Abstract
Ecological speciation occurs when reproductive isolation evolves as a consequence of divergent natural selection among environments. A direct prediction of this process is that ecologically divergent pairs of populations will exhibit greater reproductive isolation than ecologically similar pairs of populations. By comparing allopatric populations of the cynipid gall wasp Belonocnema treatae infesting Quercus virginiana and Quercus geminata, we tested the role that divergent host use plays in generating ecological divergence and sexual isolation. We found differences in body size and gall structure associated with divergent host use, but no difference in neutral genetic divergence between populations on the same or different host plant. We observed significant assortative mating between populations from alternative host plants but not between allopatric populations on the same host plant. Thus, we provide evidence that divergent host use promotes speciation among gall wasp populations.
Keywords: ecological speciation, host race, Cynipidae
1. Introduction
Understanding the mechanisms contributing to speciation is a fundamental question in biology. Ecological speciation, a process in which reproductive isolation evolves as a by-product of divergent natural selection among environments, may be an important mechanism of speciation [1,2]. Geographically separated (allopatric) pairs of populations in ecologically similar habitats may diverge owing to genetic drift, sexual conflict and certain forms of sexual selection, whereas pairs of populations in ecologically different habitats may diverge owing to any of these processes as well as divergent natural selection [3–5]. Thus, ecological speciation theory predicts that ecologically divergent pairs of populations will exhibit greater reproductive isolation than ecologically similar pairs of populations, ceteris paribus [3,4]. This experimental framework isolates the role of natural selection in generating reproductive isolation while controlling for other host-independent factors and assumes populations of a similar age, yet explicit tests of hypotheses using the powerful same-environment/different-environment comparative method remain rare ([3,5–7]; reviewed in [8]). Much of the support for ecological speciation comes from studies of herbivorous insects inhabiting different host plants where pairs of populations on the same host plant species offer ecologically similar comparisons (hereafter, ‘same-host’ pairs) and pairs of populations on different host plants offer ecologically different comparisons (hereafter, ‘different-host pairs’) [3–5,9].
In the southeastern United States, the cynipid gall-forming wasp, Belonocnema treatae, attacks two sister species of live oak within the series Virentes, Quercus virginiana and Quercus geminata, which overlap in geographical range, but occupy different habitats. Specifically, Q. virginiana occurs in moister, more nutrient-rich, and higher pH sites than Q. geminata [10]. The two species differ in leaf morphology, flowering time, and growth and photosynthetic rates [10]. Thus, populations of B. treatae on these two oak species may experience divergent natural selection.
Here we test a main prediction of ecological speciation theory: allopatric populations of B. treatae inhabiting different host plant species will exhibit greater differences in ecologically important traits (e.g. body size [11] and gall morphology [12]) and express greater sexual isolation than do allopatric populations on the same host plant. To conduct our same-host and different-host comparisons, we used gall wasps from six allopatric populations (three Q. geminata and three Q. virginiana), yielding 15 pairwise comparisons (nine different-host and six same-host). Patterns of divergence among populations are best understood within a historical context addressing the relative age of geographical separation and restricted gene flow. Here, we estimate relative divergence times among populations using sequence divergence from mtDNA collected from each study population.
2. Material and methods
(a). Study system and sampling
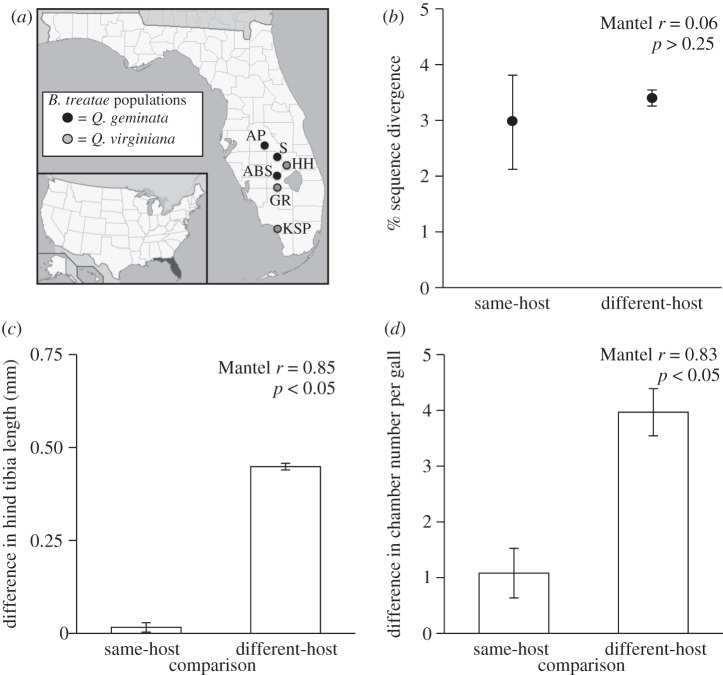
Belonocnema treatae exhibits a heterogonous life cycle [13]. The asexual generation develops within leaf galls during the summer and autumn and emerges in the winter, whereas the sexual generation develops within root galls and emerges during the spring. We collected root galls from six allopatric populations (three Q. geminata and three Q. virginiana) in April 2010 (figure 1a). Galls were husbanded under common conditions (12 L : 12 D, 23°C), and upon emergence, virgin males and females were separated. Mating assays were performed within 48 h.
Figure 1.
(a) Map of B. treatae gall wasp study populations in FL, USA. Average distance among populations was 67.8 km (±10.3 s.e.) and ranged from 20.5 to 135.7 km. (b) Neutral genetic differentiation between pairs of populations estimated as mean (±s.e.) pairwise sequence divergence. (c) Mean (±s.e.) of pairwise differences among populations for body size. (d) Mean (±s.e.) of pairwise differences among populations for gall structure.
(b). Body size and gall morphology
The body size of sexual generation adults reared from each collection site was indexed by measuring the length of the right hind tibia. Sexes were pooled, as tibia lengths did not differ between males and females (ANOVA: F1,183 = 1.9, p > 0.15). Root galls of B. treatae are typically multi-chambered, with one adult developing per chamber [13]. Following emergence, root galls were dissected to count the number of chambers per gall on each host plant.
(c). Sexual isolation assays
No-choice mating trials were conducted to assess assortative mating as a function of host plant and collection site. Trials took place within 25 × 8 cm clear plastic cups provisioned with a dried, defoliated twig from a neutral host (Prunus serotina). For each trial, one male and one female each from a different collecting site were aspirated into the cup and then observed at 5 min intervals for 1 h (∑ = 12 observations, sample sizes in table 1 and figure 2). For each observation, we recorded whether the pair was copulating.
Table 1.
Sexual isolation among B. treatae populations (population host association: Qv, Q. virginiana; Qg, Q. geminata; pop. label subscripted as in figure 1a. IPSI, index of sexual isolation; n, sample size per comparison; χ2, test statistic from likelihood-ratio test of male pop. × female pop. interaction in logistic regression).
| population comparisons | IPSI | n | χ2 | p-value |
|---|---|---|---|---|
| different-host | ||||
| QgABS × QvGR | 0.19 | 39 | 3.15 | 0.076 |
| QgABS × QvHH | 0.27 | 39 | 4.53 | 0.033 |
| QgABS × QvKSP | 0.70 | 39 | 14.54 | <0.001 |
| QgAP × QvGR | 0.39 | 34 | 5.50 | 0.019 |
| QgAP × QvHH | 0.47 | 34 | 7.69 | 0.006 |
| QgAP × QvKSP | 0.29 | 34 | 2.78 | 0.096 |
| QgS × QvGR | 0.05 | 39 | 0.23 | 0.631 |
| QgS × QvHH | 0.28 | 39 | 4.92 | 0.027 |
| QgS × QvKSP | 0.15 | 36 | 1.25 | 0.264 |
| same-host | ||||
| QgABS × QgAP | −0.03 | 35 | 1.90 | 0.168 |
| QgABS × QgS | 0.01 | 50 | 0.01 | 0.993 |
| QgAP × QgS | 0.22 | 35 | 1.12 | 0.289 |
| QvGR × QvHH | −0.02 | 49 | 0.09 | 0.758 |
| QvGR × QvKSP | −0.09 | 49 | 3.42 | 0.064 |
| QvHH × QvKSP | −0.03 | 51 | 0.17 | 0.677 |
Figure 2.
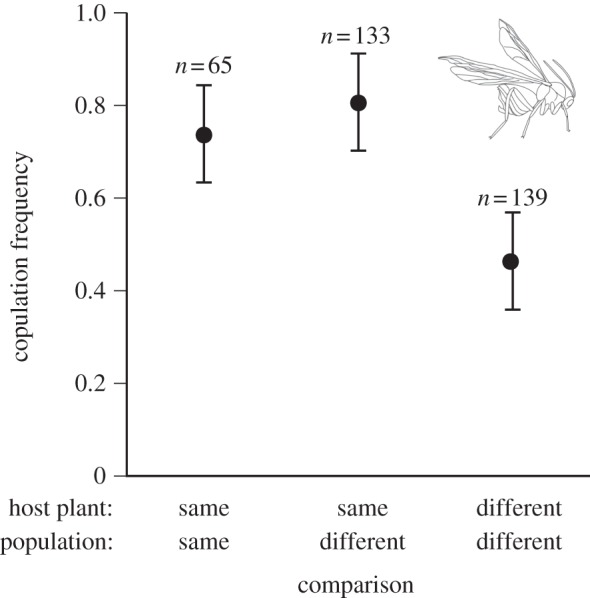
Mean (±s.e.) of copulation frequency among B. treatae populations. Mating pairs from same-host and different-host allopatric populations were used to test the role of divergent host use in the evolution of sexual isolation; pairings from the same population were used as a control to calculate the sexual isolation index, IPSI, for each pairwise comparison (table 1; n denotes sample size per comparison).
(d). Genetic analysis
To control for the confounding effects of time since divergence among allopatric populations, sequence divergence among B. treatae populations was evaluated for, in total, 44 individual wasps (equally divided by sex) based on two putatively neutrally evolving mtDNA genes, a 416 bp long segment of the cytochrome b (cyt b) gene and a 593 bp segment of the cytochrome oxidase I (COI). Sample sizes for the six collecting sites were S = 7, ABS = 8, AP = 7, HH = 9, KSP = 7 and GR = 6 (figure 1a). See electronic supplementary material for sequencing methods. Sequences were deposited in GenBank (accession nos. JQ438777–JQ438824). Cyt b and COI sequences were concatenated, and mean pairwise sequence divergences among populations were calculated in MEGA v. 5 [14] with a Kimura two-parameter model [15]. We also sequenced a 257 bp nuclear ITS2 region, but found no evidence of variation at this marker, as one might expect for recently diverging lineages (see electronic supplementary material).
(e). Statistical analysis
Our study design generated datasets in the form of matrices of pairwise comparisons. To compare elements between matrices, Mantel tests [16] were run in the ‘ecodist’ package in R v. 2.11.1 with 10 000 randomizations and one-tailed hypothesis testing. Additionally, we used logistic regression to examine the effects of male host plant, female host plant and their interaction across all populations on copulation frequency, and then separately for each pairwise comparison. Finally, we estimated the degree of sexual isolation for each pairwise population comparison by using a standard metric of sexual isolation, IPSI (0, random mating, +1, complete assortative mating) [17].
3. Results
Pairwise mtDNA sequence divergence among populations ranged from 1.0 to 5.1 per cent but was not associated with host use, as same-host pairs exhibited similar patterns of divergence as different-host pairs (Mantel r = 0.06, p > 0.25; figure 1b). Isolation-by-distance was not detected (Mantel r = 0.18, p = 0.32). However, consistent differences in both body size (Mantel r = 0.85, p < 0.05; figure 1c) and gall structure (Mantel r = 0.83, p < 0.05; figure 1d) were associated with differences in host-plant use. Larger adults and more chambers per root gall were evident for B. treatae populations on Q. geminata (figure 1c,d).
Most importantly, patterns of copulation frequency (no. of copulations/no. of mating trials) revealed strong evidence of host-associated sexual isolation. Overall, copulation was more likely if paired individuals were from the same host plant (logistic regression: male host plant × female host plant interaction, likelihood-ratio test = 37.84, d.f. = 1, p < 0.0001; figure 2) and this was consistent across host plant origin. Indeed, the mean IPSI value for sexual isolation between pairs of allopatric populations using different host plants (0.31 ± 0.06 s.e.) was significantly greater than the mean value between allopatric pairs of populations using the same host plant (0.01 ± 0.04 s.e.; Mantel r = 0.76, p < 0.05; table 1).
4. Discussion
We have demonstrated that divergent host-plant use is associated with differences in trait values and prezygotic reproductive isolation between B. treatae populations (table 1 and figure 2). Our results are consistent with partial sexual isolation evolving in this gall wasp species as a by-product of adaptation to different hosts [18].
Reproductive isolation often increases with time, where time is estimated using genetic distance at neutral loci [18]. However, time since divergence among the six allopatric B. treatae populations examined is unlikely to confound the observed associations between host-plant use and reproductive isolation for two reasons. First, allopatric different-host pairs are not more genetically divergent than allopatric same-host pairs. Second, direct comparison of sexual isolation with genetic distance among populations shows that the degree of between-population sexual isolation is not correlated with genetic distance (Mantel r = −0.07, p > 0.50). Collectively, these analyses provide evidence that host use, rather than genetic drift, underlies differences in reproductive isolation.
We found differences in body size and gall morphology associated with host use. Ecological divergence in these traits may represent adaptations to the host-plant environment. The two hosts differ in leaf morphology, the critical location for oviposition and gall induction. The large-bodied B. treatae populations were found on Q. geminata, the leaves of which have thicker midveins, denser trichomes, and are ‘thick and leathery,’ as measured by mass per area [10]. Thus, larger body size may be advantageous for females ovipositing into Q. geminata leafs. As well, soil moisture and pH [10] also differ between the two oak species and environments may select for different root gall morphologies.
Non-genetic effects of the larval host environment cannot be ruled out as contributing to the observed patterns [8]. All individuals used in the present study were reared from their native host. Yet, given our results, phenotypic divergence in B. treatae caused by plasticity would still be expected to induce reproductive isolation. Male and female B. treatae were averse to mating with individuals from the alternative host plant and chose to copulate twice as often when paired with individuals from their own host plant. Thus, regardless of the underlying basis of variation for mate preference, our results support a critical role for host-plant use in promoting reproductive isolation among populations of B. treatae. Our results are consistent with those of other studies and with theory supporting a role for initial environmentally induced differences (i.e. plasticity) promoting divergence and speciation [19], including divergent host use among herbivorous insects [8,20].
In conclusion, the same-host/different-host comparisons produced evidence consistent with divergent selection promoting speciation for B. treatae populations on alternative host plants and hints at the possible general role of host plant-driven diversification in cynipid oak systems. Our study provides the first evidence that supports predictions of ecological speciation theory for the species-rich and ecologically diverse Cynipidae.
Acknowledgements
We thank M. Deyrup and Archibold Biological Station for support, C. Brown for illustration and E. Silverfine for editorial comments.
References
- 1.Mayr E. 1947. Ecological factors in speciation. Evolution 1, 263–288 10.2307/2405327 (doi:10.2307/2405327) [DOI] [Google Scholar]
- 2.Schluter D. 2001. Ecology and the origin of species. Trends Ecol. Evol. 16, 372–380 10.1016/S0169-5347(01)02198-X (doi:10.1016/S0169-5347(01)02198-X) [DOI] [PubMed] [Google Scholar]
- 3.Funk D. J. 1998. Isolating a role for natural selection in speciation: host adaptation and sexual isolation in Neochlamisus bebbianae leaf beetles. Evolution 52, 1744–1759 10.2307/2411347 (doi:10.2307/2411347) [DOI] [PubMed] [Google Scholar]
- 4.Nosil P. 2007. Divergent host-plant adaptation and reproductive isolation between ecotypes of Timema cristinae walking-sticks. Am. Nat. 169, 151–162 10.1086/510634 (doi:10.1086/510634) [DOI] [PubMed] [Google Scholar]
- 5.Nosil P., Crespi B. J., Sandoval C. P. 2002. Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature 417, 440–443 10.1038/417440a (doi:10.1038/417440a) [DOI] [PubMed] [Google Scholar]
- 6.Vines T. H., Schluter D. 2006. Strong assortative mating between allopatric sticklebacks as a by-product of adaptation to different environments. Proc. R. Soc. B 273, 911–916 10.1098/rspb.2005.3387 (doi:10.1098/rspb.2005.3387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langerhans R. B., Gifford M. E., Joseph E. O. 2007. Ecological speciation in Gambusia fishes. Evolution 61, 2056–2074 10.1111/j.1558-5646.2007.00171.x (doi:10.1111/j.1558-5646.2007.00171.x) [DOI] [PubMed] [Google Scholar]
- 8.Rundle H., Nosil P. 2005. Ecological speciation. Ecol. Lett. 8, 336–352 10.1111/j.1461-0248.2004.00715.x (doi:10.1111/j.1461-0248.2004.00715.x) [DOI] [Google Scholar]
- 9.Funk D. J., Filchak K. E., Feder J. L. 2002. Herbivorous insects: model systems for the comparative study of speciation ecology. Genetica 116, 251–267 10.1023/A:1021236510453 (doi:10.1023/A:1021236510453) [DOI] [PubMed] [Google Scholar]
- 10.Cavender-Bares J., Pahlich A. 2009. Molecular, morphological, and ecological differentiation of sympatric sister oak species, Quercus virginiana and Q. geminata (Fagaceae). Am. J. Bot. 96, 1690–1702 10.3732/ajb.0800315 (doi:10.3732/ajb.0800315) [DOI] [PubMed] [Google Scholar]
- 11.Honek A. 1993. Intraspecific variation in body size and fecundity in insects: a general relationship. Oikos 66, 483–492 10.2307/3544943 (doi:10.2307/3544943) [DOI] [Google Scholar]
- 12.Stone G. N., Schönrogge K. 2003. The adaptive significance of insect gall morphology. Trends Ecol. Evol. 18, 512–522 10.1016/S0169-5347(03)00247-7 (doi:10.1016/S0169-5347(03)00247-7) [DOI] [Google Scholar]
- 13.Lund J. N., Ott J. R., Lyons R. 1998. Heterogony in Belonocnema treatae Mayr (Hymenoptera: Cynipidae). P. Entomol. Soc. Wash. 100, 755–763 [Google Scholar]
- 14.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 (doi:10.1093/molbev/msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura M. 1980. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 10.1007/BF01731581 (doi:10.1007/BF01731581) [DOI] [PubMed] [Google Scholar]
- 16.Goslee S. C., Urban D. L. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22, 1–19 [Google Scholar]
- 17.Rolán-Alvarez E., Caballero A. 2000. Estimating sexual selection and sexual isolation effects from mating frequencies. Evolution 54, 30–36 10.1554/0014-3820(2000)054[0030:ESSASI]2.0.CO;2 (doi:10.1554/0014-3820(2000)054[0030:ESSASI]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 18.Coyne J. A., Orr H. A. 2004. Speciation. Sunderland, MA: Sinauer [Google Scholar]
- 19.Pfennig D. W., Wund M. A., Snell-Rood E. C., Cruickshank T., Schlichting C. D., Moczek A. P. 2010. Phenotypic plasticity's impacts on diversification and speciation. Trends Ecol. Evol. 25, 459–467 10.1016/j.tree.2010.05.006 (doi:10.1016/j.tree.2010.05.006) [DOI] [PubMed] [Google Scholar]
- 20.Papaj D. R., Prokopy R. J. 1989. Ecological and evolutionary aspects of learning in phytophagous insects. Ann. Rev. Entomol. 34, 315–350 10.1146/annurev.en.34.010189.001531 (doi:10.1146/annurev.en.34.010189.001531) [DOI] [Google Scholar]