Abstract
In mammals, bitter taste is mediated by TAS2R genes, which belong to the large family of seven transmembrane G protein-coupled receptors. Because TAS2Rs are directly involved in the interaction between mammals and their dietary sources, it is likely that these genes evolved to reflect species-specific diets during mammalian evolution. Here, we investigated the sensitivities of TAS2R16s of various primates by using a cultured cell expression system, and found that the sensitivity of each primate species varied according to the ligand. Especially, the sensitivity of TAS2R16 of Japanese macaques to salicin was much lower than that of human TAS2R16, which was supported by behavioural tests. These results suggest the possibility that bitter-taste sensitivities evolved independently by replacing specific amino acid residues of TAS2Rs in different primate species to adapt to food items they use.
Keywords: old world monkey, bitter taste receptor, chimpanzee
1. Introduction
Mammals can perceive and distinguish five basic taste qualities: sweet, bitter, sour, salty and umami (the taste of glutamate) [1]. Among these, bitter sensitivity is particularly important, as can be seen from the fact that many naturally poisonous or bioactive substances taste bitter to humans, and virtually all animals show an aversive response to such tastants. This suggests that bitter transduction might have evolved as a key defence against the ingestion of harmful substances.
Bitter taste in mammals is mediated by TAS2Rs (or T2Rs), which are seven transmembrane G protein-coupled receptors that are expressed in specialized taste bud cells [2–7]. Genomic analyses have revealed the repertoires of TAS2R genes, with species-specific and intra-species variations [8,9] that probably reflect dietary changes during mammalian evolution.
Of these genes, human TAS2R16 is one of the best-studied at the molecular and population levels. For example, many β-glucopyranosides have been identified as TAS2R16 ligands by human behavioural or functional assays using expressed proteins [5]. Human-specific amino acid substitution at position 172 results in high sensitivity to harmful cyanogenic glycosides, which may have been advantageous for avoidance of toxic compounds in early human diets [10]. Ligand-binding sites were identified by mutational studies of human TAS2R16 [11]. Mutation of amino acid resides in helices 3–6 causes decreases in sensitivity for natural and artificial ligands. Notably, various primates conserve TAS2R16 largely intact with only a few amino acid substitutions. These facts prompted us to examine inter-species variations of TAS2R genes in non-human primates to better understand the biological significance of bitter perception.
Here, we report analysis of the function of TAS2R16 of some representative primate species: human, chimpanzee, macaque, langur and marmoset (figure 1a). We found that TAS2R16s of different species show different patterns of sensitivities for bitter compound repertories.
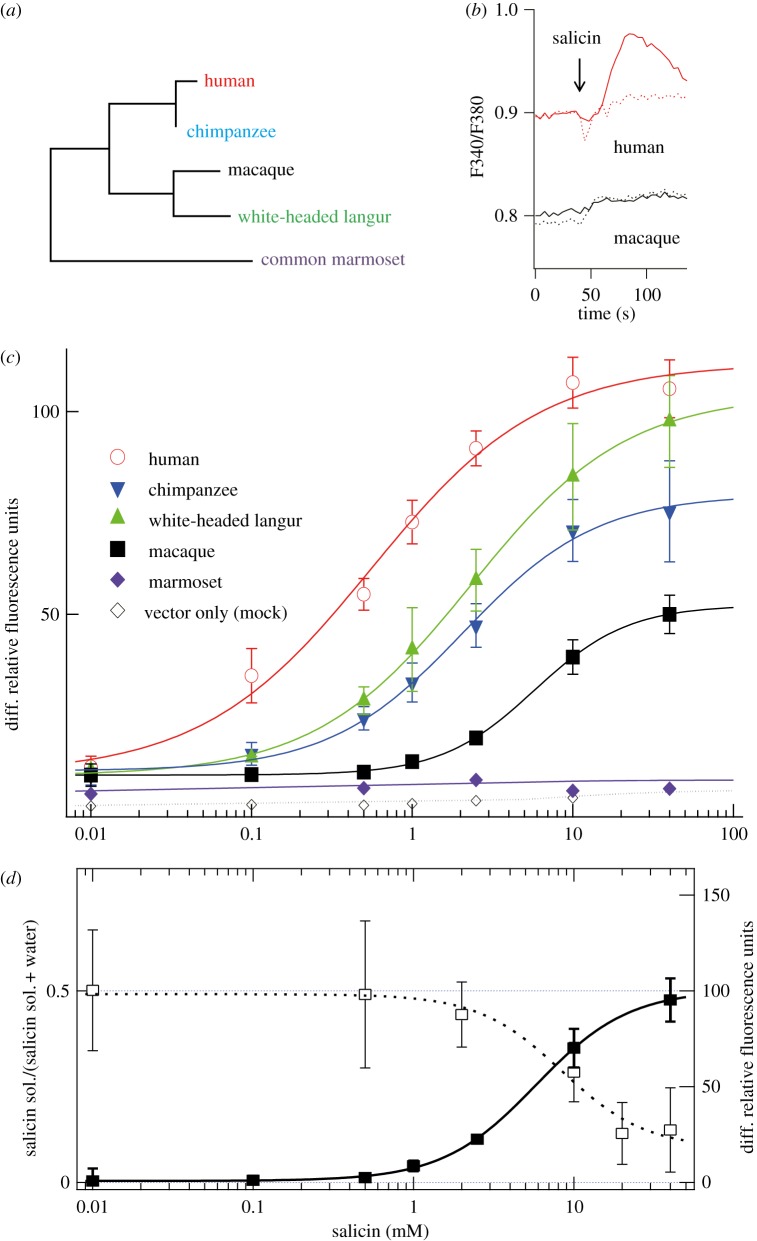
Figure 1.
The responses of TAS2R16s of various primates to salicin. (a) The phylogenetic relationships of primate TAS2R16s. (b) The averaged responses of TAS2R16s of various primates to salicin. HEK 293T cells transiently expressing TAS2R16 and G16gust44 were assayed using an intracellular calcium indicator. Representative responses of human (upper trace) and macaque (lower trace) TAS2R16s to 1 mM salicin using Fura-2 as calcium indicator are shown. Broken traces are responses of cells without transfection of TAS2Rs. (c) The responses of TAS2R16s of various primates to salicin using calcium-4 as an indicator. After adding various concentrations of salicin to cells, the increases of fluorescence were measured. Maximal responses for each time course are plotted versus the concentration of salicin (n = 4). (d) Avoidant behaviour of Japanese macaques for salicin (left axis, open squares: EC50 = 8.45 ± 2.95 mM, n = 3 for three monkeys) and relative response of TAS2R16 (right axis, filled squares: EC50 = 7.48 ± 2.20 mM, n = 4) in cell assay.
2. Material and methods
Salicin, arbutin, amygdalin and phenyl-d-glucopyranoside were purchased from Sigma. Genomic DNA of chimpanzee, macaque as well as marmoset was isolated from blood using a DNeasy blood and tissue kit (Qiagen GmbH, Hilden, Germany). Genomic DNA of the white-headed langur was isolated from faeces using a QiaAmp stool kit.
Standard PCR was used to amplify the TAS2R16 gene from genomic DNA (10 ng per 25 μl reaction). Products were subcloned into the EcoRV site of expression vector pEAK 10 with the sequences of the first 45 amino acids of rat somatostatin receptor type 3 and the last eight amino acids of bovine rhodopsin tags at the N- and C-terminal ends, respectively. We used major haplotypes human A and chimpanzee A, because their TAS2R16 genes are polymorphic ([9,10] and the electronic supplementary material, table S2). All sequences of PCR products were confirmed to be intact using standard BigDye Terminator chemistry (Applied Biosystems, CA, USA). Mutant vectors were constructed using QuikChange (Agilent Technology, CA, USA) as described previously [12].
Cell culture, transfection and cell-based assays were performed as described previously [11–13]. To calculate half maximal effective concentration (EC50) values, plots of amplitude versus concentration were prepared in IGOR Pro (WaveMetrics). Nonlinear regression of the plots produced the function, f(x) = Imin + (Imax − Imin)/(1 + (x/EC50)h), where x is the ligand concentration and h is the Hill coefficient, which was used to calculate the EC50 values for ligand–receptor interactions.
Behavioural tests were performed using a two-bottle system approved by the animal ethics committee of the Primate Research Institute, Kyoto University (no. 2011-093). Briefly, we set two bottles each in front of three Japanese macaques: one contained salicin solution and the other contained distilled water as a control. After a 2 × 2 h session with position switch of the bottles, we recorded the amounts of liquid consumed. We repeated this trial at least three times for various salicin concentrations and used the same formula as in the cell assay for calculation of the EC50 value.
3. Results
First, we compared the responses of TAS2R16 of various primates with salicin, a bitter compound contained in the bark of Salicaceae (willow) plants (figure 1). Human TAS2R16 responded to salicin with EC50 of 0.48 ± 0.09 mM, in agreement with reported values [5,10,11]. The TAS2R16s of non-human primates showed various responses to salicin. Among the responsive TAS2R16s, macaque TAS2R16 showed a remarkably reduced response to salicin compared to langur, chimpanzee and human TAS2R16s (figure 1b,c). EC50 of macaque TAS2R16 for salicin was 7.5 ± 2.2 mM, which was higher than the EC50s for langur (2.3 ± 0.2 mM), chimpanzee (1.0 ± 0.3 mM) and human TAS2R16s. This was reflected by the behaviour of macaques, as shown by a two-bottle test: captive Japanese macaques showed avoidant behaviour to salicin solution with an EC50 of 8.5 ± 3.0 mM (figure 1d), which was about 10 times higher than the EC50 of human behavioural detection and cell assays [5]. Thus, macaques are less sensitive to salicin than humans, apparently owing to the decreased sensitivity of TAS2R16.
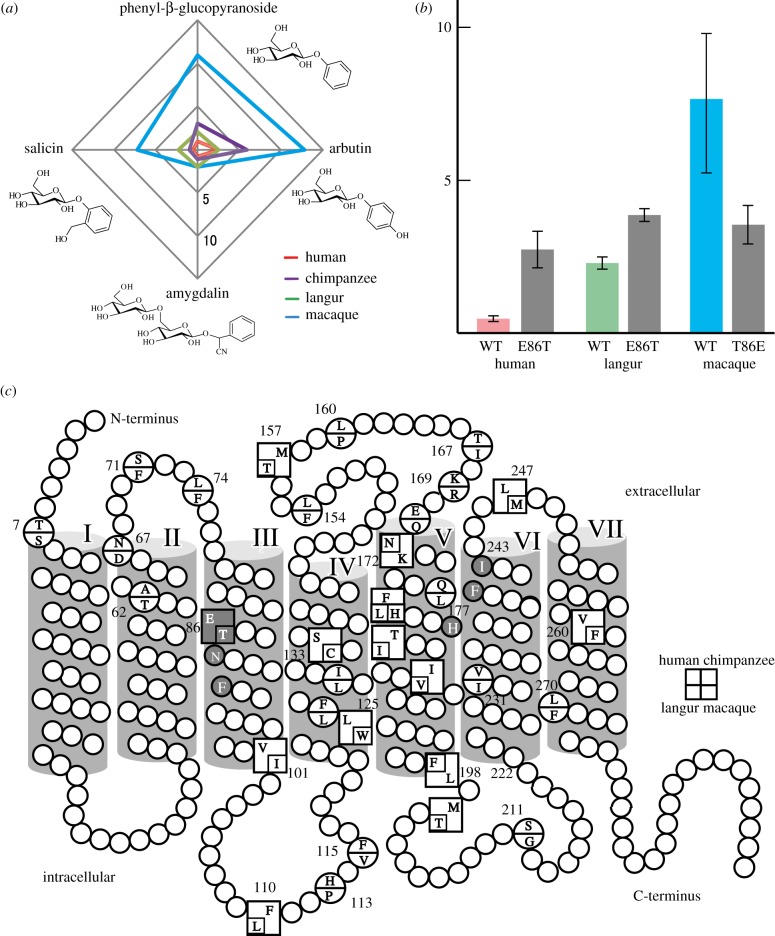
Next, we compared the responses of TAS2R16 of primates with various ligands (figure 2a and electronic supplementary material, table S1). For phenyl-β-glucopyranoside (electronic supplementary material, figure S1b), in which the hydroxymethyl group of salicin is replaced by hydrogen, the response patterns of primate TAS2R16s were similar to those for salicin. For arbutin (electronic supplementary material, figure S1c), in which a hydroxyl group is added to the 4′ position of phenyl-β-glucopyranoside, the responses of human and white-headed langur TAS2R16s showed similar EC50 values (2.3 ± 0.5 and 2.5 ± 0.3, respectively), whereas the chimpanzee and macaque TAS2R16s showed reduced sensitivity (EC50 = 6.0 ± 0.6 and 12.9 ± 2.5). For amygdalin (electronic supplementary material, figure S1d), a cyanogenic glucoside contained in Prunus plants, all of the TAS2R16s showed responses with similar low EC50 values (0.75 ± 0.04 in human ∼ 2.0 ± 0.5 in macaque). Thus, the differences of responsiveness of TAS2R16 among these primate species depend on the ligand (figure 2a) possibly owing to particular amino acid substitutions in some of these species (figure 2c). The human–chimpanzee and macaque–langur pairs, which differ by two and 12 amino acid residues respectively, showed different response patterns depending on the ligand. We replaced these sites and found that certain amino acid residues affected the sensitivities of these pairs. Comparing the human–chimpanzee pair, the amino acid residues at positions 172 and 198 additively changed the EC50 for each ligand (electronic supplementary material, table S2). When we replaced the amino acid residues specific for macaque TAS2R16, one of the six amino acid residues markedly changed the EC50 (figure 2b). Thus, the differences in affinity for each ligand are owing to differences in several amino acid residues.
Figure 2.
The sensitivities of primate TAS2R16s and their mutants to various ligands. (a) EC50 values of each primate species are plotted for each compound. (b) EC50 values of mutant TAS2R16s of humans, langurs and macaques at amino acid position 86. (c) Amino acid sequences of TAS2R16 of primates. The transmembrane topology is shown according to the structure of bovine rhodopsin. Grey circles and squares represent the putative binding site for salicin [12].
4. Discussion
Mutation of E86 to Q or D in human TAS2R16 strongly reduces the response to salicin, suggesting it is located in the binding site for salicin [11]. Therefore, one of the mechanisms of the lower sensitivity to salicin of macaque TAS2R16 compared with TAS2R16s of other species is probably the change of this residue to T in the macaque lineage (figure 2b), which presumably alters the binding site structure. Because langur TAS2R16 showed lower sensitivity than human TAS2R16, some additional residues must contribute to the higher sensitivity of human TAS2R16. In chimpanzee TAS2R16, the differences are limited to N172K and F198L compared with human TAS2R16. The K172 genotype of human TAS2R16 is about two times less sensitive than N172 [10]. In accord with this, our results showed that mutation of the residues at positions 172 and 198 changed the sensitivities additively for some natural ligands (electronic supplementary material, table S2). Because these residues are positioned at the membrane surface facing the extracellular and intracellular compartments, respectively, they are expected to change the ligand-binding site or pathway, and G-protein signalling or coupling, respectively.
Ecologically, some of the differences in the sensitivity of TAS2R16s of primates should be reflected in inter-species differences in bitter taste perception. Our demonstration of a relationship between macaques’ behaviour and the response of TAS2R16 to salicin, as previously shown in humans, suggests that such a relationship probably occurs over a wide range of primate species. Because bitter taste is a sense for avoidance, differences in sensitivity of TA2R16 should be related to the avoidance versus non-avoidance of some accessible food items. Japanese macaques often ingest willow tree bark [14], containing salicin, whereas there is no report of such behaviour in other primate species. Japanese macaques usually use this bark particularly in winter in the snowy region of Japan [14], where there are few nutritious food items other than bark; therefore, the lower sensitivity to the bitterness of salicin might have some advantage for their survival. It is possible that it is the neutral reduction of sensitivity in TAS2R16 that allows the Japanese macaques to eat tree bark. Alternatively, it is possible that a broad loss of taste sensitivity owing to a reduction of the number of taste buds or the number of bitter taste receptor cells, or to some difference of neuronal properties could contribute to the reduction of the bitter-taste sensitivity in macaques. Further investigation to address these possibilities would be necessary to fully understand the species differences in feeding behaviour. The close correlation between the EC50 value of the receptor sensitivity and behavioural discrimination could provide a clue to resolve this issue.
In conclusion, changes in the sensitivity of bitter taste receptors resulting from amino acid replacements would be helpful for the ingestion of food items that are bitter but not poisonous in some species-specific environments. This is similar to adaptive pseudogenization [15], and such a functional change would be advantageous if the replacement occurred at certain sites identified here.
Acknowledgements
Behavioural tests were performed using a two-bottle system approved by the animal ethics committee of the Primate Research Institute, Kyoto University (no. 2011-093).
We thank Drs Y. Go, T. Sugawara and A. Matsui, and Mr T. Hayakawa for helpful discussions, and Mrs Y. Hirai and M. Hakukawa for sample preparation. We also thank Dr E. Nakajima for English correction, and Dr T. Ueda for providing the expression vector of G16/gust44. This work was financially supported by Global COE A06 and by grants-in-aid from the MEXT (2137009, 22247036), and from the Ministry of the Environment (D-1007), Japan, and grants from the Takeda Foundation for Science and the Suzuken Memorial Foundation to H.I.
References
- 1.Yarmolinsky D. A., Zuker C. S., Ryba N. J. 2009. Common sense about taste: from mammals to insects. Cell 139, 234–244 10.1016/j.cell.2009.10.001 (doi:10.1016/j.cell.2009.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler E., Hoon M. A., Mueller K. L., Chandrashekar J., Ryba N. J., Zuker C. S. 2000. A novel family of mammalian taste receptors. Cell 100, 693–702 10.1016/S0092-8674(00)80705-9 (doi:10.1016/S0092-8674(00)80705-9) [DOI] [PubMed] [Google Scholar]
- 3.Chandrashekar J., Mueller K. L., Hoon M. A., Adler E., Feng L., Guo W., Zuker C. S., Ryba N. J. 2000. T2Rs function as bitter taste receptors. Cell 100, 703–711 10.1016/S0092-8674(00)80706-0 (doi:10.1016/S0092-8674(00)80706-0) [DOI] [PubMed] [Google Scholar]
- 4.Matsunami H., Montmayeur J. P., Buck L. B. 2000. A family of candidate taste receptors in human and mouse. Nature 404, 601–604 10.1038/35007072 (doi:10.1038/35007072) [DOI] [PubMed] [Google Scholar]
- 5.Bufe B., Hofmann T., Krautwurst D., Raguse J. D., Meyerhof W. 2002. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat. Genet. 32, 397–401 10.1038/ng1014 (doi:10.1038/ng1014) [DOI] [PubMed] [Google Scholar]
- 6.Mueller K. L., Hoon M. A., Erlenbach I., Chandrashekar J., Zuker C. S., Ryba N. J. 2005. The receptors and coding logic for bitter taste. Nature 434, 225–229 10.1038/nature03352 (doi:10.1038/nature03352) [DOI] [PubMed] [Google Scholar]
- 7.Behrens M., Meyerhof W. 2006. Bitter taste receptors and human bitter taste perception. Cell Mol. Life Sci. 63, 1501–1509 10.1007/s00018-006-6113-8 (doi:10.1007/s00018-006-6113-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Go Y. 2006. Lineage-specific expansions and contractions of the bitter taste receptor gene repertoire in vertebrates. Mol. Biol. Evol. 23, 964–972 10.1093/molbev/msj106 (doi:10.1093/molbev/msj106) [DOI] [PubMed] [Google Scholar]
- 9.Sugawara T., Go Y., Udono T., Morimura N., Tomonaga M., Hirai H., Imai H. 2011. Diversification of bitter taste receptor gene family in western chimpanzees. Mol. Biol. Evol. 28, 921–931 10.1093/molbev/msq279 (doi:10.1093/molbev/msq279) [DOI] [PubMed] [Google Scholar]
- 10.Soranzo N., Bufe B., Sabeti P. C., Wilson J. F., Weale M. E., Marguerie R., Meyerhof W., Goldstein D. B. 2005. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr. Biol. 15, 1257–1265 10.1016/j.cub.2005.06.042 (doi:10.1016/j.cub.2005.06.042) [DOI] [PubMed] [Google Scholar]
- 11.Sakurai T., et al. 2010. Characterization of the β-d-glucopyranoside binding site of the human bitter taste receptor hTAS2R16. J. Biol. Chem. 285, 28 373–28 378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai H., Kojima D., Oura T., Tachibanaki S., Terakita A., Shichida Y. 1997. Single amino acid residue as a functional determinant of rod and cone visual pigments. Proc. Natl Acad. Sci. USA 94, 2322–2326 10.1073/pnas.94.6.2322 (doi:10.1073/pnas.94.6.2322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueda T., Ugawa S., Yamamura H., Imaizumi Y., Shimada S. 2003. Functional interaction between T2R taste receptors and G-protein alpha subunits expressed in taste receptor cells. J. Neurosci. 23, 7376–7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki A. 1965. An ecological study of wild Japanese monkeys in snowy areas. Primates 6, 31–72 10.1007/BF01794458 (doi:10.1007/BF01794458) [DOI] [Google Scholar]
- 15.Wang X., Grus W. E., Zhang J. 2006. Gene losses during human origins. PLoS Biol. 4, e52. 10.1371/journal.pbio.0040052 (doi:10.1371/journal.pbio.0040052) [DOI] [PMC free article] [PubMed] [Google Scholar]