., Hayward B., Camm N., Lindsay H.A., Mattocks C.J., Markham A.F., Bonthron D.T., Taylor G.R. Genetic diagnosis of familial breast cancer using clonal sequencing. Human Mutat. 2010;31:484–491. doi: 10.1002/humu.21216. [DOI] [PubMed] [Google Scholar]
- 14.Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 15.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nord A.S., Lee M., King M.C., Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics. 2011;12:184. doi: 10.1186/1471-2164-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh T., Casadei S., Coats K.H., Swisher E., Stray S.M., Higgins J., Roach K.C., Mandell J., Lee M.K., Ciernikova S., Foretova L., Soucek P., King M.C. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295:1379–1388. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- 19.Walsh T., Casadei S., Lee M.K., Pennil C.C., Nord A.S., Thornton A.M., Roeb W., Agnew K.J., Stray S.M., Wickramanayake A., Norquist B., Pennington K.P., Garcia R.L., King M.C., Swisher E.M. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohonen-Corish M.R., Macrae F., Genuardi M., Aretz S., Bapat B., Bernstein I.T., Burn J., Cotton R.G., den Dunnen J.T., Frebourg T., Greenblatt M.S., Hofstra R., Holinski-Feder E., Lappalainen I., Lindblom A., Maglott D., Moller P., Morreau H., Moslein G., Sijmons R., Spurdle A.B., Tavtigian S., Tops C.M., Weber T.K., de Wind N., Woods M.O. Deciphering the colon cancer genes–report of the InSiGHT-Human Variome Project Workshop. Human Mutat. 2011;32:491–494. doi: 10.1002/humu.21450. UNESCO, Paris 2010. [DOI] [PubMed] [Google Scholar]
- 21.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 23.Schwarz J.M., Rodelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 24.Rowan A.J., Lamlum H., Ilyas M., Wheeler J., Straub J., Papadopoulou A., Bicknell D., Bodmer W.F., Tomlinson I.P. APC mutations in sporadic colorectal tumors: a mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci U S A. 2000;97:3352–3357. doi: 10.1073/pnas.97.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gayet J., Zhou X.P., Duval A., Rolland S., Hoang J.M., Cottu P., Hamelin R. Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines. Oncogene. 2001;20:5025–5032. doi: 10.1038/sj.onc.1204611. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe Y., Haugen-Strano A., Umar A., Yamada K., Hemmi H., Kikuchi Y., Takano S., Shibata Y., Barrett J.C., Kunkel T.A., Koi M. Complementation of an hMSH2 defect in human colorectal carcinoma cells by human chromosome 2 transfer. Mol Carcinog. 2000;29:37–49. [PubMed] [Google Scholar]
- 27.Deng G., Chen A., Hong J., Chae H.S., Kim Y.S. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999;59:2029–2033. [PubMed] [Google Scholar]
- 28.Pinol V., Castells A., Andreu M., Castellvi-Bel S., Alenda C., Llor X., Xicola R.M., Rodriguez-Moranta F., Paya A., Jover R., Bessa X. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 29.Yuen S.T., Chan T.L., Ho J.W., Chan A.S., Chung L.P., Lam P.W., Tse C.W., Wyllie A.H., Leung S.Y. Germline, somatic and epigenetic events underlying mismatch repair deficiency in colorectal and HNPCC-related cancers. Oncogene. 2002;21:7585–7592. doi: 10.1038/sj.onc.1205968. [DOI] [PubMed] [Google Scholar]
- 30.Hendriks Y.M., Wagner A., Morreau H., Menko F., Stormorken A., Quehenberger F., Sandkuijl L., Moller P., Genuardi M., Van Houwelingen H., Tops C., Van Puijenbroek M., Verkuijlen P., Kenter G., Van Mil A., Meijers-Heijboer H., Tan G.B., Breuning M.H., Fodde R., Wijnen J.T., Brocker-Vriends A.H., Vasen H. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology. 2004;127:17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 31.Chao E.C., Velasquez J.L., Witherspoon M.S., Rozek L.S., Peel D., Ng P., Gruber S.B., Watson P., Rennert G., Anton-Culver H., Lynch H., Lipkin S.M. Accurate classification of MLH1/MSH2 missense variants with multivariate analysis of protein polymorphisms-mismatch repair (MAPP-MMR) Human Mutat. 2008;29:852–860. doi: 10.1002/humu.20735. [DOI] [PubMed] [Google Scholar]
- 32.Stella A., Wagner A., Shito K., Lipkin S.M., Watson P., Guanti G., Lynch H.T., Fodde R., Liu B. A nonsense mutation in MLH1 causes exon skipping in three unrelated HNPCC families. Cancer Res. 2001;61:7020–7024. [PubMed] [Google Scholar]
- 33.Agostini M., Tibiletti M.G., Lucci-Cordisco E., Chiaravalli A., Morreau H., Furlan D., Boccuto L., Pucciarelli S., Capella C., Boiocchi M., Viel A. Two PMS2 mutations in a Turcot syndrome family with small bowel cancers. Am J Gastroenterol. 2005;100:1886–1891. doi: 10.1111/j.1572-0241.2005.50441.x. [DOI] [PubMed] [Google Scholar]
- 34.Goodfellow P.J., Buttin B.M., Herzog T.J., Rader J.S., Gibb R.K., Swisher E., Look K., Walls K.C., Fan M.Y., Mutch D.G. Prevalence of defective DNA mismatch repair and MSH6 mutation in an unselected series of endometrial cancers. Proc Natl Acad Sci U S A. 2003;100:5908–5913. doi: 10.1073/pnas.1030231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zighelboim I., Powell M.A., Babb S.A., Whelan A.J., Schmidt A.P., Clendenning M., Senter L., Thibodeau S.N., de la Chapelle A., Goodfellow P.J. Epitope-positive truncating MLH1 mutation and loss of PMS2: implications for IHC-directed genetic testing for Lynch syndrome. Fam Cancer. 2009;8:501–504. doi: 10.1007/s10689-009-9276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.AL-Tassan N., Chmiel N.H., Maynard J., Fleming N., Livingston A.L., Williams G.T., Hodges A.K., Davies D.R., David S.S., Sampson J.R., Cheadle J.P. Inherited variants of MYH associated with somatic G: C–>T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 37.Aretz S., Uhlhaas S., Sun Y., Pagenstecher C., Mangold E., Caspari R., Moslein G., Schulmann K., Propping P., Friedl W. Familial adenomatous polyposis: aberrant splicing due to missense or silent mutations in the APC gene. Human Mutat. 2004;24:370–380. doi: 10.1002/humu.20087. [DOI] [PubMed] [Google Scholar]
- 38.Friedl W., Aretz S. Familial adenomatous polyposis: experience from a study of 1164 unrelated german polyposis patients. Hered Cancer Clin Pract. 2005;3:95–114. doi: 10.1186/1897-4287-3-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa H., Lockman J.C., Frankel W.L., Hampel H., Steenblock K., Burgart L.J., Thibodeau S.N., de la Chapelle A. Mismatch repair gene PMS2: disease-causing germline mutations are frequent in patients whose tumors stain negative for PMS2 protein, but paralogous genes obscure mutation detection and interpretation. Cancer Res. 2004;64:4721–4727. doi: 10.1158/0008-5472.CAN-03-2879. [DOI] [PubMed] [Google Scholar]
- 40.Syngal S., Fox E.A., Eng C., Kolodner R.D., Garber J.E. Sensitivity and specificity of clinical criteria for hereditary non-polyposis colorectal cancer associated mutations in MSH2 and MLH1. J Med Genet. 2000;37:641–645. doi: 10.1136/jmg.37.9.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayward B.E., De Vos M., Valleley E.M., Charlton R.S., Taylor G.R., Sheridan E., Bonthron D.T. Extensive gene conversion at the PMS2 DNA mismatch repair locus. Human Mutat. 2007;28:424–430. doi: 10.1002/humu.20457. [DOI] [PubMed] [Google Scholar]
- 42.Karow J. 23andMe, Ambry Genetics Start Offering Exome Sequencing for Individuals. Genome Web. 2011 [Google Scholar]
- 43.Karow J. Baylor Whole Genome Laboratory Launches Clinical Exome Sequencing Test. Genome Web. 2011 [Google Scholar]
- 44.Ou J., Niessen R.C., Vonk J., Westers H., Hofstra R.M., Sijmons R.H. A database to support the interpretation of human mismatch repair gene variants. Human Mutat. 2008;29:1337–1341. doi: 10.1002/humu.20907. [DOI] [PubMed] [Google Scholar]
- 45.Hoppman-Chaney N., Peterson L.M., Klee E.W., Middha S., Courteau L.K., Ferber M.J. Evaluation of oligonucleotide sequence capture arrays and comparison of next-generation sequencing platforms for use in molecular diagnostics. Clin Chem. 2010;56:1297–1306. doi: 10.1373/clinchem.2010.145441. [DOI] [PubMed] [Google Scholar]
- 46.Crockett D.K., Lyon E., Williams M.S., Narus S.P., Facelli J.C., Mitchell J.A. Utility of gene-specific algorithms for predicting pathogenicity of uncertain gene variants. J Am Med Inform Assoc. 2012;19:207–211. doi: 10.1136/amiajnl-2011-000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crockett D.K., Piccolo S.R., Ridge P.G., Margraf R.L., Lyon E., Williams M.S., Mitchell J.A. Predicting phenotypic severity of uncertain gene variants in the RET proto-oncogene. PLoS One. 2011;6:e18380. doi: 10.1371/journal.pone.0018380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adey A., Morrison H.G., Asan X., Kitzman J.O., Turner E.H., Stackhouse B., MacKenzie A.P., Caruccio N.C., Zhang X., Shendure J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Receiver operating characteristic analysis. The sensitivity of the assay to detect HapMap-characterized single nucleotide polymorphisms is plotted versus 1-specificity at different minimum fold-coverage (number of reads) cutoffs. Even at the lowest reads cutoff, the specificity was >99%, which is why 1-specificity does not go to 0.
Assay reproducibility for introns, nonunique, and repetitive DNA. Between-run (black squares) and within-run (gray circles) reproducibility is shown as a function of minimum threshold for calling SNV and indel variants. Variants were considered reproducible if they met or exceeded the specified cutoff value in the index sample and appeared in the replicate sample regardless of cutoff value. All variants (including in deeply intronic, nonunique, and repetitive DNA) were included in this analysis. Nonreproducible variant calls were not in clinically important regions and were most prevalent in segmentally duplicated introns of PMS2.